Abstract
For centuries, philosophers and scientists have been fascinated by the principles and implications of regeneration in lower vertebrate species. Two features have made zebrafish an informative model system for determining mechanisms of regenerative events. First, they are highly regenerative, able to regrow amputated fins, as well as a lesioned brain, retina, spinal cord, heart, and other tissues. Second, they are amenable to both forward and reverse genetic approaches, with a research toolset regularly updated by an expanding community of zebrafish researchers. Zebrafish studies have helped identify new mechanistic underpinnings of regeneration in multiple tissues, and in some cases have served as a guide for contemplating regenerative strategies in mammals. Here, we review the recent history of zebrafish as a genetic model system for understanding how and why tissue regeneration occurs.
Keywords: zebrafish, regeneration, stem cells, fin, spinal cord, heart, retina, brain
A versatile model system
Zebrafish are native to river basins in and surrounding East India and were established as a laboratory model system first by Streisinger and colleagues in the 1970’s, as a potential means to apply genetic analysis to vertebrate development [1, 2]. In the decades that have followed, zebrafish have become a valuable tool to dissect embryogenesis. Experimental advantages of zebrafish for this use include large clutches, rapid external development, amenability to mutagenesis, a relatively small genome, and a reasonably short generation time. By utilizing these advantages researchers have uncovered key factors in myriad developmental events, from early germ layer patterning to how tissues derived from these layers acquire form and function [3, 4]. Recently, zebrafish have been employed more and more to investigate additional aspects of biology, including behavior, stem cells, and disease [5–9].
In this review, we provide an overview of how in the past decade, zebrafish have become a primary model system for vertebrate tissue regeneration. For this review, we have focused on their remarkable regeneration of fins, heart, and central nervous system structures, although they also regenerate jaw, hair cells (lateral line), pancreas, liver, and kidney [10–20]. We summarize what is known about mechanisms of regeneration in different tissues and contexts, and describe how new discoveries and approaches in zebrafish are impacting the field of tissue regeneration.
Zebrafish fin regeneration
Zebrafish fins are complex appendages that quickly and reliably regenerate after amputation, restoring both size and shape. The key regenerative units are their many rays of dermal bone, which are segmented and lined by osteoblasts. Rays are cylindrical and hollowed, with two concave hemirays surrounding an inner mesenchymal tissue that is innervated, vascularized, and comprised primarily of fibroblasts. An amputated fin ray is covered within the first several hours by epidermis, and within one to two days, a regeneration blastema forms. The blastema is a proliferative mass of morphologically similar cells, formed through disorganization and distal migration of fibroblasts and osteoblasts (or scleroblasts) proximal to the amputation plane. As with blastemas in other classical regenerating systems like the salamander limb and planarian head, the fin ray blastema is the major source of new structures.
The ability of teleost fish to regenerate amputated fins was first reported in 1786, in pectoral fins of goldfish, by Broussonet [21]. Although Thomas Hunt Morgan was fascinated by fin regeneration at the turn of the 20th century [22], it took nearly an additional century for fin regeneration to reach the genetic era. In 1995, Johnson and Weston described a screen for mutations that disrupt regeneration of tailfins in adult zebrafish [23], arguably the first experiments to demonstrate a technical advantage of studying regeneration in zebrafish. This screen was novel not only in its application of genetics to vertebrate regeneration, but also in its use of temperature-sensitive (TS) mutations in zebrafish. As regeneration is expected in most cases to re-employ genes used in early development, a TS screen enables identification of mutations in adults that would be lethal during early development. Over the next decade, genetic screening uncovered a handful of mutations that inhibited fin regeneration and could be localized to specific molecular defects by positional cloning [24–27]. These discoveries have contributed to the molecular models described below; yet, there has been a large time gap since the most recent identification of a regeneration gene by mutagenesis. New advances in high-throughput genome, exome, and transcriptome sequencing are likely to reboot forward genetic approaches to studying regeneration [28–33].
To best understand regeneration in any system, one must conclusively know the sources of the different cell types that are restored after injury. Only recently have modern genetic fate-mapping approaches been applied to address this question, including Cre recombinase-based technology used routinely in mice for lineage analysis. Multiple recent studies used transgenic Cre lines to focus on bone-forming osteoblasts. Their results indicated that differentiated osteoblasts transiently downregulate the osteogenic program, or dedifferentiate, as they contribute to the blastema. After this, resident osteoblasts contribute only osteoblasts to new regenerated structures [34–37]. This idea of lineage restriction was extended to other cell types like endothelium, epidermis, and fibroblasts by other studies [38]. These findings agree with similar lineage restriction observed in axolotl (Mexican salamander) limbs and mouse digit tips [39–41]. However, these studies could not exclude rare transdifferentiation events, nor do most of them address possible ancillary mechanisms under different injury contexts. For instance, osteoblasts form and bone regenerates efficiently even when resident osteoblasts are potently ablated, indicating that other cell types are capable of differentiating into osteoblasts and supporting bone regeneration [35].
Many groups have examined the molecular mechanisms underlying the formation and proliferation of the blastema. In response to injury, increased expression of key signaling components of Wnt/β-catenin and Activin-βA pathways are detectable by 3 hours post amputation (hpa) [42, 43], followed by upregulation of retinoic acid (RA), Insulin-like growth factor (Igf), and Fibroblast-like growth factor (Fgf) signaling pathway components by 6 hpa [24, 44, 45]. Although more complete functional testing is needed, one model for blastema formation is that increases in RA synthesis in response to injury induce expression of igf2b and wnt10b. These ligands then signal through canonical Wnt and Igf pathways to induce expression of fgf20a, a marker and critical regulator of blastema formation [43, 44]. Independent of this signaling cascade, activin-βA is upregulated in the interray region and is involved in re-organization of the underlying mesenchyme during blastema formation [42]. Blockade of these signaling pathways results in improper wound healing and blastema formation, implicating them in initiation of the blastema.
Blastema formation is only one step in zebrafish fin regeneration, and fins must then grow to the appropriate size. Regenerative outgrowth occurs by two processes: maintenance of a proliferative compartment at the distal end of the regenerate, and differentiation of more proximal cells. The proliferative compartment is maintained by signaling interactions between the mesenchyme and basal epidermis [46]. In addition to regulating blastema formation, RA, Fgf and canonical Wnt signaling positively regulate blastemal proliferation and outgrowth, while non-canonical Wnt signaling inhibits these events [43, 45, 47]. Inhibition of Igf receptors or the Tgf-β receptor alk4 also block blastemal proliferation during outgrowth, further indicating continued requirements for these pathways [42, 44]. Interestingly, inhibition or ectopic activation of the Notch signaling pathway results in a regenerative block, leading authors to propose models in which Notch signaling, through an unknown mechanism, enhances blastemal proliferation while suppressing osteoblast differentiation during regeneration [48, 49].
