Abstract
Poly(ADP-ribosyl)ation is a posttranslational modification catalyzed by the poly(ADP-ribose) polymerases (PARPs). These enzymes covalently modify glutamic, aspartic and lysine amino acid side chains of acceptor proteins by the sequential addition of ADP-ribose (ADPr) units. The poly(ADP-ribose) (pADPr) polymers formed alter the physico-chemical characteristics of the substrate with functional consequences on its biological activities. Recently, non-covalent binding to pADPr has emerged as a key mechanism to modulate and coordinate several intracellular pathways including the DNA damage response, protein stability and cell death. In this review, we describe the basis of non-covalent binding to pADPr that has led to the emerging concept of pADPr-responsive signaling pathways. This review emphasizes the structural elements and the modular strategies developed by pADPr-binding proteins to exert a fine-tuned control of a variety of pathways. Poly(ADP-ribosyl)ation reactions are highly regulated processes, both spatially and temporally, for which at least four specialized pADPr-binding modules accommodate different pADPr structures and reprogram protein functions. In this review, we highlight the role of well-characterized and newly discovered pADPr-binding modules in a diverse set of physiological functions.
Keywords: PARP, PARG, Poly(ADP-ribose), WWE, PBZ, Macro domain
1. Introduction
It was almost 50 years ago that poly(ADP-ribose) (pADPr) was discovered as an adenine-containing RNA-like polymer (Chambon et al., 1963) and early on, there were indications that pADPr turnover is very tightly regulated in mammalian cells (Nishizuka et al., 1967, Reeder et al., 1967, Ueda et al., 1972). Cellular levels of pADPr are governed by the finely tuned balance of the synthetic poly(ADP-ribose) polymerases (PARPs) and degrading poly(ADP-ribose) glycohydrolase (PARG) enzyme activities. In the human genome, 17 proteins share a PARP signature sequence homologous to the catalytic domain of the founding and most described member PARP1. In the context of DNA damage, PARP1 generates within minutes approximately 90% of all pADPr, preferentially on itself (automodification). However, pADPr accumulation is transient, as it is rapidly degraded by PARG (Davidovic et al., 2001). Notably, the polymerase activity has been demonstrated for only six of the PARP family members (PARP1, PARP2, PARP3, PARP4/vPARP, Tankyrases 1 and 2). Based on experimental and structural examinations, it has been proposed that the other PARP family members are either inactive (PARP9/BAL and PARP13/ZAP) or carry a mono-ADP-ribosyl transferase activity (PARP6, TiPARP, PARP8, PARP10, PARP11, PARP12, PARP14/BAL2, PARP15/BAL3, PARP16) (Goenka et al., 2007, Hottiger et al., 2010, Kleine et al., 2008). Their functions are only starting to emerge, but suggest an important role for these poorly studied proteins.
1.1. Poly(ADP-ribosyl)ation
The seminal work by Benjamin and Gill, 1980a, Benjamin and Gill, 1980b showed that PARP1 activity is highly stimulated by the presence of DNA containing single- and double-strand breaks, a discovery that has been followed up by a succession of studies that linked pADPr metabolism to maintenance of genome stability (Meyer-Ficca et al., 2005). Mono- and poly-(ADP-ribosyl)ation are reversible and phylogenetically ancient posttranslational protein modifications, for which the list of acceptor/target proteins is still expanding. Poly(ADP-ribosyl)ation can be achieved covalently or non-covalently (Fig. 1 A). The covalent posttranslational modification (PTM) occurs on glutamic, aspartic or lysine residues, while non-covalent interactions between proteins and pADPr add another level for modulating proteins biological activity. This PTM has profound physico-chemical implications, as poly(ADP-ribose) bears two negatively charged phosphate groups per ADP-ribose (ADPr) residue, i.e. twice as many charges than DNA or RNA (Fig. 1A). The size and flexibility of pADPr polymers render them capable of mediating multiple contacts with protein surfaces, thus providing a significant enhancement in binding efficiency. Such physico-chemical attributes of the pADPr are capable of contributing to the creation of a scaffold for the assembly of multiprotein complexes. Accumulating evidence indicates that pADPr actually conveys a broad spectrum of cellular signals through direct binding of a variety of protein motifs to pADPr, such as the DNA damage response, replication, chromatin structure, transcription, telomere homeostasis and cell death (Krishnakumar and Kraus, 2010).
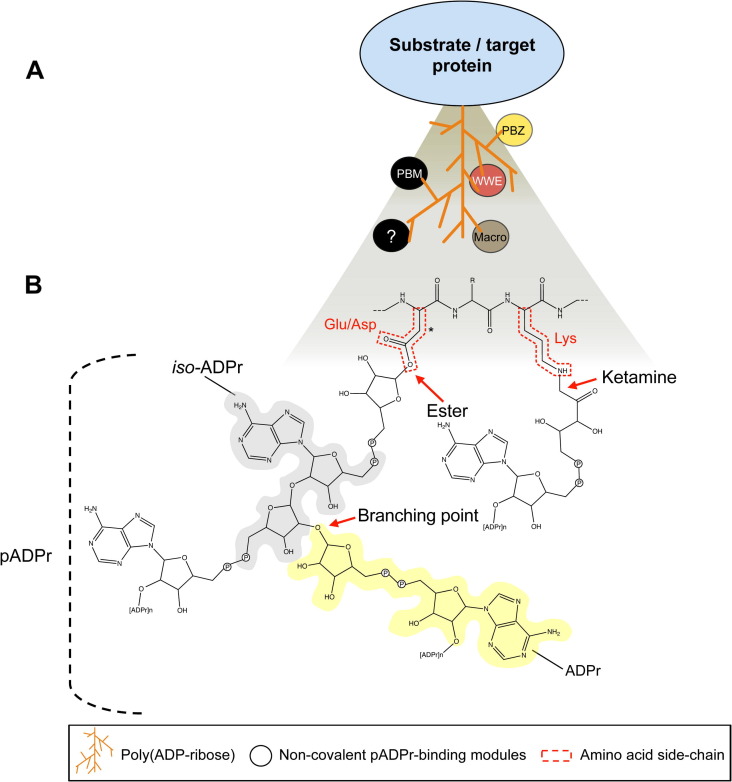
Fig. 1.
Covalent and non-covalent mechanisms of protein regulation by poly(ADP-ribosyl)ation. (A) The posttranslational modification of a protein substrate by enzymatic covalent attachment of poly(ADP-ribose) (pADPr) to specific amino acid side chains is represented. On the pADPr structure, a collection of modular protein domains non-covalently binds to pADPr through different recognition mechanisms. There are currently four pADPr-binding protein modules that have been experimentally characterized: the pADPr-binding motif (PBM); pADPr-binding zinc finger motif (PBZ); the macro domain (Macro) and the WWE domain (WWE). Experimental evidence suggests that other protein modules and sequence motifs can read this modification (see Section 3). (B) Detailed view of the covalent poly(ADP-ribosyl)ation. The proximal ADP-ribose (ADPr) is bound by an ester linkage to glutamic (Glu) and aspartic (Asp) amino acid side chains (the asterisk indicates one or two CH2 units to represent the respective side chains of Asp or Glu) or to lysine (Lys) side chains via a ketamine linkage. The mechanism that determines selective modification of specific residues and the functional significance of this heterogeneity are not known. Additional ADP-ribose units are subsequently attached by O-glycosidic linkages to form linear or branched pADPr. Some components of the pADPr chemical structure recognized by pADPr-binding modules are shown: iso-ADPr, grey shadow, ADPr, yellow shadow.
Given the structural complexity of the pADPr polymers, it is not surprising to observe that several evolutionary conserved protein domains have emerged to accomplish unique functions through interactions with pADPr. Indeed, the average chain length of pADPr synthesized by the PARP family members can range from very short and linear oligomers to extended molecules of up to 200 units and branched at every 20–50 residues (Fig. 1B) (Alvarez-Gonzalez and Jacobson, 1987, D’Amours et al., 1999, Kiehlbauch et al., 1993, Kleine et al., 2008, Tanaka et al., 1977). There are very limited investigations conducted on the physico-chemical properties of pADPr, such as flexibility and conformation, but the formation of helical pADPr structures was postulated upon protein binding (Minaga and Kun, 1983a, Minaga and Kun, 1983b, Schultheisz et al., 2009).
1.2. The emergence of non-covalent pADPr recognition motifs
The first experimental lines of evidence for proteins that bind pADPr in a non-covalent, yet specific, manner were given in the late 1960s and early 1970s when it was shown that histones possess high affinity for pADPr (Honjo et al., 1968, Nakazawa et al., 1968, Otake et al., 1969). However, whether the chemical nature of the bond was covalent or non-covalent was a highly debated topic (Adamietz et al., 1975). At that time, the prevailing view was that poly(ADP-ribosyl)ation was a covalent protein posttranslational modification on histones as is phosphorylation or methylation. This concept persisted for years until the 1980s when studies began to report interactions with PARP1-bound pADPr (Ohashi et al., 1983) and free pADPr (Sauermann and Wesierska-Gadek, 1986, Wesierska-Gadek and Sauermann, 1988). In the last decade, rapid progress has been achieved in the identification of pADPr-binding proteins among some of the chromatin-associated non-histone proteins and proteins involved in extranuclear signaling networks. Remarkably, more than four specialized pADPr-binding modules that recognize different structural features of the pADPr are responsible for the functional diversification of the pADPr-responsive cellular pathways. This paper first provides an inventory of the predominant techniques currently used to detect and measure non-covalent protein–pADPr interactions, then turns to an in-depth description of the specialized pADPr-binding modules that recognize different structural features of the pADPr, and finally, presents the functional consequences of this association in pADPr-responsive cellular pathways.
2. Affinity of pADPr-binding domains to ADP-ribose metabolites
Over the years, several different techniques have been developed to characterize the interaction of pADPr with proteins (Gagne et al., 2011). Polymer-blot and electrophoretic mobility shift assays (EMSA) are currently used to determine whether there is binding or not, while isothermal titration calorimetry (ITC), surface plasmon resonance (SPR) and a variation of the EMSA method allow to measure the affinity for pADPr by determining an affinity constant. The polymer-blot assay has also been exploited in saturation binding experiments to determine binding affinity (Wang et al., 2011).
Owing to its simplicity, the polymer-blot assay is the most frequently used method to study pADPr-binding proteins (Gagne et al., 2011). Proteins of interest are either hand-spotted, vacuum-aspirated or separated on a polyacrylamide gel prior to being transferred and renatured onto a nitrocellulose membrane. The interaction with pADPr is subsequently revealed by incubating the membrane with purified label-free pADPr, [32P]-radiolabeled pADPr or biotin-labeled pADPr. Bound pADPr is detected with anti-pADPr antibodies, by autoradiographic exposure or streptavidin conjugates, respectively. This technique is also used with peptide arrays to map the pADPr-binding regions of a protein. In this assay, peptides are chemically synthesized to represent the sequence of putative pADPr-binding sites, such as those predicted in silico based on consensus motifs (e.g. PBM, see Section 3.1). EMSA was developed to characterize pADPr–protein non-covalent interactions in solution (Fahrer et al., 2007). Incubations are made in the presence of pADPr and increasing concentrations of a purified protein. Protein-pADPr complexes are subsequently resolved by gel electrophoresis. Free and bound pADPr are detected using a streptavidin conjugate. A shift in the mobility of the protein indicates the formation of a protein–ligand complex. Since all measurements are made at equilibrium, the binding affinities can be calculated using a sigmoidal dose–response curve.
ITC and SPR are elegant label-free biophysical methods specifically designed to study the interaction kinetics between a ligand (such as pADPr) and a target protein. ITC measures the binding affinity and thermodynamics between two biomolecules in solution. In this method, a solution that contains a ligand is titrated into a solution of its binding partner until saturation is reached. A complete thermodynamic profile of the molecular interaction as well as the binding affinity (K D) are calculated from the heat released or absorbed over time during the interaction. SPR measures in real-time the refractive index changes near a sensor surface. A ligand (such as biotinylated pADPr) is immobilized on the surface of a solid support (chip) and the analyte (protein of interest) is passed over the surface to make contacts with the ligand. Interactions induce a change in the refractive index proportional to the mass on the surface. The data are fitted to a kinetic model to calculate the rate of association (ka), the rate of dissociation (kd) and the binding affinity (K D = kd/ka).
Each of these methods may also be conducted using fractionated pADPr, allowing further characterization of the binding specificity of a protein for long or short pADPr. In addition, SPR and ITC methods are amenable to determine the critical amino acid residues implicated in the binding of pADPr. Site-specific mutagenesis of critical residues that mediate pADPr-binding typically leads to at least a 10-fold reduction in binding affinity. Collectively, these methods have therefore been critical in characterizing the pADPr binding modules that are described in the following Section 3.
3. Protein modules involved in non-covalent interactions with poly(ADP-ribose)
3.1. PBM: The poly(ADP-ribose)-binding motif
The notion of non-covalent pADPr-binding was originally demonstrated with histones (Sauermann and Wesierska-Gadek, 1986, Wesierska-Gadek and Sauermann, 1988) and later better characterized using pADPr of different lengths and branching frequencies (Panzeter et al., 1992, Panzeter et al., 1993). This concept was further extended to non-histone proteins such as p53, DNA-PK or KU70/80 and led to the definition of a common polymer-binding domain of 22–26 amino acids that conveyed the specific affinity for pADPr (Table 1 ) (Althaus et al., 1999, Malanga et al., 1998). Notably, the Althaus group had a strong intuition when they raised the innovative hypothesis that “PARP-associated polymers may recruit signal proteins to sites of DNA breakage and reprogram their functions” (Althaus et al., 1999).
Table 1.
