Abstract
To unravel the mechanism of CPO-catalyzed regioselective oxidation of indole, the structure of the CPO-indole complex was studied using NMR relaxation measurements and computational techniques. The dissociation constant (KD) of the CPO-indole complex was calculated to be approximately 21 mM. The distances (r) between protons of indole and the heme iron calculated from NMR relaxation measurements and molecular docking revealed that the pyrrole ring of indole is oriented toward the heme with its 2-H pointing directly at the heme iron. Both KD and r values are independent of pH in the range of 3.0–6.5. The stability and structure of the CPO-indole complex are also independent of the concentration of chloride/iodide ion. Molecular docking suggests the formation of a hydrogen bond between the N–H of indole and the carboxyl O of Glu 183 in the binding of indole to CPO. Simulated annealing of the CPO-indole complex using r values from NMR experiments as distance restraints reveals that the van der Waals interactions were much stronger than the Coulomb interactions in indole binding to CPO, indicating that the association of indole with CPO is primarily governed by hydrophobic rather than electrostatic interactions. This work provides the first experimental and theoretical evidence for the long-sought mechanism that leads to the “unexpected” regioselectivity of CPO-catalyzed oxidation of indole. The structure of the CPO-indole complex will serve as a lighthouse in guiding the design of CPO mutants with tailor-made activities for biotechnological applications.
Keywords: NMR, docking, simulated annealing, chloroperoxidase, indole, regioselective
Chloroperoxidase (CPO), a heme-thiolate protein secreted by the marine fungus Caldariomyces fumago (1), is a versatile enzyme (2) capable of catalyzing a broad spectrum of chemical reactions including halogenation (3), dehalogenation (4), N-demethylation (5), dismutation (1), epoxidation (6), and oxidation (7–11). Therefore, CPO has been the subject of numerous biotechnological investigations due to its potential applications in synthetic chemistry as well as pharmaceutical industry (12–15). Most interestingly, CPO-catalyzed oxidation and epoxidation reactions proceed with high regio- and enantio-selectivity (6, 8, 16). Tremendous efforts have been made during the past few decades to understand the mechanism of CPO’s regio- and enantio-selectivity (17–19). Most of such studies are focused on the structures of CPO-substrate complexes. The structural feature of CPO-substrate complexes will shed light on the structure-function relationships of heme proteins in general and the structural basis of the diverse catalytic activity, strict substrate specificity, and high regio- and enatio-selectivity displayed by CPO in particular. The definitive structure of CPO-substrate complexes will also provide pivotal information regarding the topology of the heme active center of CPO, the residues involved in the catalytic cycle, the location/orientation of substrate binding, and the mechanisms of CPO-catalyzed reactions. Such information will serve as a lighthouse for designing more efficient CPO mutants that target specific substrates and produce desired products that are difficult to obtain under mild conditions (20). Unfortunately, the structural information on CPO-substrate complexes remains scarce despite the great efforts made with the application of the most powerful techniques available including X-ray crystallography and NMR spectroscopy.
X-ray crystal crystallography is one of the most powerful techniques for elucidating the three dimensional structure of enzymes and their complexes with substrates. The crystal structure of CPO has demonstrated the presence of a substrate-binding site with an opening above the heme that enables organic substrates to approach the oxoferryl oxygen of CPO compound I (2). It is generally expected that substrate binding would result in noticeable structural changes to the protein. Surprisingly, the structures of CPO-ligand complexes are indistinguishable from that of the ligand-free protein (21). On the other hand, the crystal structure of CPO complexed with its natural substrate, 1,3-cyclopentanedione (CPDO), (PDB code: 2CIX) revealed another surprising picture in which the active methylene group of CPDO is not oriented towards the heme iron (22). This is in good agreement with the results reported from an NMR relaxation study of CPO-CPDO complex in solution (23). To date, 2CIX is the only crystal structure of CPO complexed with a substrate. The limited crystal structural data on CPO-substrate complexes is mainly attributed to the difficulty of diffusing organic substrates into CPO crystals (22).
Nuclear magnetic resonance (NMR) spectroscopy is an alternative technique to X-ray crystallography for probing the structural information of proteins and their complexes with substrates in solution. Particularly, in heme proteins, the enhanced nuclear relaxation induced by the paramagnetism of the heme (24, 25) was often used to establish the structure and stability of enzyme-substrate complex (24, 26, 27). NMR relaxation studies have been successfully used in determining the structural features of CPO complexed with various substrates including phenols (28), sulfides (29), and the natural chlorination substrate, CPDO (23).
Molecular mechanics studies have become an important complement to NMR and X-ray experiments in exploring heme proteins and their interactions with substrates at the atomic level (30, 31). The NMR-determined distances between the substrate and the heme iron can be used as restraints in simulation studies utilizing a simulated annealing protocol (32, 33). This protocol has been successfully applied to studies of substrate binding to several other hemoproteins (34, 35). This success has prompted us to investigate the mechanisms of CPO-catalyzed regioselective oxidation of indole using both NMR and computational techniques.
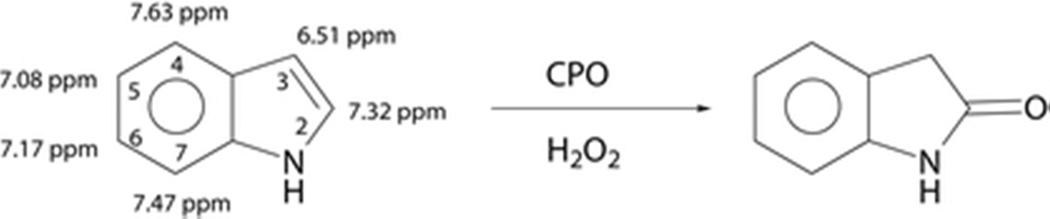
The CPO-catalyzed oxidation of indole to oxindole was first reported in 1979 (36). Van Deurzen et al. further reported that the oxidation of substituted indoles yields the corresponding oxindoles and the reactivity of the substituted indoles depends on the nature and the position of its substituent (37). However, to our best knowledge, the structural basis for CPO-catalyzed unusual regioselective oxidation of indole remains undefined. Based on the structure of indole, the oxidation would be expected to occur at the 3-position of indole, which possesses the highest electron density and is thus most susceptible to attack by oxidizing agents and by other electrophilic reagents (36). It is obvious to suspect the indolic N–H group as a structural feature controlling the position of oxidation, even though indole is an extremely weak base. However, the presence of a hydrogen-bond between the indole N–H group and an unknown group at the CPO active site has proven to be insufficient to explain the regiospecificity of indole oxidation (36). Therefore, a detailed structural characterization of CPO-indole complex would offer greater insight into the mechanisms of CPO-catalyzed oxidations and solve the long lasting puzzle of the unusual product from indole oxidation.
The aims of the present work are to reveal the orientation of indole binding at the active site of CPO, to identify the residues involved in the formation of CPO-indole complex, and to elucidate the mechanism of CPO-catalyzed regioselective oxidation of indole. The interaction of indole with CPO was probed using longitudinal NMR relaxation and two computational methods, molecular docking and simulated annealing. The dissociation constant of CPO-indole complex and the distances between the protons of indole and the heme iron of CPO are obtained. The effect of pH and halide ion on the binding of indole with CPO is presented. Simulated annealing and molecular docking of indole in the active center of CPO are provided. Our results showed that a complex with a KD of approximately 21 mM is formed between CPO and indole with indole’s 2-H pointing towards the heme iron. This structural feature satisfactorily explains the unusual product observed from CPO-catalyzed oxidation of indole. It is concluded that formation of CPO-indole complex is responsible for the escorted delivery of oxygen from CPO compound I directly to the 2-position of indole.
MATERIALS AND METHODS
Materials
Caldariomyces fumago (ATCC number: 16373) was purchased from ATCC (Manassas, VA). Unless otherwise specified, all chemicals were of analytical grade and were purchased from Sigma-Aldrich (St. Louis, MO).
Chloroperoxidase Expression and Purification
CPO was isolated and purified according to published protocols (38, 39) with slight modifications. CPO preparations with Reinheitszahl values (Rz, A398 nm/A278 nm ratios) of 1.4 or higher were used. The stock solution of CPO was prepared by repetitive (>6 times) isotope exchange of aqueous protein solution with D2O in a Centriprep-30 centrifugal filter device. The concentration of CPO was determined by measuring the absorption at 398 nm using a molar extinction coefficient of 9.12 × 104 M−1cm−1(1).
Preparation of Stock Solutions and NMR Samples
A stock solution of indole (14.0 mM) was prepared by dissolving 32.8 mg indole in 20.0 mL of warm D2O in a vortex mixer. Stock solutions of 1.0 M chloride and iodide ion were prepared from their potassium salts in D2O. Solutions of 1.0 M and 0.1 M DCl were prepared by dissolving DCl (35% w/w) in D2O to adjust the pH of all test solutions. The samples used for NMR experiments contained 2–12 mM indole and 0.1 (or 0.01) mM CPO in 100 mM phosphate buffer with 99.9% D2O. The final volume of all NMR samples was 500 µL.
Structural Characterization of Indole by NMR
Proton NMR measurements were conducted on a Bruker Avance 600 MHz NMR spectrometer at 298.0 K. Proton chemical shifts were referenced to the internal reference 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). The NMR data were processed using Topspin, version 2.1. The line width was obtained from proton spectra by fitting the proton peak to a Lorentzian line shape.