In addition to Notch signaling, other pathways have been examined for their ability to influence differentiation within the blastema. Bmp and Hedgehog signaling induce bone formation in the regenerate when ectopically activated, suggesting that the normal function of these molecules may be to drive re-differentiation of osteoblasts in the proximal blastema [50, 51].
Finally, fins provide a potentially useful model for considering the mechanisms by which an appendage regains its original shape and size after amputation. This phenomenon of positional memory, in which adult cells in the stump somehow retain and recall the correct developmental coordinates and instructions, remains in many ways a mystery. Regeneration occurs at different rates depending on the proximodistal amputation plane, regulation that involves position-dependent control of amounts of Fgf signaling [47]. Signals responsible for this, and factors that retain coordinates in adult fins and enact precise recovery, remain to be found and are likely to be broadly relevant to regeneration in other systems.
Heart regeneration
There is no significant regeneration of adult mammalian cardiac muscle after experimental injury paradigms. This deficiency is highly relevant to human disease, given that ischemic myocardial infarction (MI) and scarring is a primary cause of morbidity and mortality. Zebrafish have a high natural ability for heart regeneration, and thus can inform as to how this process occurs or might be induced [52]. There are currently several injury models that stimulate heart regeneration in zebrafish, including surgical resection of the ventricular apex, the first and most-used injury method, cryoinjury, and inducible genetic ablation [52–56]. Each of these models offers experimental advantages. For instance, whereas cryoinjury mimics aspects of MI, genetic ablation produces massive injuries, removing 60% or more of cardiomyocytes and inducing signs of end-stage heart failure. Unlike severe heart failure in humans, these signs regress within weeks and the animals typically make a full recovery concomitant with muscle regeneration [56]. Studies in zebrafish have revealed that heart regeneration involves two fundamental components: 1) proliferation of existing cardiomyocytes as the primary cellular source; and 2) an environment that stimulates muscle generation from this source. In theory, regenerated cardiomyocytes could derive directly from a progenitor pool akin to the embryonic heart fields that first create the cardiac chambers, from stem cells that populate the adult heart, or from circulating progenitor cells. However, genetic fate-mapping experiments in zebrafish have made it clear that the regenerative ability of the zebrafish heart relies mainly or exclusively on proliferation of existing cardiomyocytes [57, 58]. These source cardiomyocytes show characteristics of dedifferentiation, including a reduction in contractile structure, and can be identified after apical resection by activation of regulatory sequences of the gata4 transcription factor gene [57]. There is as of yet no definitive lineage-tracing evidence that indicates an undifferentiated progenitor cell could be anything but a minor source of heart muscle.
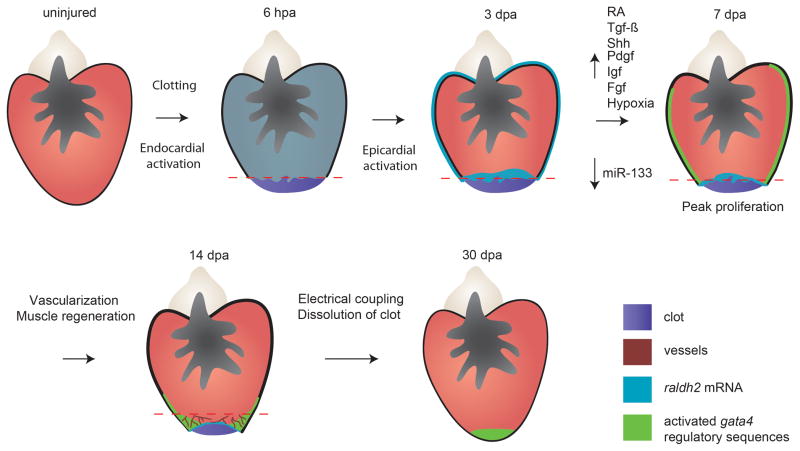
Cardiomyocyte proliferation occurs at a low rate in the adult zebrafish heart, but is sharply increased in response to tissue damage [52]. There is considerable evidence that non-muscle cells create an environment that enables this response. Injury to the zebrafish heart initiates an organ-wide reaction detectable as induced expression of raldh2 (a RA-synthesizing enzyme) as early as 1 hour post-injury in the endocardium, the endothelial lining of the lumen (Figure 1) [59]. The endocardium remains activated in the area of injury for several days adjacent to regenerating cardiomyocytes, and requires further study as a player in heart regeneration. Within a day or two of injury, the epicardium, the outer lining of the heart, shows an analogous organ-wide response of raldh2 induction [60]. Then, epicardial cells proliferate and surround the regenerating muscle, where they release signals that facilitate cardiomyocyte proliferation. RA, Tgf-β ligands, Igf2, Shh, and Platelet-derived growth factor (Pdgf) ligands all are released in the vicinity of proliferating cardiomyocytes, and have positive influences on muscle regeneration [59–64]. Epicardial cells have been fate-mapped and act as a source of vascular support cells for regeneration, just as they do during initial heart development [65]. Fgf signaling is important for vascularizing the regenerate, which ultimately aids muscle regeneration [60]. Recently developed culture techniques may allow for better characterization of epicardial cells and a greater understanding of the dynamic nature of this cell population [66]. Other potential influences on zebrafish heart regeneration have very recently been examined. Hypoxia is a general factor that appears to play a positive role in cardiomyocyte proliferation, whereas hyperoxia and the microRNA miR-133 have negative roles [67, 68]. In addition to cardiomyocyte proliferation, it has been reported that chemokine-mediated cardiomyocyte migration to the injury site is a critical step in the regenerative process [69]. New tools for manipulating gene expression in muscle, epicardium, endocardium, and other cell types like inflammatory cells will be critical for a higher resolution view of the mechanisms of heart regeneration.
Figure 1. Model for regeneration after partial resection of the cardiac ventricle.
