Abstract
Two common sources of DNA for whole exome sequencing (WES) are whole blood (WB) and immortalized lymphoblastoid cell line (LCL). However, it is possible that LCLs have a substantially higher rate of mutation than WB, causing concern for their use in sequencing studies. We compared results from paired WB and LCL DNA samples for 16 subjects, using LCLs of low passage number (<5). Using a standard analysis pipeline we detected a large number of discordant genotype calls (approximately 50 per subject) that we segregated into categories of “confidence” based on read-level quality metrics. From these categories and validation by Sanger sequencing, we estimate that the vast majority of the candidate differences were false positives and that our categories were effective in predicting valid sequence differences, including LCLs with putative mosaicism for the non-reference allele (3–4 per exome). These results validate the use of DNA from LCLs of low passage number for exome sequencing.
Keywords: graphical diagnostics, lymphoblastoid cell line, mosaicism, sequence variant call, strand bias, somatic mutation
1. Introduction
Next-generation sequencing (NGS) has become an affordable tool to probe human genomes for rare variants affecting risk for disease. Association analyses are being applied to NGS data to discover loci with rare variants that affect the risk of a wide spectrum of diseases. Nevertheless, it is becoming increasingly apparent that very large sample sizes and widespread availability will be required to attain significant results in many of these studies. To attain large samples, scientists would make use of large collections of DNA from subjects with immortalized lymphoblastoid cell lines (LCLs) in repositories such as the NIMH Center for Collaborative Genomic Disorders on Mental Disorders (http://nimhgenetics.org; hereafter the “NIMH Repository”) at the Rutgers Cell and DNA Repository (RUCDR). However, due to reports of large numbers (20 per exome) of non-germline mutations in LCLs [1], concerns now exist about use of LCL-derived DNA for sequencing studies. The concern is well-founded, because immortalization involves transformation of lymphocytes with cytomegalovirus (CMV) and this, combined with extensive serial passaging of cells, could lead to DNA sequence changes. Ultimately, mutation underlies all heritable genetic variation, but it has recently become apparent that mutations that have arisen de novo can be highly informative in identifying genetic risk factors for disorders such as autism and schizophrenia [2], [3], [4], [5], [6], [7]. Non-germline mutations (and potential artifacts) are particularly troubling in this setting, thus motivating a careful investigation of the quality of LCLs as a basis for such studies.
We sought to empirically test the degree to which mutations that arise in LCLs might impact results from WES studies. We conducted WES on paired DNA samples derived from whole blood and LCL of low passage number for 16 subjects. Cell lines were obtained from the NIMH Repository repository at RUCDR (http://www.rucdr.org/) or from the Icahn School of Medicine at Mount Sinai (ISMMS). Specifically, LCLs had been cultured through a maximum of four passages, or serial dilutions, in which a relatively small number of cells seeds a new culture to greatly expand the cell population. These cell populations are used for DNA isolation or cryopreservation of multiple new culture aliquots. Here we report (1) initial detection of genotype calls discordant between paired WB and LCL samples; (2) a novel filtering and prioritization algorithm that effectively assigns putative differences into categories of “confidence” as demonstrated by subsequent validation experiments; (3) that the vast majority of called differences are false positives and that those that do validate reflect DNA mutations that arose in the cell line; (4) in nearly all cases true positives appear as low level mosaic non-reference alleles; and finally (5) that our studies find only 3–4 valid sequence differences per subject. This work strongly supports the use of low passage number LCL samples for use in WES studies and the need for rigorous filtering algorithms that can discriminate variants most likely to be real.
2. Results
2.1. Identification of the Candidates
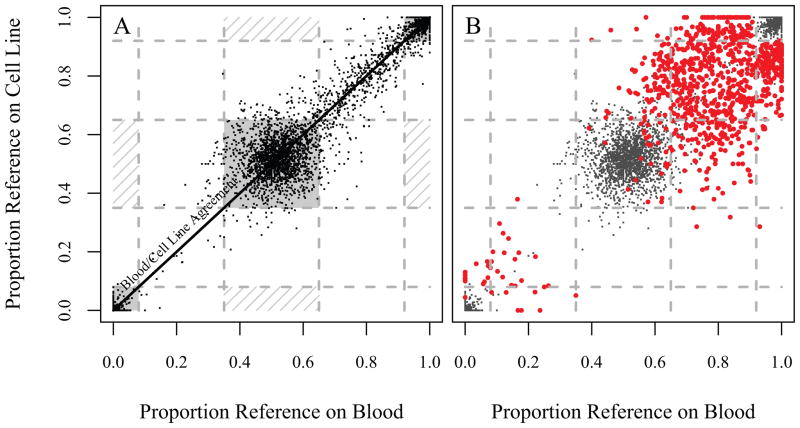
Following preliminary filtering, 15,099 call pairs were candidates for disagreement between the 16 WB and LCL pairs; further filtering reduced this count to 864 pairs (Supporting Information). Graphical inspection of the data, such as plotting the balance of reference and alternative calls, yielded clear evidence that further culling was required. First consider a random sample of variants that show agreement between blood and cell line where the proportion of the blood reads per locus that were called the reference base (horizontal axis) is plotted against the corresponding proportion of the cell line calls that were called the reference (vertical axis, Fig. 1A). Figure 1A also delineates natural (but arbitrarily chosen) regions in which one would expect the most confident homozygote matches, heterozygote matches, and mismatches would fall. The majority of matching pairs fall into regions where one would expect them (the shaded regions), but there are also some that lie close to the “Blood/Cell Line Agreement” line which are neither confident homozygotes nor heterozygotes. For these pairs, even though the variant caller was not confident that the site was either homozygote or heterozygote, the blood and cell line reads were similar enough in the proportion reference that the blood and cell line genotype calls agreed. There are no points that lie in the regions where one would expect confident Blood/Cell Line disagreements (the hashed regions).
