Abstract
The human apical sodium-dependent bile acid transporter (hASBT, SLC10A2), primarily expressed in the ileum, is involved in both the recycling of bile acids and cholesterol homeostasis. In this study, the structure-function relationship of transmembrane domain 5 (TM5) residues involved in transport is elucidated. Cysteine scanning mutagenesis of each consecutive residue on TM5 resulted in 96% of mutants with a significantly decreased transport activity although each was expressed at the cell surface. Specifically, G197 and I208 were no longer functional and G201 and G212 functioned at less than 10% upon cysteine mutation. Interestingly, each of these exists along one face of the helix. Studies suggest that neither G201 nor G212 are on the substrate pathway. Conservative alanine mutations of the four residues displayed a higher activity in all but G197A, indicating its functional importance. G197 and G201 form a GxxxG motif, which has been found to be important in helix-helix interactions. According to our model, G197 and G201 face TM4 residues G179 and P175 respectively. Similarly, G212 faces G237, which forms part of a GxxxG domain in TM6. It is possible that these GxxxG domains and their interacting partners are responsible for maintaining the structure of the helices and their interactions with one another. I205 and I208 are both in positions to anchor the GxxxG domains and direct the change in interaction of TM5 from TM4 to TM6. Combined, the results suggest that residues along TM5 are critical for ASBT function but are not directly involved in substrate translocation.
Keywords: ASBT, SCAM, GxxxG motif, transmembrane domain
The human apical sodium-dependent bile acid transporter (hASBT, SLC10A2), primarily expressed in the distal ileum, is involved in the recycling of bile acids. It is a highly efficient mechanism reclaiming 95% of the circulating bile acids daily.1 ASBT is a 39–41 kDa active co-transporter of approximately one bile acid and two sodium ions per transport cycle.2 As bile acids are synthesized de novo from cholesterol, ASBT is a potential drug target to lower cholesterol levels in hypercholesterolemia.3–5 Additionally, the efficiency of ASBT to transport structurally diverse compounds makes it a target for a prodrug approach to increase drug bioavailability.6–10 Currently there is not a crystal structure of mammalian ASBT; therefore, a combination of molecular biology and computational techniques have been used by our laboratory to elucidate hASBT’s structure-function relationships.
Previously, work form our lab has confirmed systematically that hASBT exhibits a 7-transmembrane (7TM) topology.11, 12 Further studies employed the use of cysteine scanning mutagenesis to identify structural requirements and interactions required for function. This method revealed that residues of TM613 and TM714 interact with bile acids on the exofacial half of the membrane while TM315 and TM416 interact with bile acids on the cytosolic half of the membrane. Combined, this data depicts a substrate pathway through the membrane from its entrance to its release. Additionally, residues lining one face of TM6 may impart a helical flexibility required for the function of ASBT.13 More recently, TM1 has been shown to interact with sodium ions through several key residues.17
In this study, cysteine-scanning mutagenesis and solvent-accessibility studies of each consecutive residue in transmembrane domain 5 (TM5) have been undertaken not only to understand the structure-function relationship of the residues involved in transport but also to elucidate how TM5 residues interact with nearby residues on adjacent TMs. TM5, similar to other TMs, is highly conserved amongst eukaryotic ASBT orthologs (Figure 1). Additionally, its proximity to TM4 and TM6 suggest a potential role is substrate translocation. TM5 could also interact with residues along TM6 to impart helical flexibility. In fact, the combination of results reported here indicates that TM5 plays primarily an indirect, structural role in the overall function of ASBT.
Figure 1.

ASBT TM5 sequence conservation across species. Sequences were aligned with ClustalW. Identical residues are indicated by asterisks (*); a colon (:) denotes residues with a high degree of similarity while a period (.) denotes weakly similar residues.
EXPERIMENTAL PROCEDURES
Materials
[3H]-Taurocholic acid (0.2 Ci/mmol) was purchased from PerkinElmer. Glycodeoxycholic acid (GDCA) and taurocholic acid (TCA) were purchased from Sigma. [2-(Trimethylammonium)ethyl] Methanethiosulfonate Bromide (MTSET) was purchased from Toronto Research Chemicals, Inc. Streptavidin Agarose Resin was purchased from Thermo Scientific. Synthetic oligonucleotides were purchased from Sigma-Aldrich. Plasmids were prepared using a QIAprep Spin Mini-, Midi- or Maxiprep kits from Qiagen. DNA sequencing was performed in the biopolymer laboratory at the University of Maryland, Baltimore.