After injury, the RA-synthesizing enzyme, raldh2, is induced throughout the endocardium within a few hours of amputation (hpa) and later the epicardium, before these responses localize to the wound. By 7 days post amputation (dpa), gata4 regulatory sequences are activated throughout the cortical muscle layer of the ventricle. At this point, cardiomyocyte proliferation is stimulated, under the influences of hypoxia and signaling pathways as described in the main text, and epicardial cells have begun to integrate into the wound. By 14 dpa, vascularization of the regenerating muscle begins, aided by Fgf and Pdgf signaling. By 30 dpa, a new wall of cardiac muscle is typically formed, in large part by the progeny of early cardiomyocytes that activate gata4 sequences. At this point, the myocardium is vascularized and electrically coupled with the existing muscle.
Studies of zebrafish heart regeneration have helped in considering approaches to cardiac regeneration in mammals. Because division of mature cardiomyocytes has been difficult to definitively observe in injured mammalian hearts, muscle cells have until very recently received somewhat limited attention as an endogenous target cell that could be expanded for regeneration. Arguably more consideration has instead gone to a number of potential cardiac stem cells [70], or recently to fibroblasts that can be experimentally reprogrammed into cardiomyocytes by cardiac transcription factors [71, 72]. However, recent lineage-tracing approaches have supported the idea that endogenous cardiomyocytes are a potential regenerative target. There is now evidence that adult murine cardiomyocytes proliferate at very low levels into adulthood. Some cardiomyocyte proliferation does occur after injury [73, 74], although not nearly to the extent as the injured zebrafish or neonatal mammalian heart [75]. Of potential major significance was an expression screen that reported identification of several miRNAs enhancing proliferation of adult mammalian cardiomyoyctes in vitro, with some able to stimulate significant regeneration after myocardial infarction in adult mice [76]. As cardiomyocyte proliferation as well as epicardial activation [77, 78] appear to be shared components for cardiac repair, discoveries of natural regulators of cardiac regeneration in zebrafish should continue to relate directly to mammals. The pace of the field of heart regeneration has markedly accelerated in the past few years, a development that forecasts new regenerative therapies for the injured human heart in the near future.
Neural stem cell-based regeneration
Neural regeneration
Neuronal cell loss causes visual, motor, or mental impairment in humans. This neuronal cell death often leads to glial cell hypertrophy, limited proliferation, and gliotic scarring, which prevents neuronal regeneration. Zebrafish, by contrast, have the capacity to regenerate neurons within the retina, spinal cord, and brain from resident radial glial cells. New genetic approaches have facilitated the investigation of commonalities and distinctions in the pathways necessary for regeneration of different neuronal tissues and cell types.
Retina
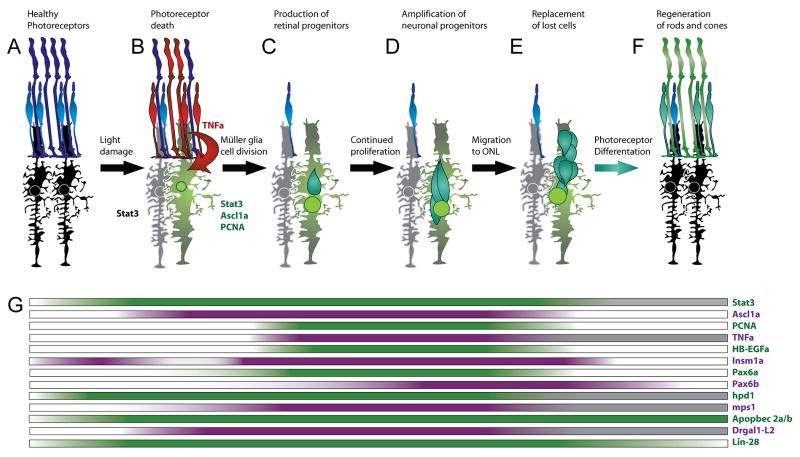
Because of the relative ease of manipulating the retina, numerous damage strategies have been employed to either destroy all or a restricted type of retinal neurons [79–90]. All of these damage models induce some Müller glia to dedifferentiate and reenter the cell cycle to produce multipotent neuronal progenitor cells (NPCs, Figure 2), which express many of the genes known to direct retinal development [91]. These NPCs proliferate, migrate to the region of damage, and differentiate into the appropriate neuronal cell type. Cre-lox-mediated lineage tracing, BrdU incorporation, and PCNA immunolabeling all demonstrated that Müller glia are the source of the new neurons in a damaged retina [79, 84, 92–94] (Figure 2B). The number of dedifferentiating and proliferating Müller glia varies according to the damage models and increases relative to the extent of damage [92, 95]. Within hours of retinal damage, all Müller glia upregulate expression of Stat3 [96], rapidly followed by Ascl1a in a subset of Müller glia [94, 96–99], both of which are required for Müller glia to reenter the cell cycle. Responder Müller glia are distinguished from non-proliferating Müller glia by their expression of Ascl1a [97].
Figure 2. Model for regeneration after light damage to photoreceptors.
A. Prior to light damage, the retina consists of healthy photoreceptors (blue) and quiescent Müller glia (black). B. Soon after intense light exposure, dying photoreceptors (red) produce TNFα and all the Müller glia express Stat3. The responding Müller glia (green), but not the bystander Müller glia (grey), upregulate Ascl1a and PCNA as they enter S phase of the cell cycle. C. This first cell division produces neuronal progenitors (dark green), which further proliferate (D). E. The new retinal progenitor cells migrate to the site of damage. F. Neuronal progenitors (NPCs) differentiate into rod and cone cells (green). G. Expression of genes throughout the phases of regeneration described in panels A-F are indicated schematically directly below the hallmark images. Colored shading represents increased expression relative to undamaged retinas. Grey shading represents times not assessed for gene expression.
Microarray studies [87, 100–104] and two-dimensional differential protein gel analysis [99] identified several candidate genes and proteins (Figure 2G) that may be required for aspects of regeneration. Recently, a technique to electroporate morpholinos into the adult retina to knockdown the expression of specific target proteins was developed to functionally validate the role of these proteins in regeneration [93, 97, 105]. This work has been complemented by the development of heat shock-inducible transgenes and temperature-sensitive zebrafish mutants, which have revealed requirements for specific genes in regeneration [103, 106].
