Abstract
The assessment of cardiac toxicity is a major challenge in both drug development and clinical trials, and numerous marketed pharmaceuticals have been removed from the market due to unpredicted cardiac effects. Serum troponins are widely-used indicators of cardiac injury; however, they are short-lived, and have not been validated in preclinical animal models. In this study, we have used filter-aided sample preparation (FASP) and tandem mass tag (TMT) labeling to investigate serum protein alterations in isoproterenol-treated African green monkeys. Our results showed that the combination of FASP and TMT labeling provided highly reproducible and efficient sample preparation, which enables us to identify and quantify serum proteins with high confidence. We focused on the proteins that exhibit long-term alteration upon isoproterenol injection and discovered nine proteins exhibiting significant changes at 48 and 72 hr post-dosing. We further chose three proteins, serum amyloid A (SAA), frutose biphosphate aldolase A (FBAA), and fetuin A, for validation using enzyme-linked immunosorbent assay (ELISA). The serum concentration of SAA showed a ~50 fold increase, while concentration of FBAA and fetuin A exhibited a significant decrease accompanying isoproterenol-induced cardiotoxicity. This work provides valuable insights for multi-marker evaluation of long-term cardiac injury.
Keywords: Tandem Mass Tag, Proteomics, Cardiotoxicity, Serum Biomarkers, African Green Monkey, Drug Safety
Introduction
Negative cardiovascular events are responsible for ~28% of drug withdrawals in the USA,1 exposing an immense need for sensitive and efficient evaluation of cardiac safety during drug development. Fenfluramine/phentermine, terfenadine, astemizole, grepafloxacin, cisapride, rofecoxib, thioridazine, pergolide, tegaserod, sibutramine, propoxyphene and rosiglitazone are all examples of marketed drugs withdrawn in the past 20 years due to adverse cardiac effects. Ideally, potential adverse cardiac effects would be detected during preclinical development, before advancing to human clinical trials; however, there are few preclinical biomarkers available for identification of potential cardiotoxicity of candidate therapeutics. Serum troponins are available, but not routinely evaluated, in preclinical toxicology studies; however, troponins are short-lived and often provide positive results only within the first 24 hr of administration, and are therefore not amenable to incorporation into routine toxicology study designs. The US Food and Drug Administration’s (FDA) Critical Path Initiative has addressed the importance of improved safety biomarkers, and has advocated for the identification of new preclinical biomarkers of cardiac safety.
Serum biomarkers have generated enormous interest for their potential as sensitive indicators of cardiotoxicity. Troponins I and T, myoglobin, creatine phosphokinase muscle and brain types (CK-MB), lactate dehydrogenase (LDH), and C-reactive proteins (CRP) have been suggested as possible cardiac safety biomarkers.2–5 The major problem with these markers is that they typically exhibit significant changes only during the first 12–24 hr, but typically return to normal ranges by 24 hr, making detection and evaluation of cardiac effects difficult. 6 Troponins I and T provide good sensitivity, making them the current standard-of-care for detection of cardiac injury;7, 8 however, recent findings demonstrate that cardiac troponins can be released due to different types of cardiac damage, limiting their value in identifying the real risk and revealing the mechanism of cardiac toxicity.9 Therefore, there is urgent need to discover more specific and sensitive cardiac biomarkers that can be detected over longer periods of time.
Isoproterenol (ISO) is a beta-adrenergic agonist used to treat bradycardia or heart block that has been widely used as a model compound to induce cardiac muscle injury with myocardial ischemia.10, 11 ISO has been shown to induce cardiac injury in a variety of preclinical animal models including non-human primates (NHP).12, 13 While various studies have focused on utilizing ISO treated animal models to analyze cardiotoxicity by testing existing markers, neither a systematic study of post-dose serum protein changes nor discovery of novel long-term markers with improved sensitivity has been carried out in ISO-induced cardiac injury models.
Quantitative proteomics, especially multiplex quantitative proteomics such as isobaric tags for relative and absolute quantitation (iTRAQ), tandem mass tag (TMT) labeling, etc., have made remarkable progress in high-throughput protein quantitation and novel biomarker discovery.14–17 In this study, we present a 2D LC-MS/MS based quantitative proteomic analysis to profile and quantify serum proteins from ISO-administrated African green monkeys at five post-dose time points, with validation of selected biomarker candidates using enzyme-linked immunosorbent assay (ELISA). A filter-aided sample preparation (FASP) strategy was applied to extract and digest proteins from individual samples, followed by TMT labeling. Labeled peptides were mixed and purified through strong cation exchange (SCX) and reverse phase (RP) chromatography. The purified peptides were then separated and analyzed by 2D-nano-LC-MS/MS. We evaluated the yield and reproducibility of each step from four biological samples with two technical replicates of each, and our results showed that a strategy combining FASP and TMT labeling can provide highly reproducible and reliable data for protein identification and quantitation. Five increased proteins and four decreased proteins were discovered with significant changes at 48 and 72 hr post-dose. We further applied an ELISA assay to validate the changes of three proteins including serum amyloid A (SAA), fructose-biphosphate aldolase A (FBAA) and fetuin A, and we detected consistent changes with results from LC-MS/MS. To our knowledge, this is the first study where a TMT-based quantitative proteomic strategy has been applied to investigate cardiotoxicity-associated protein alterations at different time points. Our results suggest that the FASP- and TMT-based strategy can provide efficient sample preparation and reproducible data for biomarker discovery, and changed proteins SAA, FBAA, and fetuin A may be potential serum biomarkers for the stratification of cardiotoxicity.
Experimental Procedures
Animal Information
Four non-naïve African green monkeys (Chlorocebus sabaeus, unknown origin, naïve to isoproterenol), with ages ranging from 3.5 to 8 years were assigned to the study. Isoproterenol hydrochloride (ISO, Sigma Aldrich, St. Louis, MO) was administered as a single subcutaneous (s.c.) dose of 4 mg/kg (concentration of 2 mg/ml) to two males and two females on Day 1 of the study. Blood for analysis of protein biomarkers was collected pre-dose, and at 1, 4, 24, 48, and 72 hr post-dose and processed to serum. Samples were snap frozen in liquid nitrogen and stored at −80°C until analysis.
Protein Extraction and Digestion
Filter-aided sample preparation (FASP), developed by Mann’s group,18 was applied to extract and digest serum proteins from 24 individual samples. Briefly, 5 µL of serum was diluted 50 times by 8 M urea in 0.1 M Tris/HCl pH 8.5 then filtered with a 0.22 µM membrane (Millipore, Billerica, MA, USA). The flow-through was collected and transferred into a 1.5 mL Microcon YM-10 centrifugal unit (Millipore, Billerica, MA, USA). Protein reduction, alkylation and tryptic digestion were performed step by step in the centrifugal unit. After overnight digestion at 37°C, the peptides were eluted twice with 150 µL 0.1% formic acid (FA). The concentration of proteins and peptides collected in each step was measured using Nano-drop 2000 (Thermo Scientific, USA). The digested peptides were then aliquoted, dried, and stored at −80°C until further use.