Site-directed Mutagenesis and Protein Expression
Cysteine and alanine point mutations were made on a hASBT C270A template in a pCMV5 vector due to the template’s MTS insensitivity.18 The mutations were generated by site-directed mutagenesis. The plasmids were sequenced, confirming the presence of each point mutation. Mutants were transiently transfected into COS-1 cells seeded at a density of 6 × 104 cells/mL on a 24 well plate using the TurboFect Transfection Reagent from Thermo Scientific. Briefly, 0.5 μg plasmid DNA and 2 μL TurboFect Reagent per well were incubated 15 minutes at room temperature in 100 μL DMEM. The mixture was then added to each well drop-wise. Cells were used for further studies 24 to 48 hours post transfection. COS-1 cells were maintained as previously described.15
In order to determine the ASBT cell surface expression levels, biotinylation of the membrane proteins was done using a membrane impermeable EZ Link Sulfo-NHS-SS-Biotin reagent (Pierce) as described previously.19 Briefly, cells were quickly washed twice with ice-cold PBS containing calcium and magnesium. The cells were incubated in 500 μL/well of 1.2 mg/mL Sulfo-NHS-SS-biotin for 30 min at 4 °C. The reaction was quenched with 300 μL/well of 1M Tris in PBS, pH 7.4. The cells were lysed in 300 μL of NP-40 lysis buffer containing a protease cocktail per well and incubated for about 1 hour at 4 °C with rocking. A fraction of the lysate was removed for analysis as a total protein control. Soluble portions of the lysate were further treated with 250 μL/well of Streptavidin Agarose Resin and incubated overnight at 4 °C on a nutator. The resin mixture was centrifuged for 1 min at 1,000 x g and the resin-attached proteins were incubated in 100 μL of 2x Laemmli buffer/well at room temperature for 40 min with rocking. The samples were boiled for 10 min and stored at 4 °C. Western blotting was also preformed as previously described.17 Briefly, samples were loaded onto precast, 15 well, 12% Tris-HCl Polyacrylamide gel (BioRad 456–1046) along with 2.5 μL Li-cor Odyssey two-color protein molecular weight marker (928-40001). A custom antibody (1:1,000) was used to visualize the glycosylated (41 kDa) and unglycoslyated (38 kDa) hASBT protein. Additionally, the selective labeling of cell surface proteins was confirmed by the absence of the 90 kDa ER protein, calnexin (mouse anti-calnexin; 1:1,000), and presence of the 140 kDa cell surface protein pan-cadherin (mouse anti-cadherin; 1:1,000).
Uptake Assay and Sodium Activation
TCA uptake experiments were conducted with slight modifications to previously reported methods.15, 17 Briefly, transiently transfected COS-1 cells were washed with warm Dulbecco’s Phosphate-Buffered (DPBS), pH 7.4 followed by equilibration in warm Modified Hanks’ balanced salt solution (MHBSS), pH 7.4 at 37°C for 15 min. Uptake was initiated by incubating cells in MHBSS containing 5.0 μM TCA, 0.2% bovine serum albumin (BSA), and 1 μCi/mL [3H] TCA for 12 min at 37°C. The reaction was halted by washing the cells in ice-cold DPBS containing 0.5 mM TCA and 0.2% BSA. Liquid scintillation counting, using a LS6500 scintillation counter (Beckmann Coulter, Inc.), was used to measure the uptake of the cells after lysis in 350 μL 1 N NaOH. The total protein concentration was obtained by Bradford assay to give a calculated rate of [3H] TCA internalization in pmol/min/mg of protein. Normalization of mutant to C270A was used as a means of comparison.
As described previously, the sensitivity of the mutants to sodium was determined by comparing the [3H] TCA transport function of the mutants at equilibrating (12 mM) versus physiological (137 mM) sodium concentrations.15 The uptake assay was conducted as described above but without the 15 min equilibration in MHBSS at 37 °C. Choline chloride was used to replace deficient sodium chloride. A ratio of 12 mM Na+ uptake to 137 mM Na+ uptake was calculated to each mutant and normalized to C270A.