A variety of experiments have demonstrated that the canonical Wnt pathway is required for zebrafish retinal regeneration [98, 107]. Wnt repression by heat-shock-induced dickkopf 1 (dkk1) misexpression [98], small-molecule drug inhibition with XAV939, which indirectly stabilizes Axin activity, or missexpression of a dominant-negative version of the Wnt target gene, T-cell transcription factor 3 (tcf3) [107] reduces the number of proliferating Müller glia in the damaged retina. Wnt activation, using intravitreally-injected lithium chloride or GSK3-β inhibitor I, results in the stimulation of Müller glial proliferation in the absence of detectable retinal damage [98], but incubation of undamaged fish with the GSK3-β inhibitor, 1-azakenpaullone, does not [107]. This suggests that Wnt is sufficient to induce Müller glia proliferation in the absence of significant cell death, but only when the retina is stressed through either blunt force trauma or increased intraocular pressure caused by intravitreal injections of large volumes. Wnt signaling, as well as EGF and Shh, can also increase the number of proliferating Müller glia in the damaged rodent retina [108–111]. In the transgenic rat S334ter model, which expresses a prematurely truncated mouse rhodopsin protein at serine 344 and results in retinal degeneration similar to rapid onset retinitis pigmentosa in humans with an analogous rhodopsin variant, overexpression of Notch and Wnt activation followed by a regimen of Shh and the Notch inhibitor DAPT is able to spare the rats from vision loss seen in the S334ter control littermates [112].
Other candidate molecules that appear to stimulate Müller glia proliferation in the zebrafish retina include Heparin-binding-Epidermal Growth Factor (HB-EGF) [113], Insulinoma associated 1a (Insm1a) [104], ADP [114], and free radicals [98], suggesting a possibly complicated process for full activation of regeneration. Additionally, TNFα expression is markedly increased specifically in dying photoreceptors following light damage, as well as in dying ganglion and amacrine cells following ouabain treatment [99]. This TNFα expression is required for Ascl1a and Stat3 expression in Müller glia [99]. Thus, several proteins have been shown to be required for Müller glia to dedifferentiate and reenter the cell cycle in the damaged retina, but only TNFα has been shown to be expressed in the dying neurons, seemingly a requirement of a de novo signal to initiate neuronal regeneration.
Zebrafish retinal regeneration is highly tuned to regenerate specifically the cell types that are damaged. For example, light-induced photoreceptor cell death results in the regeneration of only photoreceptors [86, 92]. This specificity in cell regeneration led to the identification of the molecular pathways required to regenerate rods and cones (Figure 2F). For example, Fgf receptor 1 (Fgfr1) signaling [106] and lectin, galactoside-binding, soluble, 2a (Drgal1-L2 or Lgals2a) [115] are necessary for rod regeneration, whereas Paired-box gene 6b (Pax6b) [116] and Ttk protein kinase (Ttk or Mps1) [103] are required for cone regeneration. To facilitate these studies, new transgenic tools were created, including transgenes that express E. coli nitroreductase enzyme in specific neuronal cell subtypes. Treating the different transgenic lines with the prodrug metronidazole specifically ablates the neuronal cell subtype expressing the nitroreductase enzyme, including rods [89], cones [90], or bipolar cells [88]. Although much still remains to be learned about how cell regeneration is targeted, these genetic tools make zebrafish an ideal model system to address the questions.
Spinal Cord
Damage to the human spinal cord results in irreversible loss of neurons and impaired sensory and motor functions. By contrast, zebrafish possess the ability to regrow new axonal projections from viable brain neurons across the severed spinal cord [117]. In addition to axonal growth, zebrafish can produce new neurons and interneurons at the region of damage [118, 119]. Similar to development, the type of regenerated neuron depends on dorsoventral location of its corresponding progenitor radial glial cell in the spinal cord. For example, motor neurons, characterized by Hb9, Islet-1/2, and ChAT expression, are regenerated from Olig2 radial glia cells located in the spinal cord ependyma, which surrounds the spinal ventricle [118]. The subset of Olig2-positive cells that give rise to motor neurons also coexpresses the developmental transcription factors, pax6a and nkx6.1 [120]. Additionally, motor neuron regeneration is inhibited by transgenic overexpression of the Notch intracellular domain (NICD), resulting in the increased expression of hairy-related 4.1 (her4.1) [121]. By contrast, regenerated V2 interneurons derive from more dorsally located Nkx6.1+, Pax6+, and Olig2− p2 glia, whereas serotonergic neurons derive from the ventral glia [119]. The more dorsally derived neurons show limited regeneration in zebrafish, which may be due to the lack of the expression of genes that determine dorsal biases in development, such as pax3, pax7, bmp2, bmp4, and tcf7 [119]. Inhibition of the Hh pathway reduces neurogenesis at the lesion site; interestingly, this phenotype does not impair functional recovery as assessed by swimming activity [120], suggesting that the surviving neurons may exhibit synaptic plasticity.
Axonal regeneration across the lesion site is dependent on proliferating radial glial cells that infiltrate the site. The responding glia divide soon after lesion and assume bipolar morphology. They migrate into the damaged site and connect the two sides of the lesion guiding the new axons, a process termed a “glial bridge” [122]. The early proliferation and migration of responding glia is dependent on Fgf signaling and was reduced by either global overexpression of dominant-negative Fgfr1 or by injecting SU5402, a small molecule inhibitor of Fgfr1 tyrosine kinase activity. These events were enhanced in the sprouty 4 (spry4) mutant fish [122], which is a downstream target and inhibitor of Fgf signaling. Spry4 expression is increased in activated radial glia following transection of the mouse spinal cord, and human FGF2 induces marmoset astrocytes to adopt a bipolar morphology in culture, suggesting conservation in mammals [122].
Brain
Studies of regeneration in the teleost brain were recently extensively reviewed [123]. Surgical lesion of the telencephalon causes neuronal cell death and induces radial glia to proliferate and new neurons to regenerate. Like the spinal cord, regeneration originates from radial glia that line the ventricles and express GFAP and Olig2 [124]. These glia proliferate to yield progenitor cells that then express the transcription factor Eomesa, which regulates glutamatergic neuron differentiation [124]. Recent advances using Vivo-Morpholinos to knockdown protein expression and transgenesis have allowed for functional studies of specific proteins during brain regeneration. Vivo-Morpholinos, which contain eight guanidium residues covalently attached to a trizine residue in the standard morpholino antisense-oligonucleotides [125], penetrate into the most proximal cells of the zebrafish telencephalon ventricle without electroporation or intracellular injection, after cerebroventricular microinjection (CVMI) [126]. CVMI studies indicated that Fgf signaling, the chemokine receptor, Cxcr5, and the zinc finger transcription factor Gata3, are necessary for regeneration in damaged brains, but are insufficient to stimulate proliferation in undamaged brains [127, 128]. Although Notch signaling is inhibitory in the retina [113] or motorneurons [121], it is required for brain regeneration. Her4.1+ ventricular radial glia divide in response to distal surgical injury [129], while DAPT-mediated inhibition of Notch signaling decreases the number of Neurogenin 1 (Ngn1) and T-box brain 1 (Tbr1) expressing neuronal progenitors [130], which give rise to the telencephalic cells during development. It is unclear why Notch seems to have contradictory roles in these systems.