Figure 1.
Proportion of reference allele reads for blood versus cell line. A. A random sample of blood/cell line pairs that had matched calls. These are largely high-quality calls, and hence most of the points lie in the corners (for homozygotes) or in the center (for heterozygotes). These regions are shaded, and can be interpreted as the the areas which will contain the confident blood/cell line matches. The hashed regions are those where confident mismatches would be expected. Of course, as these are matches, most of these points lie near to the axis of blood/cell line agreement. B. The red dots represent the 864 blood/cell line disagreements, the gray are a subsample of the matches. Note that the mismatches largely fall in a region outside of where high-quality homozygote and heterozygote calls would be expected: in the corners and the center, respectively.
Now consider the 864 mismatches (Fig. 1B), which clearly fall into the “unconfident” call region; there is a strong correlation between the proportion reference on both the blood and cell line, but the calls disagree. This disagreement can, in most cases, be attributed to the fact that random variation has yielded one homozygote call (in either blood or cell line DNA) and one heterozygote call; however, the evidence of a real difference is weak.
2.2. Segregation of the Candidates
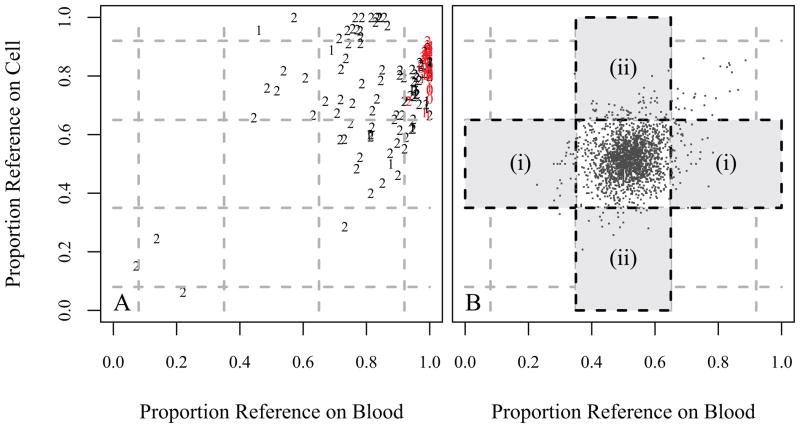
Each of the 864 candidates was segregated into 4 groups of “confidence” based on deeper inspection of the read quality, evidence for strand bias, and the relative balance of reference/alternative calls, and also by whether the heterozygote fell in blood or cell line. The candidates in each of Groups 0, 1, and 2 are shown in Section D of the Supporting Materials, along with a list of the Group 3 candidates that were tested for validation. Also, Section E of the Supporting Material includes a depiction of the reads for each of these ten cases. Plotting Groups 0 to 2 candidates in the same axis system as used previously (Fig. 2A) shows wide variation for Group 2 pairs, but many Group 0 and 1 pairs fall on the axis such that the proportion reference on blood is one.
Figure 2.
A. The 1,957 blood/cell line heterozygote matches that passed through the same filtering as applied to the mismatches. These largely fall into the central “high-quality” region. The instances in regions (i) or (ii) are potentially missed mutations or unbalanced reads. Since the count in region (i) is roughly equal to the count in region (ii) (43 and 53, respectively), the evidence suggests the majority of these observations are unbalanced reads. B. The candidates in Groups 0, 1, and 2. Note that most of the most confident candidates (“0’s” and “1’s”) are along the far right, with proportion reference on blood equal to one. The results of the validations confirmed that groups 0 and 1 consisted mostly of mosaics (shown in red). This explains why the proportion reference on blood was very close to one, while the proportion reference on cell line was between 2/3 and 1. Note that candidates in Groups 0 and 1 are largely of high quality, i.e., they do not possess other characteristics that make them of questionable quality.
A pair of “Group 1” candidates is in the upper center of the plot (Fig. 2A, Supplemental Table 5); this is the only instance among the top two groups for which a call went from heterozygous on blood to homozygous on cell line. While overall 44% of the entries in Table 1 declare blood heterozygous and cell homozygous, it is notable that only 19% of the confident differences (levels 0–2) differ in this direction. Of the high confidence entries with cell homozygous two pair are physically adjacent (Chromosome 9, Individual 2, and Chromosome 11, Individual 5) suggesting alignment problems.
Table 1.
Summary of validation results for candidate mismatches sorted by decreasing level of confidence.
| Group | Total Count | Vaid. Attempt | Valid Success | Confirmed Mismatch | Confirm. Rate | Blood Hetero. | Ti/TV Ratio |
|---|---|---|---|---|---|---|---|
| 0 | 10 | 10 | 10 | 10 | 100% | 0 | 4.00 |
|
| |||||||
| 1 | 28 | 28 | 26 | 18 | 69% | 2 | 2.11 |
|
| |||||||
| 2 | 107 | 107 | 104 | 15 | 14% | 26 | 0.91 |
|
| |||||||
| 3 (Strong) | 34 | 24 | 13 | 3 | 1.1%† | 352 | 0.47 |
| (Weak) | 685 | 9 | 9 | 0 | |||
This cell is estimated based on a weighted average.
2.3. Validation of Selected Candidates
For Group 0, all ten of candidates for blood/cell line mismatch were confirmed to be mismatches by follow-up Sanger sequencing (Table 1), which revealed small amplitude non-reference peaks on both forward and reverse strands. For Groups 1 and 2, however, the percentages drop to 69% and 14%, respectively (Table 1). There are a handful of cases for which the validation was unsuccessful due to sequencing problems; these are excluded from the count in “Validations Successful” column. Tables 4 through 7 in the Supporting Material show more detail for the validation results.