T1 Relaxation Experiments
The longitudinal relaxation time (T1) was determined using the standard inversion-recovery method with 180°-τ-90° pulse sequence (40). Eleven spectra were recorded for each sample, with the inter-pulse delay τ ranging from 0.5 to 25.0 s. For each spectrum, 32 scans were acquired. The T1 values were calculated by fitting Equation 1, where τ is the interval between 180° and 90° pulses, Mz is the Z-component of nuclear magnetization (represented by the intensity of the peak) when the interval is τ, M0 is the is the Z-component of the nuclear magnetization when the interval is infinite, and ρ is a parameter that equals to 2.0 at an exactly 180° pulse.
| (1) |
Dissociation Constants of CPO-Indole Complex
The longitudinal relaxation rate (T1obs−1) is the weighted average of the relaxation rates of the free substrate (T1f−1) and the bound substrate (T1b−1 ) (41). Thus, the relaxation-time values (T1obs−1 , T1f −1 , and T1b−1) are related as shown in Equation 2 when only one molecule of substrate binds to a molecule of enzyme, where T1obs is the relaxation time of the indole obtained from the relaxation experiment, T1f is the relaxation time of indole obtained from the relaxation experiment in the absence of CPO, T1b is the relaxation time of the CPO-bound indole, KD is the dissociation constant of the CPO-indole complex, E0 is the initial CPO concentration, and S0 is the initial indole concentration.
| (2) |
Location of Indole Binding Site
The location of indole in the active site of CPO can be determined from the distances (r) between indole protons and the heme iron of CPO calculated according to the Solomon-Bloembergen equation (Equation 3) (24, 27, 42), where µ0 is the permeability of free space, γ is the gyromagnetic ratio of the proton, g is the electronic g-factor, µB is Bohr magneton, S is the spin state of the heme iron of CPO, ωI and ωS are the nuclear and electronic Larmor frequency, respectively, r is the distance from proton nuclei to the heme iron, and τc is the correlation time that describes the dipolar interaction between the ligand and the paramagnetic iron in solution. A value of τc = 8.8×10−1 s was used for CPO (29). When the observed proton is in the extreme narrowing conditions , as is usually the case for high-spin hemoproteins (29, 43), Equation 3 is simplified to Equation 4, which was used in calculating the distances in this work.
| (3) |
| (4) |
Additionally, T1m is related to T1b through Equation 5, where T1d is the relaxation time in diamagnetic states, T1m is the relaxation time in paramagnetic states, and τM is the lifetime of the enzyme-substrate complex. Since both T1d and τM are negligible compared to T1b in our case (23, 28, 29), T1b is approximately the same as T1m.
| (5) |
Molecular Docking
Molecular docking for the preferred orientation of indole and 1-methylindole within the active site of CPO was performed with the software package AutoDock (44), version 4.2.3. The results from molecular docking studies provided the starting position of indole for distance-restrained simulated annealing. The structure of CPO for docking indole and 1-methylindole was taken from the X-ray structure (PDB code: 2CPO). Additionally, 14 of the glycosylation sites were removed from the CPO structure, and the pyroglutamic acid (Pca) residue was replaced by a proline residue, since parameters for sugars and the Pca residue are unavailable in the GROningen MOlecular Simulation (GROMOS) force field (45) that was used in our simulated annealing. The removal of sugars from CPO should have negligible effect on substrate binding, because CPO without glycosylation is still highly active (46). Furthermore, the manganese ion was removed from the CPO structure, as manganese-free CPO (47) was used in our NMR experiment. The crystallographic water in the PDB file was removed to reduce any interference from that water during docking.
Both the indole and 1-methylindole structures were built by MarvinSketch, version 5.9, in the JChem software package (ChemAxon, Ltd.) and further optimized with ORCA (48), version 2.9, using second-order Møller-Plesset perturbation theory (MP2)(49) and the def2-SVP basis set(50). AutoDockTools(51), version 1.5.4, was used to add Gasteiger charges to CPO (+1.00 was added manually on Fe), indole, and 1-methylindole, respectively. During simulation, the CPO structure was kept rigid. Indole or 1-methylindole was simulated in a box centered at the heme iron, which was confined using a grid size of 30 × 30 ×30 Å3 box with 0.375 Å spacing. Docking consisting of 60 separate simulation runs was performed with 25 million energy evaluations per run.
Distance-restrained simulated annealing
Distance-restrained simulated annealing and energy minimization for CPO-indole complex were performed with the Groningen MAchine for Chemical Simulation (GROMACS) package (52), version 4.6, using the GROMOS96 53a6 force field (45). The force field was augmented with the parameters developed in this work (see Table S1). The parameters for indole were built using the PRODRG2 server (53). The partial atomic charges of indole were assigned by analogy with the equivalent functional group, tryptophan, in the GROMOS96 53a6 force field (54). The bonded parameters and partial atomic charges for the heme ligated with cysteine were derived using Seminario’s method (55) and CHELPG (CHarges from Electrostatic Potentials using a Grid based) (56) method, respectively (see supporting information); the van der Waals parameters were those already in the force field.
The CPO-indole complex was solvated with simple point charge (SPC) waters (57). The protonation states of the titrable amino acid residues of CPO were assigned as follows: +1 for His, Lys; −1 for Glu, Asp; and neutral for Tyr. The protein has a net charge of −16, and so 16 Na+ ions were added to create a neutral system for simulation. Periodic boundary conditions were imposed using a truncated octahedral box created from an 82 × 82 × 82 Å3 cube. After deletion of overlapping waters, 12644 waters remained. A shift method with a 14 Å cutoff was used for the calculation of Lennard-Jones interactions. Electrostatic interactions were calculated with the particle mesh Ewald method (PME) (58), using a cutoff of 12 Å for real-space interactions, a Gaussian width parameter of 3.84195 Å, an FFT grid determined with the fourier spacing method using a maximum spacing of 1.2 Å, and fourth-order interpolation for reciprocal-space interactions. A relative dielectric constant (εr=1.0) was used. All bond lengths were constrained using the LINCS algorithm (59). Positional restraints were placed on the backbone α-carbon during simulated annealing. Indole was restrained with a force constant of 600 kJ mol−1 nm−2 using the NMR-derived distances between the protons of indole and the heme iron (see Table S2). A time constant of 50 ps was applied to the distance restraint, while the violation of the distance restraint was the square root of the product of the time-averaged violation and the instantaneous violation. Brief energy minimization of CPO-indole complex was performed using the conjugate gradient method to eliminate unrealistic van der Waals contacts. The system was rapidly heated to 800 K, cooled to 300 K over 150 ps, and allowed to equilibrate for 100 ps at 300 K, using a 2 fs timestep. The analysis below is based on Coulomb and Lennard-Jones interaction energies calculated during the last 50 ps, root-mean-square deviations (RMSDs) calculated with respect to the starting structure, and snapshots of the indole molecule bound to CPO during the last 50 ps of MD simulation.
RESULTS AND DISCUSSION
Proton NMR Spectrum of Indole in the Presence of CPO
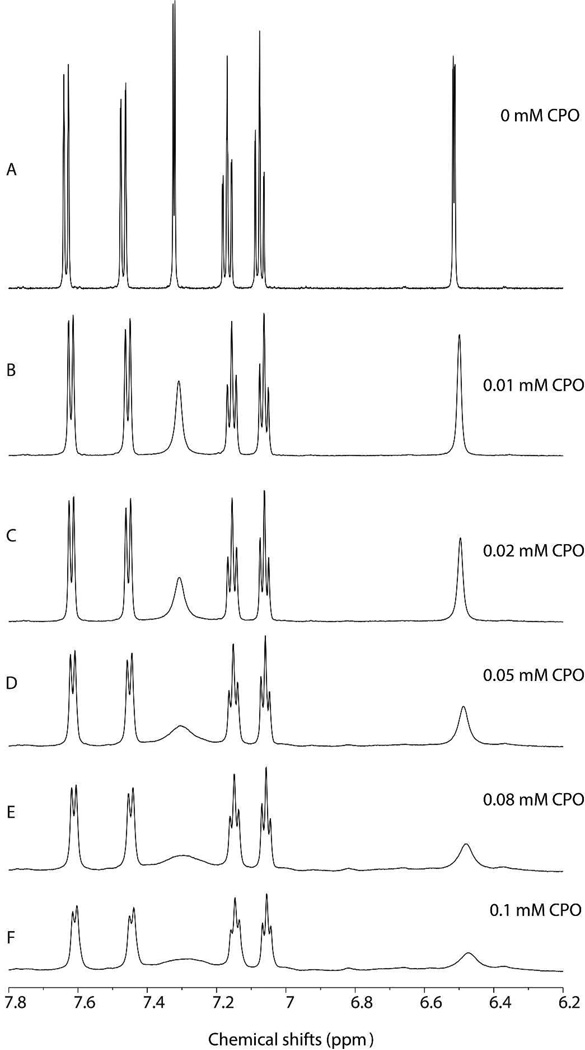
Figure 1 compares the 1H NMR spectra of 5.7 mM indole in the absence (Figure 1A) and presence of varying amount (0.01, 0.02, 0.05, 0.08, and 0.1 mM, Fig. 1B–F) of CPO at pH 6.0. The six main peaks (Figure 1A) can be easily assigned, based on the splitting pattern and shift positions, to the protons of indole shown in Scheme 1: 7.32 ppm (2-H), 6.51 ppm (3-H), 7.63 ppm (4-H), 7.08 ppm (5-H), 7.17 ppm (6-H), and 7.47 ppm (7-H). The 1-H was not observed due to its fast exchange with D2O. After addition of CPO, the doublet from 2- and 3-H (Figure 1A) was changed to two broad singlets due to the paramagnetism of the heme in CPO (Figure 1B), suggesting the close promximity of these protons to the heme iron. Further increasing the concentration of CPO led to more significant broadening of these peaks (Figure 1B–F). It is also noted that as CPO concentration was increased, all indole resonances broadened coupled with noticeable shift in their resonance positions (Figure 1B–F). For instance, the chemical shift of 4-H changed from 7.63 to 7.61 ppm. The change in chemical shifts results from the dipole-dipole interaction between indole and the heme iron (25), while line broadening of the signals is due to paramagnetic enhancements of the transverse nuclear relaxation rates (T2−1) of indole caused by the heme iron (25). The results shown in Figure 1 indicate that a reasonably stable CPO-indole complex is formed with indole bound near the heme center of the protein. More importantly, the NMR signal from all indole protons can be clearly observed, making it possible to obtain the orientation of indole in the active center of CPO because multiple distances between substrate protons and the heme iron can be experimentally determined.
Figure 1.
The effect of the concentration of CPO on the proton NMR spectra of indole. The spectra (A-F) was obtained in 10 mM deuterated phosphate buffer (pH 6.0) containing 5.7 mM indole and a fixed concentration of CPO.
Scheme 1.
The CPO-Catalyzed Oxidation of Indole to 2-oxindole Using H2O2a,b.
aThe indole structure is labeled with proton chemical shifts. bProton number assignments are shown.
As the concentration of CPO increased from 0.01 to 0.05 mM, the linewidth of the 2-proton increased from 2.47 to 54.37 Hz (Figure 1B–D). The peak of 2-H was broadened so severely that it became hardly observable when the concentration of CPO reaches 0.08 mM or higher (Figure 1E–F). The linewidth of 3-H also increased dramatically from 4.27 to 59.64 Hz (Figure 1B–F) as the concentration of CPO was increased from 0.01 to 0.1 mM, although the broadening effect was not as profound for that observed for 2-H. In contrast, other peaks were less broadened than those of the 2- and 3-H. For instance, the linewidth of 4-H increased gradually from 4.00 to 15.71 Hz in the range of the CPO concentration tested (Figure 1B–F). This suggests that protons in the pyrrole ring are closer to the heme iron than protons in the benzene ring of indole with 2-H being the closest. The systematic broadening of indole proton resonances observed here is consistent with our previous study of CPO-CPDO complex (23).