The inflammation stimulator, leukotriene C4, signals through the cysteinyl leukotriene receptor 1 (CysLT1) to induce radial glial proliferation in the undamaged zebrafish brain, whereas inhibition of CysLT1—specifically by Pranlukast or generally by dexamethasone—reduces the number of dividing radial glia in the damaged telencephalon [131]. This inflammatory response might be necessary for the regeneration of multiple zebrafish tissues, as dexamethasone-induced immunosuppression disrupts caudal fin regeneration after amputation [131]. The source of this inflammatory response, the underlying regulatory targets, and their impact in less regenerative mammalian tissues will be important to explore.
Future Advances
Zebrafish have advantages over other regenerative vertebrate model systems in regards to the relative ease and diversity by which potential factors can be manipulated (Table 1). Full utilization of emerging technologies in zebrafish will strengthen the foundation for regeneration studies.
Table 1.
A brief list of tools available for studying regeneration in zebrafish
| Category | Technique | Pros | Cons |
|---|---|---|---|
| Gene Manipulation | Gal4-UAS | modular reagents tissue specificity |
requires multiple transgenic lines non-reversible non-inducible generational or stage-specific silencing |
| hsp70 (heat-inducible promoter) | reversible tunable single transgenic line inducible |
no tissue specificity elevated temperature may affect regenerative events |
|
| Cre mediated recombination | tissue specificity inducible modular |
non-reversible requires multiple transgenic lines |
|
| ENU mutagenesis | unbiased screening technique can identify temperature-sensitive alleles | requires much time and animal space | |
| Morpholinos | rapid Versatile loss-of-function approach |
non-specific effects | |
| Zinc Finger Nucleases | site directed mutagenesis | low efficiency | |
| Transcription Activator-Like endonucleases (TALENs) | site specific mutations high efficiency can facilitate homologous recombination in zebrafish | ||
| CRISPR-Cas System | Site specific mutations Easy to produce guide RNAs High efficiency |
||
| Visualization Strategies | Reporter Lines | ease of visualization BACs enable inclusion of enhancers |
lengthy generation time potential insertional effects on expression generational or stage-specific silencing |
| Multicolor clonal analysis | High-resolution clonal analysis | Need specific inducible Cre lines | |
| Photoconvertible proteins | Can be used to trace regionalized cell subsets | Trace is not genetic or permanent |
One drawback of the zebrafish model system has been the inability to generate conditional loss-of-function alleles. Over the past few years, zinc finger nucleases (ZFNs) and, more recently, transcription activator-like effector nucleases (TALENs) and the CRISPR-Cas system have aided directed mutagenesis [132–136]. Very recently, a system was described for inducing site-specific homologous recombination in zebrafish embryos utilizing TALENs [137]. Double-stranded breaks could enable the incorporation of sequences from co-injected short single-stranded DNA oligos at a low frequency, in both somatic and germline cells. Adapting this technology, one can envision creating conditional “knock-out” alleles through the insertion of two compatible loxP sites targeting a gene of interest. This technology will enable the study of individual gene products in a tissue-specific manner during regeneration, and provide potential upgrades over current dominant-negative, pharmacologic, and antisense morpholino-based approaches for loss-of-function studies. These current strategies are less specific than genetic mutants, and the ranges in treatment conditions or phenotypic penetrance can make it difficult to connect multiple pathways and synthesize a coherent blueprint for regeneration. Conditional loss-of-function alleles when standardized will remove a key element of speed, but can provide clarity by eliminating some of the drawbacks of other approaches. In addition to loss-of-function, the ability to perform homologous recombination downstream of endogenous regulatory sequences may help circumvent transgenic silencing in adult zebrafish, a current challenge in the field [138].
In addition to using TALENs for genome editing, there is evidence that the TALE architecture can be employed as a transcriptional activator or repressor to alter expression of specific target genes in living cells [139]. Combining TALEs with Cre-Lox technology could also provide another method for altering gene expression in a conditional and tissue-specific manner.
High-throughput screening using zebrafish embryos is a powerful method to identify small molecules with the potential to enhance regeneration. For instance, transgenic zebrafish expressing cell cycle indicators specifically in cardiomyocytes were used to identify several small molecules capable of enhancing or blocking cardiomyocyte proliferation in growing embryonic or injured adult hearts [64]. In the best-case scenario, promising candidates can be directly applied to mammalian systems to assess their impact on regeneration. A most successful series of studies toward this end came from a small molecule screen for effects on hematopoietic stem cell markers in zebrafish embryos. It was found that inducers of prostaglandin E2 synthesis and prostaglandin E2 itself are capable of expanding hematopoietic stem cell (HSC) numbers in zebrafish, and that they have similar effects on the adult HSCs of mice and non-human primates [140]. Thus, studying stem cell and regenerative biology in the zebrafish system might lead to potential new therapies in humans.
Concluding remarks
Adult mammals are naturally incapable of re-growing amputated limbs, significant amounts of cardiac muscle, or recovering from traumatic injury to the brain or spinal cord. Current studies address two main options for functional recovery after these injuries: 1) introduction of an exogenous cell source, which could engraft and integrate with existing tissue; or 2) stimulation of endogenous cell populations to induce regeneration. By pairing model organism genetics with remarkable regenerative abilities, the zebrafish has a strong track record and high potential to inform methodology to activate endogenous cell populations for regeneration. Recent studies with zebrafish have used an evolving toolset to identify the cellular sources activated for regeneration, an important first step in understanding the complex events of organ regeneration. Continued insights into the molecular mechanisms regulating regeneration will provide guidance for understanding and augmenting the regenerative abilities of less naturally capable vertebrate species like humans. Advances in gene targeting, chemical screening, and visualization techniques in zebrafish should facilitate the next generation of insights and discoveries.