The results from the validation of Group 3 candidates are also shown in Table 1, divided into “Strong” and “Weak” candidates, as described in Methods. Taking a weighted average of the two subgroups, we estimate that the rate of mismatch in Group 3 is 1.1%. Assessing the Ti/Tv ratio of all candidates in each of the groups, there is a clear decreasing trend as quality decreases. Among all confirmed mismatches, the Ti/Tv ratio is 3.18.
Finally, we did not validate any candidates for which a call went from heterozygous on blood to homozygous on cell line. For each candidate, based on Sanger sequencing, we determined that WB was actually homozygous, or LCL was heterozygous leading to concordant calls. This result supports our conjecture that in the process of creating LCL from WB, errors are much more likely to result in homozygous loci becoming heterozygous.
2.4. Checking for Missed Mutations
We note that with our current filtering procedure we miss the opportunity to discover mutations when the truth is that blood is homozygous and cell line is heterozygous, but the variant call is that both are heterozygous. We can assess the probability of this occurring by inspection of the variant calls.
Figure 2B shows the 1,957 calls where WB and LCL agreed on the heterozygote call (following the filtering procedure). It is evident that most of these fall into the center region of the plot, as to be expected with high-quality heterozygote calls. Here we consider two regions away from the center, in an effort to determine if we are missing a significant number of cases where the truth is that WB is homozygous and LCL is heterozygous. Such cases, if they existed, would likely fall into one of the regions labelled (i), since such cases should have a relatively large or small proportion of WB reference calls. Region (ii) is create for comparison; if the number of cases falling into these two regions are roughly equal, then there would appear to be little evidence of missed WB-to-LCL mutations. In our data there are 43 instances that fell into region (i), but 53 that were in region (ii).
2.5. Inspection of Other Potential Mismatches
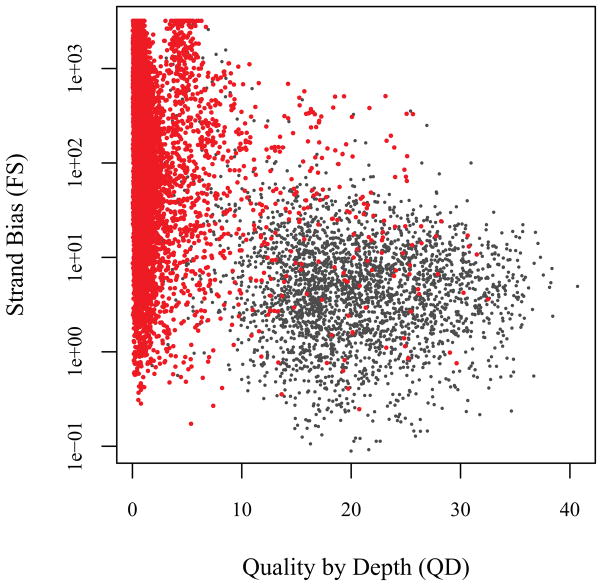
To ensure that we have not overscreened the candidate mismatches, we assess the full 15,099 variants for which LCL and WB mismatch. This examination focused on two standard quality metrics, the quality by depth (QD) and a quantification of the amount of strand bias present at the locus (Fig. 3). The strand bias measure FS is the Phred-scaled p-value from an application of Fisher’s exact test to the two-by-two table formed by the factors “direction of the strand” and “reference or non-reference base call.” Hence, the p-value will be small (and FS large) when evidence for a heterozygote call is largely present only in either the forward or reverse direction. This is a significant challenge for the remaining candidates, as there are many for which FS is large. In fact FS is large for the large number of candidates for which the reference proportion is not close to 0, 0.5, or 1 because of erroneous mapping of one of the four direction/chromosome combinations (results not shown). It is also apparent that strand bias is related to low average quality reads (low QD) providing further evidence that these calls are dubious (Fig. 3). The mismatched variants that fall into the acceptable quality region of the display (Fig. 3) exclude all but the 864 variants that were investigated further in the validation study. This analysis suggests that the mismatching variants excluded from the validation study are of even lower quality than those in the Level 3, weak variant category.
Figure 3.
Quality metrics in variants with candidate mismatches. Strand bias (FS) versus Quality by Depth (QD) is shown for each of the candidates (in red), and for a subset of the matched pairs (in gray). Note that there is a large amount of strand bias among the mismatched calls (high FS), and that these calls are largely of poor quality (low QD).