Tandem Mass Tag (TMT) labeling
Fifty micrograms of peptides from each sample were used for Amine-reactive 6-plex TMT labeling (Thermo Scientific, USA). Peptides from each time point (hr) taken from the same animal were labeled with one of the 6-plex TMT tags (Supplemental Table S1). The labeling experiment was performed essentially according to manufacturer’s protocol. The peptides from each sample were redissolved in dissolution buffer and then incubated with a specific TMT tag for 1 hr at room temperature. To each sample, 8 µL of 5% hydroxylamine was added and incubated for 15 minutes to quench the reaction. Samples with different tags were then mixed for the following peptide purification.
Excess TMTs were removed from the labeled peptides by a 3×8 mm SCX trap column (Bruker-Michrom, CA, USA). Peptides were eluted by 0.5 M NaCl in 0.1% FA. Collected peptides were then desalted by a 3×8 mm C18 RP trap column (Bruker-Michrom, CA, USA). 98% acetontrile in 0.1% FA was used to elute desalted peptides from the RP trap column. Peptide purification was performed on Shimazdu fast protein liquid chromatography (FPLC) system (Shimazdu, CA, USA) at a flow rate of 0.5 mL/min, and the absorbance was monitored at 280 and 220 nm. Sample containing purified peptides was dried and redissolved in 0.1% FA for LC-MS/MS analysis.
Nano-LC-MS/MS
Peptides labeled with TMTs were separated using a nano-LC system (Agilent 1200, Palo Alto, CA, USA) that was equipped with a self-packed 5 µm, 300Å polysulfoethyl A SCX column (100 µm × 100mm) and a 5 µm, 300Å C18 reverse phase column (75 µm × 150mm). The salt steps were 5, 15, 25, 50, 75, 100, 200, 500, and 1500 mM ammonium formate, each of which was followed by a 146-min reverse phase gradient from 2 to 97% acetonitrile in 0.1% FA at a constant flow rate of 300 nl/min.
Mass spectra were recorded on an LTQ-XL Orbitrap ETD (Thermo Fisher Scientific, San Jose, CA, USA). We applied a CID-HCD acquisition method reported by Mechtler et al.19 for separation, identification, and quantitation. For each cycle of one full mass scan (range of m/z from 350 to 1800), the four most intense ions in the spectrum were selected for CID (collision induced dissociation) and HCD (higher-energy collisional dissociation) analysis, unless they appeared in the dynamic or mass exclusion lists. Four CID scans (automated gain control [AGC] target value 1×104, maximum inject time 100 ms, minimum signal threshold 500 counts, collison energy 35%, activation time 30 ms, isolation width 2.0 m/z) were used for MS/MS identification and 4 corresponding HCD scans (AGC target value 3×105, maximum inject time 300 ms, minimum signal threshold 500 counts, collision energy 60%, activation time 30 ms, isolation width 2.0 m/z) were used for quantitation. Data were acquired in data-dependent mode using Xcalibur 2.2 software.
Peptide Identification and Quantitation
All MS/MS spectra were searched against a decoy database20 generated from Macaca (downloaded from NCBI April 18th, 2011). The search was performed using SEQUEST algorithm incorporated in Proteome Discoverer 1.3.0.339 (Thermo Finnigan, CA, USA). The search parameters were as follows: (1) tryptic digestion, allow up to 1 missed cleavage; (2) fixed modification, carbamidomethyl of C; (3) variable modification, oxidation of M; (4) variable modification, TMT of N-terminal; (5) variable modification, TMT of K and Y; (6) peptide ion mass tolerance 13 ppm; (7) fragment ion mass tolerance 0.8 Da; (8) peptide charges +1, +2, and +3. The identified peptides were searched against the decoy database and the false positive rate (FDR) was set as 0.05 using Percolator scoring with posterior error probability validation.
Peptide quantitation was also performed in Proteome Discoverer 1.3.0.339. A TMT 6-plex quantitation method was used for HCD-based quantitation. Mass tolerance was 797 ppm for reporter TMT tags. The intensity of each peptide was normalized to protein median intensity before comparing the ratio of same peptides with different tags. Both peptide identification and quantitation were performed in an overall workflow in Proteome Discoverer.
ELISA Validation
An ELISA assay was applied to validate the changes of selected proteins to confirm the MS results. Human FBAA ELISA kit was purchased from Antibody Online (GA, USA); monkey amyloid A and human fetuin A ELISA kits were purchased from Kamiya Biomedical (WA, USA). All ELISA assays were performed according to manufacturer’s protocols. Each serum sample was tested in triplicates and the average concentration was used for data analysis.
Results
Cardiotoxicity Evaluations
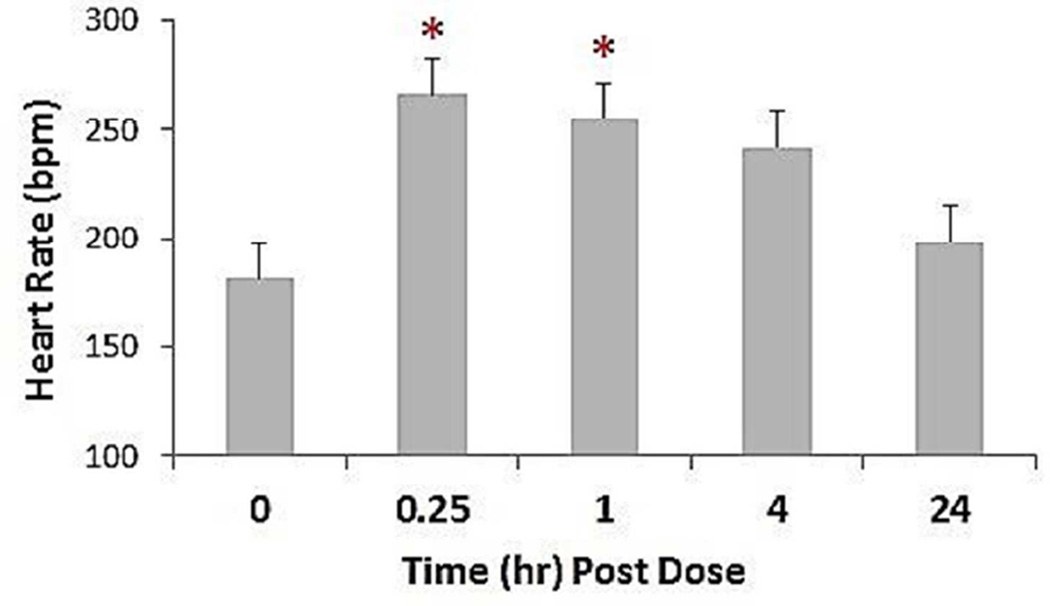
Monkeys treated with ISO demonstrated a variety of adverse cardiac effects. Heart rate, as measured by electrocardiograms (ECG), increased within 15 min post-treatment and remained elevated for up to 4 hr (Figure 1). Cardiac abnormalities observed during the first 24 hr post-dose included paroxysmal ventricular tachycardia, ST segment elevation, occasional second-degree atrioventricular block with atrioventricular junctional escape complexes, occasional second-degree atrioventricular block (Mobitz type I) with a ventricular escape complex, ventricular premature complexes, frequent atrial premature complexes, and hypotension. A majority of the treatment-related findings were noted within 15–60 min post-treatment.