Highlights.
Zebrafish are a key genetic model system for vertebrate regeneration research
Toolsets continue to evolve for studies of zebrafish appendage, heart, and neural regeneration
Regeneration concepts and mechanisms in zebrafish have implications for mammals
Acknowledgments
We thank Amy Dickson for artwork, and Gregory Nachtrab and Mayssa Mokalled for comments on the manuscript. K.D.P. acknowledges current funding from Howard Hughes Medical Institute, National Institute of General Medical Sciences, National Heart, Lung, and Blood Institute, and American Federation for Aging Research. D.R.H. acknowledges current funding from the National Eye Institute (R01-EY018417) and the University of Notre Dame Center for Zebrafish Research.
Glossary of Terms
- Regeneration
events by which lost or damaged tissue is replaced through endogenous mechanisms, restoring organ form and function
- Blastema
a proliferative mass of morphologically similar cells that accumulates in certain tissues after trauma and develops into the lost structures
- Dedifferentiation
process by which a differentiated cell reverts to a less differentiated state to enable proliferation or differentiation
- Transdifferentiation
conversion from one differentiated cell type to another
- Fate mapping
permanent labeling of a cell type to determine the contribution of these cells and their progeny during developmental and regenerative events
- Positional memory
the process by which spared adult cells retain positional information to recover only those structures lost by injury, of correct size and pattern
- Myocardial infarction
massive cardiac muscle cell death and a leading cause of morbidity and mortality in humans, typically caused by coronary artery occlusion and ischemia
- Osteoblasts
bone-depositing cells
- Epicardium
mesothelial cell type that covers the periphery of the heart and can act as progenitor tissue for fibroblasts, vascular support cells, and possibly other cells
- Genetic ablation
selective killing of a specific cell type by the expression of a toxin, pro-apoptotic factor, or pro-drug converting enzyme
- Müller glia
specialized glial cells found in the retina that act as neuronal support cells and resident stem cells after injury
- Radial glial cell
glial cells in the brain and spinal cord that act as neuronal progenitors during development and after injury
- Telencephalon
the most rostral of two subdivisions of the developing forebrain, the caudal subdivision being the diencephalon
- Transection
a precise transverse cut into the tissue that leaves much of the surrounding tissue undisturbed
- CRISPR-Cas
the Cas9 protein can be targeted through a CRISPR guide RNA to induce site-specific dsDNA breaks for targeting genome editing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Streisinger G, et al. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291(5813):293–6. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 2.Streisinger G, et al. Segregation analyses and gene-centromere distances in zebrafish. Genetics. 1986;112(2):311–9. doi: 10.1093/genetics/112.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Stainier DY. Zebrafish in the study of early cardiac development. Circ Res. 2012;110(6):870–4. doi: 10.1161/CIRCRESAHA.111.246504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolman M, Granato M. Behavioral genetics in larval zebrafish: learning from the young. Dev Neurobiol. 2012;72(3):366–72. doi: 10.1002/dneu.20872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jesuthasan S. Fear, anxiety, and control in the zebrafish. Dev Neurobiol. 2012;72(3):395–403. doi: 10.1002/dneu.20873. [DOI] [PubMed] [Google Scholar]
- 7.Kabashi E, et al. In the swim of things: recent insights to neurogenetic disorders from zebrafish. Trends Genet. 2010;26(8):373–81. doi: 10.1016/j.tig.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Leach SD. Zebrafish models for cancer. Annu Rev Pathol. 2011;6:71–93. doi: 10.1146/annurev-pathol-011110-130330. [DOI] [PubMed] [Google Scholar]
- 9.Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011;91(2):279–88. doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, et al. Two origins of blastemal progenitors define blastemal regeneration of zebrafish lower jaw. PLoS One. 2012;7(9):e45380. doi: 10.1371/journal.pone.0045380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chitnis AB, Nogare DD, Matsuda M. Building the posterior lateral line system in zebrafish. Dev Neurobiol. 2012;72(3):234–55. doi: 10.1002/dneu.20962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brignull HR, Raible DW, Stone JS. Feathers and fins: non-mammalian models for hair cell regeneration. Brain Res. 2009;1277:12–23. doi: 10.1016/j.brainres.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hata S, Namae M, Nishina H. Liver development and regeneration: from laboratory study to clinical therapy. Dev Growth Differ. 2007;49(2):163–70. doi: 10.1111/j.1440-169X.2007.00910.x. [DOI] [PubMed] [Google Scholar]
- 14.Shin D, et al. Intrinsic and extrinsic modifiers of the regulative capacity of the developing liver. Mech Dev. 2012;128(11–12):525–35. doi: 10.1016/j.mod.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Wingert RA. Regenerative medicine for the kidney: stem cell prospects & challenges. Clin Transl Med. 2013;2(1):11. doi: 10.1186/2001-1326-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson O, et al. Adenosine signaling promotes regeneration of pancreatic beta cells in vivo. Cell Metab. 2012;15(6):885–94. doi: 10.1016/j.cmet.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson RM, et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334(1):213–23. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss JB, et al. Regeneration of the pancreas in adult zebrafish. Diabetes. 2009;58(8):1844–51. doi: 10.2337/db08-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diep CQ, et al. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature. 2011;470(7332):95–100. doi: 10.1038/nature09669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pisharath H, et al. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124(3):218–29. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MB . Observations sur la regeneration de quelques parties du corps des poissons. Hist d. l’AcadRoy des Sciences; 1786. [Google Scholar]
- 22.Morgan TH. Regeneration in teleosts. Archiv Fur Entwicklungsmechanik Der Organismen. 1900;10(1):120–134. [Google Scholar]
- 23.Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141(4):1583–95. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitehead GG, et al. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310(5756):1957–60. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- 25.Poss KD, et al. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129(22):5141–9. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 26.Makino S, et al. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc Natl Acad Sci U S A. 2005;102(41):14599–604. doi: 10.1073/pnas.0507408102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nechiporuk A, et al. Positional cloning of a temperature-sensitive mutant emmental reveals a role for sly1 during cell proliferation in zebrafish fin regeneration. Dev Biol. 2003;258(2):291–306. doi: 10.1016/s0012-1606(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 28.Voz ML, et al. Fast homozygosity mapping and identification of a zebrafish ENU-induced mutation by whole-genome sequencing. PLoS One. 2012;7(4):e34671. doi: 10.1371/journal.pone.0034671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowen ME, et al. Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics. 2012;190(3):1017–24. doi: 10.1534/genetics.111.136069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obholzer N, et al. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development. 2012;139(22):4280–90. doi: 10.1242/dev.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leshchiner I, et al. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22(8):1541–8. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill JT, et al. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013;23(4):687–97. doi: 10.1101/gr.146936.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller AC, et al. RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res. 2013;23(4):679–86. doi: 10.1101/gr.147322.