2.6. Inferences
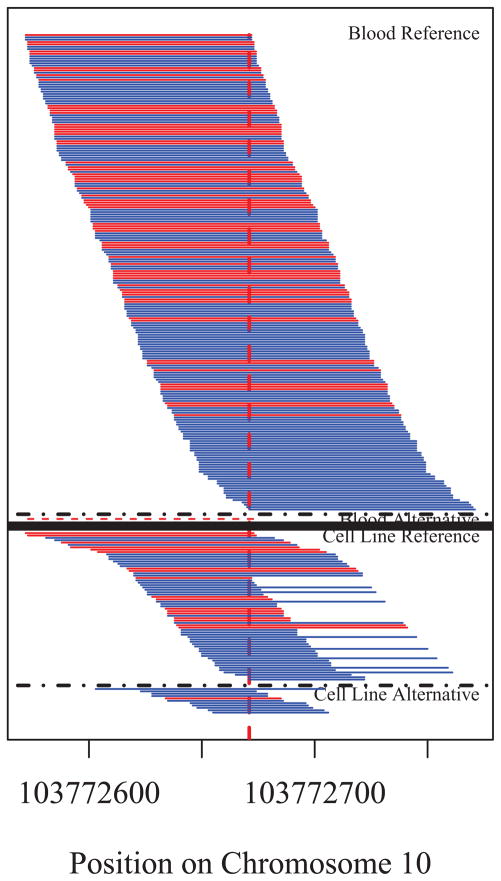
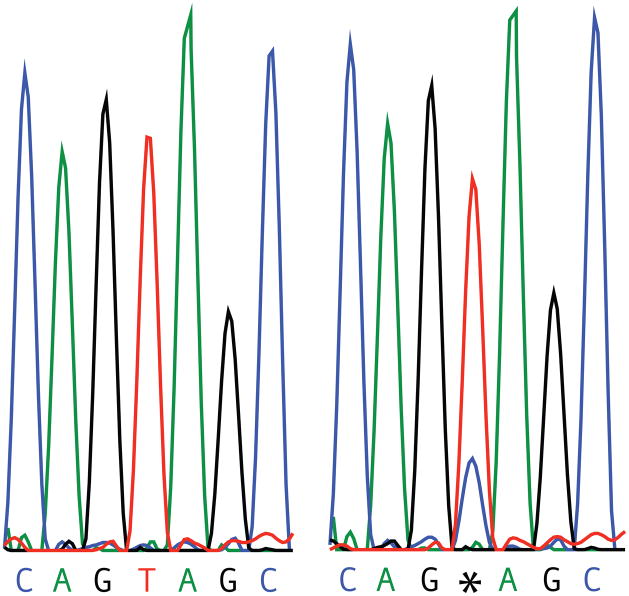
A notable feature of the validation results is that in those cases for which a blood/cell line disagreement was confirmed, the locus was homozygous for the allele on blood, but then, with only one possible exception, displayed the characteristics of mosaicism within the cell line. The mosaic nature of these loci is evident not only in the results of the targeted validation via Sanger sequencing, but also in the original WES results. For example, consider one of the candidates in Group 2: Individual 4, on Chromosome 10 at position 103772671. In a pileup diagram showing the individual NGS reads as horizontal lines (Fig. 4), all but one of the reads in the blood sample were for T, the reference allele; yet for the cell line sample, a majority (62 of 73) of the reads were for T, but a quarter returned C. This is far from the 50/50 split one would expect if the site were heterozygous. Sanger sequencing results were consistent in that the electropherogram (Fig. 5) reveals a peak for both T and C for the cell line sample. This example of relative over-representation of the reference allele is consistent with virtually all of the other instances of validated blood/cell line disagreements. The exception is a single candidate for which the Sanger trace is balanced in one direction and unbalanced in the other direction.
Figure 4.
Pileup diagram from the sequencing results of Individual 4, on Chromosome 10 at position 103772671, a confirmed mosaic. Note that although there is a large number of reads for the alternative allele on the cell line, the proportion of such reads is much less than one half of all of the reads. The dashed line under “Blood Alternative” represents a read which did not match the either the reference or alternative allele for this site.
Figure 5.
Results from Sanger sequencing of a confirmed mosaic. This is from Individual 4, on Chromosome 10 at position 103772671, as were was the results from Figure 4. Note the additional peak in the chromatogram on the right. This result confirmed that this site was a mosaic, as suggested in Figure 4.
In addition there were no confirmed mismatches among the candidates for which blood was called heterozygous and cell line was called homozygous. Hence, the overall mismatch rate per person is also our estimate of the overall rate of mosaics per person. This estimate can be obtained by taking a weighted average of the rate within each group, with weight calculated using the proportion of the candidates within that group (Table 1). Hence, the probability of a randomly chosen candidate being a mosaic is approximately 6.1%.
The estimated number of mosaics per individual is hence 0.0609 × 864/16 = 3.29. This estimate is predicated on the assumption that loci that were filtered in the initial steps of the processing consist entirely of mismatches that can be attributed to variant caller errors and poor quality reads. (Recall that the list of 864 candidates was culled from the initial set of 15,099 blood/cell line differences, see Supplemental Materials, Tables 1–3.) It is most natural to think of the 14,235 loci that were excluded from the focused, read-level analysis as forming a fifth group, one whose quality metrics are even worse than the weak candidates in Group 3, and that among the loci in this group, a very small percentage would be true blood/cell line mismatches.