Figure 1. Heart rate in African green monkeys following a single s.c. administration of 4 mg/kg isoproterenol (ISO).
Heart rate values were obtained from EKG recordings collected at the following time points: immediately prior to dose administration (time 0) during the first 15 min after dosing, then at 1, 4 and 24 hr post-dose. Results for males and females showed no significant differences and are pooled. *p<0.05
Serum sample preparation
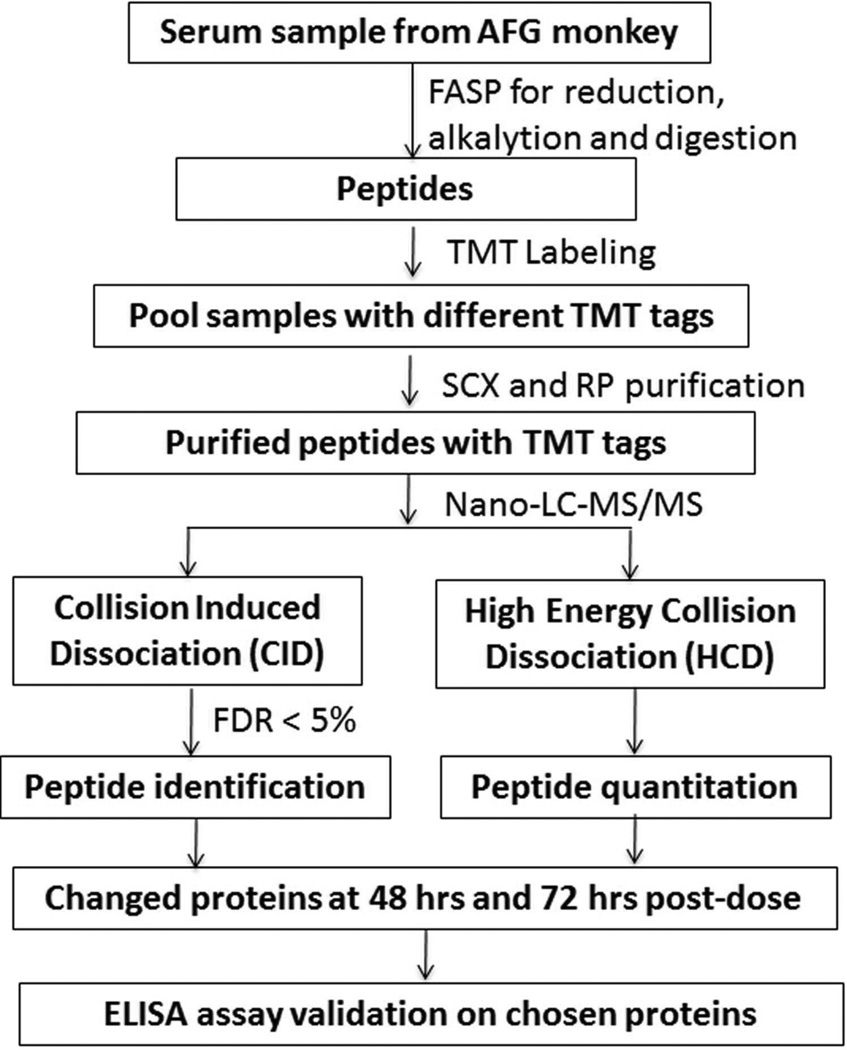
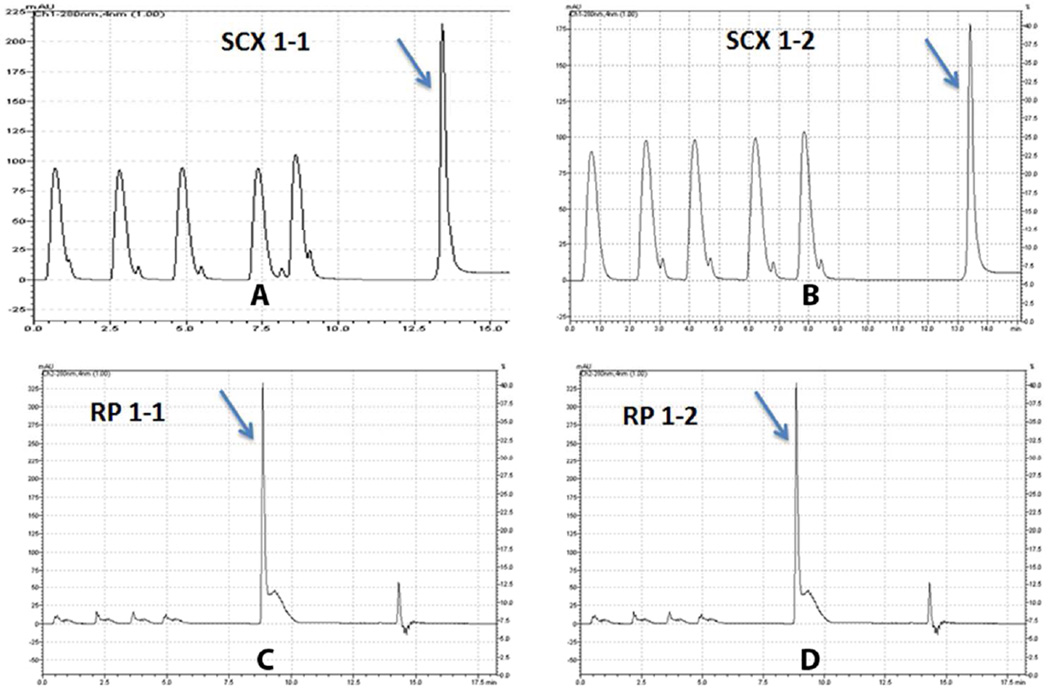
The overall workflow strategy is shown in Figure 2. We monitored sample yields and reproducibility in each step during sample preparation due to the importance of both in quantitative proteomic analysis. A total of 24 samples were processed in replicate to evaluate the reproducibility of sample handling and the subsequent mass spectrometry analysis. The protein concentration of original samples was 58.15 ± 5.44 g/L. The yields of FASP, SCX, and RP steps are 64.1% ± 9%, 63.0 ± 2.5%, and 79.0 ± 3.0%, respectively (see Supplemental Table S2). SCX and RP high performance liquid chromatography (HPLC) chromatograms from two technical replicates are shown in Figure 3.
Figure 2. Workflow of proteomic strategy.
TMT-based serum protein profiling and quantitation were applied to study long-term cardiac injury following isoproterenol administration.
Figure 3. SCX and RP chromatogram of TMT labeled peptides from two technical replicates of animal1.
(A) and (B) indicate chromatogram from two independent SCX runs and (C) and (D) indicate chromatogram from two RP runs. The absorbance was monitored at 280 nm and peptide peak is identified by arrow in the figure.