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart S, Stankunas K. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol. 2012;365(2):339–49. doi: 10.1016/j.ydbio.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22(4):879–86. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sousa S, et al. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138(18):3897–905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- 37.Knopf F, et al. Bone Regenerates via Dedifferentiation of Osteoblasts in the Zebrafish Fin. Dev Cell. 2011;20(5):713–24. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell. 2011;20(5):725–32. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehoczky JA, Robert B, Tabin CJ. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc Natl Acad Sci U S A. 2011;108(51):20609–14. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460(7251):60–5. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 41.Rinkevich Y, et al. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476(7361):409–13. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jazwinska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17(16):1390–5. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 43.Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134(3):479–89. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 44.Chablais F, Jazwinska A. IGF signaling between blastema and wound epidermis is required for fin regeneration. Development. 2010;137(6):871–9. doi: 10.1242/dev.043885. [DOI] [PubMed] [Google Scholar]
- 45.Blum N, Begemann G. Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development. 2012;139(1):107–16. doi: 10.1242/dev.065391. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y, et al. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol. 2009;331(2):270–80. doi: 10.1016/j.ydbio.2009.05.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee Y, et al. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132(23):5173–83. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- 48.Grotek B, Wehner D, Weidinger G. Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development. 2013;140(7):1412–23. doi: 10.1242/dev.087452. [DOI] [PubMed] [Google Scholar]
- 49.Munch J, Gonzalez-Rajal A, de la Pompa JL. Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development. 2013;140(7):1402–11. doi: 10.1242/dev.087346. [DOI] [PubMed] [Google Scholar]
- 50.Smith A, et al. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblasts differentiation and function. Dev Biol. 2006;299(2):438–54. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 51.Quint E, et al. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 2002;99(13):8713–8. doi: 10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Rosa JM, et al. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138(9):1663–74. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 54.Chablais F, et al. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnabel K, et al. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One. 2011;6(4):e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138(16):3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464(7288):601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20(3):397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 61.Chablais F, Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development. 2012;139(11):1921–30. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- 62.Kim J, et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci U S A. 2010;107(40):17206–10. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lien CL, et al. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4(8):e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi WY, et al. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140(3):660–6. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138(14):2895–902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J, et al. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat Protoc. 2012;7(2):247–55. doi: 10.1038/nprot.2011.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin VP, et al. Regulation of zebrafish heart regeneration by miR-133. Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jopling C, et al. Hypoxia induces myocardial regeneration in zebrafish. Circulation. 2012;126(25):3017–27. doi: 10.1161/CIRCULATIONAHA.112.107888. [DOI] [PubMed] [Google Scholar]
- 69.Itou J, et al. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development. 2012;139(22):4133–42. doi: 10.1242/dev.079756. [DOI] [PubMed] [Google Scholar]
- 70.Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: lessons from development. Genes Dev. 2011;25(4):299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–8. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–6. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bersell K, et al. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138(2):257–70. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 75.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eulalio A, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–81. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 77.Huang GN, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338(6114):1599–603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou B, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011;121(5):1894–904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernardos RL, et al. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27(26):7028–40. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fimbel SM, et al. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27(7):1712–24. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sherpa T, et al. Ganglion cell regeneration following whole-retina destruction in zebrafish. Dev Neurobiol. 2008;68(2):166–81. doi: 10.1002/dneu.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Otteson DC, Hitchcock PF. Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Res. 2003;43(8):927–36. doi: 10.1016/s0042-6989(02)00400-5. [DOI] [PubMed] [Google Scholar]
- 83.Cameron DA. Cellular proliferation and neurogenesis in the injured retina of adult zebrafish. Vis Neurosci. 2000;17(5):789–97. doi: 10.1017/s0952523800175121. [DOI] [PubMed] [Google Scholar]
- 84.Yurco P, Cameron DA. Responses of Muller glia to retinal injury in adult zebrafish. Vision Res. 2005;45(8):991–1002. doi: 10.1016/j.visres.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 85.Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26(23):6303–13. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44(3):289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 87.Kassen SC, et al. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67(8):1009–31. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- 88.Ariga J, Walker SL, Mumm JS. Multicolor time-lapse imaging of transgenic zebrafish: visualizing retinal stem cells activated by targeted neuronal cell ablation. J Vis Exp. 2010:43. doi: 10.3791/2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Montgomery JE, Parsons MJ, Hyde DR. A Novel Model of Retinal Ablation Demonstrates That the Extent of Rod Cell Death Regulates the Origin of the Regenerated Zebrafish Rod Photoreceptors. Journal of Comparative Neurology. 2010;518(6):800–814. doi: 10.1002/cne.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fraser B, et al. Regeneration of cone photoreceptors when cell ablation is primarily restricted to a particular cone subtype. PLoS One. 2013;8(1):e55410. doi: 10.1371/journal.pone.0055410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raymond PA, et al. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vihtelic TS, et al. Retinal regional differences in photoreceptor cell death and regeneration in light-lesioned albino zebrafish. Exp Eye Res. 2006;82(4):558–75. doi: 10.1016/j.exer.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 93.Thummel R, et al. Inhibition of Muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev Neurobiol. 2008;68(3):392–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramachandran R, et al. Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration. J Comp Neurol. 2010;518(20):4196–212. doi: 10.1002/cne.22448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomas JL, et al. Characterization of multiple light damage paradigms reveals regional differences in photoreceptor loss. Exp Eye Res. 2012;97(1):105–16. doi: 10.1016/j.exer.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nelson CM, et al. Stat3 defines three populations of Muller glia and is required for initiating maximal muller glia proliferation in the regenerating zebrafish retina. J Comp Neurol. 2012;520(18):4294–311. doi: 10.1002/cne.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fausett BV, Gumerson JD, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28(5):1109–17. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramachandran R, Zhao XF, Goldman D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A. 2011;108(38):15858–63. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nelson CM, et al. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013;33(15):6524–39. doi: 10.1523/JNEUROSCI.3838-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cameron DA, et al. Gene expression profiles of intact and regenerating zebrafish retina. Mol Vis. 2005;11:775–91. [PubMed] [Google Scholar]