Dissociation Constant of CPO-indole Complex
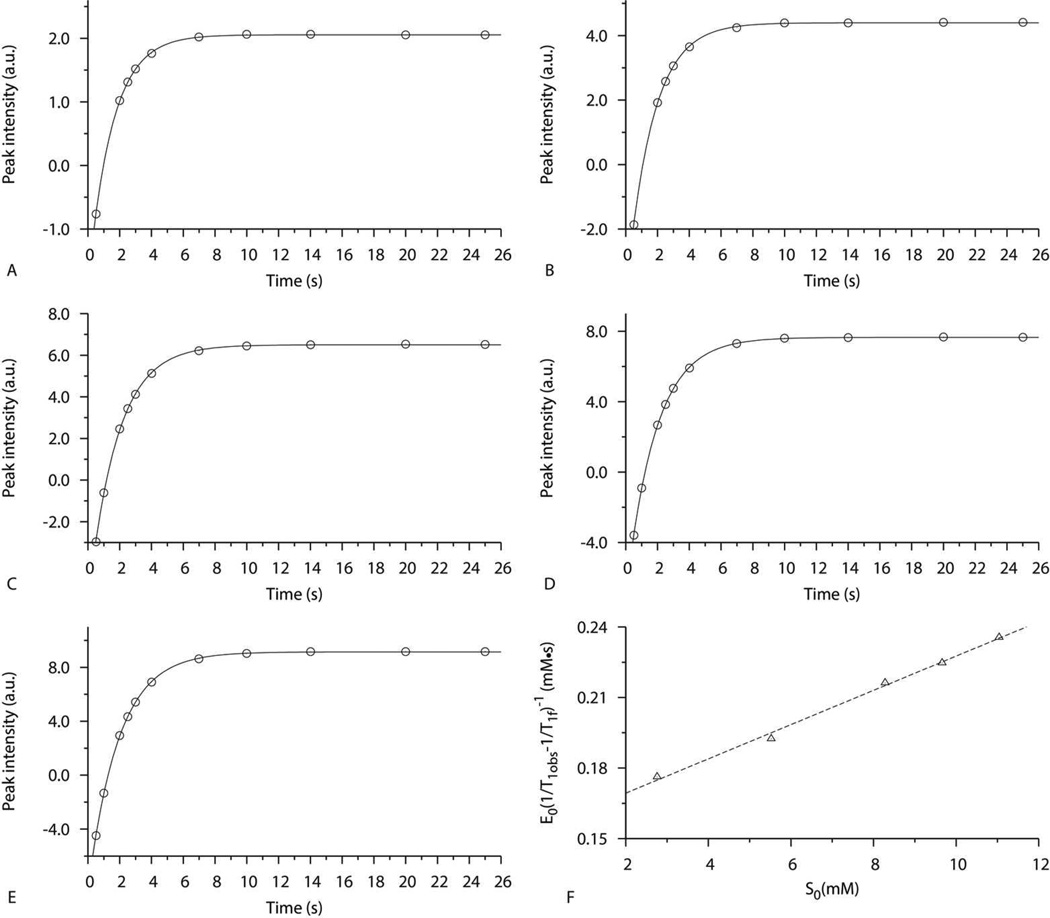
To evaluate the binding affinity between CPO and indole, the longitudinal relaxation time (T1) of indole protons was measured in the presence of 0.1 mM CPO and different concentrations of indole (2.8–11.0 mM) at pH 6.0. Figure 2 shows the nice exponential fit of signal intensities (Mz) as a function of the delay time (τ) as defined in Equation 1, using the resonance at 7.61 ppm (4-H) as an example. Similar fits are obtained for other protons of indole (data not shown). Therefore, T1obs of 4-H (Figure 2A–E) can be obtained from Equation 1 (1.51 s, 1.63 s, 1.79 s, 1.85 s, and 1.92 s, respectively). The increase in T1obs is attributed to the increase of the fraction of free indole as its concentration is increased.
Figure 2.
NMR relaxation experiments for 4-proton of indole in the presence of CPO. (A-E) The plots of peak intensities at 7.61 ppm as a function of inversion recovery time (τ). The experimental results (dots) was obtained in 100 mM deuterated phosphate buffer (pH 6.0) containing 0.1 mM CPO and (A) 2.8 mM indole, (B) 5.5 mM indole, (C) 8.3 mM indole, (D) 9.7 mM indole, and (E) 11.0 mM indole. The curve presents the exponential fit to Equation 1. (F) The plot of E0[1/T1obs−1/T1f]−1 as a function of the concentration of indole (S0). The linear line presents the least-squares fit to Equation 2 and is used to estimate the KD.
Figure 2F shows a plot of E0(1/T1obs-1/T1f)−1 versus S0 for 4-H. The fitted straight line demonstrates the reliability of the data (R2=0.9957). The KD value can be calculated as the intercept of the fitted line divided by its slope using Equation 2. The KD calculated from Figure 2F was approximately 21 mM as listed in Table 1. This value is dramatically different from the KM (3.3 mM) for CPO catalyzed oxidation of indole (60). The inconsistence between KD and KM are most probably due to the fact that KM is affected by many species involved in the entire catalytic process, while KD is soly determined by the stability of the enzyme-substrate complex. Higher KD than KM has been observed for complexes between CPO and other substrates (23). To verify the results from our NMR studies, the KD value for CPO-indole complex was also studied using difference optical absorption techniques. Result from our UV-Vis studies is in good agreement with those derived from our NMR studies (data not shown).
Table 1.
The dissociation constant (KD) of CPO-indole complex and the distances (r) between the protons of indole and CPO heme irona
| position | δ (ppm) | T1b (s)b | r (Å) | KD (mM) |
|---|---|---|---|---|
| 2-H | 7.31 | 6.8×10−5 | 4.2 | 21 |
| 3-H | 6.50 | 3.7×10−4 | 5.5 | 26 |
| 4-H | 7.61 | 7.2×10−3 | 9.1 | 21 |
| 5-H | 7.06 | 1.6×10−2 | 10.4 | 22 |
| 6-H | 7.15 | 1.8×10−2 | 10.5 | 20 |
| 7-H | 7.44 | 9.9×10−3 | 9.5 | 19 |
The relaxation experiments were performed in 10 mM deuterated phosphate buffer (pH 6.0) containing 0.01 mM CPO to measure the δ and T1 of 2- and 3-protons or 0.1 mM CPO for the other protons by changing the concentration of indole from 2.8 to 11.0 mM.
The average was from two independent experiments.
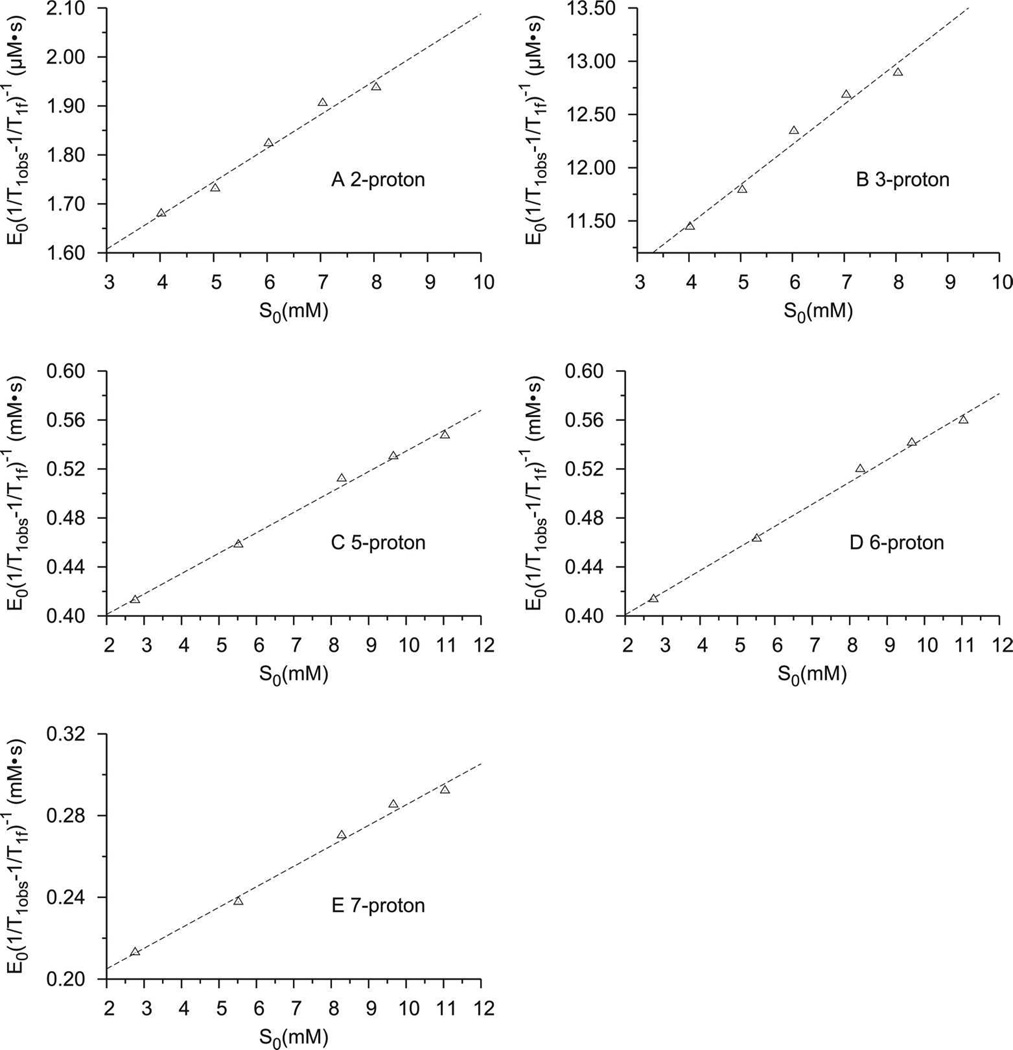
Figure 3 shows plots of E0(1/T1obs-1/T1f)−1 versus S0 for other protons. All correlation values of the fits (R2) were higher than 0.98. It should be noted that linear fits could only be obtained for 2- and 3-H in the presence of 0.01 mM CPO. Protons in the benzene ring of indole did not give reliable results in 0.01 mM CPO solution and thus do not fit into Equation 2 possibly due to the weak paramagnetic effect at low enzyme concentrations and long distances from these protons to the heme iron. Therefore, a higher concentration of CPO (0.1 mM) was used to measure the relaxation rates of protons on the benzene ring of indole. Table 1 lists the dissociation constant (KD) of the CPO-indole complex at pH 6.0. The KD values obtained from the relaxation property of different protons in indole are quite consistent, further proving the validity of the relaxation method in probing the structural properties of heme protein-substrate complexes. The average of KD calculated from all protons of indole is ~21 mM, indicating a weak binding of indole to CPO. This value is comparable to the reported KD of 33 mM for the binding of CPDO to CPO (23). The stability of CPO-indole complex also compares nicely with that of indole-P450 BM3 complex (61).
Figure 3.
The plots of E0[1/T1obs-1/T1f]−1 as a function of the concentration on indole (S0) for (A) 2-proton, (B) 3-proton, (C) 5-proton, (D) 6-proton, and (E) 7-proton of indole. The relaxation experiment were carried out (A-B) in 10 mM deuterated phosphate buffer containing 0.01 mM CPO and varying the concentration of indole, or (C-E) in 100 mM deuterated phosphate buffer containing 0.1 mM CPO and varying the concentration of indole. To plot the label on y axis clearly, E0[1/T1obs-1/T1f]−1 of (A) and (B) was plotted in the unit of µM•S.