Protein identification and quantitation
To study protein changes after ISO administration, we labeled peptides from six different time points with six TMT tags, respectively. Four monkeys (biological replicates) were tested with two technical replicates for each monkey. Labeled peptides were then separated and identified by 2D LC-MS/MS. CID spectra generated in the linear ion trap were used for identification, while the HCD spectra acquired in the Orbitrap with higher collision energy and accuracy were used for reporter ion–based quantitation. Tandem mass spectra of peptides with >95% confidence were used for protein identification and quantitation. The spectra were compared to a protein database downloaded from NCBI based on the Macaca genome, with a 5% false discovery rate21 cut-off based on the reverse database decoy search. Perculator was applied to further screen peptides with posterior error probability (PEP) of less than 0.05.22 PEP was calculated based on peptide match of each spectrum. It has been reported that if the threshold of PEP is 0.05, the actual FDR will be much smaller than 0.05.23 We applied both FDR and PEP because FDR is good for peptide identification because it measures all peptide confidence analysis, while PEP is good for peptide quantitation because it measures the confidence of each matched spectrum.
We also evaluated the protein-level FDR using a targeted-decoy search strategy24, 25. Briefly, a concatenated target decoy database of macaca was generated using Perl, where the decoy IDs have a prefix of "rev-". All raw data were searched against the target decoy database using Proteome Discoverer. The protein-level FDR was calculated in the following way: FDR = 2 × decoy hits/total hits. In our study, the protein-level FDR was ~8% (5% with PEP validation). This is consistent with many quantitative studies which use isotopic tag labeling strategy26, 27. The molecular weight of TMT tag is ~229 Da. During MS/MS analysis, the TMT tag gererates report ions through CID/HCD fragmentation while at the same time it also generates unknown peaks which will be considered as unmatched peaks during spectum analysis. To make sure the changed proteins were correctly identified and quantified, all CID and HCD spectra of these proteins were manually inspected.
The 6-plex TMT quantitation was achieved through the reporter ions in HCD mass spectra. Each of the six tags has a specific molecular weight that appears at m/z of 126.1, 127.1, 128.1, 129.1, 130.1, and 131.1, respectively. Peptides at the 0 hr time point were used as controls and the relative abundance of each peptide at each time point (1, 4, 24, 48, or 72 hr) was indicated by the ratio of the intensity of a specific tag to that of the corresponding control tag on the same peptide. The ratio of each protein was determined by the average ratio of its matched peptides. Both unique and non-unique peptides were used for the protein quantitation.28
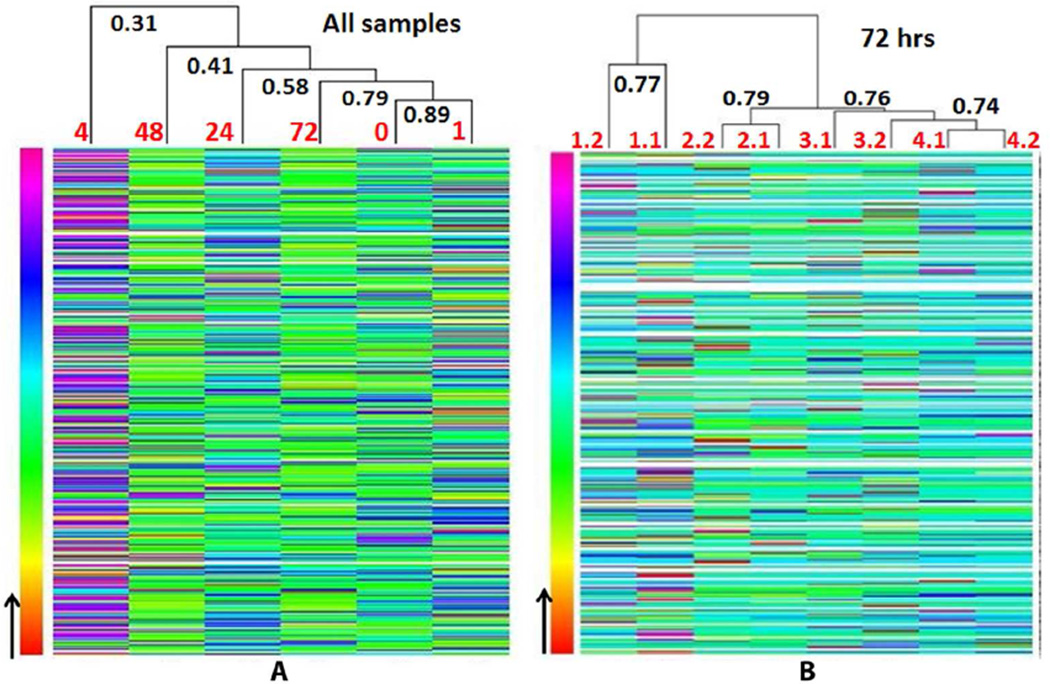
A total of 384 proteins were identified and quantitated. We first analyzed the correlation between technical replicates (independent sample processing on the same sample) and biological replicates (four monkeys). The correlation coefficient (R) between two technical replicates of the same sample was ~0.7–0.8 while R between each biological sample was ~0.3–0.4. A representative heat-map of correlation between different runs at the 72 hr time point is shown in Figure 4A. We then looked at the correlation of protein abundances between different time points (Figure 4B, analyzed by R program). The correlation between control and 1 hr or 72 hr are higher than correlation between control and other time points. The highest correlation was observed between control and 1 hr (R = 0.86) while the lowest correlation was observed between control and 4 hr (R = 0.31).
Figure 4. Heatmap of LC-MS/MS results.
(A) Correlation of protein abundance between different time points. The correlation between 0 hr and 72 hr is higher than the correlation between 0 hr and 4 hr, indicating that protein changes induced by isoproterenol peaked at 4 hr, then returned to normal ranges after 24 hr. (B) Correlation of protein abundance between technical replicates and biological replicates. 1.1 and 1.2, 2.1 and 2.2, 3.1 and 3.2, and 4.1 and 4.2 indicate two replicates from each of four biological samples, respectively. Correlation coefficient (R) is shown in bold.
To select proteins which can exhibit long-term changes after isoproterenol treatment, we focused on proteins with significant changes at 48 hr and 72 hr compared with the 0 hr time point (p < 0.05) across all technical and biological replicates. Student t-test was applied to screen the altered proteins with an average change >2-fold. All CID spectra of selected proteins were manually inspected (Supplemental Figure S1) to ensure accurate identification. We also performed manual inspection of HCD spectra of each quantified peptides of the changed proteins to ensure quantitation is highly confident (Figure 5).