List and functional classification of experimentally demonstrated pADPr-binding proteins.a
| Description | Motifb | pADPr-dependent recruitment at DNA damagesc | References |
|---|---|---|---|
| DNA damage response and checkpoint | |||
| Aprataxin | n.d | + | Harris et al. (2009) |
| Aprataxin and PNK-like factor (APLF) | PBZ | + | Eustermann et al., 2010, Li et al., 2010, Rulten et al., 2008, Rulten et al., 2011 |
| Cellular tumor antigen p53 | PBM | Fahrer et al., 2007, Pleschke et al., 2000 | |
| Cyclin-dependent kinase inhibitor 1 (p21) | PBM | Pleschke et al. (2000) | |
| DNA excision repair protein ERCC-6 | PBM | Gagne et al. (2008) | |
| DNA ligase 3 | PBM | + | Gagne et al., 2012, Leppard et al., 2003, Pleschke et al., 2000 |
| DNA mismatch repair protein MSH6 | PBM | Pleschke et al. (2000) | |
| DNA polymerase epsilon catalytic subunit A (POL ε) | PBM | Pleschke et al. (2000) | |
| DNA repair protein complementing XP-A cells | PBM | Fahrer et al., 2007, Pleschke et al., 2000 | |
| DNA repair protein complementing XP-C cells | n.d | + | Luijsterburg et al. (2012) |
| DNA repair protein XRCC1 | PBM | + | El-Khamisy et al., 2003, Gagne et al., 2012, Mortusewicz and Leonhardt, 2007, Pleschke et al., 2000 |
| DNA topoisomerase 1 (TOP1) | PBM | Malanga and Althaus (2004) | |
| DNA topoisomerase 2-alpha (TOP2A) | PBM | Malanga and Althaus (2005) | |
| DNA-dependent protein kinase catalytic subunit (DNA-PK) | PBM | Pleschke et al. (2000) | |
| Double-strand break repair protein MRE11A | PBM/GAR | + | Haince et al. (2008) |
| E3 ubiquitin-protein ligase RNF146 (Iduna) | WWE | + | Andrabi et al., 2011, Callow et al., 2011, Kang et al., 2011, Wang et al., 2012, Zhou et al., 2011 |
| Flap endonuclease 1 (FEN1) | n.d | + | Gagne et al., 2012, Kleppa et al., 2012 |
| Histone H2A | PBM | Pleschke et al. (2000) | |
| Histone H2B | PBM | Pleschke et al. (2000) | |
| Histone H3 | PBM | Pleschke et al. (2000) | |
| Histone H4 | PBM | Pleschke et al. (2000) | |
| Nibrin (NBS1) | n.d | + | Haince et al. (2008) |
| Non-POU domain-containing octamer-binding protein (NONO) | RRM1 | + | Krietsch et al. (2012) |
| RNA-binding motif protein, X chromosome (RBMX) | n.d | + | Adamson et al. (2012) |
| Serine-protein kinase ATM | PBM | Haince et al. (2007) | |
| X-ray repair cross-complementing protein 6 (XRCC6 / KU70) | PBM | Pleschke et al. (2000) | |
| Werner syndrome ATP-dependent helicase (WRN) | PBM | Popp et al. (2012) | |
| Chromatin regulation and modification | |||
| Core histone macroH2A1.1 | Macro | + | Xu et al. (2012) |
| O-acetyl-ADP-ribose deacetylase MACROD1 | Macro | Neuvonen and Ahola (2009) | |
| Chromodomain-helicase-DNA-binding protein 1-like (CHD1L/ALC1) | Macro | + | Ahel et al., 2009, Gottschalk et al., 2009 |
| Chromodomain-helicase-DNA-binding protein 4 (CHD4) | n.d. | + | Chou et al., 2010, Polo et al., 2010 |
| Chromodomain-helicase-DNA-binding protein Mi-2 homolog (dMi-2) | K/R-rich | Murawska et al. (2011) | |
| Condensin complex subunit 1 (hCAP-D2) | PBM | Gagne et al. (2008) | |
| DNA methyltransferase 1 (DNMT1) | PBM | Reale et al., 2005, Zampieri et al., 2012 | |
| E3 SUMO-protein ligase CBX4 | n.d | + | Ismail et al. (2012) |
| Metastasis-associated protein MTA1 | n.d | + | Chou et al. (2010) |
| Polycomb complex protein BMI-1 | n.d | + | Gieni et al. (2011) |
| Protein DEK | PBM | Fahrer et al. (2010) | |
| Apoptosis | |||
| Apoptosis-inducing factor 1, mitochondrial (AIF) | PBM | Wang et al. (2011) | |
| DNA fragmentation factor subunit beta (DFF40/CAD) | PBM | Pleschke et al., 2000, Reh et al., 2005, West et al., 2005 | |
| E3 ubiquitin-protein ligase RNF146 (Iduna) | WWE | + | Andrabi et al., 2011, Callow et al., 2011, Kang et al., 2011, Wang et al., 2012, Zhou et al., 2011 |
| Hexokinase domain-containing protein 1 (HKDC1) | PBM | Gagne et al. (2008) | |
| Transcription, replication and gene expression | |||
| Cellular tumor antigen p53 | PBM | Pleschke et al. (2000) | |
| DNA topoisomerase 1 (TOP1) | PBM | Malanga and Althaus, 2004 | |
| DNA topoisomerase 2-alpha (TOP2A) | PBM | Malanga and Althaus, 2005 | |
| DNA topoisomerase 2-beta (TOP2B) | PBM | Gagne et al. (2008) | |
| E3 SUMO-protein ligase PIAS4 (PIASy) | PBM | Stilmann et al. (2009) | |
| Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) | PBM | Gagne et al., 2003, Gagne et al., 2008, Ji and Tulin, 2009, Ji and Tulin, 2012 | |
| NF-kappa-B essential modulator (NEMO/IKKγ) | n.d | Stilmann et al. (2009) | |
| SARS coronavirus non-structural protein nsp3 | Macro | Egloff et al. (2006) | |
| Nuclear factor NF-kappa-B p100 subunit | PBM | Pleschke et al. (2000) | |
| Polycomb group RING finger protein 2 (MEL-18/RNF110) | n.d | + | Chou et al. (2010) |
| RNA-binding motif protein, X chromosome (RBMX) | n.d | + | Adamson et al. (2012) |
| Serine/arginine-rich splicing factor 1 (ASF/SF2) | RRM1 / SR | Malanga et al. (2008) | |
| Telomerase reverse transcriptase (TERT) | PBM | Pleschke et al. (2000) | |
| Transcriptional repressor CTCF | PBM | Caiafa et al. (2009) | |
| G3BP1 | PBM | Isabelle et al. (2012) | |
| Centromere function and cell cycle checkpoint | |||
| Aurora kinase A-interacting protein | PBM | Gagne et al. (2008) | |
| E3 ubiquitin-protein ligase CHFR | PBZ | + | Gagne et al., 2012, Isogai et al., 2010, Oberoi et al., 2010 |
| Histone H3-like centromeric protein A (CENP-A) | PBM | Gagne et al., 2008, Saxena et al., 2002 | |
| Major centromere autoantigen B (CENP-B) | PBM | Saxena et al. (2002) | |
| Mitotic checkpoint protein BUB3 | PBM | Gagne et al., 2008, Saxena et al., 2002 | |
| Others | |||
| Capsid protein viral protein 1 (VP1) | PBM | Carbone et al. (2006) | |
| Heat shock factor (HSF-1) | PBM | Fossati et al. (2006) | |
| Major vault protein | PBM | Gagne et al. (2008) | |
| Myristoylated alanine-rich C-kinase substrate (MARCKS) | PBM | Pleschke et al., 2000, Schmitz et al., 1998 | |
| Nicotinamide mononucleotide adenylyltransferase 1 (NMNAT-1) | PBM | Berger et al. (2007) | |
| Nitric oxide synthase, inducible (iNOS) | PBM | Pleschke et al. (2000) | |
Listed proteins were retrieved from studies that specifically addressed the direct non-covalent binding to pADPr.
n.d. not determined.
Proteins shown to accumulate at DNA-damage sites in a pADPr-dependent fashion.
The first defined pADPr-binding motif (PBM) was derived from a region of high similarity in a multiple sequence alignment of proteins involved in signaling pathways that control cell cycle progression and DNA damage (Pleschke et al., 2000). This PBM is composed of a property-based sequence motif harboring basic and hydrophobic residues downstream of a lysine- and arginine-rich region (Fig. 2 A). Consistent with their previous observation with histones, the authors reported that long and branched pADPr are the preferred ligands of non-chromatin proteins comprising the PBM (Panzeter et al., 1992, Panzeter et al., 1993, Pleschke et al., 2000).
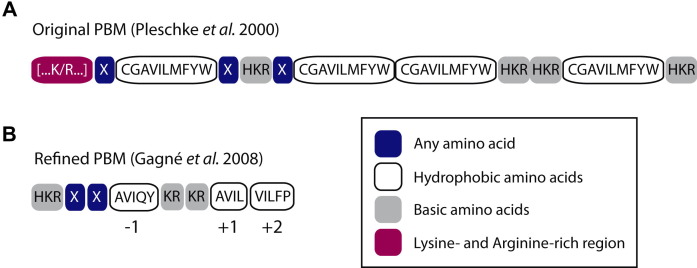
Fig. 2.
The non-covalent pADPr-binding motif (PBM). (A) The first PBM has been described by Pleschke and collaborators (2000) in a variety of DNA damage repair and checkpoint proteins. The motif is primarily composed of a hydrophobic and basic amino acid core flanked by a cluster of positively charged residues […K/R…]. Each box represents one amino acid position. (B) A refinement of the motif was proposed by Gagne et al. (2008) based on a number of PBM variations found in human proteins. The refined pADPr-binding signature confirmed the overall basic nature of the PBM but represents a minimal stand-alone version of the motif, the K/R-rich N-terminal cluster being dispensable for efficient binding. Outside the dual [KR][KR] site, there are additional preferences for hydrophobic amino acids (positions −1, +1 and +2), mostly those with alkyl side chains. The basic [KR][KR] doublet is an important requirement for the PBM since most substitutions in this region result in a substantially reduced binding affinity for pADPr.
To better define and address prediction accuracy of the PBM, we adopted a strategy based on a refinement of the consensus PBM motif (Gagne et al., 2008). We showed that restrictions to specific amino acid types exist for positions within the PBM (Fig. 2B). The previously reported preference for hydrophobic residues [ACGVILMFYW] was recovered, but there was a clear tendency for limited residue types to be allowed (mostly aliphatic residues), especially at position −1, +1 and +2 relative to the central K/R doublet (Fig. 2B). Clearly, the PBM refers to the conservation of a physical property pattern rather than a fixed sequence motif. The refined motif offers a more stringent definition of the original motif that decreases the probability of a PBM arising by chance in a protein database search. Computational PBM prediction has proven to be a powerful tool for the identification of protein regions that could mediate interaction with pADPr. They have been shown to convey important functions in animal models. Notably, the PBM discovered in the apoptosis-inducing factor (AIF) is critical for the ability of AIF to induce cell death by parthanatos (PARP1-dependent cell death) in cells and in vivo (Wang et al., 2011). A detailed PBM-pADPr complex has yet to be modeled but a study of the structural features of AIF’s PBM showed that it occupies an area on the surface of the protein which could provide stabilizing non-covalent contacts of amino acid side chains with pADPr (Wang et al., 2011). We can only speculate as to whether all PBMs possess common structural features, but a highly exposed solvent-accessible surface must be present to make contacts with pADPr molecules. Based on the helix propensity scale, positively charged amino acids (K/R) have a tendency to form α-helices (Pace and Scholtz, 1998). Since PBMs are located in lysine- and arginine-rich regions, it would be likely to find several of them in a helical conformation.
A summary of pADPr-binding proteins for which binding affinity constants have been determined is given in Table 2 . Of particular interest, several reports have shown that pADPr chain length is a crucial determinant for high affinity non-covalent interactions of PBM proteins with pADPr. The binding of the tumor suppressor protein p53, the nucleotide excision repair XPA, and the DEK oncoprotein with long (55-mer) and short (16-mer) pADPr chains were assessed by EMSA and SPR (Fahrer et al., 2007, Fahrer et al., 2010). These experiments revealed the high affinity (10−7 to 10−9 M range) of XPA and DEK to long pADPr chains but the lack of binding to short pADPr while p53 bound both short and long chains of pADPr with equivalent affinity.
Table 2.
List of pADPr-binding proteins with experimentally determined affinity constant (KD).
| Protein/peptide/domain | Method | O-acetyl-ADPr KD [M] | iso-ADPr KD [M] | ADP-ribose KD [M] | Short pADPr KD [M] | Long pADPr KD [M] | NFd pADPr KD [M] | Refs. | |
|---|---|---|---|---|---|---|---|---|---|
| PBM | XPA | EMSA | NBb | 3.2 × 10−7 | Fahrer et al. (2010) | ||||
| “ | SPR | NB | 6.5 × 10−9 | Fahrer et al. (2007) | |||||
| p53 | EMSA | 2.5 × 10−7 | 1.3 × 10−7 | Fahrer et al. (2010) | |||||
| “ | SPR | 3.4 × 10−9 | NMc | Fahrer et al. (2007) | |||||
| DEK | EMSA | NB | 6.1 × 10−8 | Fahrer et al. (2010) | |||||
| XRCC1 | SPR | 3.6 × 10−8 | Ahel et al. (2008) | ||||||
| NONO | SPR | NB | 2.01 × 10−8 | 2.32 × 10−8 | Krietsch et al. (2012) | ||||
| AIF | PBAa | 6.63 × 10−8 | Wang et al. (2011) | ||||||
| PBZ | CHFR | SPR | 5 × 10−10 | Ahel et al. (2008) | |||||
| “ | SPR | 7 × 10−9 | Oberoi et al. (2010) | ||||||
| APLF | SPR | 9 × 10−10 | Ahel et al. (2008) | ||||||
| APLF/TZF peptide | SPR | 9.5 × 10−10 | Li et al. (2010) | ||||||
| APLF/ZF1 peptide | SPR | 5.2 × 10−7 | Li et al. (2010) | ||||||
| APLF/ZF2 peptide | SPR | 8.3 × 10−6 | Li et al. (2010) | ||||||
| Macro | macroH2A1.1 | ITC | 2.6 × 10−6 | Kustatscher et al. (2005) | |||||
| MACROD1 | ITC | 9 × 10−7 | Neuvonen and Ahola (2009) | ||||||
| MACROD2 | ITC | 1.5 × 10−7 | Neuvonen and Ahola (2009) | ||||||
| WWE | Iduna/RNF146 | PBA | 1.2 × 10−8 | Andrabi et al. (2011) | |||||
| Iduna/RNF146 WWE domain | ITC | 3.7 × 10−7 | 1.7 × 10−3 | He et al., 2012, Wang et al., 2012 | |||||
| HUWE1 WWE domain | ITC | 1.3 × 10−5 | Wang et al. (2012) | ||||||
| PARP11 WWE domain | ITC | 4.0 × 10−4 | He et al. (2012) | ||||||
PBA, polymer-blot assay.
NB, no binding.
NM, no model found to describe the binding.
NF, non-fractionated pADPr.
Remarkably, PBMs are present in a marked number of proteins involved in the response to DNA damage and other chromatin transactions such as chromatin structure, replication and transcription (Table 1). Furthermore, the PBM often overlaps with important regulatory protein domains (Pleschke et al., 2000). This has triggered the idea that upon binding to pADPr, the PBM could shield a regulatory surface by steric hindrance, thus destabilizing several protein–protein or protein–ligand interactions. Alternatively, a highly extended and flexible polymer bound to a protein domain could distort it so that perturbations of the native fold may arise. Globally, molecular crowding by the pADPr provides the basis for the concept of “reprogrammation” of protein functions as suggested (Malanga and Althaus, 2005). Actually, the affinity of several DNA damage response factors for pADPr can modulate (I) the sensing of DNA lesions; (II) the dynamic chromatin remodeling events and (III) the assembly and functionality of DNA repair complexes. We believe that the transient accumulation of pADPr following DNA-dependent PARP activation can result in vast pleiotropic effects on a systems-wide scale that implicates numerous DNA damage response effectors. This is supported by the predominant presence of nucleic acid-interacting proteins in the PBM’s prediction datasets (Gagne et al., 2008). Indeed, DNA- and RNA-binding modules are significantly over-represented as putative pADPr-binding modules and thus represent a general class of pADPr-targeted proteins with potential for broad implication in the DNA damage response. However, in some proteins, the PBM is distinct from the nucleic acid binding domains, such as in AIF, providing the ability of pADPr to modulate protein function in the context of nucleic acid binding. Generally, proteins associate in multi-protein complex machineries that execute biological processes that a single protein cannot execute alone. Macromolecules that disrupt or stabilize such complexes drive a wide variety of cellular processes. pADPr possesses the biochemical and structural properties to fulfill such functions through non-covalent interactions.
3.2. Alternative PBMs
Similar to DNA and RNA, the pADPr carries a net negative charge due to its phosphate backbone. Because these three macromolecules tend to bind positively charged protein domains, some competition exists among DNA, RNA or pADPr for the same binding site in specific cellular contexts. Indeed, in addition to the classical PBM, recent studies suggest alternative PBMs located in nucleic acid-interaction domains.