Distances between the Heme Iron and the Protons of Indole
To reveal the orientation of indole in the active site of CPO, paramagnetically induced NMR relaxation of indole in the presence of CPO was used to calculate the distance between the heme iron and the protons of indole. Table 1 lists the NMR relaxation time, T1b, and the calculated distances between the protons of indole and the heme iron of CPO. These distances are consistent with those obtained for alky phenyl sulfides (8.0–10.9 Å) (29), phenols (5.6–9.3 Å) (28), and CPDO (7.1 Å) (23) binding with CPO. Examination of Table 1 revealed that the pyrrole ring of indole is closer than the benzene ring to the heme iron. The distance between the heme iron of CPO and the 2-H of indole is the shortest. This position happens to be the location at which the C–H oxidation takes place, indicating that the oxidation of indole proceeds by a direct insertion of oxygen from the oxyferryl intermediates of CPO. Furthermore, the use of these distance constraints (Table S2) in advanced computational methods such as simulated annealing (62, 63) can provide the precise orientation and position of indole in the active site of CPO, which will be presented in Distance-Restrained Model of CPO Complexed with Indole.
Effect of pH on Indole Binding to CPO
To check the effect of pH on indole binding to CPO, the KD and the distances from 2- and 3-H to the heme iron were determined in the pH range of 3.0–6.5 due to stability restriction of CPO (Table 2). It has been reported that in the absence of hydrogen peroxide, CPO is stable up to pH 6.5 (64, 65). The relaxation time of 3-H of indole cannot be determined at pH ≤ 4.0 due to acid-catalyzed tautomerization of the N-protonated species (66) that leads to the exchange of 3-H with deuterated solvent. Therefore, the KD values listed in Table 2 were derived from T1 of 2-H only. Surprisingly, both the dissociation constant, KD, and the distance, r, between the 2-H of indole and CPO heme iron were independent of pH in the pH range tested (Table 2), indicating that no ionizable group is involved in indole binding to CPO. This is in conflict with the fact that the activity of CPO catalyzed oxidation of indole is strongly pH dependent. Similar as most heme peroxidases, CPO uses compound I rather than the resting state enzyme itself to oxidize substrates. Therefore, our results imply that the effect of pH on CPO’s catalytic activity is primarily due to changes in the rate of formation of CPO compound I rather than the formation of CPO-substrate complexes, especially for hydrophobic organic substrates. It should be noted that the observed pH inertness of indole binding to CPO is also at odds with the case of CPDO where an ionizable group with a pKa between 4.5 and 6.5 is involved in the formation of CPO-CPDO complex (23). The ionizable group has been suggested to be either a carboxylate residue (Glu 183) or a histidine imidazole group (His 105) (23). Since CPDO is polar, it may require hydrogen bonding with Glu 183 or His 105 to stabilize its binding with CPO. In contrast to CPDO, indole is neutral and hydrophobic; therefore, its binding with CPO would naturally be dominated by hydrophobic interactions with the enzyme. The X-ray structure of CPO has revealed the presence of a putative hydrophobic channel for the approach of organic substrates to the heme active site (22). At the bottom of this hydrophobic channel, two phenylanalines, Phe 103 and Phe 186, serve as the gate keeper in controlling substrate selectivity of CPO. Therefore, these two residues are likely to be the principal contact sites for the hydrophobic interactions between CPO and indole.
Table 2.
The dissociation constant (KD) of CPO-indole complex and the distances (r) between the protons of indole and the CPO heme irona at different pH
| pH | T1bs (s) | r (Å) | KD (mM)c | ||
|---|---|---|---|---|---|
| 2-H | 3-Hb | 2-H | 3-Hb | ||
| 3.0 | 7.0×10−5 | n.d. | 4.2 | n.d. | 25 |
| 3.5 | 5.6×10−5 | n.d. | 4.0 | n.d. | 25 |
| 4.0 | 6.2×10−5 | n.d. | 4.1 | n.d. | 23 |
| 4.5 | 5.6×10−5 | 3.2×10−4 | 4.0 | 5.4 | 25 |
| 5.0 | 6.0×10−5 | 4.1×10−4 | 4.1 | 5.6 | 23 |
| 5.5 | 6.1×10−5 | 3.8×10−4 | 4.1 | 5.5 | 23 |
| 6.0 | 6.8×10−5 | 3.7×10−4 | 4.2 | 5.5 | 21 |
| 6.5 | 6.0×10−5 | 4.1×10−4 | 4.1 | 5.6 | 23 |
The relaxation experiment were performed in 10 mM deuterated phosphate buffer (pH 3.0~6.5) containing 0.01 mM CPO to measure T1 of 2- and 3-protons by changing the concentration of indole from 2.0 to 12.0 mM.
n.d. means not determined owing to deuterated exchanged of the 3-proton.
The dissociation constants were derived from relaxation experiments of 2-proton.
Association of Indole with CPO in the Presence of Halide Ion
It is well known that halides (including chloride, bromide, and iodide) are co-substrates of CPO-catalyzed halogenations (3). Although CPO from Caldariomyces fumago is incapable of catalyzing the halogenation of indole, halide binding sites have indeed been identified by a recent X-ray diffraction study of CPO (62). To probe the effect of chloride and iodide on the binding of indole to CPO, the KD and the distances from 2- and 3-H to the heme iron in the presence of chloride and iodide were measured. Both KD and the distances displayed negligible dependence on the presence of chloride and iodide (Table 2 and Table 3), indicating that chloride and iodide do not affect the binding affinity and orientation of indole to CPO. This is consistent with the previous report of CPO-CPDO complex in the presence of chloride ion (23) and is supported by the absence of chloride binding sites in chloride-soaked crystals of CPO (2, 22). However, the essentially identical KD of CPO-indole complex in the presence of varying concentrations of iodide is somewhat surprising as CPO does have three iodide binding sites at the distal heme pocket (22). The effect of iodide on substrate binding was confirmed by our previous study on CPO-CPDO complex, which displayed a dramatic increase in KD (from 33 to 123 mM) as the concentration of iodide is increased (23). This was later rationalized by possible steric clashes between CPDO and CPO bound iodide, since the distance between the carbonyl oxygen of CPDO and CPO bound iodide is only 1.9 Å (22). It is thus proposed that the effect of iodide ion on the binding of substrates to CPO depends on the specific structure and nature of the substrates. Substrates such as CPDO that undergo halogenation reactions are affected by the presence of iodide, while substrates undergoing other transformations are not, implying that iodide binding results in little or no structural rearrangement for the substrate binding site of CPO. For CPO-catalyzed chlorination reactions, chloride ion is captured by compound I (67, 68) without the need for a bound chloride ion at the active site of CPO. This is plausible because the physiological environment where CPO functions has sufficient chloride ion to satisfy its need for chlorination reactions.
Table 3.
The dissociation constant (KD) of CPO-indole complex and the distances (r) between the protons of indole and CPO heme irona in the presence of chloride or iodide ion
| Cions (mM) | T1bs(s) | r (Å) | KD(mM)b | ||
|---|---|---|---|---|---|
| 2-H | 3-H | 2-H | 3-H | ||
| Cl− | at pH6.0 | ||||
| 10 | 6.5×l0−5 | 4.2×l0−4 | 4.1 | 5.6 | 24 |
| 20 | 5.5×l0−5 | 3.4×l0−4 | 4.0 | 5.4 | 25 |
| 30 | 5.6×l0−5 | 3.4×l0−4 | 4.0 | 5.4 | 24 |
| 40 | 6.2×l0−5 | 3.4×l0−4 | 4.1 | 5.4 | 22 |
| at pH 4.0 | |||||
| 20 | 7.1×10−5 | 4.2 | 20 | ||
| I− | at pH 6.0 | ||||
| 10 | 6.6×l0−5 | 3.8×l0−4 | 4.1 | 5.5 | 25 |
| 20 | 6.4×l0−5 | 3.4×l0−4 | 4.1 | 5.4 | 20 |
| 30 | 5.4×l0−5 | 4.0×l0−4 | 4.0 | 5.6 | 26 |
| 40 | 6.8×l0−5 | 3.7×l0−4 | 4.1 | 5.5 | 21 |
| at pH 4.0 | |||||
| 20 | 8.6×l0−5 | 4.3 | 20 | ||
The relaxation experiment were performed in 10 mM deuterated phosphate buffer (pH 4.0 and pH 6.0) containing 0.01 mM CPO and a certain concentration (Cion) of chloride or iodide ion to measure T1 of 2- and 3-protons by changing the concentration of indole from 2.0 to 12.0 mM.
The dissociation constants were evaluated from relaxation experiments of 2-proton.
Molecular Docking Models of Indole Bound to CPO
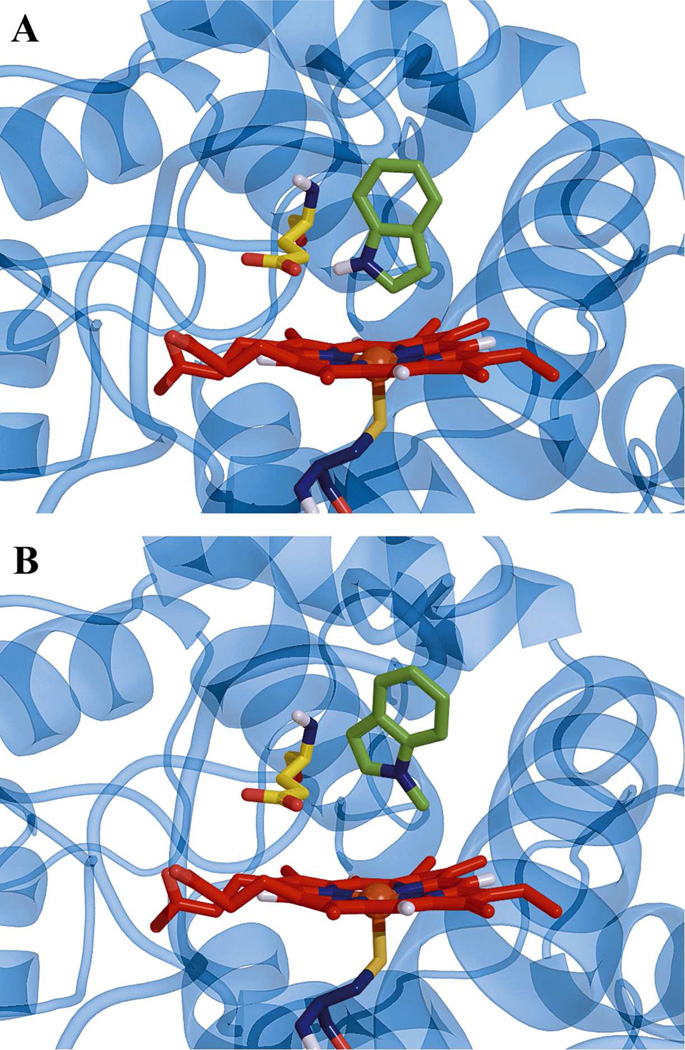
As a first step in determing the preferred orientations of indole within the active site of CPO, computer-aided docking of the molecules including indole and its N-methyl derivative into the active site of CPO was performed. During 60 Autodock runs, only one cluster of poses for indole (or its derivative) was observed. The key issue described here is the position and orientation of the substrate with respect to the heme and Glu 183 in the active site of CPO, since Glu 183 is postulated to be the acid-base catalyst in compound I formation (69, 70) and thus to tune the catalytic activity of CPO (71). Figure 4A shows the CPO-indole complex deduced from molecular docking. The pyrrole ring of indole faces the heme with the 2-H being the closest to the heme iron. The orientation of indole observed here is consistent with that derived from the distances between the protons of indole and the heme iron in our relaxation experiments (Table 1). Additionally, the N–H of indole is 1.7 Å from the γ-carboxyl group of Glu 183, indicating that, potentially, a hydrogen bond can be formed between the N–H of indole and Glu 183. This hydrogen bond, even if it does form, is not sufficient to be the major factor in keeping indole in its binding site in CPO (36). However, it is reported that the hydrogen bond between Glu 183 and the alcoholic OH group directs benzyl alcohol or 2-phenylethanol to a specific orientation (72). To further elucidate the function of a hydrogen bond formed between the N–H of indole and Glu 183, 1-methylindole was also docked into the active site (Figure 4B). It is observed that the methyl group of 1-methylindole points toward the heme, not Glu 183. Thus, the orientation of 1-methylindole is different from that of indole within the active site of CPO (Figure 4A). This is attributed to both the hindrance from a bulkier methyl group and the disruption of the putative hydrogen bond between indole and Glu 183. This also confirms that the N–H of indole does play a role in the binding of indole to CPO, since 1-methylindole is indeed hardly oxidized by CPO (36). Therefore, the observed negligible effect of pH on the KD of the CPO-indole complex from our NMR relaxation studies can be attributed to the extremely low pKa of Glu 183 within the active site of CPO.