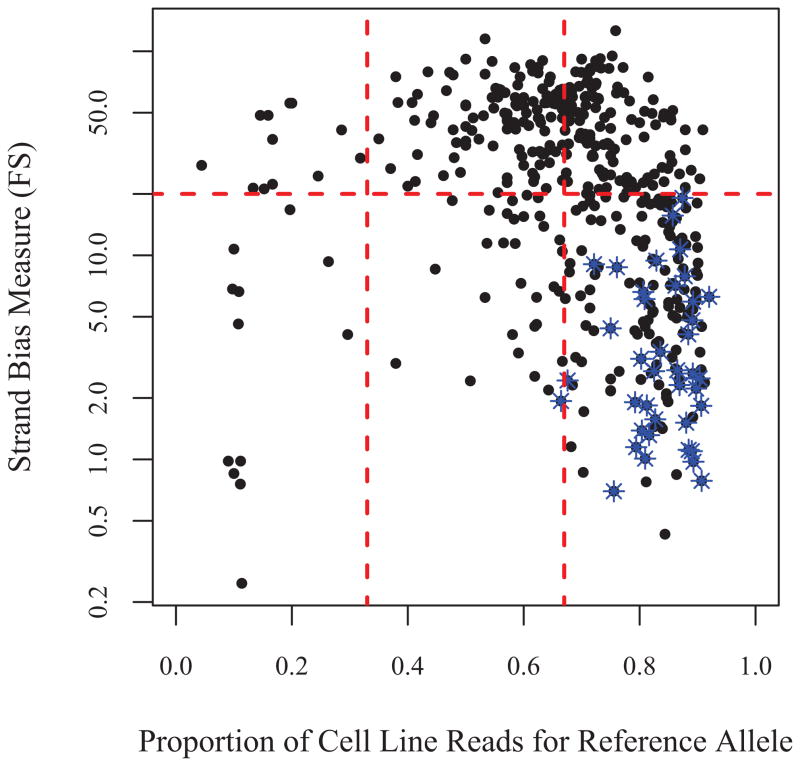
Finally, in practice with only cell line data available one could not assess the suspicion flags to discover false calls. Here we investigate whether or not the 864 false heterozygote calls could have been successfully filtered out using only the meta information available in the output produced by GATK from a single vcf. In Figure 6 we plot the 484 candidate mismatched LCL heterozygotes as a function of two variables by which to assess the quality of these calls. By requiring the proportion of reads for reference (or alternative) allele to be less than 67%, all but one of the validated mosaics is eliminated. In addition, imposing an additional filter for strand bias (FS < 20) we remove the majority of the apparent false heterozygotes. After imposing these 2 filters jointly we retain only 33 calls; that is about 2 mismatches per individuals. Our previous analysis suggests that none of the mismatched calls are true non-mosaic mismatches. Hence we conclude that these simple filters can eliminate most false positives. Moreover, it is interesting to note that all of the validated mosaics passed the strand bias filter, and that about 19% of the calls remaining after imposing this FS-filter are mosaics rather than simple false heterozygotes.
Figure 6.
Plot of strand bias (FS) versus proportion reference reads for the 484 heterozygous LCL calls among the candidates. Since each of these was called as homozygous on blood, these are all assumed to be either incorrect calls or mosaics on cell line. The mosaics are indicated by the blue stars. Most of these calls could have been filtered by constraining the proportion reference to be between 1/3 and 2/3, and ensuring that FS, the strand bias measure returned by GATK, is less than 20.
3. Discussion
To identify mutations in LCLs we compared results from WES of DNA from both whole blood and LCLs for 16 subjects. All LCLs were at low passage (< 5) and, therefore, the samples are much more likely to be representative of those in widely accessed public repositories. After filtering based on standard quality metrics, we identified 864 discordant genotype calls between blood and LCL samples (approximately 50/subject). These candidates were further prioritized based on read-level analyses of sequence data, including evaluation of read depth, base-call, mapping quality and read direction. This permitted segregation of candidate blood/LCL differences into four categories of decreasing “confidence.” We used Sanger sequencing to evaluate each candidate variant in the top 3 confidence levels (N=145 variants), as well as some of the lower level candidates. The results of this validation study yielded few instances of confirmed blood/LCL differences, and, notably, in virtually every instance the difference manifested as low-level mosaicism in the LCL. Aside from these apparently mosaic loci, there was only one instance of a sequence variant arising in the cell line and showing approximately equal allelic representation. The evidence for mosaicism was present not only in the Sanger sequencing, but also in the WES results. Hence, most of the 864 candidate differences were false positives, and the remainder had characteristics that would cause them to be filtered by standard variant calling procedures.
There are two possible sources for the mosaicism. First, there is a possibility that mutations accumulate in the cell lines during transformation and subsequent culturing. Second, lymphocytes represent a mixture of different cells that have undergoing genomic changes from which clonal progeny are derived. Any one of these clones could have a point mutation, which could be amplified during subsequent culturing. In either case, low passage of the LCLs and the careful analysis of sequencing traces for mosaicism would help identify such sites.
Although we have not tested higher passage LCLs, our results do point towards an elevated risk of using such samples. We identified 46 mosaic LCL mutations in the exomes of 16 subjects and predict fewer than ten remain in the unvalidated sample suggesting a rate of 3.3 per person per exome. Prior studies [5] predict a rate of 1 germline mutation per exome per person. Our estimated ratio of 3.3:1 non-germline to germline de novo mutation comports well with a prior estimate [2] of 1:1 and differs sharply from the estimate of 20:1 reported from HapMap samples [1]. As noted [1], a difference in mutation rate could be caused by the number of passages of the cell line as well as other factors, including external factors affecting mutation rate. The reduced and tractable number of mutations we observed likely reflects the focus on lower passage cells.
In earlier research, the suitability of LCL for use in genetic studies was evaluated in the context of single nucleotide polymorphism (SNP) arrays [8]. To test for genotypic errors potentially induced by the Epstein-Barr Virus transformation process, this study compared SNP genotype calls in WB and LCL from the same individuals using cells that were not passaged after immortalization. Genotypic discrepancies found in the matched WB and LCL pairs were not notable relative to differences observed among control pairs, suggesting that most genotypic discrepancies were due to technical artifacts rather than the transformation process. Prior studies also supported the conclusion that LCL have a minor effect on genomic structural variation [9, 10, 11, 12]. Even in the HapMap samples, putative LCL-specific genomic errors accounted for less than 0.5% of observed deletions [9].