3.2.1. The glycine- and arginine-rich domain (GAR)
The glycine- and arginine-rich domain (GAR) lacks hydrophobic amino acids commonly found in PBMs. This region rather accumulates a very high positive charge owing to the presence of a repetition of arginine residues, thus making it an ideal binding surface for a polymer with a high negative charge density such as the pADPr. The GAR, also referred to as RGG box and the RG domain, is a protein module typically found in proteins involved in RNA metabolism (Burd and Dreyfuss, 1994) as well as in some chromatin associated proteins (Bernstein and Allis, 2005, Kornblihtt et al., 2009). Selected examples include fibrillarin (FBL), heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), fragile X mental retardation protein 1 (FMRP), small nuclear ribonucleoprotein Sm D1 (SNRPD1), chromatin target of PRMT1 (Protein arginine N-methyltransferase 1) protein (CHTOP), bromodomain and WD repeat-containing protein 3 (BRWD3), cell death and ATM (serine-protein kinase ATM) regulator AVEN, Ras GTPase-activating protein-binding protein 1 (G3BP1), double-strand break repair protein MRE11 and tumor suppressor p53-binding protein 1 (53BP1).
Evidence that the GAR is a pADPr-binding module came from the study of MRE11 (Haince et al., 2008). MRE11, a core component of the MRN complex (MRE11, RAD50 and NBS1), is responsible for the initial recognition of DNA double-strand breaks (DSBs), mediates end-resection by its exonuclease activity and together with 53BP1 and other DNA damage response factors, facilitates repair. The GAR domain of MRE11 is required for its DNA binding activity (Boisvert et al., 2005, Dery et al., 2008) but also mediates its interaction with pADPr as well as its rapid accumulation at DNA strand breaks (Haince et al., 2008). The MRE11 exonuclease activity is inhibited by pADPr in vitro, suggesting that pADPr may regulate MRE11-dependent end-resection at DSBs or at stalled replication forks, as recently reported (Ying et al., 2012). Interestingly, several other GAR-containing proteins participate in the DNA damage response and genome surveillance. In view of the high pADPr level that transiently accumulates at sites of damage, it is suspected that the function of some of these GAR-bearing proteins might be regulated by pADPr. 53BP1 regulates repair of DSBs, while the nuclear DNA helicase II (RNA Helicase A) interacts and regulates the DSBs biomarker γ-H2AX (Mischo et al., 2005) and the Werner syndrome helicase (WRN) (Friedemann et al., 2005). The nucleosome remodeling and histone deacetylase (NuRD) complex comprises several core components with affinity for pADPr. Methyl-CpG-binding domain protein 2 (MBD2) has a GAR motif, while CHD4 (chromodomain helicase DNA binding protein 4) and MTA1 (metastasis associated 1) interact with pADPr through a still undefined motif (Chou et al., 2010, Lai and Wade, 2011, Polo et al., 2010). The latter two proteins are involved in the recruitment of the NuRD complex to DNA strand breaks in a pADPr-dependent manner (Lai and Wade, 2011). Our current understanding suggests that the presence of pADPr acts as a recruitment module for the organization of the PARP1-associated DNA repair and chromatin remodeling machinery at DNA lesions. On the other hand, pADPr binding to the GAR domain could be considered as a DNA displacement mechanism required to reconfigure the DNA repair protein complexes and provide access to damaged DNA. It may also interfere with other DNA damage-induced posttranslational modifications, such as PRMT1-dependent arginine methylation in the GAR domain (Bedford and Richard, 2005). This view supports a concept where pADPr is a key orchestrator of the DNA damage response.
Recently, it has been suggested that pADPr regulates post-transcriptional gene regulation in the cytoplasm, notably through the assembly of cytoplasmic stress granules (Leung et al., 2012). In support of this, we showed that the stress granule effector G3BP1 binds pADPr by its GAR domain (Isabelle et al., 2012). Importantly, G3BP1-mediated stress granule assembly is impaired by PARP inhibition during genotoxic insult, suggesting that pADPr is critical for the reprogrammation of messenger ribonucleoparticles in cellular stress responses. This result adds to the experimental evidence that the pADPr-binding protein AIF functions as a negative regulator of stress granules (Cande et al., 2004). These results emphasize the fact that pADPr can perform various functions in several different DNA damage-processing pathways and can enable a crosstalk between the regulation of the early and late steps of the DNA damage response.
3.2.2. The RNA recognition motif (RRM) and Serine/Arginine repeats (SR)
The RNA recognition motif (RRM), also referred to as RNA-binding domain (RBD) or ribonucleoprotein domain (RNP), is the most abundant nucleic acid-binding motif in the human genome (Clery et al., 2008). RRMs are found in a wide variety of RNA and ssDNA-binding proteins. RRMs may be found in conjunction with GAR-containing proteins. One prominent example is hnRNP A1 that possesses two RRMs in addition to its GAR motif. hnRNPs are highly versatile proteins that can participate in various aspects of nucleic acid metabolism: mRNA stability and splicing, DNA replication, chromatin remodeling, telomere maintenance, DNA repair and genome stability (Han et al., 2010). Based on a proteome-wide screen to identify pADPr-binding proteins, our laboratory was the first to identify hnRNPs as a family of proteins with affinity for pADPr (Gagne et al., 2003). More recently, Ji and Tulin (2009) validated this finding by providing evidence that hrp38 (the Drosophila melanogaster homologue of human hnRNP A1) binds pADPr in a non-covalent way in vivo, with the consequence of reduced RNA-binding ability. RNA processing factors (such as NONO and RBMX) recently emerged as guardians of genomic stability with widespread involvement in preventing DNA damage (Adamson et al., 2012, Krietsch et al., 2012, Paulsen et al., 2009). Both NONO (Non-POU domain-containing octamer-binding protein) and RBMX (RNA-binding motif protein, X chromosome) comprise RRMs and are recruited in a pADPr-dependent manner to DNA damage (Adamson et al., 2012, Krietsch et al., 2012). We have recently reported the binding of pADPr to the RRM1 domain of NONO. As it was observed for hnRNPA1, pADPr interfered with the interaction of NONO with RNA in vitro. Notably, the binding of NONO to pADPr by RRM1 is crucial for the recruitment of NONO to DNA damage sites and influences the outcome of DNA DSB repair. The high binding affinity of pADPr to NONO was assessed by SPR (Table 2). Similar to DEK and XPA described above, NONO showed a strong affinity for non-fractionated and long pADPr chains while no binding was detected for short pADPr chains. These observations therefore provide further support for RRMs as biologically relevant pADPr-binding modules (Krietsch et al., 2012). Given the frequent occurrence of RRM-containing proteins in the human proteome, interactions with pADPr are likely to have a significant impact on cell signaling through a complex network of biochemical pathways.
Another large group of RRM-containing proteins that bind to RNAs are the SR (Serine/Arginine repeats) proteins (Long and Caceres, 2009) that, along with hnRNPs, contribute to the formation of messenger ribonucleoprotein particles (mRNPs). It has been shown that the SR protein ASF/SF2 binds pADPr with high affinity (Malanga et al., 2008). Two domains in ASF/SF2 can mediate the interaction with pADPr: (I) a N-terminal fragment that contains a RRM1 and (II) a C-terminal SR domain (Malanga et al., 2008). Given that the SR domain carries an excess positive charge with the predominance of arginine residues, this pADPr-binding feature resembles that of the GAR which also harbors a basic arginine-rich cluster expected to interact with the phosphate backbone of pADPr. Similarly, strong pADPr-binding was observed in lysine- and arginine-rich (K/R-rich) motifs located in the nucleosome remodeler dMi-2 (Murawska et al., 2011). It remains to be determined whether the presence of a basic electrostatic patch on a protein surface could be considered as a general pADPr-protein interface or if additional structural determinants are required (such as the helical conformation of the SR domain (Sellis et al., 2012)).
3.3. PBZ: a poly(ADP-ribose)-binding zinc finger
Zinc fingers specifically interacting with pADPr rather than DNA or RNA were discovered in a subset of proteins related directly or indirectly with pADPr metabolism (Ahel et al., 2008). This newly identified C2H2-type “pADPr-binding zinc finger” (PBZ) has a consensus sequence defined as [K/R]xxCx[F/Y]GxxCxbbxxxxHxxx[F/Y]xH where b denotes a basic residue and x any residue (Ahel et al., 2008). PBZ domains have been observed only in eukaryotic proteins, excluding yeast. The absence of PBZ motifs in prokaryote and yeast proteins parallels the absence of pADPr metabolism in those, suggesting a co-evolution of the PBZ motif with the presence of PARPs. Only three human proteins appear to carry a PBZ motif: the aprataxin and PNK-like factor (APLF, also called XIP1, PALF), the checkpoint protein with FHA and RING domains (CHFR), and the DNA cross-link repair 1A protein (DCLRE1A/SNM1A) (Fig. 3 ) (Ahel et al., 2008). Interestingly, the PBZ module was found in some non-human proteins involved in the maintenance of genome integrity or DNA repair: Ku70, Rad17, Parp and Chk2 in Dictyostelium discoideum and DNA Ligase in Caenorhabditis elegans corresponding to human DNA Ligase III. However, the human orthologues do not contain any PBZ domain (Ahel et al., 2008, Isogai et al., 2010).
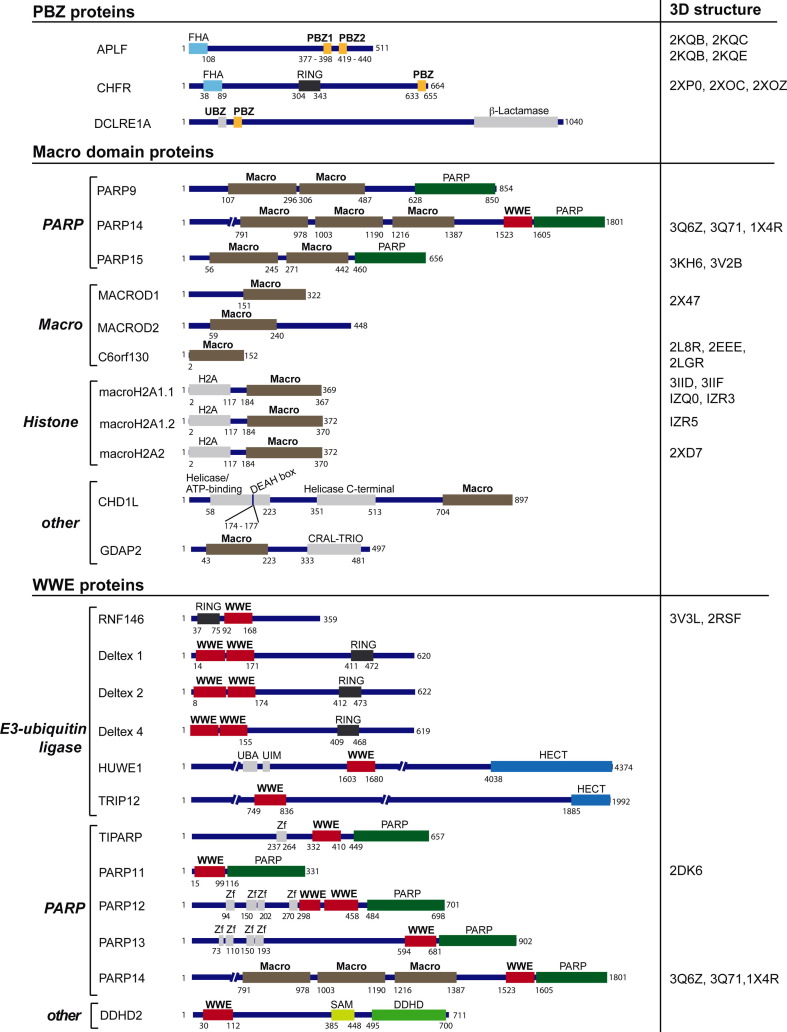
Fig. 3.
Schematic representation of human proteins harboring a PBZ, Macro domain or WWE binding module. The three protein folds currently recognized to confer high-affinity to pADPr are listed with their individual protein members. When available, 3D structure accession numbers (Protein Data Bank (PDB)) are given. The domain organization is schematized and drawn to scale according to the Uniprot database. Binding to pADPr remains to be formally demonstrated for some of the listed proteins while binding was undetectable for others. See Sections 3.3, 3.4, 3.5 for more details. FHA, forkhead-associated domain; β-Lactamase, beta-Lactamase domain; RING, RING finger; H2A, domain with similarity to histone H2A; Helicase ATP-binding, helicase superfamily 1/2 ATP-binding domain; DEAH box, DEAH box motif; Helicase C-terminal; Helicase conserved C-terminal domain; CRAL-TRIO, domain named after cellular retinaldehyde-binding protein (CRALBP) and TRIO guanine exchange factor, this domain binds lipophilic molecules; UBA, ubiquitin associated domain; UIM, ubiquitin interaction motif; HECT, homologous to E6-AP carboxyl terminus domain (has E3-ubiquitin ligase activity); Zf, zinc finger; SAM, sterile α motif domain; DDHD, domain named after the conserved residues DDHD.
Human CHFR and DCLRE1A contain a unique PBZ while APLF has two PBZ placed in tandem (PBZ1 and PBZ2 – Fig. 3). Structural studies of the PBZ domains with small molecules that mimic the features of pADPr have revealed that besides the cysteines and histidines coordinating the Zn ion, critical aromatic residues mediate interactions with the adenine ring of ADPr (Ahel et al., 2008, Eustermann et al., 2010, Isogai et al., 2010, Li et al., 2010, Oberoi et al., 2010). The lack of conservation of most of these critical binding residues in the PBZ sequence of DCLRE1A suggests that it may not bind pADPr, but this has not been experimentally assessed (Oberoi et al., 2010). The affinity of CHFR (5 × 10−10 M) and APLF (9.5 × 10−10 M) for pADPr measured by SPR indicates that this module has the highest affinity for pADPr of all pADPr-binding domains (Table 2). Interestingly, the affinity of APLF for pADPr is in the same range than that of CHFR, despite the fact that it has two PBZ. Each PBZ of APLF binds independently pADPr, but with reduced affinity relative to the tandem PBZ and the full length protein (Table 2) (Ahel et al., 2008, Eustermann et al., 2010, Li et al., 2010, Rulten et al., 2008). Moreover, the affinity of PBZ1 for pADPr is 10-fold higher than that of PBZ2 (Table 2) (Eustermann et al., 2010, Li et al., 2010). These observations are in line with the structural details of CHFR and APLF, which strongly suggested that the CHFR PBZ and the PBZ1 domain of APLF are able to interact with two consecutive ADPr moieties in pADPr while the second PBZ of APLF probably binds only one (Oberoi et al., 2010). These observations are also consistent with the more deleterious effects of mutations in PBZ1 than in PBZ2 for the recruitment of APLF to UV-induced DNA strand breaks (Li et al., 2010, Rulten et al., 2008). PBZ1 may also interact with PARP1 as well, providing further affinity of PBZ1 for automodified PARP1 (Eustermann et al., 2010, Macrae et al., 2008).
The role of APLF in the DNA damage response and repair via the non-homologous end-joining (NHEJ) pathway has been recognized by several research groups (Bekker-Jensen et al., 2007, Iles et al., 2007, Kanno et al., 2007, Li et al., 2010, Macrae et al., 2008, Rulten et al., 2008). APLF comprises a N-terminal FHA domain and displays apurinic-apyrimidinic (AP) endonuclease and 3′-5′ exonuclease activities in vitro. APLF is rapidly recruited to DNA strand breaks introduced by UV irradiation via both its FHA and PBZ domains. While the FHA domain mediates interactions with the repair proteins XRCC1 and XRCC4, the tandem PBZ domain directs pADPr-dependent recruitment of APLF to DNA strand breaks, where APLF facilitates NHEJ. Both PARP1 and PARP3-dependent poly(ADP-ribosyl)ation have been shown to promote APLF responses to DNA strand breaks (Rulten et al., 2008, Rulten et al., 2011).