Figure 4.
Ribbon and stick representations of (A) indole and (B) 1-methylindole docking in the active site of CPO. There was only one orientation of indole and 1-methylindole found in docking. The indole and 1-methylindole are colored green. Heme is shown as red sticks, and Glu 183 is shown as yellow sticks.
The binding energy, which is the free energy released by the formation of the complex between the substrate and the enzyme, is an important parameter for evaluating the bioaffinity of the substrate for the enzyme (73). The docking study resulted in binding energies of −23.2 kJ/mol for the CPO-indole complex and −21.9 kJ/mol for the CPO-1-methylindole complex. Compared to the binding of substrates to P450, such as indole (−53.5 kJ/mol) (74) and ticlopidine (−32.6 kJ/mol) (75) , the binding of indole to CPO is apparently weaker. This agrees with the conclusion from our NMR relaxation experiments (Table 1). The slight smaller (in magnitude) binding energy of 1-methylindole to CPO relative to that of indole, can be partly attributed to loss of a hydrogen bond with Glu 183.
Distance-Restrained Model of CPO Complexed with Indole
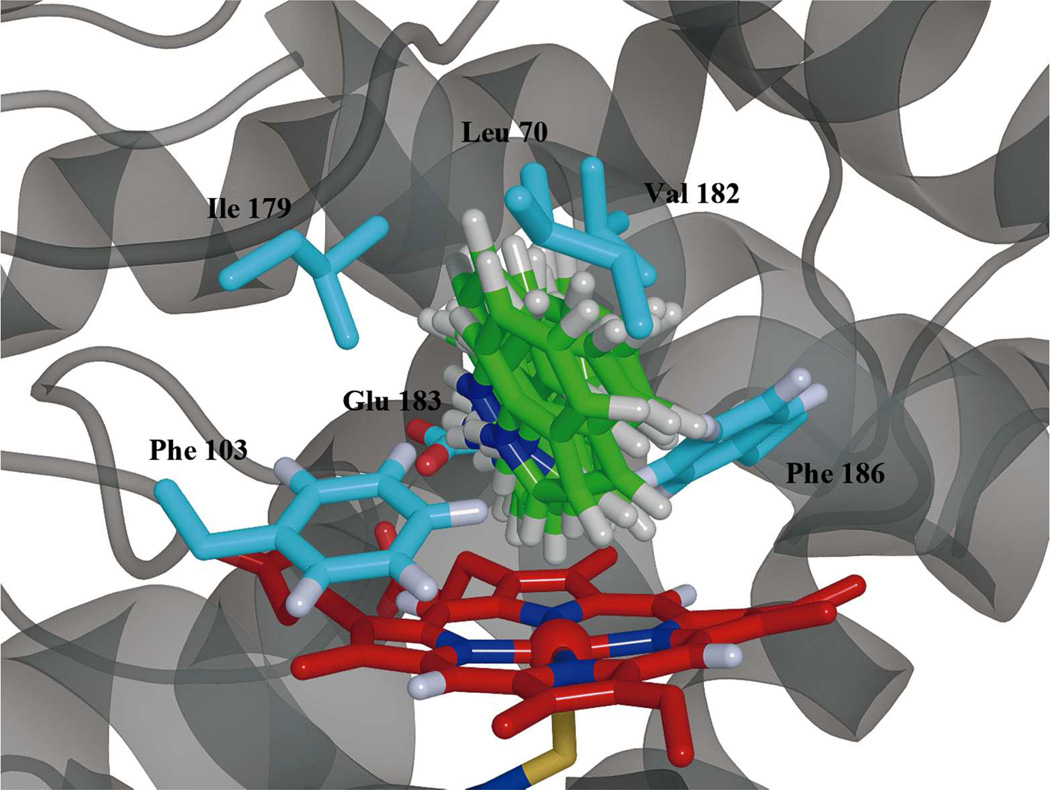
To further investigate the interaction between indole and CPO, simulated annealing of the CPO-indole complex was performed using the distances calculated from our NMR relaxation studies at pH 5.0 (Table S2). Mean interaction energies of CPO with indole were calculated for the last 50 ps of the simulatedannealing. The root-mean-square deviation (RMSD) of the coordinates of CPO in the CPO-indole complex relative to the crystal structure reaches a constant values (~1.4 Å) in the last 50 ps, indicating stability of the CPO-indole complex. Figure 5 depicts the structural model of indole in the active site of CPO deduced from simulated annealing. Ten equally-spaced snapshots from the last 50 ps of simulation are overlaid. The orientation of the indole is seen to be quite stable. The distances between the protons of indole and the heme iron observed in Figure 5 are ~2 Å smaller than those of NMR results (Table S2). This is consistent with previous results in which the distances between the protons of CPDO and the heme iron of the crystal structure of a CPO-CPDO complex (22) (4.9 Å) are much smaller than those of a relaxation measurement (23) (7.1 Å). Furthermore, differences between distances obtained with the distancerestrained model and the NMR results are also observed in a study of the CYP1A1-phenacetin complex (76). Such differences may be attributed to the fact that NMR results reflect an ensemble of multiple orientations of the substrate in the active site (75). In spite of such an ensemble, there should be only one major indole orientation as indicated by the significant broadness of NMR peaks of 2- and 3-protons of indole compared to other protons (Figure 1). Therefore, the distance-restrained model investigated here should be sufficient for studying the mechanism of regioselective oxidation catalyzed by CPO. It is observed that residues within 5 Å radius of indole are Val 67, Ile 68, Leu 70, Ala 71, Asn 74, Phe 103, Ile 179, Val 182, Glu 183, Phe 186, and Ala 267. The side-chain RMSDs of these residues, except for Glu 183, averaged over the last 50 ps are comparable to the corresponding root-mean-square fluctuations (RMSFs) (see Table S3). The RMSD of Glu 183 turned out to be much larger than the corresponding RMSF, suggesting that the substantial adjustment of the orientation of the charged Glu 183 side chain is a prerequisite for indole binding at the active site. This is consistent with a previous study (71) showing that Glu 183 mobility is critical for the catalytic reactivity of CPO. Furthermore, the residues in the active site strongly interacting with indole are of critical importance; identification of these residues can serve as a lighthouse for designing better CPO mutants for the oxidation of not only indole but also similar types of organic substrates. Therefore, the interaction energies between the active-site residues and the indole molecule were examined (Table S4).
Figure 5.
The distance-restrained model of indole in the active site of CPO. Residues within a 5 Å radius of indole are Val 67, Ile 68, Leu 70, Ala 71, Asn 74, Phe 103, Ile 179, Val 182, Glu 183, Phe 186, and Ala 267. The side chains of Leu 70, Phe 103, Ile 179, Val 182, Glu 183, and Phe 186 are shown for the sake of clarity. To differentiate the protein residues from the substrate, the indole molecule are colored green, the heme is colored red, and the carbons of the residues are colored blue.
The calculated Coulomb and Lennard-Jones potential energies of indole bound to CPO (Table S3) can be interpreted as representing electrostatic and hydrophobic interactions, respectively. A strong Coulomb interaction (−25.7 kJ/mol) was found between indole and Glu 183, indicating the presence of a hydrogen bond between bound substrate and Glu 183. This agrees with our molecular docking studies of this system. His 105 in CPO has been postulated to form a hydrogen bond with and modulate the acidity of Glu 183 (77, 78). The interaction between His 105 and Glu 183 helps hold Glu 183 in its place to interact with iron-bound hydrogen peroxide that facilitates the formation of compound I. It has been demonstrated that chemical modification of His 105 is detrimental to CPO-catalyzed oxidations (79). In order to learn whether His 105 can affect indole binding to CPO, the interaction between His 105 and indole was evaluated. It was found that only a small repulsive Coulomb interaction (4.3 kJ/mol) exists between His 105 and indole, indicating that His 105 has little direct effect on indole binding to CPO, although His 105 can indirectly influence indole binding via its effect on the orientation and pKa of Glu 183. Additionally, a weak Coulomb interaction (−2.1 kJ/mol) was found between indole and Val 67. Surprisingly, the Coulomb interaction between indole and Asn74 was also weak (−1.2 kJ/mol), indicating that the interaction between Asn 74 and indole is not essential for substrate binding. This is quite different from the case of polar substrates such as ethylene glycol and dimethyl sulfoxide, which interact with Asn 74 extensively (22).
The van der Waals interaction (−103 kJ/mol) was much higher than the Coulomb interaction between CPO and indole (−30 kJ/mol) from our simulated annealing studies. This indicates that the formation of the CPO-indole complex is mainly governed by hydrophobic interaction, rather than hydrogen bond or electrostatic interactions. Specifically, indole interacts strongly with several key hydrophobic residues including Leu 70, Phe 103, Ile 179, Val 182, and Phe 186 at the active center of CPO. These residues contribute −6.6 to −10.4 kJ/mol per residue to the formation of the CPO-indole complex (see Table S4). This agrees with reports that these hydrophobic residues interact significantly with hydrophobic substrates such as cis-β-methylstyrene (71, 80) and anthracene (81).