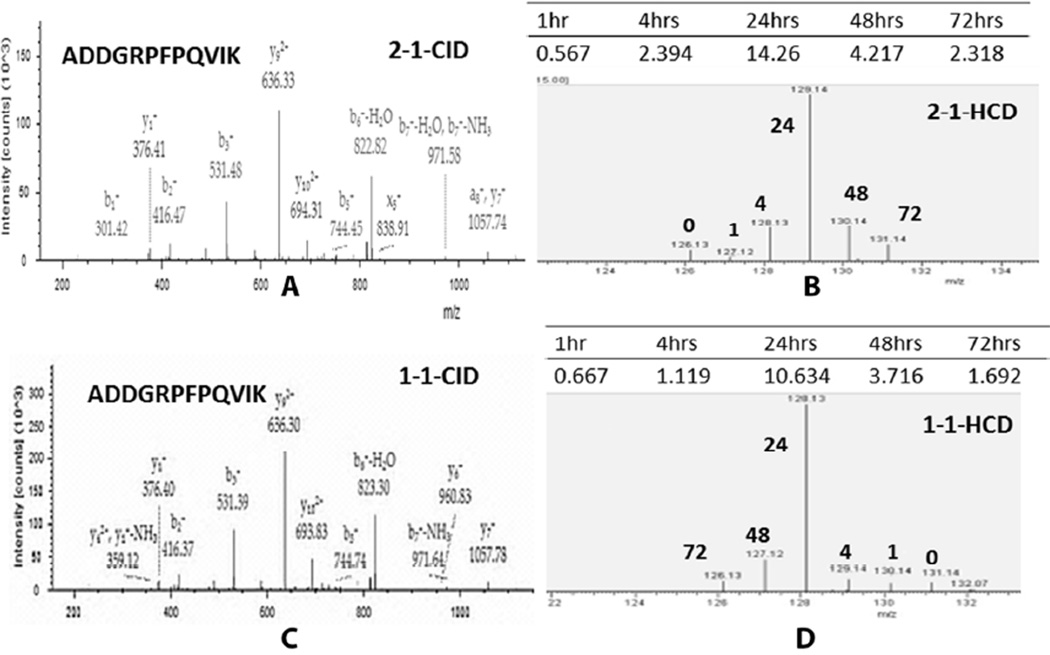
Figure 5. MS/MS spectra of “ADDGRPFPQVIK” of FBAA collected by CID and HCD.
(A) and (C) are CID spectra from the same peptide from two different biological samples. The CID spectra were used for peptide identification. (B) and (D) are corresponding HCD spectra of TMTs. The intensity of each tag was used for peptide quantitation. Forward and reverse labeling was applied on different samples and the corresponding time point of each tag is indicated in bold.
Five proteins, including beta-enolase–like protein, FBAA, glycogen phosphorylase, SAA, and peroxiredoxin-2 isoform 5 were significantly up-regulated, while another four proteins, including fetuin A, apolipoprotein A-I, apolipoprotein A-II and InaD-like protein, were down regulated at 48 and 72 hr after ISO treatment (Table 1 and Supplemental Table S3 & Figure S1). We also investigated the protein changes at the other time points. Six increased proteins and four decreased proteins were discovered from 4 hr post-dose (Table 2 and Supplemental Table S4 & Figure S2) but their expression levels reverted to baseline after 48 hr.
Table 1.
Proteins demonstrating significant changes 48 and 72 hr after treatment with isoproterenol. Values shown represent fold-increases over 0 hr values.
| Accession | Description | 0 hr | 1hr | 4hr | 24hr | 48hr | 72hr |
|---|---|---|---|---|---|---|---|
| 109113005 | beta-enolase-like | 1.00 | 1.61 | 5.19 | 19.63 | 10.39 | 6.11 |
| 109128138 | fructose-bisphosphate aldolaseA isoform1 | 1.00 | 0.83 | 1.47 | 8.87 | 6.08 | 3.61 |
| 297267491 | glycogen phosphorylase, muscle form isoform 1 | 1.00 | 1.15 | 1.70 | 3.98 | 3.72 | 3.26 |
| 109107123 | serum amyloid A protein-like isoform 1 | 1.00 | 0.77 | 0.78 | 3.33 | 7.61 | 5.99 |
| 109123563 | peroxiredoxin-2 isoform 5 | 1.00 | 0.83 | 1.01 | 17.50 | 3.21 | 6.25 |
| 109042277 | fetuin A | 1.00 | 1.06 | 1.10 | 0.60 | 0.40 | 0.38 |
| 109108768 | apolipoprotein A-I isoform 1 | 1.00 | 1.22 | 1.02 | 0.48 | 0.34 | 0.34 |
| 109017814 | apolipoprotein A-II isoform 2 | 1.00 | 0.94 | 1.13 | 1.09 | 0.44 | 0.39 |
| 297278839 | inaD-like protein-like | 1.00 | 0.20 | 0.15 | 0.46 | 0.51 | 0.49 |
Table 2.
Proteins demonstrating significant changes 4 hr after treatment with isoproterenol. Values shown represent fold-increases over 0 hr values.
| Accession | Description | 0 hr | 1hr | 4hr | 24hr | 48hr | 72hr |
|---|---|---|---|---|---|---|---|
| 109017524 | c-reactive protein isoform 2 | 1.00 | 1.44 | 2.39 | 4.20 | 2.26 | 1.04 |
| 109093377 | heparin cofactor 2 isoform 2 | 1.00 | 6.14 | 4.59 | 3.10 | 0.88 | 0.89 |
| 109126335 | alpha-1B-glycoprotein | 1.00 | 13.12 | 13.32 | 5.57 | 1.89 | 2.22 |
| 109110480 | alpha-1-acid glycoprotein 1-like isoform 2 | 1.00 | 3.32 | 2.52 | 4.38 | 1.45 | 1.36 |
| 109022951 | complement factor H-like isoform 2 | 1.00 | 6.46 | 4.49 | 4.62 | 1.13 | 0.98 |
| 109019000 | complement factor H-like isoform 3 | 1.00 | 3.09 | 2.47 | 2.50 | 1.35 | 1.39 |
| 113461945 | vitamin K-dependent protein S | 1.00 | 0.47 | 0.46 | 0.64 | 1.13 | 0.87 |
| 189011659 | prothrombin | 1.00 | 0.44 | 0.52 | 0.61 | 0.83 | 0.77 |
| 297278313 | apolipoprotein C-II isoform 2 | 1.00 | 0.45 | 0.48 | 0.34 | 0.91 | 0.98 |
ELISA Validation
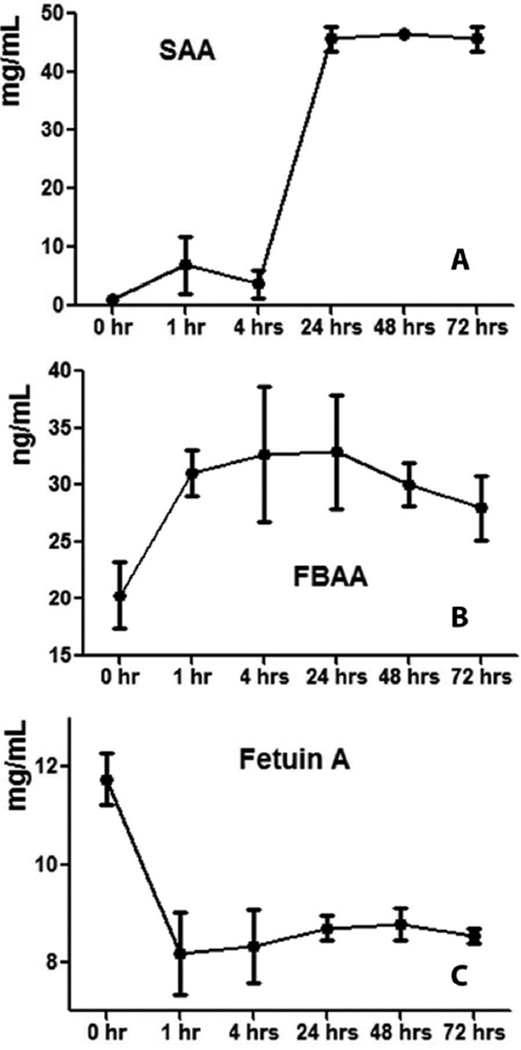
Three proteins, FBAA, SAA, and fetuin A, were validated by ELISA assay. The ELISA results were consistent with the LC-MS/MS data. FBAA and SAA were significantly up regulated, while fetuin A was down-regulated at 48 and 72 hr post-dose (Figure 6 and Supplemental Table S5). The concentrations of FBAA at 0, 48, and 72 hr after ISO administration were 20.3 ± 2.9, 30.0 ± 2.0 and 28.0 ± 2.8 ng/mL, respectively. Serum concentrations of SAA at 48 and 72 hr post-dose were 46.6 ± 0.3 and 45.7 ± 2.2 mg/mL, which are significantly higher than the level at 0 hr (1.0 ± 0.2 mg/mL). The expression level of fetuin A was significantly lower at 48 hr (8.8 ± 0.3 mg/mL) and 72 hr (8.5 ± 0.1 mg/mL) compared with that at 0 hr (11.7 ± 0.5 mg/mL).