The results of our work could be used to identify mosaic loci in studies that make use of LCL DNA. In particular, the evidence for mosaicism is most clearly defined by significant imbalance in the traces showing the reference and variant allele, but it is also frequently apparent in WES statistics. Even without further filtering, the number of mosaics is sufficiently modest such that LCL-derived DNA is appropriate for gene discovery using case-control analyses of rare variants. If the mosaic loci were called as variant alleles, there would be minimal loss of power in gene discovery studies in complex genetics, because many samples are used and multiple independent hits are required for statistical evidence of association. For analyses of de novo variants in families, the statistics are more sensitive to errors. Nevertheless, multiple hits in the same gene are required for implicating a novel gene as one affecting risk [4], providing some measure of protection against false discoveries. Moreover, it is standard practice to validate de novo calls and this would typically identify mosaics. Important findings can be further validated in blood samples, when available, as well as independent samples. If validation in blood is not feasible, observations derived from individuals with numerous it de novo mutations could be downweighted in analysis. In summary, our results, which are restricted to LCL of low passage, support the use of LCL-derived DNA for NGS sequencing in research.
4. Material and Methods
4.1. Whole Exome Sequencing
The sixteen samples comprising this study correspond to ten subjects (autism probands) recruited at Vanderbilt University (VU) and another six subjects recruited at ISMMS. WB DNA was extracted at either site using Qiagen Puregene kit. LCLs for VU samples were established at and obtained from the NIMH Repository at RUCDR, and those from ISMMS were transformed locally using standard procedures.. Exome capture and sequencing was performed at VU and ISMMS using very similar methods. Genomic DNA (~3 ug) was sheared to 200–300 bp using a Covaris Acoustic Adaptor, and (VU) DNA purified using Agencourt’s AMPure XP Solid Phase Reversible Immobilization paramagnetic (SPRI) beads. Fragments were end-repaired, dA-tailed, and sequencing adaptor oligonucleotides ligated using reagents from New England BioLabs (Beverly, MA; http://neb.com/). Libraries were barcoded using the Illumina index read strategy, which uses six-base sequences within the adapter that are sequenced separately from the genomic DNA insert. Ligated products were size selected with gel electrophoresis (ISMMS) or purified using SPRI beads (VU). The DNA library was subsequently enriched for sequences with 5 and 3 adapters by PCR amplification using with primers complementary to the adapter sequences (ligation-mediated PCR, LM-PCR). Exons were captured using the Agilent 38Mb SureSelect v2 exon enrichment reagents (http://genomics.agilent.com/). After capture, another round of LM-PCR was performed to generate sufficient DNA for sequencing. Libraries were sequenced to produce 50- to 100-bp paired end reads using Illumina GAIIx or HiSeq2000 instruments (http://illumina.com). Over 99% of targets were hit, with over 82% covered at 10x and over 70% at 20x. Average coverage was approximately 50x across the exome. Paired samples were all sequenced together to minimize batch effects.
Sequence data were processed with Picard (http://picard.sourceforge.net/), which utilizes base quality-score recalibration and local realignment at known indels [described in [13], and alignment of raw sequence reads was performed using a fast light-weighted Burrows-Wheeler Alignment Tool (BWA) [14] for mapping reads to hg19, followed by genotype calling by Genome Analysis Toolkit (GATK) [15] as detailed below.
4.2. Overview of Our Approach to Filtering
Our primary goal was to determine which loci were mutations. To identify point mutations we focused on observations in which the LCL sample was called heterozygous, but WB DNA was called homozygous corresponding to reference sequence, and designed validation experiments (see below) to determine which of those observations indeed came from mutations. To avoid missing any true differences between the samples, we also examined observations in which the WB sample was called heterozygous, but LCL DNA was called homozygous. To formalize our investigation, we establish some notation. Let f+ denote the probability of a truly homozygous locus being called heterozygous (a false positive), and let f− denote the probability of a truly heterozygous locus being called homozygous (a false negative). The probability of a homozygous locus being called homozygous, but for an incorrect allele, is considered negligible. Hence these are the two error probabilities of interest.
There are four cases into which all loci can be divided based on these considerations, shown as the rows of Table 2. The table illustrates, for each of the possible true genotypes, the probabilities for each of the four possible calls. We adopted a principled approach to reducing the different error probabilities and discovering evidence of mutations. We utilized the UnifiedGenotyper of GATK with settings that were set at liberal levels, so that a heterozygote call would be made even in cases where the evidence is not overwhelming. Thus, f− would be limited, even if it is at the expense of a larger value of f+. This choice facilitates filtering of false mismatches in the next stage. For instance, when there is a true difference and LCL is heterozygous, the variant caller is unlikely to report both WB and LCL are homozygous using this strategy. If both are homozygous, however, LCL might be falsely called heterozygous. The latter call is will be based on poor quality data and hence it will be possible to remove this false mismatch at the next stage of filtering. Alternatively, employing a strict filtering strategy leads to a types of error that is difficult to reconcile – mismatches of true heterozygotes that are due to very small differences in the quality of the reads.
Table 2.
Probabilities of different possibilities.
| Variant Call | ||||
|---|---|---|---|---|
| Truth | Both hom. | Only Cell Line het. | Only Blood het. | Both het. |
| Both hom. | (1 − f+)(1 − f+) | f+(1 − f+) | f+(1 − f+) | f+2 |
| Only Cell Line het. | (1 − f+)f− | (1 − f+)(1 − f−) | f−f+ | f+(1− f−) |
| Only Blood het. | (1 − f+)f− | f−f+ | (1 − f+)(1 − f−) | f+(1 − f−) |
| Both het. | f−2 | f − (1 − f−) | f − (1 − f−) | (1 − f−)(1 − f−) |
Comparisons were made between the blood and cell line variant calls, and differences were found; however, many of the discrepant loci in this initial set were a result of false positives variant calls (see Supplemental Materials). Segregation of these loci based on quality metrics began by applying the GATK Variant Quality Score Recalibration (VQSR) procedure. VQSR fits a Gaussian mixture model to both known and unknown variants based on various features of the variant calls including quality by depth (QD), strand bias (SB), and haplotype score (HapScore). VQSR then assigns a score proportional to the false positive rate; this score (VQSLOD) can be interpreted as the log odds of the variant being real. We only retain those loci for which this log odds is larger than zero. This is still a generous cutoff, and, although the total count of loci was reduced greatly, there are still many false differences among the remaining loci.