This structural model (Figure 5) is in good agreement with the fact that the catalytically active form of CPO is compound I intermediate with O as the axial ligand. Assuming an Fe-O bond of 1.65 Å in CPO compound I (82) and the structure of CPO-indole complex derived from distancerestrained model remain essentially unchanged after formation of compound I, the distance between oxoferryl oxygen and 2-C of indole would be ~2.6 Å. This short distance agrees with the direct transfer of oxygen to the oxidation site (2-C) of indole. Although the flexibility of CPO’s distal pocket is important to tune the catalytic reactivity of CPO (71), the assumption of the same structure may hold as the crystal structure of CPO-ligand adduct (21) and the hydroperoxoferric intermediate, compound 0 (83), are indistinguishable from that of CPO. Furthermore, the binding of substrates should weaken the Fe-O bond in the transition state, ensuring the efficient transfer of O from compound I to substrate, as demonstrated by the elongation of Fe-O distance in the oxidation of dimethyl sulfide by compound I of cytochrome P450 (84).
Mechanism of CPO-Catalyzed Oxidation of Indole
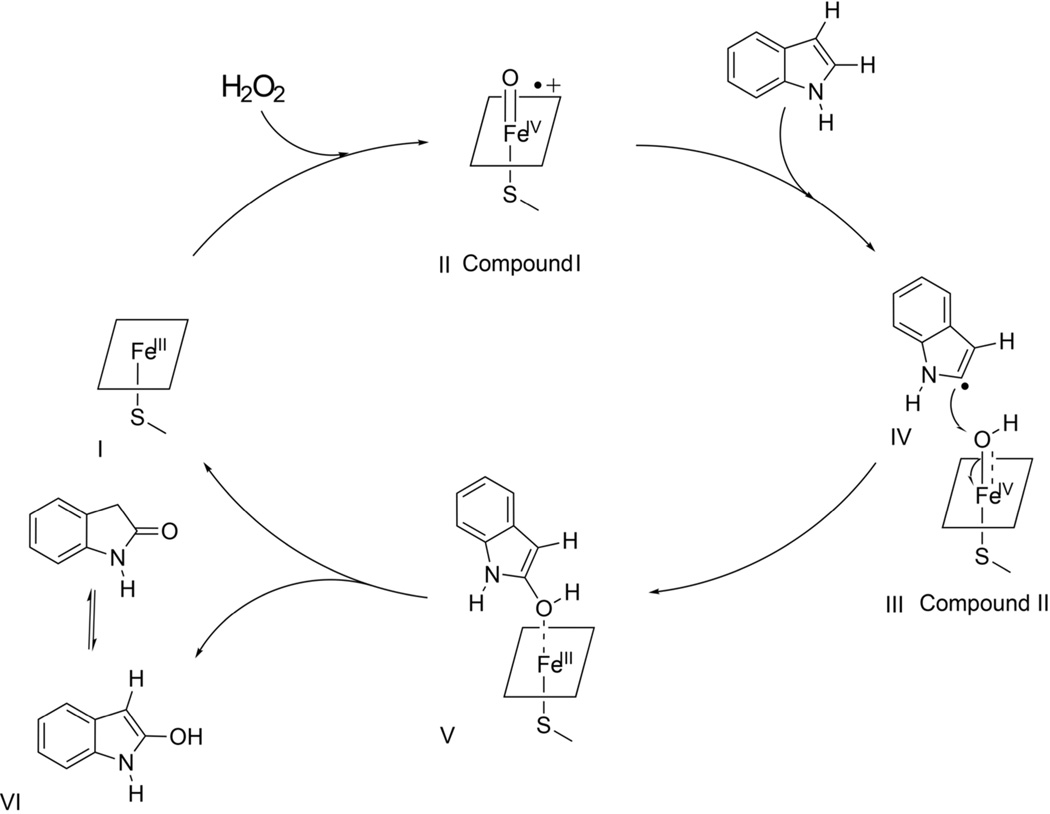
On the basis of our NMR relaxation measurements and the distance-restrained models presented in Figure 5, a mechanism for CPO-catalyzed regioselective oxidation of indole is proposed (Scheme 2). As for all CPO catalyzed reactions, the oxidation of indole is initiated by the binding of a neutral hydrogen peroxide molecule to the heme iron of CPO (FeIII, Scheme 2, I). Binding of H2O2 to the heme iron is facilitated by the transfer of one of the peroxide hydrogens to the carboxyl oxygen of Glu 183(2). The hydrogen ion is then delivered to the distal oxygen of the peroxide as the peroxide bond is heterolytically cleaved to produce an oxoferryl π-cation radical intermediate (FeIV=O•+, Scheme 2, II) known as compound I. Formation of compound I has been observed and characterized through many different techniques including electron paramagnetic resonances (EPR) (85), Mössbauer (85), X-ray absorption (82), and electron nuclear double resonance spectroscopy (86). Single electron reduction of compound I by indole converts compound I to compound II (Scheme 2, III, (87)) and generates an indole radical intermediate (Scheme 2, IV). The presence of substrate intermediates as carbon-centered radicals has been established in P450-catalyzed reactions (88). Additional evidence in favor of such a radical intermediate is provided by EPR studies of lactoperoxidase in the sulfoxidation (89) and by 1H NMR and EPR studies of a synthetic heme system in the epoxidation of styrene (90). In addition, the hydrogen-atom abstraction of the substrate is also presumed to be the mechanism in CPO-catalyzed demethylation of N,N-dimethylanilines (91). Green and Bukowski et al. suggested that the hydrogen-atom abstraction from reducing substrates is facilitated by the axial thiolate ligand (92, 93). Reduction of compound II results in the transfer of the ferryl oxygen to the substrate and the regeneration of the initial ferric resting state of CPO. Consequently, 2-hydroxyindole is formed (Scheme 2, V), which spontaneously tautomerizes to the stable oxindole (Scheme 2, VI). Similar mechanisms have been reported in CPO-catalyzed oxidative dehalogenation (94). Furthermore, our results clearly indicate that the consecutive electron transfers occur between the 2-position of indole and CPO intermediates, since 2-H of indole is pointing directly to the heme iron. Therefore, CPO-catalyzed oxidation of indole has a regioselectivity at the 2-position.
Scheme 2.
Proposed Mechanism of CPO-Catalzyed Oxidation of Indole.
CONCLUSION
The work presented here provides insights into the mechanism of CPO-catalzyed regioselective oxidation of indole. Association of indole with CPO was confirmed from both linewidth and longitudinal relaxation time measurements of the proton NMR signals of indole. Significantly, the precise orientation of indole in the CPO heme cavity was deduced from our results. The dissociation constant of the CPO-indole complex was calculated to be ~21 mM, and the distances between protons of indole (2-, 3-, 4-, 5-, 6-, and 7-H) and the heme iron of CPO were 4.3 Å, 5.6 Å, 9.2 Å, 10.5 Å, 10.7 Å, and 9.7 Å, respectively. In the pH range of 3.0–6.5, the dissociation constant of the CPO-indole complex and the position of indole in the active site are independent of solution pH. It was also found that the presence of halide (chloride and iodide) ion negligibly affect the stability of the CPO-indole complex and the binding geometry of indole in the active site of CPO at pH 6. Molecular docking of indole into the active site of CPO revealed that the pyrrole ring of indole points to the heme and the 2-H of indole is the closest to the heme iron, confirming the results from NMR studies of the CPO-indole complex. This indicates a direct-insertion mechanism of oxygen into the C–H bond at the 2-posion of indole and provides the first experimental explanation for the “unusual” regioselectivity of CPO catalyzed oxidation of indole. On the other hand, molecular docking of 1-methylindole reveals that the methyl group points toward the heme iron, not Glu 183 as in the case of indole. These docking studies suggest that the N–H of indole plays an important role in the binding of indole to CPO. The observed pH independence of indole binding to CPO in our pH effect experiments may be attributed to the low pKa of Glu 183, which remains fully deprotonated within the pH range tested in our experiments. Furthermore, simulated annealing of the CPO-indole complex indicates that the Coulombic interaction between indole and Glu 183 is much stronger than that between Val 67, Asn 74, and His 105. Finally, our study demonstrated that the association of CPO with indole is mainly governed by hydrophobic interactions rather than electrostatic interactions. This work provides the first experimental and theoretical explanation for the observed “unexpected” regioselectivity of CPO catalyzed oxidation of indole. Our results will also serve as a lighthouse in guiding the engineering of CPO into an efficient biocatalyst for synthetic and pharmaceutical applications.
Supplementary Material
ACKNOWLEDGMENT
The authors wish to acknowledge FIU’s NMR facility for the use of its Bruker Avance 600 MHz spectrometer sponsored by a grant from DOD (ARO:W911NF0411-0022).
Funding Sources
This research is supported by the National Science Foundation under Award No CHE-0540763 to X.W. (CAREER Award), a start-up fund from Florida International University, and the NIH under award number SC3GM83723 to D.C.