Figure 6. ELISA results of (A) serum amyloid A (SAA), (B) fructose-biphosphate aldolase A (FBAA) and (C) fetuin A.
SAA and FBAA showed significant increases at 48 and 72 hr after isoproterenol administration while fetuin A showed a significant decrease. Serum samples of four monkeys from six different time points were tested. Each sample and protein standards were measured in triplicates.
Discussion
Improved prediction of cardiac injury during preclinical testing of pharmaceuticals would provide a significant improvement in the prediction of drug safety prior to testing in humans.29 Preclinical and clinical serum biomarkers offer a promising method for predicting cardiotoxicity before drugs can produce cardiac injury and death.30 In this study, we applied a TMT 6-plex–based proteomic approach to investigate the serum protein changes in monkeys at time points ranging from 1 to 72 hr following administration of ISO, a model cardiotoxicant. Serum proteins were extracted and digested into peptides using a FASP strategy, and then the resulting peptides were labeled with TMTs to allow simultaneous identification and quantitation by LC-MS/MS. The changes of proteins of interest were further validated using a traditional ELISA strategy.
To confirm the overall quality of the serum biomarker candidates for monitoring cardiotoxicity, we evaluated reproducibility and yield, the two most crucial factors in biomarker discovery, using quantitative proteomics.31, 32 We applied FASP to process 48 serum samples collected from six time points of four monkeys, with two technical replicates of each. The average yield of FASP is ~64.1% with 9% variation among all 48 samples. Our results show that FASP allows us to achieve better consistency than other methods (acetone and trichloroacetic acid precipitation, data not shown) in sample preparation, and thus improves the reliability of the proteomics-based protein quantitation.
In this study, we focused on biomarker discovery from whole serum proteins instead of low-abundance proteins only. We also tested the application of the depletion procedure using a IgY-14 depletion column (Sigma-Aldrich); however, after depletion the reproducibility decreased to ~40%, resulting in difficulties for the biomarker discovery (data not shown). Our results also showed that two high-abundance proteins apolipoprotein A1and fetuin A exihibited significant expression changes in 48 hrs and 72 hrs post dose. Therefore, the low reproducibility of depletion could be induced by the changes of high-abundance and medium abundance proteins.
For the TMT labeling step, we used HPLC-based SCX and RP chromatography to purify labeled peptides. The SCX and RP steps have average yields of 63% and 79%, respectively. The technical variation of TMT labeling and peptide purification is less than 10% among all samples. We further evaluated the reproducibility of TMT-based quantitation in LC-MS/MS analysis. Correlation between each of the two technical replicates is ~0.7–0.8 (see Figure 3a), which is consistent with the Dayon et al. report of 20% technical variation in relative quantitation with the TMT method.33 Our data showed that through the combination of FASP and TMT-CID-HCD quantitative strategy, we are able to obtain accurate and reproducible results for biomarker discovery in multiple complicated serum samples. Note that the correlation of protein quantitation between different biological replicates is only ~0.3–0.4, indicating a larger variation among individual monkeys than that among all technical replicates.
Serum proteins provide a convenient source for us to discover changed proteins from the same individual at different post-dose time points. As illustrated in Figure 3, our results show that the overall protein expression before ISO injection (0 hr) showed the highest correlation with that of 1 hr and 72 hr samples (R = 0.79) and the lowest correlation with that of 4 hr samples (R = 0.31). This observation is consistent with many reports that serum levels of most cardiac biomarkers peak at 2–12 hr and return to baseline after 24 hr although chronic cardiac myodegeneration may still exist. In order to discover proteins relevant to long-term cardiac injury, we focused on the protein changes at 48 and 72 hr and discovered nine significantly changed proteins (Table 1). Three proteins of interest, FBAA, SAA, and fetuin A, were measured by ELISA assay, and their changes were consistent with LC-MS/MS. Monkey SAA ELISA kits were used for SAA while human FBAA and fetuin A kits were used, due to the lack of availability of NHP reagents. We detected a ~50-fold increase of SAA concentration at 72 hr post-dose using ELISA and a ~6-fold increase using proteomics. We also detected significant changes of serum FBAA and fetuin A using ELISA, but the ratios analyzed from ELISA results were not as significant as those of proteomic results. The manufacturer has reported a 10% cross-reactivity of human FBAA kit on monkey FBAA. Therefore, the ratio differences between ELISA assay and proteomic strategy may result from the low reactivity of human-specific antibodies against monkey antigens.
Glycogen phosphorylase has been known as a marker for both short-term and long-term cardiac injury.34 It releases early, within 2–4 h, as an indicator of irreversible myocardial damage,35 and the late release of glycogen phosphorylase has been linked to cardiac toxicity and inflammatory damage.36 Glycogen phosphorylase is known as the primary enzyme in glycogen breakdown.37 We detected significant elevation of glycogen phosphorylase from 4 to 72 hr, suggesting an up-regulated glycogen uptake. Our results from LC-MS/MS and the ELISA assay showed that FBAA, another protein involved in glycogen metabolism, was significantly up regulated at 48 and 72 hr upon ISO administration. Since ISO can induce a decrease in cardiac glycogen level,38 our findings suggest that the decrease of glycogen may be due to elevated glycogen uptake and that glycogen phosphorylase and FBAA may serve as indicators of cardiac injury at time points later than 24 hr.
We also discovered a decreased expression level of fetuin A and an increased level of protein SAA in serum from 48 and 72 hr post-dose and further validated the changes of the two proteins by ELISA assay. Nordfors et al. have reported that low fetuin A levels are associated with inflammation as well as with increased cardiovascular and all-cause mortality.39, 40 Fetuin A has also been reported to have lower expression in subclinical coronary artery calcification, stable angina and myocardial infarction.41–43 Other studies have indicated that SAA is a sensitive indicator of inflammation and cardiovascular risk.44 Elevated SAA has been linked to high mortality in acute coronary syndromes; more importantly, Braunwald et al. has discovered that SAA can identify a population of patients without elevation of cardiac-specific troponin T (cTnT), and thus can provide additional prognostic information for predicting cardiac necrosis.45 Our findings suggest that the changes of fetuin A and SAA may be useful in detecting cardiac injury and further evaluation on the significance of fetuin A and SAA could be helpful in assessing cardiovascular risk.