Similar to APLF, CHFR comprises a phospho-binding FHA module but also a RING finger domain with E3 ubiquitin ligase activity that plays an essential role in the antephase checkpoint, delaying mitotic entry under certain stress conditions (Chaturvedi et al., 2002, Scolnick and Halazonetis, 2000). This function of CHFR is dependent on its interaction with PARP1 and pADPr, because mutations in the CHFR PBZ that disrupts pADPr-binding as well as PARP inhibitors both abolish the CHFR-dependent mitotic checkpoint (Ahel et al., 2008, Kashima et al., 2012, Oberoi et al., 2010). Recently, a mechanistic interplay behind CHFR and pADPr interactions inducing a mitotic entry delay was uncovered (Kashima et al., 2012). The activation of PARP1 following mitotic stress promotes the pADPr-dependent ubiquitylation of PARP1 by CHFR and its proteasomal degradation (Fig. 4A). This finding further revealed that PARP1 levels must be critically controlled during cell proliferation.
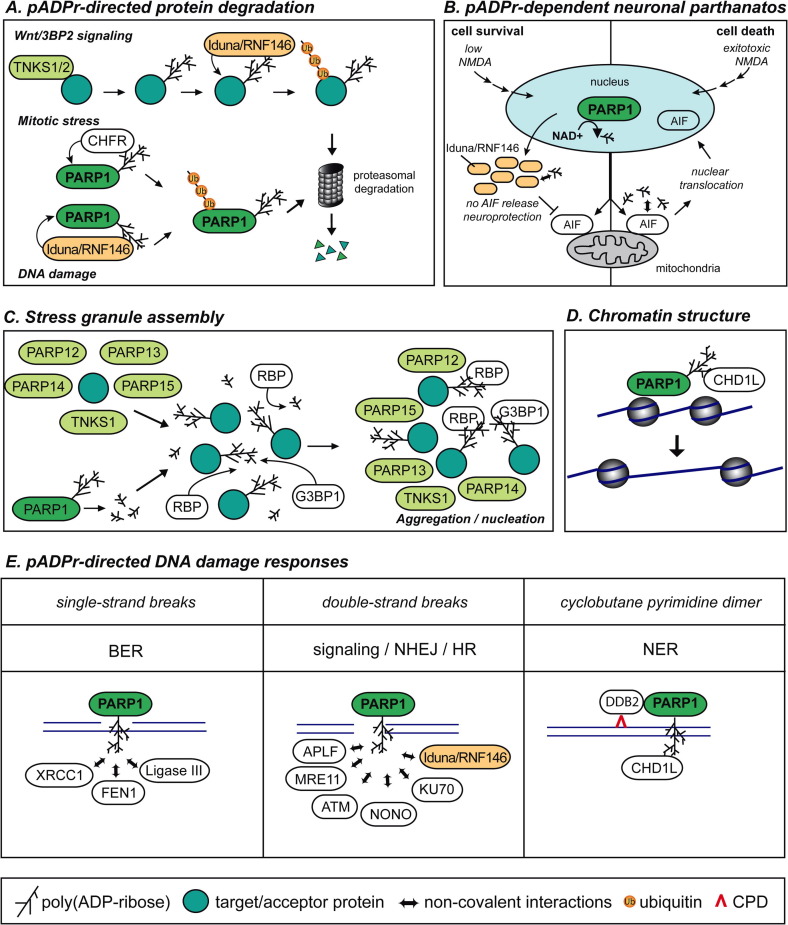
Fig. 4.
Schematic models of pADPr regulatory functions. (A) Protein stability can be regulated via pADPr-directed recruitment of ubiquitin-conjugating enzymes. Some protein substrates of tankyrases (TNKS1/2) (3BP2 and axin) or automodified PARP1 undergo proteasomal degradation after ubiquitylation by the WWE-containing E3 ubiquitin ligase Iduna/RNF146 or by the PBZ-containing E3 ubiquitin ligase CHFR (see Sections 3.3, 4.1 for details). (B) Neuronal cell fate after toxic stress. The excitation of glutamate receptor by N-methyl-d-aspartate (NMDA) triggers PARP activation. Non-toxic NMDA activation (left panel) induces the expression of the cell survival factor Iduna/RNF146 and its cytoplasmic accumulation. Interactions of Iduna/RNF146 with pADPr prevent apoptosis inducing factor (AIF) translocation to the nucleus and parthanatos. Excitotoxic activation of glutamate receptors (right panel) fail to induce Iduna/RNF146 expression. The accumulation of cytoplasmic pADPr promotes the release of apoptosis-inducing factor (AIF) from the mitochondria. AIF subsequently translocates to the nucleus and induces parthanatos. See Sections 3.5, 4.2 for details. (C) pADPr-dependent assembly of stress granules. Two models have been proposed (see Section 4.3 for details). In the first view, cytoplasmic ADPr and pADPr are synthesized by tankyrases and PARP12-15 upon stress exposure. This triggers the aggregation of RNA-binding proteins (RBP) to ADPr/pADPr and stress granule formation. In the second view, the nucleo-cytoplasmic trafficking of pADPr is responsible for its accumulation into the cytoplasm. By virtue of its endoglycosidic activity, PARG releases free and protein-bound pADPr following genotoxic stress and PARP activation. pADPr translocated into the cytoplasm is targeted by G3BP1 and RNA-binding proteins to initiate the aggregation of stress granules. In both views, pADPr in ribonucleoparticles acts as a scaffold for the recruitment of RNA-binding proteins. (D) pADPr plays regulatory roles in the dynamics of chromatin structure. Automodification of PARP1 and poly(ADP-ribosy)ation of histones induce chromatin relaxation. This also involves the chromatin remodeling factor CHD1L, which is recruited to specific sites by pADPr. The ATPase activity of CHD1L is stimulated by pADPr and triggers nucleosome sliding. This possibly facilitates access of the DNA repair machineries. (E) Functions of pADPr in DNA damage responses. DNA strand breaks as well as other types of altered DNA structures and DNA adducts activate PARP1. pADPr triggers the recruitment of proteins and enzymes involved in DNA damage signaling, in base excision repair (BER), non-homologous end joining (NHEJ), homologous recombination (HR) and nucleotide excision repair (NER). See Section 4.5 for details.
The binding of DCLRE1A to pADPr has not been studied. This protein has an endonuclease function and it is involved in the repair of DNA interstrand cross-links (Hazrati et al., 2008, Yang et al., 2010). However, the putative PBZ does not comprise the aromatic residues needed to contact pADPr, suggesting that its functions are independent of poly(ADP-ribosyl)ation.
3.4. The macro domain: an ADP-ribose binding module
The macro domain was first described in a variant of the histone H2A, as a 130–190 amino acid module on the C-terminal side of the histone domain (Pehrson and Fried, 1992). It was soon recognized as an important protein domain, well conserved throughout all living organisms, as well as in a small subset of single-stranded RNA viruses that infect mammalian cells, including coronaviruses, alphaviruses and hepatitis E virus. Determination of the 3D structure of the thermophile Archaeoglobus fulgidus macro protein Af1521 provided the first clues to the potential function of this domain. It revealed an organization of helices and sheets reminiscent of the P-loop found in nucleotide hydrolases, suggesting a related enzymatic function for the macro fold (Allen et al., 2003). Additional studies with Af1521 and with the yeast macro domain protein YBR022 W supported this observation as they revealed the ability of these macro domains to hydrolyze ADP-ribose-1”-phosphate into ADPr and inorganic phosphate (Pi) (Karras et al., 2005, Martzen et al., 1999). Subsequent studies indicated that some macro domains could not only interact with ADPr but also with pADPr, making it a novel pADPr-interaction module (Ahel et al., 2009, Gottschalk et al., 2009, Karras et al., 2005, Neuvonen and Ahola, 2009, Timinszky et al., 2009).
Based on primary amino acid sequence comparisons, 11 human proteins comprise a macro domain (Fig. 3). This macro domain may exist on its own (macroD1, also called MDO1 and LRP16, macroD2 also called MDO2 and C6orf130) or in association with the histone fold (macrohistones H2A), the PARP catalytic domain (PARP9, 14 and 15), the SNF2/helicase ATPase domain (ALC1/CHD1L), or the Sec14p/CRAL-TRIO protein–lipid interaction module (GDAP2). Intriguingly, macroPARPs (also called BAL PARPs) have the unique feature of bearing two (PARP9 and PARP15) or even three (PARP14) macro domains in tandem, which, in PARP14, are further associated with two more putative pADPr binding modules, a RRM and a WWE (Fig. 3).
The affinity of most human macro domain proteins for NAD+-derived metabolites permits the assessment of the importance of this module for pADPr binding and metabolism. It should be stressed that, out of nine macro domain-containing human proteins so far tested for pADPr binding (PARP14 and PARP15 have not been examined), only four bind pADPr (namely macroH2A1.1, CHD1L, macroD1 and PARP9). The others have affinity only for ADPr or O-acetyl-ADP-ribose (Fig. 1; Table 2). Furthermore, the detailed structural analysis of macroH2A1.1 revealed that it is able to bind solely the terminal ADPr of the polymer, indicating that its macro domain is in practice an ADPr binding module (Timinszky et al., 2009). Therefore, structural details critically affect the ability of the macro fold binding pocket to accommodate ligands such as ADPr. For instance, the macroH2A1.2 and macroH2A2 variants as well as GDAP2 do not interact with ADPr (Kustatscher et al., 2005, Till and Ladurner, 2009). While the structural characteristics of macroH2A1.2 differ only slightly from those of the macroH2A1.1 variant, three additional amino acid residues in macroH2A1.2 critically fall in the ADPr binding pocket, thereby hindering the interaction (Kustatscher et al., 2005). The structure of some viral macro domains (also called X-domain) has also been investigated as well as their ability to interact with ADPr and pADPr (Egloff et al., 2006, Malet et al., 2009, Neuvonen and Ahola, 2009, Piotrowski et al., 2009). As it is the case for human proteins, some but not all tested present affinity for ADPr or pADPr. The binding of viral macro domains to ADPr is at least 10-fold lower (K D of the severe acute respiratory syndrome (SARS) coronavirus is 24 μM, that of hepatitis E virus above 50 μM) but interactions with pADPr have been shown by polymer blot assays (Egloff et al., 2006, Neuvonen and Ahola, 2009). It remains to be determined whether this interaction is critical for viral host infections. Collectively, these observations indicate that the presence of a macro fold hints to a possible interaction with ADPr-related metabolites, which however needs to be experimentally addressed.
3.4.1. Hidden macro domains in pADPr-binding proteins
Recent structural analysis of poly(ADP-ribose) glycohydrolase (PARG) revealed the striking finding that its pADPr-interaction module folds in a macro domain-like structure. First identified in the distantly PARG-related bacterial protein DUF2263, this finding was then confirmed with the structural analysis of a protozoan PARG and rat PARG (Dunstan et al., 2012, Kim et al., 2012a, Slade et al., 2011). Therefore, despite minimal sequence similarity between the typical macro domain and PARG, part of the PARG catalytic domain adopts this characteristic macro fold organization in which the PARG sequence signature GGG-X6-8-QEE lines the ADP-ribose binding pocket. However, in the mammalian PARG, additional essential sequences extending beyond the macro domain adopt structural conformations around the macro fold that specify the catalytic pocket and the glycohydrolase activity. Importantly, a unique “tyrosine clasp” near the catalytic pocket offers an explanation for the exoglycosidic and endoglycosidic activities of mammalian PARG towards pADPr (Brochu et al., 1994), the latter endoglycosidic activity lacking in the bacterial PARG (Kim et al., 2012a). These recent findings have thus highlighted that some macro domains may only be revealed once the 3D structure is determined, indicating that there may be other pADPr-binding macro proteins in mammalian cells awaiting identification.
Many of the macro domain-ADPr/pADPr interactions have been examined in vitro, using purified macro domains or proteins and purified ADPr/pADPr. Because PARPs catalyze the addition of ADPr onto protein substrates, it will be critical to investigate whether the macro-ADPr interaction can be extended to ADPr covalently attached to acceptor proteins. A recent study using synthetic peptides corresponding to mono-ADP-ribosylated histone H2B tail showed that it could be the case, as macroH2A1.1 did bind such peptides (Moyle and Muir, 2010).
Recent detailed analysis of the enzymatic activity of the macro domain indicates that some deacetylate O-acetyl-ADP-ribose. This molecule is produced by the sirtuin deacetylases, which uses NAD+ as a co-factor to deacetylate proteins. The three stand-alone macro domain proteins, namely macroD1, macroD2, and C6orf130, appear to form a subgroup of macro domain proteins showing this deacetylase activity by cleaving the ester bond between the acetyl group and ADPr. One could envision that some macro domain proteins possessing O-acetyl-ADPr deacetylase activity may be able to hydrolyze the protein-ADPr ester bond (Fig. 1B). Until now, the ability of PARG and ARH3 to fulfill this function has been questioned. The existence of a distinct enzyme (an ADP-ribose lyase) able to hydrolyze the ester bond between the glutamic or aspartic acid residue of the acceptor protein and ADPr has been proposed nearly 30 years ago (Oka et al., 1984), but remains to be characterized. However, the recent indications that lysines could also constitute ADPr acceptor sites on PARP1 and histones, forming a ketamine in this case (Fig. 1B) (Altmeyer et al., 2009, Messner et al., 2010), suggest that there may be more than one “lyase”.
Biological functions of macro domain proteins remain to be examined in details. Macrohistones and CHD1L contribute to the structure of chromatin (see Sections 4.4, 4.5) and regulatory transcriptional functions have been ascribed to PARP9, PARP14, macroD1 and macroH2A variants. The latter macrohistones have been generally linked to transcriptional repression as they induce a more condensed chromatin state and impede access to transcription factors, although in some specific cases they can also promote transcription (reviewed by (Gamble and Kraus, 2010)) (Muthurajan et al., 2011). PARP9 was also shown to repress transcription via its macro domains (Aguiar et al., 2005). In contrast, PARP14, also named CoaSt6 because of its co-activator function for the transcription factor Stat6, promotes interleukin-4 dependent gene activation in a manner dependent on its macrodomains as well on its mono-ADP-ribosyl-transferase activity (Goenka and Boothby, 2006, Goenka et al., 2007). MacroD1, originally named leukemia related protein 16 (LRP16), was identified as a co-activator of androgen and estrogen receptor transcriptional activity as well as of NF-κB (Han et al., 2007, Wu et al., 2011, Yang et al., 2009). This transcriptional co-activation of macroD1 is dependent on its macro domain. Collectively, these examples suggest that pADPr-macro domain interactions contribute to transcriptional regulation.
3.5. The WWE domain
The most recently discovered pADPr-binding motif, the so-called WWE domain, is named after its most conserved amino acids (tryptophan (W) and glutamate (E)), which are flanked by an otherwise rather low degree of sequence conservation (Wang et al., 2012). The 12 human proteins that contain a WWE domain (Fig. 3) belong mostly to two functional classes of proteins, namely those associated with ubiquitylation and those associated with poly(ADP-ribosyl)ation (Wang et al., 2012).