ABBREVIATIONS
- CPO
chloroperoxidase
- CPDO
1,3-cyclopentanedione
- CYP1A1
cytochrome P450, family 1, member A1
- DSS
4,4-dimethyl-4-silapentane-1-sulfonic acid
- KD
the dissociation constant
- MD
molecular dynamic
- MP2
second-order Møller–Plesset perturbation theory
- NMR
nuclear magnetic resonance
- RZ
Reinheitszahl
- RMSD
root-mean-square deviation
- SPC
simple point charge
- T1
longitudinal relaxation time
- P450
cytochrome P450, P450 BM3, Cytochrome P450 BM3
- PDB
protein data bank
- Pca
pyroglutamic acid
- PME
particle mesh Ewald method
Footnotes
ASSOCIATED CONTENT
Supporting Information. Details of the parameterization of the ferric heme for use with the GROMOS96 force field (Table S1), table of distances between protons of indole and the heme iron for the CPO-indole complex obtained from relaxation experiments at pH 5.0 and the corresponding distances observed in the simulation (Table S2), table of RMSDs and RMSFs of the active-site residues (Table S3), table of the energies averaged over last 50 ps of the simulation (Table S4), figure showing the distances between protons of indole and the heme iron as a function of the simulation time (Figure S1), and figure of RMSDs of the active-site residues during the last 50 ps of the simulation (Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Morris DR, Hager LP. Chloroperoxidase I. Isolation and properties of the crystalline glycoprotein. J. Biol. Chem. 1966;241:1763–1768. [PubMed] [Google Scholar]
- 2.Sundaramoorthy M, Terner J, Poulos TL. The crystal structure of chloroperoxidase: a heme peroxidase-cytochrome P450 functional hybrid. Structure (Cambridge, MA, U. S.) 1995;3:1367–1378. doi: 10.1016/s0969-2126(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 3.Hager LP, Morris DR, Brown FS, Eberwein H. Chloroperoxidase II. Utilization of halogen anions. J. Biol. Chem. 1966;241:1769–1777. [PubMed] [Google Scholar]
- 4.Osborne RL, Raner GM, Hager LP, Dawson JH. C. fumago chloroperoxidase is also a dehaloperoxidase: oxidative dehalogenation of halophenols. J. Am. Chem. Soc. 2006;128:1036–1037. doi: 10.1021/ja056213b. [DOI] [PubMed] [Google Scholar]
- 5.Kedderis GL, Koop DR, Hollenberg PF. N-demethylation reactions catalyzed by chloroperoxidase. J. Biol. Chem. 1980;255:10174–10182. [PubMed] [Google Scholar]
- 6.Allain EJ, Hager LP, Deng L, Jacobsen EN. Highly enantioselective epoxidation of disubstituted alkenes with hydrogen peroxide catalyzed by chloroperoxidase. J. Am. Chem. Soc. 1993;115:4415–4416. [Google Scholar]
- 7.Carmichael R, Fedorak PM, Pickard MA. Oxidation of phenols by chloroperoxidase. Biotechnol. Lett. 1985;7:289–294. [Google Scholar]
- 8.Colonna S, Gaggero N, Manfredi A, Casella L, Gullotti M, Carrea G, Pasta P. Enantioselective oxidations of sulfides catalyzed by chloroperoxidase. Biochemistry. 1990;29:10465–10468. doi: 10.1021/bi00498a006. [DOI] [PubMed] [Google Scholar]
- 9.Corbett MD, Baden DG, Chipko BR. Arylamine oxidations by chloroperoxidase. Bioorg. Chem. 1979;8:91–95. [Google Scholar]
- 10.Geigert J, Dalietos DJ, Neidleman SL, Lee TD, Wadsworth J. Peroxide oxidation of primary alcohols to aldehydes by chloroperoxidase catalysis. Biochem. Biophys. Res. Commun. 1983;114:1104–1108. doi: 10.1016/0006-291x(83)90676-9. [DOI] [PubMed] [Google Scholar]
- 11.Kiljunen E, Kanerva LT. Chloroperoxidase-catalysed oxidation of alcohols to aldehydes. J. Mol. Catal. B: Enzym. 2000;9:163–172. [Google Scholar]
- 12.Hager LP. A lifetime of playing with enzymes. J. Biol. Chem. 2010;285:14852–14860. doi: 10.1074/jbc.X110.121905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hager LP, Lakner FJ, Basavapathruni A. Chiral synthons via chloroperoxidase catalysis. J. Mol. Catal. B: Enzym. 1998;5:95–101. [Google Scholar]
- 14.Lakner FJ, Cain KP, Hager LP. Enantioselective epoxidation of ω-Bromo-2-methyl-1-alkenes catalyzed by chloroperoxidase. Effect of chain length on selectivity and efficiency. J. Am. Chem. Soc. 1997;119:443–444. [Google Scholar]
- 15.Lakner FJ, Hager LP. Chloroperoxidase as enantioselective epoxidation catalyst: an efficient synthesis of (R)-(−)-mevalonolactone. J. Org. Chem. 1996;61:3923–3925. doi: 10.1021/jo960074m. [DOI] [PubMed] [Google Scholar]
- 16.Zaks A, Dodds DR. Chloroperoxidase-catalyzed asymmetric oxidations: substrate specificity and mechanistic study. J. Am. Chem. Soc. 1995;117:10419–10424. [Google Scholar]
- 17.Dawson JH. Probing structure-function relations in heme-containing oxygenases and peroxidases. Science (Washington DC) 1988;240:433–439. doi: 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- 18.Dawson JH, Sono M. Cytochrome P-450 and chloroperoxidase: thiolateligated heme enzymes. Spectroscopic determination of their active-site structures and mechanistic implications of thiolate ligation. Chem. Rev. (Washington, DC U. S.) 1987;87:1255–1276. [Google Scholar]
- 19.Woggon WD, Wagenknecht HA, Claude C. Synthetic active site analogues of heme-thiolate proteins: Characterization and identification of intermediates of the catalytic cycles of cytochrome P450cam and chloroperoxidase. J. Inorg. Biochem. 2001;83:289–300. doi: 10.1016/s0162-0134(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 20.Torres E, Ayala M. Biocatalysis based on heme peroxidases: peroxidases as potential industrial biocatalysts. New York: Springer; 2010. Challenges in the application of peroxidases; pp. 209–352. [Google Scholar]
- 21.Sundaramoorthy M, Terner J, Poulos TL. Stereochemistry of the chloroperoxidase active site: crystallographic and molecular-modeling studies. Chem. Biol. (Cambridge, MA U. S.) 1998;5:461–473. doi: 10.1016/s1074-5521(98)90003-5. [DOI] [PubMed] [Google Scholar]
- 22.Kühnel K, Blankenfeldt W, Terner J, Schlichting I. Crystal structures of chloroperoxidase with its bound substrates and complexed with formate, acetate, and nitrate. J. Biol. Chem. 2006;281:23990–23998. doi: 10.1074/jbc.M603166200. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Goff HM. A nuclear paramagnetic relaxation study of the interaction of the cyclopentanedione substrate with chloroperoxidase. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1997;1339:88–96. doi: 10.1016/s0167-4838(96)00216-6. [DOI] [PubMed] [Google Scholar]
- 24.Bakhmutov VI. Practical nuclear magnetic resonance relaxation for chemists. Wiley; 2005. Paramagnetic NMR Relaxation; pp. 179–193. [Google Scholar]
- 25.Otting G. Protein NMR using paramagnetic ions. Annu. Rev. Biophys. 2010;39:387–405. doi: 10.1146/annurev.biophys.093008.131321. [DOI] [PubMed] [Google Scholar]
- 26.Modi S, Primrose WU, Boyle JMB, Gibson CF, Lian LY, Roberts GCK. NMR studies of substrate binding to cytochrome P450 BM3: comparisons to cytochrome P450 cam. Biochemistry. 1995;34:8982–8988. doi: 10.1021/bi00028a006. [DOI] [PubMed] [Google Scholar]
- 27.Solomon I. Relaxation processes in a system of two spins. Phys. Rev. 1955;99:559. [Google Scholar]
- 28.Casella L, Gullotti M, Selvaggini C, Poli S, Beringhelli T, Marchesini A. The chloroperoxidase-catalyzed oxidation of phenols. Mechanism, selectivity, and characterization of enzyme-substrate complexes. Biochemistry. 1994;33:6377–6386. doi: 10.1021/bi00187a001. [DOI] [PubMed] [Google Scholar]
- 29.Casella L, Gullotti M, Ghezzi R, Poli S, Beringhelli T, Colonna S, Carrea G. Mechanism of enantioselective oxygenation of sulfides catalyzed by chloroperoxidase and horseradish peroxidase. Spectral studies and characterization of enzyme-substrate complexes. Biochemistry. 1992;31:9451–9459. doi: 10.1021/bi00154a018. [DOI] [PubMed] [Google Scholar]
- 30.Bikiel DE, Boechi L, Capece L, Crespo A, De Biase PM, Di Lella S, Lebrero MCG, Martí MA, Nadra AD, Perissinotti LL. Modeling heme proteins using atomistic simulations. Phys. Chem. Chem. Phys. 2006;8:5611–5628. doi: 10.1039/b611741b. [DOI] [PubMed] [Google Scholar]
- 31.van Gunsteren WF, Dolenc J, Mark AE. Molecular simulation as an aid to experimentalists. Curr. Opin. Struct. Biol. 2008;18:149–153. doi: 10.1016/j.sbi.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Brünger AT, Adams PD, Rice LM. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure (Cambridge, MA, U. S.) 1997;5:325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 33.Clore GM, Schwieters CD. Theoretical and computational advances in biomolecular NMR spectroscopy. Curr. Opin. Struct. Biol. 2002;12:146–153. doi: 10.1016/s0959-440x(02)00302-0. [DOI] [PubMed] [Google Scholar]
- 34.Roberts AG, A SSE, Fomina N, Vu KT, Almutairi A, Halpert JR. NMR-derived models of amidopyrine and its metabolites complexed to rabbit cytochrome P450 2B4 reveal a structural mechanism of sequential N-dealkylation. Biochemistry. 2011;50:2123–2134. doi: 10.1021/bi101797v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts AG, Yang J, Halpert JR, Nelson SD, Thummel KT, Atkins WM. The structural basis for homotropic and heterotropic cooperativity of midazolam metabolism by human cytochrome P450 3A4. Biochemistry. 2011;50:10804–10818. doi: 10.1021/bi200924t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbett MD, Chipko BR. Peroxide oxidation of indole to oxindole by chloroperoxidase catalysis. Biochem. J. 1979;183:269–276. doi: 10.1042/bj1830269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Deurzen MPJ, van Rantwijk F, Sheldon RA. Synthesis of substituted oxindoles by chloroperoxidase catalyzed oxidation of indoles. J. Mol. Catal. B: Enzym. 1996;2:33–42. [Google Scholar]
- 38.Hallenberg PF, Hager LP. Purification of chloroperoxidase from Caldariomyces fumago . Methods Enzymol. 1978;52:521–529. doi: 10.1016/s0076-6879(78)52057-0. [DOI] [PubMed] [Google Scholar]
- 39.Hashimoto A, Pickard MA. Chloroperoxidases from caldariomyces (= leptoxyphium) cultures: glycoproteins with variable carbohydrate content and isoenzymic forms. J. Gen. Microbiol. 1984;130:2051–2058. [Google Scholar]
- 40.Vold RL, Waugh JS, Klein MP, Phelps DE. Measurement of spin relaxation in complex systems. J. Chem. Phys. 1968;48:3831–3832. [Google Scholar]
- 41.Rea V, Kolkman AJ, Vottero E, Stronks EJ, Ampt KAM, Honing M, Vermeulen NPE, Wijmenga SS, Commandeur JNM. Active site substitution A82W improves the regioselectivity of steroid hydroxylation by cytochrome P450 BM3 mutants as rationalized by spin relaxation nuclear magnetic resonance studies. Biochemistry. 2012;51:750–760. doi: 10.1021/bi201433h. [DOI] [PubMed] [Google Scholar]
- 42.Sakurada J, Takahashi S, Hosoya T. Nuclear magnetic resonance studies on the spatial relationship of aromatic donor molecules to the heme iron of horseradish peroxidase. J. Biol. Chem. 1986;261:9657–9662. [PubMed] [Google Scholar]