In summary, we have applied a TMT labeling-based CID-HCD method to identify serum proteins from African green monkeys at different post-dose timepoints upon injection of the model cardiotoxicant ISO. A FASP strategy was used to extract and digest proteins from serum samples and the peptides were then labeled using different TMT tags, purified by SCX and RP trap columns, and identified and quantified by 2D LC-MS/MS. To our knowledge, this is the first study that combines FASP and TMT labeling for sample processing and data acquisition. Our strategy provides highly reproducible data, which are essential for discovering biomarkers of cardiac injury. Five increased proteins and four decreased proteins were identified from the serum drawn at 48 and 72 hr after ISO treatment. We further applied an ELISA assay to confirm the alteration of three proteins, FBAA, SAA, and fetuin A, which are involved in glycogen metabolism, inflammation and cardiac necrosis. These results suggest that these nine proteins have potential for inclusion in multi-marker analysis of long-term cardiac injury. We are currently conducting similar evaluations in Cynomolgus macaques and Rhesus macaques, and are comparing the protein markers presented here with other commercially available and experimental markers of cardiac injury.
Supplementary Material
Acknowledgements
This work was supported, in part, by funding under contract N01-AI-70043, awarded by the National Institute of Allergy and Infectious Diseases (NIAID). The authors gratefully acknowledge Dr. Hao Zhang, NIAID, for his support and helpful suggestions. Technical assistance in the conduct of the animal studies was provided by Maya Patrick, Janet Gahagen, Dr. Karen Tinajero, and Dr. Ken Lopez.
Abbreviations
- TMT
Tandem Mass Tag
- FASP
Filter-aided sample preparation
- NHP
Non-human primates
- SCX
Strong cation exchange
- RP
Reverse phase
- ECG
Electrocardiograms
References
- 1.Gwathmey JK, Tsaioun K, Hajjar RJ. Cardionomics: a new integrative approach for screening cardiotoxicity of drug candidates. Expert opinion on drug metabolism & toxicology. 2009;5(6):647–660. doi: 10.1517/17425250902932915. [DOI] [PubMed] [Google Scholar]
- 2.Lott JA, Stang JM. Differential diagnosis of patients with abnormal serum creatine kinase isoenzymes. Clinics in laboratory medicine. 1989;9(4):627–642. [PubMed] [Google Scholar]
- 3.Ohman EM, Casey C, Bengtson JR, Pryor D, Tormey W, Horgan JH. Early detection of acute myocardial infarction: additional diagnostic information from serum concentrations of myoglobin in patients without ST elevation. British heart journal. 1990;63(6):335–338. doi: 10.1136/hrt.63.6.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazawa K, Fukuyama H, Yamaguchi I, Kobayashi M, Washio M, Oda J. Serial determinations of serum enzymes following aorta-coronary bypass surgery and acute myocardial infarction. Japanese heart journal. 1985;26(1):45–52. doi: 10.1536/ihj.26.45. [DOI] [PubMed] [Google Scholar]
- 5.Koenig W, Sund M, Frohlich M, Fischer HG, Lowel H, Doring A, Hutchinson WL, Pepys MB. C-Reactive protein, a sensitive marker of inflammation, predicts future risk of coronary heart disease in initially healthy middle-aged men: results from the MONICA (Monitoring Trends and Determinants in Cardiovascular Disease) Augsburg Cohort Study, 1984 to 1992. Circulation. 1999;99(2):237–242. doi: 10.1161/01.cir.99.2.237. [DOI] [PubMed] [Google Scholar]
- 6.Rajappa M, Sharma A. Biomarkers of cardiac injury: an update. Angiology. 2005;56(6):677–691. doi: 10.1177/000331970505600605. [DOI] [PubMed] [Google Scholar]
- 7.Maynard SJ, Menown IB, Adgey AA. Troponin T or troponin I as cardiac markers in ischaemic heart disease. Heart. 2000;83(4):371–373. doi: 10.1136/heart.83.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebell MH, Flewelling D, Flynn CA. A systematic review of troponin T and I for diagnosing acute myocardial infarction. The Journal of family practice. 2000;49(6):550–556. [PubMed] [Google Scholar]
- 9.Blich M, Sebbag A, Attias J, Aronson D, Markiewicz W. Cardiac troponin I elevation in hospitalized patients without acute coronary syndromes. The American journal of cardiology. 2008;101(10):1384–1388. doi: 10.1016/j.amjcard.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 10.York M, Scudamore C, Brady S, Chen C, Wilson S, Curtis M, Evans G, Griffiths W, Whayman M, Williams T, Turton J. Characterization of troponin responses in isoproterenol-induced cardiac injury in the Hanover Wistar rat. Toxicologic pathology. 2007;35(4):606–617. doi: 10.1080/01926230701389316. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman PL. Isoproterenol dose-outflow facility response relationships in the vervet monkey. Current eye research. 1985;4(8):877–883. doi: 10.3109/02713688509095255. [DOI] [PubMed] [Google Scholar]
- 12.Hasic S, Jadric R, Kiseljakovic E, Valjevac A, Mornjakovic Z, Winterhalter-Jadric M. Time-dependent responses of rat troponin I and cardiac injury following isoproterenol administration. Medicinski glasnik : official publication of the Medical Association of Zenica-Doboj Canton, Bosnia and Herzegovina. 2011;8(1):140–145. [PubMed] [Google Scholar]
- 13.Acikel M, Buyukokuroglu ME, Erdogan F, Aksoy H, Bozkurt E, Senocak H. Protective effects of dantrolene against myocardial injury induced by isoproterenol in rats: biochemical and histological findings. International journal of cardiology. 2005;98(3):389–394. doi: 10.1016/j.ijcard.2003.10.054. [DOI] [PubMed] [Google Scholar]
- 14.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7(3):340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 15.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical chemistry. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 16.Tiberti N, Hainard A, Lejon V, Robin X, Ngoyi DM, Turck N, Matovu E, Enyaru J, Ndung'u JM, Scherl A, Dayon L, Sanchez JC. Discovery and verification of osteopontin and Beta-2-microglobulin as promising markers for staging human African trypanosomiasis. Molecular & cellular proteomics : MCP. 2010;9(12):2783–2795. doi: 10.1074/mcp.M110.001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armah HB, Wilson NO, Sarfo BY, Powell MD, Bond VC, Anderson W, Adjei AA, Gyasi RK, Tettey Y, Wiredu EK, Tongren JE, Udhayakumar V, Stiles JK. Cerebrospinal fluid and serum biomarkers of cerebral malaria mortality in Ghanaian children. Malaria journal. 2007;6:147. doi: 10.1186/1475-2875-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 19.Pichler P, Kocher T, Holzmann J, Mohring T, Ammerer G, Mechtler K. Improved precision of iTRAQ and TMT quantification by an axial extraction field in an Orbitrap HCD cell. Analytical chemistry. 2011;83(4):1469–1474. doi: 10.1021/ac102265w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 21.Thingholm TE, Bak S, Beck-Nielsen H, Jensen ON, Gaster M. Characterization of human myotubes from type 2 diabetic and nondiabetic subjects using complementary quantitative mass spectrometric methods. Molecular & cellular proteomics : MCP. 2011;10(9):M110 006650. doi: 10.1074/mcp.M110.006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kall L, Storey JD, MacCoss MJ, Noble WS. Posterior error probabilities and false discovery rates: two sides of the same coin. Journal of proteome research. 2008;7(1):40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- 23.Choi H, Nesvizhskii AI. False discovery rates and related statistical concepts in mass spectrometry-based proteomics. Journal of proteome research. 2008;7(1):47–50. doi: 10.1021/pr700747q. [DOI] [PubMed] [Google Scholar]