The best studied example is the RING finger protein 146 (RNF146) also called Iduna. pADPr binding of Iduna/RNF146 was first ascribed to a PBM (Kang et al., 2011) which was further defined as part of the WWE domain that mediates the interaction with pADPr (Wang et al., 2012). The structural characteristics of the Iduna/RNF146 WWE domain and its interaction with pADPr were thoroughly investigated by polymer blot assays, NMR and crystallography (Andrabi et al., 2011, Wang et al., 2012). Iso-ADPr rather than ADPr is the smallest unit that can be bound by the Iduna/RNF146 domain (Fig. 1B). The lack of interaction between WWE and ADPr was explained by the need for phosphate groups on either side of the adenine-ribose structure to make extensive contacts with the binding pocket. This supported the idea that the WWE domain is a pADPr-binding module because at least two ADPr units are needed to generate the iso-ADPr ligand. The proposed structure is compatible with interactions with iso-ADPr within longer pADPr chains (Wang et al., 2012), consistent with the co-purification of Iduna/RNF146 with pADPr (Andrabi et al., 2011). Four residues have been identified as crucial for iso-ADPr-binding in the Iduna/RNF146 WWE domain. These residues are well conserved throughout most human WWE domains (Wang et al., 2012), including the putative or demonstrated ubiquitin ligases Deltex 1,2,4, HUWE1 and TRIP12, which have been shown by SPR to also bind pADPr (Table 2). The WWE domains of several PARP family members (PARP11, PARP13 and the first WWE of PARP12) also comprise the conserved residues, suggesting that they bind pADPr. Only the binding of PARP11 has been examined, and showed interactions with ADPr and pADPr with rather low affinity (Table 2) (He et al., 2012, Wang et al., 2012). In contrast, two of the residues are not conserved in the second WWE of PARP12, in TiPARP, PARP14 and the putative phospholipase DDHD2, suggesting that these may not bind pADPr, as shown for DDHD2 (DDHD domain containing 2) in in vitro binding experiments (Wang et al., 2012).
A common theme among WWE containing proteins is the association with domains of the E3 ligase type, strongly suggesting a functional link between ubiquitylation and poly(ADP-ribosyl)ation. This link was uncovered in the regulation of Wnt/β-catenin signaling pathway by tankyrases and Iduna/RNF146 (Huang et al., 2009, Zhang et al., 2011). This pathway, essential for embryonic development and adult tissue homeostasis, is tightly regulated by the concentration of axin. It turns out that axin is a substrate for tankyrase such that its poly(ADP-ribosyl)ation allows its recognition by Iduna/RNF146. Binding of Iduna/RNF146 to poly(ADP-ribosyl)ated axin triggers axin ubiquitylation and proteasomal degradation (Fig. 4A). Interestingly, we concurrently showed that Iduna/RNF146 plays a prominent role in the context of DNA damage through its pADPr-dependent E3 ligase activity as it also ubiquitylates several DNA repair factors in a way that depends on pADPr. PARP1/2, KU70, XRCC1 and DNA ligase III were identified as Iduna/RNF146 substrates when poly(ADP-ribosyl)ated by PARP1 (see Section 4.2 and Fig. 4B and E) (Kang et al., 2011).
The cross-talk between ubiquitylation and poly(ADP-ribosyl)ation may not be restricted to Iduna/RNF146 but awaits further experimental examination. For instance, the E3 ligases Deltex1, Deltex2 and Deltex3 play a role in notch signaling. The N-terminus of these proteins contains tandem WWE motifs mediating interactions with the ankyrin repeats of Notch intracellular domain. Several studies performed in vivo and in cell culture have shown that Notch ubiquitylation is promoted by Deltex expression (Baron, 2012, Hori et al., 2004, Matsuno et al., 2002, Mukherjee et al., 2005, Wilkin et al., 2008).
In the context of DNA damage responses, HUWE1 (also called Mule, ARF-BP1, LASU1 and HectH9) and TRIP12 (also called E3 ubiquitin ligase for Arf (ULF)) both belong to the family of HECT domain (homologous to E6-AP carboxyl terminus) E3 ligases. HUWE1 participates in the DNA damage response by tightly regulating steady state levels of XRCC1, DNA polymerase β, and DNA ligase III (Khoronenkova and Dianov, 2011, Parsons et al., 2009). Several substrates of HUWE1 for ubiquitylation/proteosomal degradation have been identified: Cdc6, the c-Myc oncogene, histones, as well as p53 (Adhikary et al., 2005, Chen et al., 2005, Hall et al., 2007). TRIP12 is a key regulator of the DNA damage response (Gudjonsson et al., 2012) by acting as a guard against excessive spreading of ubiquitylated chromatin at DNA damage sites as it functionally suppresses RNF168, another E3 ligase, which promotes the concerted accumulation of H2A and H2AX at DNA damage site. The importance of pADPr-binding by the WWE motif of HUWE1 and TRIP12 for their recruitment to DNA strand breaks remains to be addressed.
It is interesting to note that the members of the PARP family that carry WWE domains are most likely mono(ADP-ribosyl)transferases and unable to produce the iso-ADPr moiety bound by WWE. Little is known about these proteins and their functions. One aspect that may be of further functional relevance is the presence of classical zinc fingers associated with TiPARP, PARP12, PARP13 (Fig. 3). Of these, PARP13 has been examined as an antiviral protein (also named zinc antiviral protein ZAP). PARP13 inhibits the replication of viruses by recruiting the cellular RNA degradation machineries to degrade the viral mRNAs. It targets RNA viruses such as the retroviridae human immunodeficiency virus type 1 (HIV-1) (Chen et al., 2012, Zhu et al., 2011).
4. Cellular processes influenced by protein-pADPr interactions and its clinical applications
4.1. Regulation of protein stability
The mechanistic link between poly(ADP-ribosyl)ation and the regulation of protein degradation is one of the most surprising aspects of the recent advances on poly(ADP-ribosyl)ation. Three research groups identified independently the E3-ligase Iduna/RNF146 as being a key regulator of protein stability in a pADPr-dependent manner (Andrabi et al., 2011, Callow et al., 2011, Kang et al., 2011, Zhang et al., 2011). Remarkably, this pathway appears to function in several biological contexts, regulated not only by PARP1 but also by the tankyrases 1 and 2 (Fig. 4 A). In the context of DNA damage, PARP1 automodification triggers its ubiquitylation by Iduna/RNF146 and subsequent degradation by the proteasome (Kang et al., 2011). In the context of Wnt signaling, it is tankyrase-dependent poly(ADP-ribosyl)ation of axin that induces its Iduna/RNF146-dependent proteasomal degradation and subsequent β-catenin-dependent transcription (Callow et al., 2011, Zhang et al., 2011). Iduna/RNF146 binds to pADPr of varying lengths (Andrabi et al., 2011) and ubiquitylates substrates poly(ADP-ribosyl)ated with short chains as occurs with tankyrases (Zhang et al., 2011) and substrates poly(ADP-ribosyl)ated with long chains that occurs with PARP1 (Kang et al., 2011). Thus Iduna/RNF146’s dynamic range of protein quality control in the setting of poly(ADP-ribosyl)ation may be extensive. Because of this, there are likely to be multiple checkpoints that control Iduna/RNF146’s activity and biological actions that require further investigation. Interestingly, regulation of protein stability in a pADPr-dependent manner is not restricted to WWE-containing E3-ligases because the PBZ-bearing CHFR E3-ligase has been shown recently to ubiquitylate PARP1 to target it for proteasomal degradation in the context of mitotic stress (see Section 3.3; Fig. 4A) (Kashima et al., 2012).
The clinical importance of the tankyrase-Iduna/RNF146-dependent regulation of protein stability was revealed recently in studies focusing on the cherubism-mutated protein 3BP2 (Guettler et al., 2011, Levaot et al., 2011). Cherubism is a rare autosomal dominant human disorder characterized by inflammatory destructive bone lesions resulting in abnormal fibrous tissue growth in the lower part of the face. Approximately 80% of all cherubism patients carry a mutation in the Sh3bp2 gene, which encodes the Src homology 3 domain-binding protein-2 (3BP2). Interestingly, most of these mutations lie within a six amino acid sequence (RSPPDG) that corresponds to the tankyrase substrate-recognition motif (Levaot et al., 2011). Upon successful binding of tankyrase 2 to the latter motif in 3BP2 of healthy cells, 3BP2 is poly(ADP-ribosyl)ated, which serves as a signal for its Iduna/RNF146-directed ubiquitylation and proteasome-mediated degradation (Fig. 4A) (Guettler et al., 2011, Levaot et al., 2011). The regulated degradation of 3BP2 is profoundly disturbed in cherubism patients because the interaction and hence poly(ADP-ribosyl)ation of mutated 3BP2 by tankyrase 2 is impaired, abolishing 3BP2 ubiquitylation by Iduna/RNF146. The abnormal accumulation of 3BP2 within cells appears to alter the signaling balance of SRC kinase multiprotein complex to which it is associated, causing systematic inflammation, and leading to the cherubism phenotype (Guettler et al., 2011, Levaot et al., 2011). Hence, understanding the concerted action of poly(ADP-ribosyl)ation and ubiquitylation might improve therapeutic approaches targeting cherubism.
It is tempting to speculate that the tankyrase-dependent poly(ADP-ribosyl)ation coupled to Iduna/RNF146 ubiquitylation/degradation pathway is a widespread process to regulate protein steady states. By in silico searches for putative tankyrase interacting proteins, Guettler et al. (2011) have identified a very large list of proteins carrying the tankyrase interaction sequence RXX(G/P)DG that could constitute potential targets. For instance, the stability of the centrosomal associated protein (CPAP), important for centriole duplication during mitosis, is regulated by tankyrase-dependent poly(ADP-ribosyl)ation and proteasomal degradation (Kim et al., 2012b).
4.2. Cell death – Parthanatos
Massive activation of PARP1 following a genotoxic stress has been long recognized as a critical event in the induction of cell death. However, it is only in recent years that pADPr has been recognized as a death signaling molecule after the identification of pADPr as the trigger of apoptosis inducing factor (AIF)-dependent cell death. This insight came from studies attempting to determine the linkage between PARP1 activation during genotoxic stress and translocation of AIF from the mitochondria to the nucleus (Yu et al., 2002). Experiments to determine the mechanism of how PARP1 activation was intimately coupled to AIF translocation led to the discovery that pADPr translocates from the nucleus to mitochondria where it acts as an AIF releasing factor to cause the mitochondrial-nuclear translocation of AIF, initiating cell death (Fig. 4B) (Andrabi et al., 2006, Yu et al., 2006). This type of cell death, called parthanatos, occurs in neuronal cells following excitotoxicity as well as other cell types in which DNA damage is induced by specific genotoxic agents (Fig. 4B) (Wang et al., 2009). As noted above, the PBM within AIF is required for the release of AIF after PARP1 activation (Wang et al., 2011). It is important to note that the pADPr binding site in AIF is distinct from its DNA binding site. Thus, agents could be designed to block this interaction serving as inhibitors of parthanatos or to enhance the release of AIF for cancer chemotherapeutics. In a screen for cell survival factors, we identified the E3-ligase Iduna/RNF146 as a pADPr-dependent pro-survival factor, triggered by the same pADPr signal, in this case during non-toxic neuronal stress (Fig. 4B) (Andrabi et al., 2011). Iduna/RNF146 protects against parthanatos (pADPr-dependent) cell death mediated by glutamate excitotoxicity both in vitro and in vivo and against middle cerebral artery occlusion-induced stroke. Iduna/RNF146’s protective properties are due to its ability to bind pADPr, consistent with the notion that pADPr can act as a death signal during parthanatos. PARG also inhibits parthanatos via degradation of the pADPr (Koh et al., 2004), whereas Iduna/RNF146 interferes with pADPr death signaling. Together both PARG and Iduna/RNF146 function as inhibitors of parthanatos during genotoxic stress providing endogenous controllers of cell death initiated by activation of PARP1.
Iduna/RNF146 also protects against the N-methyl-N-nitro-N-nitrosoguanidine (MNNG), a DNA damaging agent, and after γ-irradiation rescues cells from G1 arrest and promotes cell survival. Iduna/RNF146 reduces purinic/apyrimidinic (AP) sites after MNNG exposure and it also facilitates DNA repair following γ-irradiation. Iduna/RNF146’s protective function not only requires a functional pADPr-binding domain, but its ubiquitin E3 ligase activity as well (Kang et al., 2011). Thus, Iduna/RNF146 links poly(ADP-ribosyl)ation and protein stability in the DNA damage response by controlling the levels of proteins important in this process through modulating the levels of poly(ADP-ribosyl)ated proteins via ubiquitin proteasomal degradation. Identification of Iduna/RNF146’s substrates that play roles in parthanatos and DNA repair are the critical next steps.
4.3. Assembly of stress granules
A novel function of pADPr has been recently described in the regulation of stress granules (SG) (Leung et al., 2011, Leung et al., 2012). These structures are composed of cytoplasmic aggregates of stalled pre-initiation mRNA complexes. In addition to a subset of ribosomal proteins and translation initiation factors, the SG contains a variety of RNA-binding proteins that promote its nucleation and participate in the reprogrammation of translation during stress. The localization of several PARPs in SG (PARP12, PARP13, PARP14, PARP15 and tankyrase, called here SG-PARPs) uncovered a connection between poly(ADP-ribosyl)ation reactions and posttranscriptional regulation of gene expression. It has been shown that the overexpression of the SG-PARPs promotes the poly(ADP-ribosyl)ation of mRNA-associated proteins and the assembly of SGs, while PARG overexpression inhibits their appearance supporting a key role for pADPr in the assembly of SG (Fig. 4C). This notion was further supported by the identification of G3BP1, one of the primary nucleator of SGs (Kedersha and Anderson, 2007), as a pADPr-binding protein (Isabelle et al., 2012). In contrast to the cytoplasm-restricted poly(ADP-ribosyl)ation reactions of the SG-PARPs, the G3BP1-associated SG are assembled with a pADPr-dependent mechanism that originates from the nuclear activation of PARP1/2. These two possibilities are schematized in Fig. 4C.
4.4. Chromatin structure
PARP activation results in chromatin relaxation (reviewed in Beneke, 2012, Krishnakumar and Kraus, 2010). The mechanism through which this occurs remains to be characterized. One model stipulates that histones, which are poly(ADP-ribosyl)ated after PARP activation, are less tightly bound to DNA, thereby inducing relaxation (Ball and Yokomori, 2011, Poirier et al., 1982). Another mechanism has arisen from studies of the non-covalent interaction of ALC1/CHD1L with pADPr. PARP1 activation not only recruits this protein to sites of DNA damage, it also stimulates its ATPase activity and induces CHD1L-dependent nucleosome repositioning (Fig. 4D) (Ahel et al., 2009, Gottschalk et al., 2009). This PARP-dependent function of CHD1L may be of importance during DNA damage signaling and repair of DNA strand breaks, as well as of cyclobutane pyrimidine dimers by nucleotide excision repair (Fig. 4E) (Luijsterburg et al., 2012, Pines et al., 2012).
4.5. DNA damage response
Poly(ADP-ribosyl)ation in the context of DNA damage responses has long been recognized, but the concept is emerging that it is not involved in the DNA repair processes per se. pADPr synthesis at sites of DNA lesions triggers the recruitment of many DNA damage mediators and repair factors as well as chromatin remodeling and may serve as a scaffold for the assembly of repair complexes (Table 1; Fig. 4E). Regardless of the type of DNA damage, be it single-strand breaks repaired by base excision repair, double-strand breaks repaired by non-homologous end joining or homologous recombination, or cyclobutane pyrimidine dimers repaired by nucleotide excision repair, the recent elucidation of large sets of pADPr-binding proteins and associated complexes supports critical functions for pADPr in the early sensing and signaling of DNA damage (Gagne et al., 2012).