- 101.Craig SE, Calinescu AA, Hitchcock PF. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebra fish. J Ocul Biol Dis Infor. 2008;1(2–4):73–84. doi: 10.1007/s12177-008-9011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Calinescu AA, et al. Cellular expression of midkine-a and midkine-b during retinal development and photoreceptor regeneration in zebrafish. J Comp Neurol. 2009;514(1):1–10. doi: 10.1002/cne.21999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qin Z, Barthel LK, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009;106(23):9310–5. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ramachandran R, Zhao XF, Goldman D. Insm1a-mediated gene repression is essential for the formation and differentiation of Muller glia-derived progenitors in the injured retina. Nat Cell Biol. 2012;14(10):1013–23. doi: 10.1038/ncb2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thummel R, Bailey TJ, Hyde DR. In vivo electroporation of morpholinos into the adult zebrafish retina. J Vis Exp. 2011;(58):e3603. doi: 10.3791/3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qin Z, et al. FGF signaling regulates rod photoreceptor cell maintenance and regeneration in zebrafish. Exp Eye Res. 2011;93(5):726–34. doi: 10.1016/j.exer.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meyers JR, et al. beta-catenin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural Dev. 2012;7:30. doi: 10.1186/1749-8104-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Osakada F, et al. Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007;27(15):4210–9. doi: 10.1523/JNEUROSCI.4193-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Das AV, et al. Neural stem cell properties of Muller glia in the mammalian retina: regulation by Notch and Wnt signaling. Dev Biol. 2006;299(1):283–302. doi: 10.1016/j.ydbio.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 110.Wan J, et al. Preferential regeneration of photoreceptor from Muller glia after retinal degeneration in adult rat. Vision Res. 2008;48(2):223–34. doi: 10.1016/j.visres.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 111.Close JL, et al. Epidermal growth factor receptor expression regulates proliferation in the postnatal rat retina. Glia. 2006;54(2):94–104. doi: 10.1002/glia.20361. [DOI] [PubMed] [Google Scholar]
- 112.Del Debbio CB, et al. Notch and Wnt signaling mediated rod photoreceptor regeneration by Muller cells in adult mammalian retina. PLoS One. 2010;5(8):e12425. doi: 10.1371/journal.pone.0012425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wan J, Ramachandran R, Goldman D. HB-EGF Is Necessary and Sufficient for Muller Glia Dedifferentiation and Retina Regeneration. Dev Cell. 2012;22(2):334–47. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Battista AG, et al. Extracellular ADP regulates lesion-induced in vivo cell proliferation and death in the zebrafish retina. J Neurochem. 2009;111(2):600–13. doi: 10.1111/j.1471-4159.2009.06352.x. [DOI] [PubMed] [Google Scholar]
- 115.Craig SE, et al. The zebrafish galectin Drgal1-l2 is expressed by proliferating Muller glia and photoreceptor progenitors and regulates the regeneration of rod photoreceptors. Invest Ophthalmol Vis Sci. 2010;51(6):3244–52. doi: 10.1167/iovs.09-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thummel R, et al. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp Eye Res. 2010;90(5):572–82. doi: 10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Becker T, et al. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol. 1997;377(4):577–95. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 118.Reimer MM, et al. Motor neuron regeneration in adult zebrafish. J Neurosci. 2008;28(34):8510–6. doi: 10.1523/JNEUROSCI.1189-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kuscha V, et al. Lesion-induced generation of interneuron cell types in specific dorsoventral domains in the spinal cord of adult zebrafish. J Comp Neurol. 2012;520(16):3604–16. doi: 10.1002/cne.23115. [DOI] [PubMed] [Google Scholar]
- 120.Reimer MM, et al. Sonic hedgehog is a polarized signal for motor neuron regeneration in adult zebrafish. J Neurosci. 2009;29(48):15073–82. doi: 10.1523/JNEUROSCI.4748-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dias TB, et al. Notch signaling controls generation of motor neurons in the lesioned spinal cord of adult zebrafish. J Neurosci. 2012;32(9):3245–52. doi: 10.1523/JNEUROSCI.6398-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Goldshmit Y, et al. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J Neurosci. 2012;32(22):7477–92. doi: 10.1523/JNEUROSCI.0758-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kizil C, et al. Adult neurogenesis and brain regeneration in zebrafish. Dev Neurobiol. 2012;72(3):429–61. doi: 10.1002/dneu.20918. [DOI] [PubMed] [Google Scholar]
- 124.Marz M, et al. Regenerative response following stab injury in the adult zebrafish telencephalon. Dev Dyn. 2011;240(9):2221–31. doi: 10.1002/dvdy.22710. [DOI] [PubMed] [Google Scholar]
- 125.Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques. 2008;45(6):613–4. 616. doi: 10.2144/000113005. 618 passim. [DOI] [PubMed] [Google Scholar]
- 126.Kizil C, Brand M. Cerebroventricular microinjection (CVMI) into adult zebrafish brain is an efficient misexpression method for forebrain ventricular cells. PLoS One. 2011;6(11):e27395. doi: 10.1371/journal.pone.0027395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kizil C, et al. The chemokine receptor cxcr5 regulates the regenerative neurogenesis response in the adult zebrafish brain. Neural Dev. 2012;7:27. doi: 10.1186/1749-8104-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kizil C, et al. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev Cell. 2012;23(6):1230–7. doi: 10.1016/j.devcel.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 129.Kroehne V, et al. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138(22):4831–41. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- 130.Kishimoto N, Shimizu K, Sawamoto K. Neuronal regeneration in a zebrafish model of adult brain injury. Dis Model Mech. 2012;5(2):200–9. doi: 10.1242/dmm.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kyritsis N, et al. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012;338(6112):1353–6. doi: 10.1126/science.1228773. [DOI] [PubMed] [Google Scholar]
- 132.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26(6):702–8. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meng X, et al. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26(6):695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29(8):697–8. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang P, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29(8):699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 136.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bedell VM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491(7422):114–8. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thummel R, Burket CT, Hyde DR. Two different transgenes to study gene silencing and re-expression during zebrafish caudal fin and retinal regeneration. Scientific World Journal. 2006;6(Suppl 1):65–81. doi: 10.1100/tsw.2006.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 140.North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–11. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]