- 24.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nature methods. 2005;2(9):667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 25.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Molecular & cellular proteomics : MCP. 2009;8(11):2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohr MJ, Aponte A, Sun J, Gucek M, Steenbergen C, Murphy E. Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: short communication. Circulation research. 2012;111(10):1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur P, Rizk NM, Ibrahim S, Younes N, Uppal A, Dennis K, Karve T, Blakeslee K, Kwagyan J, Zirie M, Ressom HW, Cheema AK. iTRAQ-based quantitative protein expression profiling and MRM verification of markers in type 2 diabetes. Journal of proteome research. 2012;11(11):5527–5539. doi: 10.1021/pr300798z. [DOI] [PubMed] [Google Scholar]
- 28.Dost B, Bandeira N, Li X, Shen Z, Briggs SP, Bafna V. Accurate Mass Spectrometry Based Protein Quantification via Shared Peptides. Journal of computational biology : a journal of computational molecular cell biology. 2012;19(4):337–348. doi: 10.1089/cmb.2009.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piccini JP, Whellan DJ, Berridge BR, Finkle JK, Pettit SD, Stockbridge N, Valentin JP, Vargas HM, Krucoff MW. Current challenges in the evaluation of cardiac safety during drug development: translational medicine meets the Critical Path Initiative. American heart journal. 2009;158(3):317–326. doi: 10.1016/j.ahj.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Walker DB. Serum chemical biomarkers of cardiac injury for nonclinical safety testing. Toxicologic pathology. 2006;34(1):94–104. doi: 10.1080/01926230500519816. [DOI] [PubMed] [Google Scholar]
- 31.Silberring J, Ciborowski P. Biomarker discovery and clinical proteomics. Trends in analytical chemistry : TRAC. 2010;29(2):128. doi: 10.1016/j.trac.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annual review of biomedical engineering. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 33.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Analytical chemistry. 2008;80(8):2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 34.Kemp M, Donovan J, Higham H, Hooper J. Biochemical markers of myocardial injury. British journal of anaesthesia. 2004;93(1):63–73. doi: 10.1093/bja/aeh148. [DOI] [PubMed] [Google Scholar]
- 35.Rabitzsch G, Mair J, Lechleitner P, Noll F, Hofmann V, Krause EG, Dienstl F, Puschendorf B. Isoenzyme BB of glycogen phosphorylase b and myocardial infarction. Lancet. 1993;341(8851):1032–1033. doi: 10.1016/0140-6736(93)91129-a. [DOI] [PubMed] [Google Scholar]
- 36.Horacek JM, Tichy M, Pudil R, Jebavy L. Glycogen phosphorylase BB could be a new circulating biomarker for detection of anthracycline cardiotoxicity. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2008;19(9):1656–1657. doi: 10.1093/annonc/mdn414. [DOI] [PubMed] [Google Scholar]
- 37.Krause EG, Rabitzsch G, Noll F, Mair J, Puschendorf B. Glycogen phosphorylase isoenzyme BB in diagnosis of myocardial ischaemic injury and infarction. Molecular and cellular biochemistry. 1996;160–161:289–295. doi: 10.1007/BF00240061. [DOI] [PubMed] [Google Scholar]
- 38.Todd GL, Pieper GM, Clayton FC, Eliot RS. Heterogeneity in distribution of cardiac glycogen following isoproterenol infusions in the dog. The Histochemical journal. 1979;11(4):425–434. doi: 10.1007/BF01002770. [DOI] [PubMed] [Google Scholar]
- 39.Stenvinkel P, Wang K, Qureshi AR, Axelsson J, Pecoits-Filho R, Gao P, Barany P, Lindholm B, Jogestrand T, Heimburger O, Holmes C, Schalling M, Nordfors L. Low fetuin-A levels are associated with cardiovascular death: Impact of variations in the gene encoding fetuin. Kidney international. 2005;67(6):2383–2392. doi: 10.1111/j.1523-1755.2005.00345.x. [DOI] [PubMed] [Google Scholar]
- 40.Marechal C, Schlieper G, Nguyen P, Kruger T, Coche E, Robert A, Floege J, Goffin E, Jadoul M, Devuyst O. Serum fetuin-A levels are associated with vascular calcifications and predict cardiovascular events in renal transplant recipients. Clinical journal of the American Society of Nephrology : CJASN. 2011;6(5):974–985. doi: 10.2215/CJN.06150710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tousoulis D, Siasos G, Maniatis K, Oikonomou E, Vlasis K, Papavassiliou AG, Stefanadis C. Novel biomarkers assessing the calcium deposition in coronary artery disease. Current medicinal chemistry. 2012;19(6):901–920. doi: 10.2174/092986712799034833. [DOI] [PubMed] [Google Scholar]
- 42.Ix JH, Barrett-Connor E, Wassel CL, Cummins K, Bergstrom J, Daniels LB, Laughlin GA. The associations of fetuin-A with subclinical cardiovascular disease in community-dwelling persons: the Rancho Bernardo Study. Journal of the American College of Cardiology. 2011;58(23):2372–2379. doi: 10.1016/j.jacc.2011.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bilgir O, Kebapcilar L, Bilgir F, Bozkaya G, Yildiz Y, Pinar P, Tastan A. Decreased serum fetuin-A levels are associated with coronary artery diseases. Internal medicine. 2010;49(13):1281–1285. doi: 10.2169/internalmedicine.49.3223. [DOI] [PubMed] [Google Scholar]
- 44.Stettler C, Witt N, Tapp RJ, Thom S, Allemann S, Tillin T, Stanton A, O'Brien E, Poulter N, Gallimore JR, Hughes AD, Chaturvedi N. Serum amyloid A, C-reactive protein, and retinal microvascular changes in hypertensive diabetic and nondiabetic individuals: an Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT) substudy. Diabetes care. 2009;32(6):1098–1100. doi: 10.2337/dc08-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrow DA, Rifai N, Antman EM, Weiner DL, McCabe CH, Cannon CP, Braunwald E. Serum amyloid A predicts early mortality in acute coronary syndromes: A TIMI 11A substudy. Journal of the American College of Cardiology. 2000;35(2):358–362. doi: 10.1016/s0735-1097(99)00574-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.