Hence, the above process yielded a shorter list of candidates more likely to be “real” blood/cell line differences. These candidates were further segregated into groups based on deeper inspection of the read quality, evidence for strand bias, and the relative balance of reference/alternative calls. Secondary validation experiments were conducted using Sanger sequencing to confirm the suspected differences. The within-group rate of confirmation, along with the estimated proportion of loci within each group, was then used to estimate the overall mutation and deletion rate.
In what follows, we present more details regarding these steps.
4.2.1. Identification and Segregation of the Candidates
The GATK Unified Genotyper was utilized to call variants; a single call was made for both of the sites and then the calls were filtered using standard metrics (Supporting Information). The 864 candidates with VQSLOD larger than zero are the starting point for the next stage in the process. Here, these 864 were segregated into groups defined by criteria described below. These criteria were based on properties of the individual reads that cover a given locus for an individual.
To begin, individual reads were assigned a Phred-scaled score to assess the quality of the mapping. Each base call was also assigned a Phred-scaled score to quantify the quality of the call. A read was defined to be of good quality if both the mapping quality and the base quality at the locus of interest were at least 15. Also, in what follows, there will be several applications of Fisher’s exact test. In each case, the categories that comprise the two-by-two table were the same: Each read was classified as either “Reference” or “non-Reference” and as either from “Blood” or “Cell Line.” By finding the p-value found for these tests, one directly assesses the evidence in support of discordance between the blood and cell line bases. Small p-values corresponded to the conclusion that there is a true difference between blood and cell line. For this reason we flag instances where the p-value is larger than 0.1 as evidence of a false mismatch.
Suspicion Flags
Inspection of the candidates led to the creation of seven “suspicion flags,” so-named because these are not, by themselves, justification for eliminating a site from consideration as being the location of a true blood/cell line mismatch, but these did raise concerns as to the reliability of the called difference. For each of these criteria, a restriction was made to only those reads classified as “good quality.”
Strand Direction Imbalance. The initial filtering may not be completely effective in removing strand bias. Consequently, some of the heterozygous calls could be a result of reads in one direction or the other, but not both. Here, to assess this, the aforementioned Fisher’s exact test was applied separately to the reads that were in the forward direction and the reads in the reverse direction. If at least one of the two resulting p-values is larger than 0.1, this would be evidence that the difference between blood and cell line is not apparent in both strands weakening the case for a true difference. Hence, a candidate for which either of these p-values is large was flagged for strand bias.
Skewed Reads. At a high-quality site, one would expect to see reads that have the target locus at a range of positions, going from the far left to the far right. This skew is evidence of a mapping problem that is present only when the non-reference base is at the locus. As was the case with strand bias, to assess this, Fisher’s exact test was applied separately to the reads that had the target locus to the left of center, and then to the reads that had the target locus to the right of center. If either of these two p-values was larger than 0.1, then the site was flagged for evidence of skew.
Insertions and Deletions. Reads with called insertions and/or deletions could be the result of mapping errors, i.e., the mapping software trying to “fit” a read in a position it should not be in. To eliminate this issue, Fisher’s exact test was applied only to those reads for which the mapping procedure found no insertions or deletions. If the p-value resulting from this test was greater 0.1, then this flag was set.
Proper Pairing of Reads. Fisher’s exact test was applied only to those reads for which a “proper pair” was found during the mapping process. If this p-value was greater than 0.1, then this flag was set.
Extreme Proportion Reference. If the proportion of reads called as the reference exceeded 0.85 for both blood and cell line, this was taken as some evidence that both sites are indeed homozygous reference. Likewise, if both of these proportions were less than 0.15, there was basic evidence that the site is homozygous in the alternative.
Low VQSLOD. If the VQSLOD for the site was less than 1.5, then this flag was set. As described above, low VQSLOD called into question the overall quality of the site.
Large Depth. If there is excessive depth at a locus, this was indicative of a potential problem with mapping reads to this location. Here, this flag was set if the total number of reads (on both blood and cell line) exceeded 400.
Once determined, sites were classified based on the number of these flags that are raised. Sites for which there are no flags were assigned to Group 0; sites for which there was a single flag to Group 1; sites for which there were two flags to Group 2; and sites for which there were more than two flags were placed in Group 3.
4.2.2. Validation of Selected Candidates
Validation of the indicated variant calls was carried out using standard Sanger dye-terminator sequencing of amplimers for regions containing putative WB/LCL sequence differences. Primers were designed using Primer 3 software (http://frodo.wi.mit.edu/primer3/) and subjected to a BLAST-Like Alignment Tool search to ensure amplification specificity. PCR products were amplified from ~20 ng DNA using either AmpliTaq Gold PCR Mastermix (Life Technologies; Carlsbad, CA) or AccuPrime Pfx (Invitrogen, Carlsbad, CA, USA) with individually optimized PCR conditions, in a total volume of 10 l. Genomic and LCL DNA from the same subject were amplified simultaneously. Amplified DNA fragments were then purified using either MultiScreen PCR96 filtration plates (Millipore; Billerica, MA) or QIAquick PCR purification columns (QIAGEN, Valencia, CA, USA) and yields were determined using a NaroDrop spectrophotometer ND-1000 (Thermo Scientific, Wilmington, DE). Sequencing reactions were carried out using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and reactions were subjected to analysis on an ABI 3730xl DNA Analyzer. Sequence elecropherogram data were viewed to examine sites of putative difference using Sequencher v5.0.1 (Gene Codes; Ann Arbor, MI), the Assembler module of MacVector v12.6 (Cary, NC), FinchTV (Geospiza Inc., Seattle, WA) and ABI Sequencing Analysis Software v5.2.