4.6. Clinical implications
A number of cancer cells are crucially dependent on the DNA repair pathways regulated by PARP1. BRCA1 and 2 are the major breast and ovarian susceptibility genes reported. Mutations in the latter genes render cells deficient for DSB repair and exquisitely sensitive to PARP inhibitors. This concept is now being extended to other DNA repair factors, including mutations in ATM, p53 and the MRE11-RAD50-NBS1 complex (Oplustilova et al., 2012, Williamson et al., 2012). Interestingly, the exact mechanism of action of PARP inhibitors is still a matter of ongoing debate (Helleday, 2011, Patel et al., 2011). Large-scale sequencing projects of human genomes, such as the ENCODE project consortium, may help to reveal novel sequence variants or mutations in proteins involved in the maintenance of genomic stability with critical implications in the development of human cancers (Dunham et al., 2012). It is also reasonable to think that a critical mutation in a pADPr-binding motif might have deleterious consequences in signaling pathways that comprise the DNA damage response. Such information might be positively exploited clinically.
While defective DNA damage repair pathways are one type of susceptibility to PARP inhibitors, there appear to be others for which mechanistic basis are failing to be explained at the moment. For instance, the susceptibility of human epidermal growth factor 2 positive (HER2+) breast cancer cells to PARP inhibitors alone was recently shown, independent of defects in HR (Nowsheen et al., 2012). Similarly intriguing is the sensitivity of cancers and cells bearing gene fusions with ETS transcription factors (mainly ERG), including prostate cancer and Ewing’s sarcoma, to PARP inhibitors (Brenner et al., 2011, Brenner et al., 2012, Garnett et al., 2012). In this case, both the transcriptional and DNA damage signaling functions of PARP1 may be involved to explain the sensitivity (Brenner et al., 2011, Garnett et al., 2012, Schiewer et al., 2012). Based on the latter findings and on the fact that PARP inhibitors have minimal side-effects, they have been in clinical trials for almost a decade, either in combination with chemotherapeutic drugs, as a single-agent or very recently in combination with phosphoinositide 3-kinase (PI3K) inhibitors, that further expand the treatment options for cancer patients (Ibrahim et al., 2012, Javle and Curtin, 2011, Juvekar et al., 2012, Rouleau et al., 2010).
With these examples in mind, and as the list of pADPr-binding proteins and pathways using poly(ADP-ribosyl)ation as signaling mechanisms is still expanding, it becomes crucial to investigate the broad spectrum of biological implications of pADPr-protein interactions. It will undoubtedly lead to a better understanding of more applications of PARP inhibitors as single-agent and combination therapies. For instance, several pADPr-binding proteins have been linked to cancer progression or aggressiveness. PARP9, initially referred to as “BAL PARP”, was originally identified as a risk-related gene in diffuse large B-cell aggressive lymphoma, being over-expressed in such cancer cells (Aguiar et al., 2000, Takeyama et al., 2003). Similarly, CHD1L was originally named “amplified in liver cancer 1” (ALC1) because it was discovered as a candidate oncogene in hepatocellular carcinoma (Chen et al., 2009, Chen et al., 2010, Ma et al., 2008).
Further investigation of the mechanistic roles of pADPr in the regulation of cancer-associated protein networks and signaling pathways will be fundamental for the development of innovative treatment strategies, and to overcome resistance to such treatments.
5. Concluding remarks
Our understanding of the role of pADPr has remarkably expanded since the original observation that DNA strand breaks activate PARP1. As described above, in humans, there are at least four pADPr-binding modules (and others perhaps waiting to be discovered), coupled to a discrete number of additional domains linking poly(ADP-ribosyl)ation to ubiquitylation and chromatin structure in several cellular contexts such as protein degradation, DNA damage responses and cell death. Although sufficient to ensure binding to pADPr, these pADPr-binding domains vary widely in their degree of ligand specificity. While some seems to have a general affinity for the polyanionic backbone of biomolecules (i.e. DNA, RNA and pADPr) due to the presence of basic patches of amino acid residues, others evolved to perform specialized functions and exhibit a high degree of target specificity towards pADPr. Currently, the PBZ domain, which forms a divergent C2H2-type zinc finger fold with specialized functions, possesses the highest affinity for pADPr. The C2H2 zinc finger fold is amongst the most prevalent protein motifs in the human proteome and comprises the largest family of regulatory proteins in mammals. With this knowledge, we can speculate that uncharacterized variations in finger-like protrusions might provide the specificity required to recognize different pADPr structures.
Over 60 human proteins have been shown to interact with pADPr (Table 1), but based on in silico predictions of the occurrence of the PBM sequence, there may be over 500 of them, and many more if we consider that proteins in complexes with pADPr-binding proteins are (indirectly) affected by pADPr. Because the PBM represents a short contiguous protein segment, examination of other context criteria, such as protein surface accessibility, evolutionary conservation as well as the determination of three-dimensional PBM-bound pADPr complexes will help to establish the local structural environment required for pADPr-binding. Because of this potential that poly(ADP-ribosyl)ation affects a significant portion of the proteome, it is crucial to pursue the extensive examination of the pADPr-protein interactions.
In this regard, it is very intriguing to consider the high number of pADPr-binding proteins working in a single pathway. One may envision that there is a “pADPr code” where the length, complexity, and conformation adopted by pADPr covalently linked to a particular protein target, all contribute to favor some non-covalent interactions relative to others. The remarkably high affinity and processivity of PARG for long pADPr will certainly have a role to play in the regulation of the non-covalent interactions. Equally intriguing is the presence of several pADPr-binding modules within single proteins (i.e. Deltex, macroPARPs, APLF, etc.). Does it confer higher specificity, stronger interactions, or preference for pADPr in a particular conformation? Or does it organize pADPr in a scaffold as proposed for the formation of stress granules?
The functions of pADPr in pathological conditions (i.e. DNA damage, mitotic stress, etc.) are now better understood. Still, the mechanisms underlying the relocalization of pADPr to the cytoplasm after specific stresses are largely unknown. The physiological aspects of poly(ADP-ribosyl)ation are only starting to emerge as they are more difficult to grasp. pADPr levels often fall below the threshold of detectability using current methods, especially in the cytoplasm. The triggers of pADPr synthesis by the damage-independent PARPs, such as tankyrases and SG-PARPs, are undefined. Nonetheless, in view of their important functional outcomes in regulating protein stability and posttranslational gene expression in the cytoplasm, no doubt that it must be finely regulated. Considering the critical functions of Iduna/RNF146 in directing poly(ADP-ribosyl)ated proteins towards proteasomal degradation, and the high catabolic activity of PARG, it is conceivable that some functional aspects of pADPr-binding proteins may consist in protecting pADPr from degradation, or in shielding the pADPr from Iduna/RNF146/CHFR to protect target proteins from degradation. We can also wonder whether there may be pADPr-dependent deubiquitylases that further regulate this process. It is now important to critically examine the regulation of pADPr degradation by PARG, ARH3 and possibly some macro domain proteins, in cellular contexts where pADPr-binding proteins also operate, to truly understand the extent of signaling afforded by pADPr. Some pathways appear to rely on the generation of free pADPr molecules, requiring an endoglycosidic activity that so far, only PARG is known to display. Our understanding of these fundamental questions now depends on the development of sensitive and quantitative methods for the detection of nuclear and cytoplasmic pADPr structures.
Acknowledgements
We apologize to authors of relevant publications that were not cited in this review due to space limitations. We thank John D. Chapman (Washington University, WA) for help with the drawings of chemical structures. JYM and GGP acknowledge the Canadian Institutes of Health Research for supporting their research. GGP holds a Canadian Research Chair in Proteomics. VLC and TMD acknowledge that their work is supported by grants from the NIH/NINDS NS067525 and NIDA DA000266. TMD is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. JYM is a FRSQ Senior scholar. JK is supported by a scholarship from the FQRNT.
References
- Adamietz P., Oldekop M., Hilz H. Proceedings: is poly(ADP-ribose) covalently bound to nuclear proteins? J. Biochem. 1975;77:4p. [PubMed] [Google Scholar]
- Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Kogel U., Scheffner M., Helin K., Eilers M. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Aguiar R.C., Takeyama K., He C., Kreinbrink K., Shipp M.A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- Aguiar R.C., Yakushijin Y., Kharbanda S., Salgia R., Fletcher J.A., Shipp M.A. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96:4328–4334. [PubMed] [Google Scholar]
- Ahel D., Horejsi Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P., Owen-Hughes T., Boulton S.J. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Allen M.D., Buckle A.M., Cordell S.C., Lowe J., Bycroft M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- Althaus F.R., Kleczkowska H.E., Malanga M., Muntener C.R., Pleschke J.M., Ebner M., Auer B. Poly ADP-ribosylation: a DNA break signal mechanism. Mol. Cell. Biochem. 1999;193:5–11. [PubMed] [Google Scholar]
- Altmeyer M., Messner S., Hassa P.O., Fey M., Hottiger M.O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R., Jacobson M.K. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- Andrabi S.A., Kang H.C., Haince J.F., Lee Y.I., Zhang J., Chi Z., West A.B., Koehler R.C., Poirier G.G., Dawson T.M., Dawson V.L. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat. Med. 2011;17:692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C., Hurn P.D., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball A.R., Jr., Yokomori K. Damage site chromatin: open or closed? Curr. Opin. Cell Biol. 2011;23:277–283. doi: 10.1016/j.ceb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M. Endocytic routes to Notch activation. Semin. Cell Dev. Biol. 2012;23:437–442. doi: 10.1016/j.semcdb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Fugger K., Danielsen J.R., Gromova I., Sehested M., Celis J., Bartek J., Lukas J., Mailand N. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem. 2007;282:19638–19643. doi: 10.1074/jbc.C700060200. [DOI] [PubMed] [Google Scholar]
- Beneke S. Regulation of chromatin structure by poly(ADP-ribosyl)ation. Front. Genet. 2012;3:169. doi: 10.3389/fgene.2012.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin R.C., Gill D.M. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J. Biol. Chem. 1980;255:10493–10501. [PubMed] [Google Scholar]
- Benjamin R.C., Gill D.M. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J. Biol. Chem. 1980;255:10502–10508. [PubMed] [Google Scholar]
- Berger F., Lau C., Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc. Natl. Acad. Sci. USA. 2007;104:3765–3770. doi: 10.1073/pnas.0609211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Allis C.D. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., Hendzel M.J., Masson J.Y., Richard S. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle. 2005;4:981–989. doi: 10.4161/cc.4.7.1830. [DOI] [PubMed] [Google Scholar]
- Brenner J.C., Ateeq B., Li Y., Yocum A.K., Cao Q., Asangani I.A., Patel S., Wang X., Liang H., Yu J., Palanisamy N., Siddiqui J., Yan W., Cao X., Mehra R., Sabolch A., Basrur V., Lonigro R.J., Yang J., Tomlins S.A., Maher C.A., Elenitoba-Johnson K.S., Hussain M., Navone N.M., Pienta K.J., Varambally S., Feng F.Y., Chinnaiyan A.M. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J.C., Feng F.Y., Han S., Patel S., Goyal S.V., Bou-Maroun L.M., Liu M., Lonigro R., Prensner J.R., Tomlins S.A., Chinnaiyan A.M. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. 2012;72:1608–1613. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochu G., Duchaine C., Thibeault L., Lagueux J., Shah G.M., Poirier G.G. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim. Biophys. Acta. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Caiafa P., Guastafierro T., Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 2009;23:672–678. doi: 10.1096/fj.08-123265. [DOI] [PubMed] [Google Scholar]
- Callow M.G., Tran H., Phu L., Lau T., Lee J., Sandoval W.N., Liu P.S., Bheddah S., Tao J., Lill J.R., Hongo J.A., Davis D., Kirkpatrick D.S., Polakis P., Costa M. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One. 2011;6:e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande C., Vahsen N., Metivier D., Tourriere H., Chebli K., Garrido C., Tazi J., Kroemer G. Regulation of cytoplasmic stress granules by apoptosis-inducing factor. J. Cell Sci. 2004;117:4461–4468. doi: 10.1242/jcs.01356. [DOI] [PubMed] [Google Scholar]
- Carbone M., Reale A., Di Sauro A., Sthandier O., Garcia M.I., Maione R., Caiafa P., Amati P. PARP-1 interaction with VP1 capsid protein regulates polyomavirus early gene expression. J. Mol. Biol. 2006;363:773–785. doi: 10.1016/j.jmb.2006.05.077. [DOI] [PubMed] [Google Scholar]
- Chambon P., Weill J.D., Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P., Sudakin V., Bobiak M.L., Fisher P.W., Mattern M.R., Jablonski S.A., Hurle M.R., Zhu Y., Yen T.J., Zhou B.B. Chfr regulates a mitotic stress pathway through its RING-finger domain with ubiquitin ligase activity. Cancer Res. 2002;62:1797–1801. [PubMed] [Google Scholar]
- Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Chen L., Chan T.H., Yuan Y.F., Hu L., Huang J., Ma S., Wang J., Dong S.S., Tang K.H., Xie D., Li Y., Guan X.Y. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these processes in human patients. J. Clin. Invest. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Hu L., Chan T.H., Tsao G.S., Xie D., Huo K.K., Fu L., Ma S., Zheng B.J., Guan X.Y. Chromodomain helicase/adenosine triphosphatase DNA binding protein 1-like (CHD1l) gene suppresses the nucleus-to-mitochondria translocation of nur77 to sustain hepatocellular carcinoma cell survival. Hepatology. 2009;50:122–129. doi: 10.1002/hep.22933. [DOI] [PubMed] [Google Scholar]
- Chen S., Xu Y., Zhang K., Wang X., Sun J., Gao G., Liu Y. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat. Struct. Mol. Biol. 2012;19:430–435. doi: 10.1038/nsmb.2243. [DOI] [PubMed] [Google Scholar]
- Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clery A., Blatter M., Allain F.H. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Davidovic L., Vodenicharov M., Affar E.B., Poirier G.G. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res. 2001;268(1):7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- D’Amours D., Desnoyers S., D’Silva I., Poirier G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- Dery U., Coulombe Y., Rodrigue A., Stasiak A., Richard S., Masson J.Y. A glycine–arginine domain in control of the human MRE11 DNA repair protein. Mol. Cell Biol. 2008;28:3058–3069. doi: 10.1128/MCB.02025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R., Khatun J., Lajoie B.R., Landt S.G., Lee B.K., Pauli F., Rosenbloom K.R., Sabo P., Safi A., Sanyal A., Shoresh N., Simon J.M., Song L., Trinklein N.D., Altshuler R.C., Birney E., Brown J.B., Cheng C., Djebali S., Dong X., Ernst J., Furey T.S., Gerstein M., Giardine B., Greven M., Hardison R.C., Harris R.S., Herrero J., Hoffman M.M., Iyer S., Kelllis M., Kheradpour P., Lassman T., Li Q., Lin X., Marinov G.K., Merkel A., Mortazavi A., Parker S.C., Reddy T.E., Rozowsky J., Schlesinger F., Thurman R.E., Wang J., Ward L.D., Whitfield T.W., Wilder S.P., Wu W., Xi H.S., Yip K.Y., Zhuang J., Bernstein B.E., Green E.D., Gunter C., Snyder M., Pazin M.J., Lowdon R.F., Dillon L.A., Adams L.B., Kelly C.J., Zhang J., Wexler J.R., Good P.J., Feingold E.A., Crawford G.E., Dekker J., Elinitski L., Farnham P.J., Giddings M.C., Gingeras T.R., Guigo R., Hubbard T.J., Kellis M., Kent W.J., Lieb J.D., Margulies E.H., Myers R.M., Starnatoyannopoulos J.A., Tennebaum S.A., Weng Z., White K.P., Wold B., Yu Y., Wrobel J., Risk B.A., Gunawardena H.P., Kuiper H.C., Maier C.W., Xie L., Chen X., Mikkelsen T.S., Gillespie S., Goren A., Ram O., Zhang X., Wang L., Issner R., Coyne M.J., Durham T., Ku M., Truong T., Eaton M.L., Dobin A., Lassmann T., Tanzer A., Lagarde J., Lin W., Xue C., Williams B.A., Zaleski C., Roder M., Kokocinski F., Abdelhamid R.F., Alioto T., Antoshechkin I., Baer M.T., Batut P., Bell I., Bell K., Chakrabortty S., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M.J., Gao H., Gonzalez D., Gordon A., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Li G., Luo O.J., Park E., Preall J.B., Presaud K., Ribeca P., Robyr D., Ruan X., Sammeth M., Sandu K.S., Schaeffer L., See L.H., Shahab A., Skancke J., Suzuki A.M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Hayashizaki Y., Reymond A., Antonarakis S.E., Hannon G.J., Ruan Y., Carninci P., Sloan C.A., Learned K., Malladi V.S., Wong M.C., Barber G.P., Cline M.S., Dreszer T.R., Heitner S.G., Karolchik D., Kirkup V.M., Meyer L.R., Long J.C., Maddren M., Raney B.J., Grasfeder L.L., Giresi P.G., Battenhouse A., Sheffield N.C., Showers K.A., London D., Bhinge A.A., Shestak C., Schaner M.R., Kim S.K., Zhang Z.Z., Mieczkowski P.A., Mieczkowska J.O., Liu Z., McDaniell R.M., Ni Y., Rashid N.U., Kim M.J., Adar S., Zhang Z., Wang T., Winter D., Keefe D., Iyer V.R., Sandhu K.S., Zheng M., Wang P., Gertz J., Vielmetter J., Partridge E.C., Varley K.E., Gasper C., Bansal A., Pepke S., Jain P., Amrhein H., Bowling K.M., Anaya M., Cross M.K., Muratet M.A., Newberry K.M., McCue K., Nesmith A.S., Fisher-Aylor K.I., Pusey B., DeSalvo G., Parker S.L., Balasubramanian S., Davis N.S., Meadows S.K., Eggleston T., Newberry J.S., Levy S.E., Absher D.M., Wong W.H., Blow M.J., Visel A., Pennachio L.A., Elnitski L., Petrykowska H.M., Abyzov A., Aken B., Barrell D., Barson G., Berry A., Bignell A., Boychenko V., Bussotti G., Davidson C., Despacio-Reyes G., Diekhans M., Ezkurdia I., Frankish A., Gilbert J., Gonzalez J.M., Griffiths E., Harte R., Hendrix D.A., Hunt T., Jungreis I., Kay M., Khurana E., Leng J., Lin M.F., Loveland J., Lu Z., Manthravadi D., Mariotti M., Mudge J., Mukherjee G., Notredame C., Pei B., Rodriguez J.M., Saunders G., Sboner A., Searle S., Sisu C., Snow C., Steward C., Tapanan E., Tress M.L., van Baren M.J., Washieti S., Wilming L., Zadissa A., Zhengdong Z., Brent M., Haussler D., Valencia A., Raymond A., Addleman N., Alexander R.P., Auerbach R.K., Bettinger K., Bhardwaj N., Boyle A.P., Cao A.R., Cayting P., Charos A., Cheng Y., Eastman C., Euskirchen G., Fleming J.D., Grubert F., Habegger L., Hariharan M., Harmanci A., Iyenger S., Jin V.X., Karczewski K.J., Kasowski M., Lacroute P., Lam H., Larnarre-Vincent N., Lian J., Lindahl-Allen M., Min R., Miotto B., Monahan H., Moqtaderi Z., Mu X.J., O’Geen H., Ouyang Z., Patacsil D., Raha D., Ramirez L., Reed B., Shi M., Slifer T., Witt H., Wu L., Xu X., Yan K.K., Yang X., Struhl K., Weissman S.M., Tenebaum S.A., Penalva L.O., Karmakar S., Bhanvadia R.R., Choudhury A., Domanus M., Ma L., Moran J., Victorsen A., Auer T., Centarin L., Eichenlaub M., Gruhl F., Heerman S., Hoeckendorf B., Inoue D., Kellner T., Kirchmaier S., Mueller C., Reinhardt R., Schertel L., Schneider S., Sinn R., Wittbrodt B., Wittbrodt J., Jain G., Balasundaram G., Bates D.L., Byron R., Canfield T.K., Diegel M.J., Dunn D., Ebersol A.K., Frum T., Garg K., Gist E., Hansen R.S., Boatman L., Haugen E., Humbert R., Johnson A.K., Johnson E.M., Kutyavin T.M., Lee K., Lotakis D., Maurano M.T., Neph S.J., Neri F.V., Nguyen E.D., Qu H., Reynolds A.P., Roach V., Rynes E., Sanchez M.E., Sandstrom R.S., Shafer A.O., Stergachis A.B., Thomas S., Vernot B., Vierstra J., Vong S., Weaver M.A., Yan Y., Zhang M., Akey J.A., Bender M., Dorschner M.O., Groudine M., MacCoss M.J., Navas P., Stamatoyannopoulos G., Stamatoyannopoulos J.A., Beal K., Brazma A., Flicek P., Johnson N., Lukk M., Luscombe N.M., Sobral D., Vaquerizas J.M., Batzoglou S., Sidow A., Hussami N., Kyriazopoulou-Panagiotopoulou S., Libbrecht M.W., Schaub M.A., Miller W., Bickel P.J., Banfai B., Boley N.P., Huang H., Li J.J., Noble W.S., Bilmes J.A., Buske O.J., Sahu A.O., Kharchenko P.V., Park P.J., Baker D., Taylor J., Lochovsky L. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan M.S., Barkauskaite E., Lafite P., Knezevic C.E., Brassington A., Ahel M., Hergenrother P.J., Leys D., Ahel I. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat. Commun. 2012;3:878. doi: 10.1038/ncomms1889. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy S.F., Masutani M., Suzuki H., Caldecott K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S., Brockmann C., Mehrotra P.V., Yang J.C., Loakes D., West S.C., Ahel I., Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat. Struct. Mol. Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J., Kranaster R., Altmeyer M., Marx A., Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J., Popp O., Malanga M., Beneke S., Markovitz D.M., Ferrando-May E., Burkle A., Kappes F. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49:7119–7130. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati S., Formentini L., Wang Z.Q., Moroni F., Chiarugi A. Poly(ADP-ribosyl)ation regulates heat shock factor-1 activity and the heat shock response in murine fibroblasts. Biochem. Cell Biol. 2006;84:703–712. doi: 10.1139/o06-083. [DOI] [PubMed] [Google Scholar]
- Friedemann J., Grosse F., Zhang S. Nuclear DNA helicase II (RNA helicase A) interacts with Werner syndrome helicase and stimulates its exonuclease activity. J. Biol. Chem. 2005;280:31303–31313. doi: 10.1074/jbc.M503882200. [DOI] [PubMed] [Google Scholar]
- Gagne J.P., Haince J.F., Pic E., Poirier G.G. Affinity-based assays for the identification and quantitative evaluation of noncovalent poly(ADP-ribose)-binding proteins. Methods Mol. Biol. 2011;780:93–115. doi: 10.1007/978-1-61779-270-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.P., Hunter J.M., Labrecque B., Chabot B., Poirier G.G. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.P., Isabelle M., Lo K.S., Bourassa S., Hendzel M.J., Dawson V.L., Dawson T.M., Poirier G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.P., Pic E., Isabelle M., Krietsch J., Ethier C., Paquet E., Kelly I., Boutin M., Moon K.M., Foster L.J., Poirier G.G. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012;40:7788–7805. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble M.J., Kraus W.L. Multiple facets of the unique histone variant macroH2A: from genomics to cell biology. Cell Cycle. 2010;9:2568–2574. doi: 10.4161/cc.9.13.12144. [DOI] [PubMed] [Google Scholar]
- Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W., Greninger P., Thompson I.R., Luo X., Soares J., Liu Q., Iorio F., Surdez D., Chen L., Milano R.J., Bignell G.R., Tam A.T., Davies H., Stevenson J.A., Barthorpe S., Lutz S.R., Kogera F., Lawrence K., McLaren-Douglas A., Mitropoulos X., Mironenko T., Thi H., Richardson L., Zhou W., Jewitt F., Zhang T., O’Brien P., Boisvert J.L., Price S., Hur W., Yang W., Deng X., Butler A., Choi H.G., Chang J.W., Baselga J., Stamenkovic I., Engelman J.A., Sharma S.V., Delattre O., Saez-Rodriguez J., Gray N.S., Settleman J., Futreal P.A., Haber D.A., Stratton M.R., Ramaswamy S., McDermott U., Benes C.H. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieni R.S., Ismail I.H., Campbell S., Hendzel M.J. Polycomb group proteins in the DNA damage response: a link between radiation resistance and “stemness”. Cell Cycle. 2011;10:883–894. doi: 10.4161/cc.10.6.14907. [DOI] [PubMed] [Google Scholar]
- Goenka S., Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc. Natl. Acad. Sci. USA. 2006;103:4210–4215. doi: 10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka S., Cho S.H., Boothby M. Collaborator of Stat6 (CoaSt6)-associated poly(ADP-ribose) polymerase activity modulates Stat6-dependent gene transcription. J. Biol. Chem. 2007;282:18732–18739. doi: 10.1074/jbc.M611283200. [DOI] [PubMed] [Google Scholar]
- Gottschalk A.J., Timinszky G., Kong S.E., Jin J., Cai Y., Swanson S.K., Washburn M.P., Florens L., Ladurner A.G., Conaway J.W., Conaway R.C. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grofte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S., Mailand N., Neumann B., Heriche J.K., Shearer R., Saunders D., Bartek J., Lukas J., Lukas C. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- Guettler S., LaRose J., Petsalaki E., Gish G., Scotter A., Pawson T., Rottapel R., Sicheri F. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- Haince J.-F., Kozlov S., Dawson V.L., Dawson T.M., Hendzel M.J., Lavin M.F., Poirier G.G. Ataxia Telangiectasia Mutated (ATM) Signaling Network Is Modulated by a Novel Poly(ADP-ribose)-dependent Pathway in the Early Response to DNA-damaging Agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- Haince J.F., McDonald D., Rodrigue A., Dery U., Masson J.Y., Hendzel M.J., Poirier G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- Hall J.R., Kow E., Nevis K.R., Lu C.K., Luce K.S., Zhong Q., Cook J.G. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol. Biol. Cell. 2007;18:3340–3350. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.P., Tang Y.H., Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- Han W.D., Zhao Y.L., Meng Y.G., Zang L., Wu Z.Q., Li Q., Si Y.L., Huang K., Ba J.M., Morinaga H., Nomura M., Mu Y.M. Estrogenically regulated LRP16 interacts with estrogen receptor alpha and enhances the receptor’s transcriptional activity. Endocr. Relat. Cancer. 2007;14:741–753. doi: 10.1677/ERC-06-0082. [DOI] [PubMed] [Google Scholar]
- Harris J.L., Jakob B., Taucher-Scholz G., Dianov G.L., Becherel O.J., Lavin M.F. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum. Mol. Genet. 2009;18:4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- Hazrati A., Ramis-Castelltort M., Sarkar S., Barber L.J., Schofield C.J., Hartley J.A., McHugh P.J. Human SNM1A suppresses the DNA repair defects of yeast pso2 mutants. DNA Repair (Amsterdam) 2008;7:230–238. doi: 10.1016/j.dnarep.2007.09.013. [DOI] [PubMed] [Google Scholar]
- He F., Tsuda K., Takahashi M., Kuwasako K., Terada T., Shirouzu M., Watanabe S., Kigawa T., Kobayashi N., Guntert P., Yokoyama S., Muto Y. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586:3858–3864. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T., Nishizuka Y., Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 1968;243:3553–3555. [PubMed] [Google Scholar]
- Hori K., Fostier M., Ito M., Fuwa T.J., Go M.J., Okano H., Baron M., Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- Hottiger M.O., Hassa P.O., Luscher B., Schuler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C.J., Mickanin C., Myer V., Fazal A., Tomlinson R., Serluca F., Shao W., Cheng H., Shultz M., Rau C., Schirle M., Schlegl J., Ghidelli S., Fawell S., Lu C., Curtis D., Kirschner M.W., Lengauer C., Finan P.M., Tallarico J.A., Bouwmeester T., Porter J.A., Bauer A., Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Ibrahim Y.H., Garcia-Garcia C., Serra V., He L., Torres-Lockhart K., Prat A., Anton P., Cozar P., Guzman M., Grueso J., Rodriguez O., Calvo M.T., Aura C., Diez O., Rubio I.T., Perez J., Rodon J., Cortes J., Ellisen L.W., Scaltriti M., Baselga J. PI3K Inhibition Impairs BRCA1/2 Expression and Sensitizes BRCA-Proficient Triple-Negative Breast Cancer to PARP Inhibition. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles N., Rulten S., El-Khamisy S.F., Caldecott K.W. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell Biol. 2007;27:3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabelle M., Gagne J.P., Gallouzi I.E., Poirier G.G. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 2012 doi: 10.1242/jcs.106963. [DOI] [PubMed] [Google Scholar]
- Ismail I.H., Gagne J.P., Caron M.C., McDonald D., Xu Z., Masson J.Y., Poirier G.G., Hendzel M.J. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40:5497–5510. doi: 10.1093/nar/gks222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S., Kanno S., Ariyoshi M., Tochio H., Ito Y., Yasui A., Shirakawa M. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- Javle M., Curtin N.J. The role of PARP in DNA repair and its therapeutic exploitation. Br. J. Cancer. 2011;105:1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Tulin A.V. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Tulin A.V. Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nat. Commun. 2012;3:760. doi: 10.1038/ncomms1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvekar A., Burga L.N., Hu H., Lunsford E.P., Ibrahim Y.H., Balmana J., Rajendran A., Papa A., Spencer K., Lyssiotis C.A., Nardella C., Pandolfi P.P., Baselga J., Scully R., Asara J.M., Cantley L.C., Wulf G.M. Combining a PI3K Inhibitor with a PARP Inhibitor Provides an Effective Therapy for BRCA1-Related Breast Cancer. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagne J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G., Dawson V.L., Dawson T.M. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S., Kuzuoka H., Sasao S., Hong Z., Lan L., Nakajima S., Yasui A. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., Bycroft M., Ladurner A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima L., Idogawa M., Mita H., Shitashige M., Yamada T., Ogi K., Suzuki H., Toyota M., Ariga H., Sasaki Y., Tokino T. CHFR protein regulates mitotic checkpoint by targeting PARP-1 protein for ubiquitination and degradation. J. Biol. Chem. 2012;287:12975–12984. doi: 10.1074/jbc.M111.321828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Khoronenkova S.V., Dianov G.L. The emerging role of Mule and ARF in the regulation of base excision repair. FEBS Lett. 2011;585:2831–2835. doi: 10.1016/j.febslet.2011.06.015. [DOI] [PubMed] [Google Scholar]
- Kiehlbauch C.C., Aboul-Ela N., Jacobson E.L., Ringer D.P., Jacobson M.K. High resolution fractionation and characterization of ADP-ribose polymers. Anal. Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- Kim I.K., Kiefer J.R., Ho C.M., Stegeman R.A., Classen S., Tainer J.A., Ellenberger T. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nat. Struct. Mol. Biol. 2012;19:653–656. doi: 10.1038/nsmb.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.K., Dudognon C., Smith S. Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. 2012;13:724–732. doi: 10.1038/embor.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H., Poreba E., Lesniewicz K., Hassa P.O., Hottiger M.O., Litchfield D.W., Shilton B.H., Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Kleppa L., Mari P.O., Larsen E., Lien G.F., Godon C., Theil A.F., Nesse G.J., Wiksen H., Vermeulen W., Giglia-Mari G., Klungland A. Kinetics of endogenous mouse FEN1 in base excision repair. Nucleic Acids Res. 2012;40:9044–9059. doi: 10.1093/nar/gks673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh D.W., Lawler A.M., Poitras M.F., Sasaki M., Wattler S., Nehls M.C., Stoger T., Poirier G.G., Dawson V.L., Dawson T.M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A.R., Schor I.E., Allo M., Blencowe B.J. When chromatin meets splicing. Nat. Struct. Mol. Biol. 2009;16:902–903. doi: 10.1038/nsmb0909-902. [DOI] [PubMed] [Google Scholar]
- Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R., Kraus W.L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., Wade P.A. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppard J.B., Dong Z., Mackey Z.B., Tomkinson A.E. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A., Todorova T., Ando Y., Chang P. Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol. 2012;9:542–548. doi: 10.4161/rna.19899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levaot N., Voytyuk O., Dimitriou I., Sircoulomb F., Chandrakumar A., Deckert M., Krzyzanowski P.M., Scotter A., Gu S., Janmohamed S., Cong F., Simoncic P.D., Ueki Y., La Rose J., Rottapel R. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.Y., McCulloch R.D., Fenton A.L., Cheung M., Meng L., Ikura M., Koch C.A. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. USA. 2010;107:9129–9134. doi: 10.1073/pnas.1000556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.C., Caceres J.F. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., Lindh M., Acs K., Vrouwe M.G., Pines A., van Attikum H., Mullenders L.H., Dantuma N.P. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 2012;197:267–281. doi: 10.1083/jcb.201106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N.F., Hu L., Fung J.M., Xie D., Zheng B.J., Chen L., Tang D.J., Fu L., Wu Z., Chen M., Fang Y., Guan X.Y. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47:503–510. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- Macrae C.J., McCulloch R.D., Ylanko J., Durocher D., Koch C.A. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amsterdam) 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Malanga M., Althaus F.R. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and Induces DNA strand break resealing. J. Biol. Chem. 2004;279:5244–5248. doi: 10.1074/jbc.C300437200. [DOI] [PubMed] [Google Scholar]
- Malanga M., Althaus F.R. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- Malanga M., Atorino L., Tramontano F., Farina B., Quesada P. Poly(ADP-ribose) binding properties of histone H1 variants. Biochim. Biophys. Acta. 1998;1399:154–160. doi: 10.1016/s0167-4781(98)00110-9. [DOI] [PubMed] [Google Scholar]
- Malanga M., Czubaty A., Girstun A., Staron K., Althaus F.R. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008;283:19991–19998. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- Malet H., Coutard B., Jamal S., Dutartre H., Papageorgiou N., Neuvonen M., Ahola T., Forrester N., Gould E.A., Lafitte D., Ferron F., Lescar J., Gorbalenya A.E., de Lamballerie X., Canard B. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzen M.R., McCraith S.M., Spinelli S.L., Torres F.M., Fields S., Grayhack E.J., Phizicky E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- Matsuno K., Ito M., Hori K., Miyashita F., Suzuki S., Kishi N., Artavanis-Tsakonas S., Okano H. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development. 2002;129:1049–1059. doi: 10.1242/dev.129.4.1049. [DOI] [PubMed] [Google Scholar]
- Messner S., Altmeyer M., Zhao H., Pozivil A., Roschitzki B., Gehrig P., Rutishauser D., Huang D., Caflisch A., Hottiger M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca M.L., Meyer R.G., Jacobson E.L., Jacobson M.K. Poly(ADP-ribose) polymerases: managing genome stability. Int. J. Biochem. Cell Biol. 2005;37:920–926. doi: 10.1016/j.biocel.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Minaga T., Kun E. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. J. Biol. Chem. 1983;258:5726–5730. [PubMed] [Google Scholar]
- Minaga T., Kun E. Spectral analysis of the conformation of polyadenosine diphosphoribose. Evidence indicating secondary structure. J. Biol. Chem. 1983;258:725–730. [PubMed] [Google Scholar]
- Mischo H.E., Hemmerich P., Grosse F., Zhang S. Actinomycin D induces histone gamma-H2AX foci and complex formation of gamma-H2AX with Ku70 and nuclear DNA helicase II. J. Biol. Chem. 2005;280:9586–9594. doi: 10.1074/jbc.M411444200. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O., Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol. Biol. 2007;8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle P.M., Muir T.W. Method for the synthesis of mono-ADP-ribose conjugated peptides. J. Am. Chem. Soc. 2010;132:15878–15880. doi: 10.1021/ja1064312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Veraksa A., Bauer A., Rosse C., Camonis J., Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nat. Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- Murawska M., Hassler M., Renkawitz-Pohl R., Ladurner A., Brehm A. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet. 2011;7:e1002206. doi: 10.1371/journal.pgen.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthurajan U.M., McBryant S.J., Lu X., Hansen J.C., Luger K. The linker region of macroH2A promotes self-association of nucleosomal arrays. J. Biol. Chem. 2011;286:23852–23864. doi: 10.1074/jbc.M111.244871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa K., Ueda K., Honjo T., Yoshihara K., Nishizuka Y., Hayaishi O. Nicotinamide adenine dinucleotide glycohydrolases and poly adenosine diphosphate ribose synthesis in rat liver. Biochem. Biophys. Res. Commun. 1968;32:143–149. doi: 10.1016/0006-291x(68)90360-4. [DOI] [PubMed] [Google Scholar]
- Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y., Ueda K., Nakazawa K., Hayaishi O. Studies on the polymer of adenosine diphosphate ribose. I. Enzymic formation from nicotinamide adenine dinuclotide in mammalian nuclei. J. Biol. Chem. 1967;242:3164–3171. [PubMed] [Google Scholar]
- Nowsheen S., Cooper T., Bonner J.A., Lobuglio A.F., Yang E.S. HER2 overexpression renders human breast cancers sensitive to PARP inhibition independently of any defect in homologous recombination DNA repair. Cancer Res. 2012;72:4796–4806. doi: 10.1158/0008-5472.CAN-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi J., Richards M.W., Crumpler S., Brown N., Blagg J., Bayliss R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING domains (CHFR) J. Biol. Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Ueda K., Kawaichi M., Hayaishi O. Activation of DNA ligase by poly(ADP-ribose) in chromatin. Proc. Natl. Acad. Sci. USA. 1983;80:3604–3607. doi: 10.1073/pnas.80.12.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka J., Ueda K., Hayaishi O., Komura H., Nakanishi K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J. Biol. Chem. 1984;259:986–995. [PubMed] [Google Scholar]
- Oplustilova L., Wolanin K., Mistrik M., Korinkova G., Simkova D., Bouchal J., Lenobel R., Bartkova J., Lau A., O’Connor M.J., Lukas J., Bartek J. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle. 2012;11:3837–3850. doi: 10.4161/cc.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otake H., Miwa M., Fujimura S., Sugimura T. Binding of ADP-ribose polymer with histone. J. Biochem. 1969;65:145–146. [PubMed] [Google Scholar]
- Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeter P.L., Realini C.A., Althaus F.R. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992;31:1379–1385. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- Panzeter P.L., Zweifel B., Malanga M., Waser S.H., Richard M., Althaus F.R. Targeting of histone tails by poly(ADP-ribose) J. Biol. Chem. 1993;268:17662–17664. [PubMed] [Google Scholar]
- Parsons J.L., Tait P.S., Finch D., Dianova I.I., Edelmann M.J., Khoronenkova S.V., Kessler B.M., Sharma R.A., McKenna W.G., Dianov G.L. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 2009;28:3207–3215. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.G., Sarkaria J.N., Kaufmann S.H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl. Acad. Sci. USA. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E., Meyer T., Cimprich K.A. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Pines A., Vrouwe M.G., Marteijn J.A., Typas D., Luijsterburg M.S., Cansoy M., Hensbergen P., Deelder A., de Groot A., Matsumoto S., Sugasawa K., Thoma N., Vermeulen W., Vrieling H., Mullenders L. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 2012;199:235–249. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski Y., Hansen G., Boomaars-van der Zanden A.L., Snijder E.J., Gorbalenya A.E., Hilgenfeld R. Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property. Protein Sci. 2009;18:6–16. doi: 10.1002/pro.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- Poirier G.G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S.E., Kaidi A., Baskcomb L., Galanty Y., Jackson S.P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp O., Veith S., Fahrer J., Bohr V.A., Burkle A., Mangerich A. Site-specific non-covalent interaction of the biopolymer poly(ADP-ribose) with the Werner syndrome protein regulates protein functions. ACS Chem. Biol. 2012 doi: 10.1021/cb300363g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale A., Matteis G.D., Galleazzi G., Zampieri M., Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- Reeder R.H., Ueda K., Honjo T., Nishizuka Y., Hayaishi O. Studies on the polymer of adenosine diphosphate ribose. II. Characterization of the polymer. J. Biol. Chem. 1967;242:3172–3179. [PubMed] [Google Scholar]
- Reh S., Korn C., Gimadutdinow O., Meiss G. Structural basis for stable DNA complex formation by the caspase-activated DNase. J. Biol. Chem. 2005;280:41707–41715. doi: 10.1074/jbc.M509133200. [DOI] [PubMed] [Google Scholar]
- Rouleau M., Patel A., Hendzel M.J., Kaufmann S.H., Poirier G.G. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten S.L., Cortes-Ledesma F., Guo L., Iles N.J., Caldecott K.W. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell Biol. 2008;28:4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten S.L., Fisher A.E., Robert I., Zuma M.C., Rouleau M., Ju L., Poirier G., Reina-San-Martin B., Caldecott K.W. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Sauermann G., Wesierska-Gadek J. Poly(ADP-ribose) effectively competes with DNA for histone H4 binding. Biochem. Biophys. Res. Commun. 1986;139:523–529. doi: 10.1016/s0006-291x(86)80022-5. [DOI] [PubMed] [Google Scholar]
- Saxena A., Saffery R., Wong L.H., Kalitsis P., Choo K.H. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- Schiewer M.J., Goodwin J.F., Han S., Brenner J.C., Augello M.A., Dean J.L., Liu F., Planck J.L., Ravindranathan P., Chinnaiyan A.M., McCue P., Gomella L.G., Raj G.V., Dicker A.P., Brody J.R., Pascal J.M., Centenera M.M., Butler L.M., Tilley W.D., Feng F.Y., Knudsen K.E. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A.A., Pleschke J.M., Kleczkowska H.E., Althaus F.R., Vergeres G. Poly(ADP-ribose) modulates the properties of MARCKS proteins. Biochemistry. 1998;37:9520–9527. doi: 10.1021/bi973063b. [DOI] [PubMed] [Google Scholar]
- Schultheisz H.L., Szymczyna B.R., Williamson J.R. Enzymatic synthesis and structural characterization of 13C, 15N-poly(ADP-ribose) J. Am. Chem. Soc. 2009;131:14571–14578. doi: 10.1021/ja903155s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick D.M., Halazonetis T.D. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- Sellis D., Drosou V., Vlachakis D., Voukkalis N., Giannakouros T., Vlassi M. Phosphorylation of the arginine/serine repeats of lamin B receptor by SRPK1-insights from molecular dynamics simulations. Biochim. Biophys. Acta. 2012;1820:44–55. doi: 10.1016/j.bbagen.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilmann M., Hinz M., Arslan S.C., Zimmer A., Schreiber V., Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol. Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Takeyama K., Aguiar R.C., Gu L., He C., Freeman G.J., Kutok J.L., Aster J.C., Shipp M.A. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 2003;278:21930–21937. doi: 10.1074/jbc.M301157200. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Miwa M., Hayashi K., Kubota K., Matsushima T. Separation of oligo(adenosine diphosphate ribose) fractions with various chain lengths and terminal structures. Biochemistry. 1977;16:1485–1489. doi: 10.1021/bi00626a037. [DOI] [PubMed] [Google Scholar]
- Till S., Ladurner A.G. Sensing NAD metabolites through macro domains. Front Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- Timinszky G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E.H., Scheffzek K., Hottiger M.O., Ladurner A.G. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Ueda K., Oka J., Naruniya S., Miyakawa N., Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 1972;46:516–523. doi: 10.1016/s0006-291x(72)80169-4. [DOI] [PubMed] [Google Scholar]
- Wang Y., Dawson V.L., Dawson T.M. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp. Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kim N.S., Haince J.F., Kang H.C., David K.K., Andrabi S.A., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci. Signal. 2011;4 doi: 10.1126/scisignal.2000902. ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesierska-Gadek J., Sauermann G. The effect of poly(ADP-ribose) on interactions of DNA with histones H1, H3 and H4. Eur. J. Biochem. 1988;173:675–679. doi: 10.1111/j.1432-1033.1988.tb14051.x. [DOI] [PubMed] [Google Scholar]
- West J.D., Ji C., Marnett L.J. Modulation of DNA fragmentation factor 40 nuclease activity by poly(ADP-ribose) polymerase-1. J. Biol. Chem. 2005;280:15141–15147. doi: 10.1074/jbc.M413147200. [DOI] [PubMed] [Google Scholar]
- Wilkin M., Tongngok P., Gensch N., Clemence S., Motoki M., Yamada K., Hori K., Taniguchi-Kanai M., Franklin E., Matsuno K., Baron M. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev. Cell. 2008;15:762–772. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Williamson C.T., Kubota E., Hamill J.D., Klimowicz A., Ye R., Muzik H., Dean M., Tu L., Gilley D., Magliocco A.M., McKay B.C., Bebb D.G., Lees-Miller S.P. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol. Med. 2012;4:515–527. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Li Y., Li X., Ti D., Zhao Y., Si Y., Mei Q., Zhao P., Fu X., Han W. LRP16 integrates into NF-kappaB transcriptional complex and is required for its functional activation. PLoS One. 2011;6:e18157. doi: 10.1371/journal.pone.0018157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Xu Y., Gursoy-Yuzugullu O., Price B.D. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 2012;586:3920–3925. doi: 10.1016/j.febslet.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhao Y.L., Wu Z.Q., Si Y.L., Meng Y.G., Fu X.B., Mu Y.M., Han W.D. The single-macro domain protein LRP16 is an essential cofactor of androgen receptor. Endocr. Relat. Cancer. 2009;16:139–153. doi: 10.1677/ERC-08-0150. [DOI] [PubMed] [Google Scholar]
- Yang K., Moldovan G.L., D’Andrea A.D. RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J. Biol. Chem. 2010;285:19085–19091. doi: 10.1074/jbc.M109.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S., Hamdy F.C., Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72:2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- Yu S.W., Andrabi S.A., Wang H., Kim N.S., Poirier G.G., Dawson T.M., Dawson V.L. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.W., Wang H., Poitras M.F., Coombs C., Bowers W.J., Federoff H.J., Poirier G.G., Dawson T.M., Dawson V.L. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Zampieri M., Guastafierro T., Calabrese R., Ciccarone F., Bacalini M.G., Reale A., Perilli M., Passananti C., Caiafa P. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem. J. 2012;441:645–652. doi: 10.1042/BJ20111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G.A., Schirle M., Shi X., Hild M., Bauer A., Myer V.E., Finan P.M., Porter J.A., Huang S.M., Cong F. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- Zhou Z.D., Chan C.H., Xiao Z.C., Tan E.K. Ring finger protein 146/Iduna is a poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase. Cell Adh. Migr. 2011;5:463–471. doi: 10.4161/cam.5.6.18356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Chen G., Lv F., Wang X., Ji X., Xu Y., Sun J., Wu L., Zheng Y.T., Gao G. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Nat.l Acad. Sci. USA. 2011;108:15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]