- 43.Cameron MD, Wen B, Allen KE, Roberts AG, Schuman JT, Campbell AP, Kunze KL, Nelson SD. Cooperative binding of midazolam with testosterone and α-naphthoflavone within the CYP3A4 active site: a NMR T 1 paramagnetic relaxation study. Biochemistry. 2005;44:14143–14151. doi: 10.1021/bi051689t. [DOI] [PubMed] [Google Scholar]
- 44.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 45.Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 46.Zong Q, Osmulski PA, Hager LP. High-pressure-assisted reconstitution of recombinant chloroperoxidase. Biochemistry. 1995;34:12420–12425. doi: 10.1021/bi00038a040. [DOI] [PubMed] [Google Scholar]
- 47.Pickard MA. A defined growth medium for the production of chloroperoxidase by Caldariomyces fumago . Can. J. Microbiol. 1981;27:1298–1305. doi: 10.1139/m81-199. [DOI] [PubMed] [Google Scholar]
- 48.Neese F, Wennmohs F. ORCA- an ab initio, DFT and semiempirical SCF-MO package [Google Scholar]
- 49.Head-Gordon M, Pople JA, Frisch MJ. MP2 energy evaluation by direct methods. Chem. Phys. Lett. 1988;153:503–506. [Google Scholar]
- 50.Weigend F, Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005;7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 51.Sanner MF. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 52.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 53.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr Sect. D: Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 54.Lemkul JA, Allen WJ, Bevan DR. Practical considerations for building GROMOS-compatible small-molecule topologies. J. Chem. Inf. Model. 2010;50:2221–2235. doi: 10.1021/ci100335w. [DOI] [PubMed] [Google Scholar]
- 55.Seminario JM. Calculation of intramolecular force fields from second-derivative tensors. Int. J. Quantum Chem. 1996;60:1271–1277. [Google Scholar]
- 56.Breneman CM, Wiberg KB. Determining atom-centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. J. Comput. Chem. 1990:361–373. [Google Scholar]
- 57.Berendsen HJC, Postma JPM, van Gunstetren WF, Hermans J. Intermolecular forces. Dordrecht: D. Reidel Publishing Company; 1981. Interaction models for water in relation to protein hydration; pp. 331–342. [Google Scholar]
- 58.Darden T, York D, Pedersen L. Particle mesh Ewald: an N·log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 59.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 60.van Deurzen MPJ, Remkes IJ, Van Rantwijk F, Sheldon RA. Chloroperoxidase catalyzed oxidations in t-butyl alcohol/water mixtures. J. Mol. Catal. A: Chem. 1997;117:329–337. [Google Scholar]
- 61.Huang WC, Westlake AC, Mar\'e cJD, Joyce MG, Moody PC, Roberts GC. Filling a hole in cytochrome P450 BM3 improves substrate binding and catalytic efficiency. J. Mol. Biol. 2007;373:633–651. doi: 10.1016/j.jmb.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 62.Constantine KL. Evaluation of site-directed spin labeling for characterizing protein-ligand complexes using simulated restraints. Biophys. J. 2001;81:1275–1284. doi: 10.1016/S0006-3495(01)75785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yue SY. Distance-constrained molecular docking by simulated annealing. Protein Eng. 1990;4:177–184. doi: 10.1093/protein/4.2.177. [DOI] [PubMed] [Google Scholar]
- 64.Pickard MA, Kadima TA, Carmichael RD. Chloroperoxidase, a peroxidase with potential. J. Ind. Microbiol. Biotechnol. 1991;7:235–241. [Google Scholar]
- 65.Manoj KM, Hager LP. Utilization of peroxide and its relevance in oxygen insertion reactions catalyzed by chloroperoxidase. Biochim. Biophys. Acta. 2001;1547:408–417. doi: 10.1016/s0167-4838(01)00210-2. [DOI] [PubMed] [Google Scholar]
- 66.Hinman RL, Whipple EB. The protonation of indoles: position of protonation. J. Am. Chem. Soc. 1962;84:2534–2539. [Google Scholar]
- 67.Butler A, Sandy M. Mechanistic considerations of halogenating enzymes. Nature (London) 2009;460:848–854. doi: 10.1038/nature08303. [DOI] [PubMed] [Google Scholar]
- 68.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Nature's inventory of halogenation catalysts: oxidative strategies predominate. Chem. Rev. (Washington, DC, U. S.) 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 69.Filizola M, Loew GH. Probing the role of protein environment in compound I formation of chloroperoxidase (CPO) J. Am. Chem. Soc. 2000;122:3599–3605. [Google Scholar]
- 70.Yi X, Conesa A, Punt PJ, Hager LP. Examining the role of glutamic acid 183 in chloroperoxidase catalysis. J. Biol. Chem. 2003;278:13855–13859. doi: 10.1074/jbc.M210906200. [DOI] [PubMed] [Google Scholar]
- 71.Morozov AN, Chatfield DC. Chloroperoxidase-catalyzed epoxidation of cis-β-methylstyrene: distal pocket flexibility tunes catalytic reactivity. J. Phys. Chem. B. 2012;116:12905–12914. doi: 10.1021/jp302763h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baciocchi E, Fabbrini M, Lanzalunga O, Manduchi L, Pochetti G. Prochiral selectivity in H2O2-promoted oxidation of arylalkanols catalysed by chloroperoxidase. Eur. J. Biochem. 2001;268:665–672. doi: 10.1046/j.1432-1327.2001.01924.x. [DOI] [PubMed] [Google Scholar]
- 73.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 74.Li W, Tang Y, Hoshino T, Neya S. Molecular modeling of human cytochrome P450 2W1 and its interactions with substrates. J. Mol. Graphics Modell. 2009;28:170–176. doi: 10.1016/j.jmgm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 75.Gay SC, Roberts AG, Maekawa K, Talakad JC, Hong WX, Zhang Q, Stout CD, Halpert JR. Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel. Biochemistry. 2010;49:8709–8720. doi: 10.1021/bi100914z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Q, Deshmukh RS, Ericksen SS, Tu Y, Szklarz GD. Preferred binding orientations of phenacetin in CYP 1A1 and 1A2 are associated with isoform-selective metabolism. Drug Metab. Dispos. 2012;40:2324–2331. doi: 10.1124/dmd.112.047308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen H, Hirao H, Derat E, Schlichting I, Shaik S. Quantum mechanical/molecular mechanical study on the mechanisms of compound I formation in the catalytic cycle of chloroperoxidase: an overview on heme enzymes. J. Phys. Chem. B. 2008;112:9490–9500. doi: 10.1021/jp803010f. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Tachikawa H, Yi X, Manoj KM, Hager LP. Two-dimensional NMR study of the heme active site structure of chloroperoxidase. J. Biol. Chem. 2003;278:7765–7774. doi: 10.1074/jbc.M209462200. [DOI] [PubMed] [Google Scholar]
- 79.Manoj KM, Hager LP. Chloroperoxidase, a Janus Enzyme? Biochemistry. 2008;47:2997–3003. doi: 10.1021/bi7022656. [DOI] [PubMed] [Google Scholar]
- 80.Morozov AN, D'Cunha C, Alvarez CA, Chatfield DC. Enantiospecificity of chloroperoxidase-catalyzed epoxidation: biased molecular dynamics study of a cis-β-methylstyrene/chloroperoxidase-compound I complex. Biophys. J. 2011;100:1066–1075. doi: 10.1016/j.bpj.2010.12.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aburto J, Correa-Basurto J, Torres E. Atypical kinetic behavior of chloroperoxidase-mediated oxidative halogenation of polycyclic aromatic hydrocarbons. Arch. Biochem. Biophys. 2008;480:33–40. doi: 10.1016/j.abb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 82.Stone KL, Behan RK, Green MT. X-ray absorption spectroscopy of chloroperoxidase compound I: insight into the reactive intermediate of P450 chemistry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16563–16565. doi: 10.1073/pnas.0507069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuhnel K, Derat E, Terner J, Shaik S, Schlichting I. Structure and quantum chemical characterization of chloroperoxidase compound 0, a common reaction intermediate of diverse heme enzymes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:99–104. doi: 10.1073/pnas.0606285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Porro CS, Sutcliffe MJ, de Visser SP. Quantum mechanics/molecular mechanics studies on the sulfoxidation of dimethyl sulfide by compound I and compound 0 of cytochrome P450: which is the better oxidant? J. Phys. Chem. A. 2009;113:11635–11642. doi: 10.1021/jp9023926. [DOI] [PubMed] [Google Scholar]
- 85.Rutter R, Hager LP, Dhonau H, Hendrich M, Valentine M, Debrunner P. Chloroperoxidase compound I: electron paramagnetic resonance and Mössbauer studies. Biochemistry. 1984;23:6809–6816. doi: 10.1021/bi00321a082. [DOI] [PubMed] [Google Scholar]
- 86.Kim SH, Perera R, Hager LP, Dawson JH, Hoffman BM. Rapid freeze-quench ENDOR study of chloroperoxidase compound I: the site of the radical. J. Am. Chem. Soc. 2006;128:5598–5599. doi: 10.1021/ja060776l. [DOI] [PubMed] [Google Scholar]
- 87.Green MT, Dawson JH, Gray HB. Oxoiron (IV) in chloroperoxidase compound II is basic: implications for P450 chemistry. Science (Washington, DC) 2004;304:1653–1656. doi: 10.1126/science.1096897. [DOI] [PubMed] [Google Scholar]
- 88.Cooper HLR, Groves JT. Molecular probes of the mechanism of cytochrome P450. Oxygen traps a substrate radical intermediate. Arch. Biochem. Biophys. 2011;507:111–118. doi: 10.1016/j.abb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tuynman A, Vink MKS, Dekker HL, Schoemaker HE, Wever R. The sulphoxidation of thioanisole catalysed by lactoperoxidase and Coprinus cinereus peroxidase: evidence for an oxygen-rebound mechanism. Eur. J. Biochem. 2001;258:906–913. doi: 10.1046/j.1432-1327.1998.2580906.x. [DOI] [PubMed] [Google Scholar]
- 90.Gross Z, Nimri S. Seeing the long-sought intermediate in the reaction of oxoiron (IV) porphyrin cation radicals with olefins. J. Am. Chem. Soc. 1995;117:8021–8022. [Google Scholar]
- 91.Takahashi A, Kurahashi T, Fujii H. Redox potentials of oxoiron (IV) porphyrin π-cation radical complexes: participation of electron transfer process in oxygenation reactions. Inorg. Chem. 2011;50:6922–6928. doi: 10.1021/ic102564e. [DOI] [PubMed] [Google Scholar]
- 92.Bukowski MR, Koehntop KD, Stubna A, Bominaar EL, Halfen JA, E M, Nam W, Que L., Jr A thiolate-ligated nonheme oxoiron (IV) complex relevant to cytochrome P450. Science (Washington, DC) 2005;310:1000–1002. doi: 10.1126/science.1119092. [DOI] [PubMed] [Google Scholar]
- 93.Green MT. CH bond activation in heme proteins: the role of thiolate ligation in cytochrome P450. Curr. Opin. Chem. Biol. 2009;13:84–88. doi: 10.1016/j.cbpa.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 94.Osborne RL, Coggins MK, Terner J, Dawson JH. Caldariomyces fumago chloroperoxidase catalyzes the oxidative dehalogenation of chlorophenols by a mechanism involving two one-electron steps. J. Am. Chem. Soc. 2007;129:14838–14839. doi: 10.1021/ja0746969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.