All variants in Groups 0, 1, and 2 were sequenced. In addition, we also pursued validation of a subset of the Group 3 candidates. Instead of validating a random subset of Group 3, results from the first three groups were exploited to determine what, if any, features of the candidates increased their chances of being real mismatches. It was determined that among the confirmed mismatches, almost all shared two characteristics: First, the proportion of the WB DNA calls that corresponded to the reference allele was either very close to zero or very close to one. Second, the cell line reads for both alleles were well-represented on both the forward and reverse strands. Hence, a filter was applied that labeled a candidate as “Strong” if (1) for the homozygous call the proportion of reads for the reference allele was either larger than 0.97 or smaller than 0.03 and (2) the proportion of reads in a single direction did not exceed 0.95 for either allele present in the cell line. The remaining candidates were labeled “Weak.” Group 3 validation was therefore pursued chiefly for “Strong” candidates.
Supplementary Material
Highlights.
We compare variant calls in matched DNA from 16 blood and cell lines.
Read-level characteristics are useful for detecting true genotype mismatches.
Sanger sequencing reveals a small number of actual differences.
Actual variant differences exhibit mosaicism.
Predicted rate of mosaicism is 3–4 per exome.
Acknowledgments
This work was directly supported by the ARRA Autism Sequencing Consortium-grant (R01MH089208, R01MH089025 - J.D.B., R01MH089004, R01MH089175 and R01 MH089482-J.S.S.), and supported in part by NIH grants RO1 MH057881 (B.D.) and R01 MH061009 (J.S.S.). J.D.B. is supported by the Seaver Foundation. We thank T. Lehner, A. Felsenfeld and P. Bender for their support and contribution to the project. We acknowledge the assistance of M. Potter, A. McGrew and G. Crockett without whom these studies would not be possible, and Center for Human Genetics Research resources: Computational Genomics Core, Genetic Studies Ascertainment Core and DNA Resources core, supported in part by NIH NCRR grant UL1 RR024975, and the Vanderbilt Kennedy Center for Research on Human Development (P30 HD015052). We acknowledge the clinicians and organizations that contributed to samples used in this study and the particular support of families and clinicians of the Icahn School of Medicine at Mount Sinai and Vanderbilt University. M.J.D., R.A.G., G.D.S., B.D, J.S.S, J.D.B. and K.R., are lead investigators in the Autism Sequencing Consortium (ASC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conrad DF, Keebler JEM, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, Zilversmit M, Cartwright R, Rouleau GA, Daly M, Stone EA, Hurles ME, Awadalla P. 1000 Genomes Project, Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–4. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR, Côté M, Henrion E, Spiegelman D, Tarabeux J, Piton A, Yang Y, Boyko A, Bustamante C, Xiong L, Rapoport JL, Addington AM, DeLisi JLE, Krebs MO, Joober R, Millet B, Fombonne E, Mottron L, Zilversmit M, Keebler J, Daoud H, Marineau C, Roy-Gagnon MH, Dubé MP, Eyre-Walker A, Drapeau P, Stone EA. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87:316–24. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gauthier J, Champagne N, Lafrenière RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Côté M, Noreau A, Hamdan FF, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Joober R, Fathalli F, Mouaffak F, Haghighi AP, Néri C, Dubé MP, Samuels ME, Marineau C, Stone EA, Awadalla P, Barker PA, Carbonetto S, Drapeau P, Rouleau GA. S2D Team, De novo mutations in the gene encoding the synaptic scaffolding protein shank3 in patients ascertained for schizophrenia. Proc Natl Acad Sci U S A. 2010;107:7863–8. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders S, Murtha M, Gupta A, Murdoch J, Raubeson M, Willsey A, Ercan-Sencicek A, DiLullo N, Parikshak N, Stein J, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:82–93. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neale B, Kou Y, Liu L, Ma’ayan A, Samocha K, Sabo A, Lin C, Stevens C, Wang L, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Roak B, Vives L, Girirajan S, Karakoc E, Krumm N, Coe B, Levy R, Ko A, Lee C, Smith J, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iossifov I, Zheng T, Baron M, Gilliam TC, Rzhetsky A. Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network. Genome Res. 2008;18:1150–62. doi: 10.1101/gr.075622.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbeck JT, Gottlieb GS, Wong K, Detels R, Phair JP, Rinaldo CR, Jacobson LP, Margolick JB, Mullins JI. Fidelity of snp array genotyping using epstein barr virus-transformed b-lymphocyte cell lines: implications for genome-wide association studies. PLoS One. 2009;4:e6915. doi: 10.1371/journal.pone.0006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon JP, Shim SM, Nam HY, Baik SY, Kim JW, Han BG. Copy number increase of 1p36.33 and mitochondrial genome amplification in epstein-barr virus-transformed lymphoblastoid cell lines. Cancer Genet Cytogenet. 2007;173:122–30. doi: 10.1016/j.cancergencyto.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, Crawley A, Keen JC, Nash J, Borgaonkar D, Hardy J, Singleton A. Genome-wide snp assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 12.McElroy JP, Nelson MR, Caillier SJ, Oksenberg JR. Copy number variation in african americans. BMC Genet. 2009;10:15. doi: 10.1186/1471-2156-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation dna sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a mapreduce framework for analyzing next-generation dna sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.