TCA and Sodium Kinetics
TCA inhibition kinetics were determined for a select group of mutants using a range of TCA concentrations from 0 – 200 μM, 137 mM NaCl and 1 μCi/mL (0.2 Ci/mmol) [3H] TCA. Sodium kinetics were determined for sodium sensitive mutants using a range of sodium chloride concentrations from 0–200 mM, 5 μM TCA and 1 μCi/mL (0.2 Ci/mmol) [3H] TCA. Data was normalized to cell surface expression levels. Kinetic parameters were determined using GraphPad 5.0 software as described previously.19
MTSET Inhibition and Substrate Protection
Transiently transfected COS-1 cells were incubated in 1 mM MTSET for 10 min at room temperature prior to undergoing the uptake assay as described above.15, 17, 19 Control cells, treated in MHBSS buffer alone, were run in parallel. The ratio of uptake for the MTSET treated cells versus the control cells was calculated to determine the inhibition of transport. This ratio was then compared to that of C270A. Additionally, as previously described, the transiently transfected cells were incubated in 1 mM MTSET with or without sodium and with or without 200 μM GDCA for 10 min at room temperature prior to undergoing the [3H] TCA uptake experiments as described above.15, 17 Again, ratios were calculated comparing each treatment to untreated (buffer only) cells and these ratios were normalized to those of C270A.
Data Analysis
The data from each mutant are shown as the mean value with bars indicating the standard deviation (SD) for n≥3. Data analysis was done using GraphPad 5.0 or Kaleidagraph 4.0 using either a one-way analysis of variance (ANOVA) with Dunnett’s post-hoc test or a two-tailed unpaired Student’s t-test as appropriate. Differences were considered statistically significant at p≤0.05.
RESULTS
Membrane Expression and Uptake Activity of TM5 Cysteine Mutants
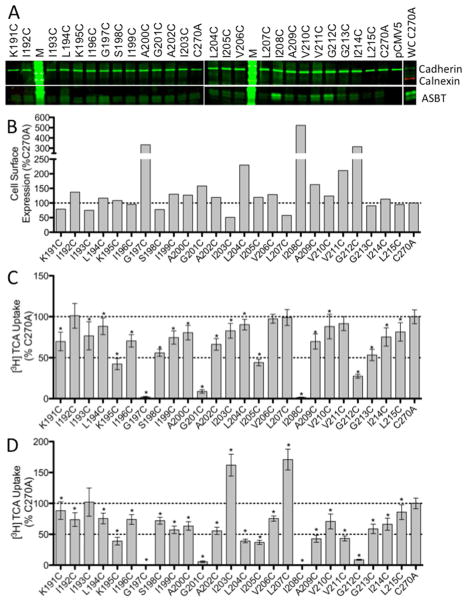
Each residue along the TM5 helix (Lys191 – Leu215) was individually mutated to a cysteine residue in the C270A hASBT scaffold. The mutant DNA was then transiently transfected into COS-1 cells followed by studies on protein expression and function. Expression data indicate that each mutant generates protein that is present on the cell surface (Figure 2a).
Figure 2.
Protein expression and [3H] TCA uptake in TM5 cysteine mutants. A. Western Blot of TM5 cysteine mutants expressed on the cell surface of COS-1 cells. ASBT is seen a two bands, glycosylated (41 kDa) and unglycosylated (38 kDa). Cadherin (140 kDa), expressed at the cell surface, was used as a positive loading control while Calnexin (90 kDa), expressed in the ER, was used as a negative control and was only observed in whole cell (WC) fractions (C270A is shown). Blots are representative of five independent experiments. B. ASBT cell surface expression normalized to C270A. C. Uptake of [3H] TCA expressed as a percentage of the parental transporter, C270A, in pmol [3H] TCA transported per minute per milligram of protein. D. Uptake expressed as a percentage of C270A and normalized to ASBT cell surface expression. Bars represent the S.D. of at least three separate experiments performed in triplicate. A significant difference was determined by an ANOVA calculation with p < 0.05 represented with an asterisk (*).
After normalizing to membrane expression, results of activity assays of the cysteine mutants show that 88% of the mutants demonstrated a significant decrease in the rate of TCA uptake in comparison to C270A (Figure 2c). The activities for residues G197C and I208C were close to empty vector, pCMV5, indicating a near total loss of function. Additionally, G201C maintained only about 5.6% of the activity of C270A and G212C maintained approximately 8.8%. The direct neighbors of each of these residues maintained at least 40% of the C270A activity.
Sodium Activation
Bile acid substrate is transported by ASBT through the use of the energy provided by the cotransport of sodium through a sodium gradient.2 By altering the sodium concentration outside the cell from a physiological concentration (137 mM) to a concentration equal to that inside the cell, an equilibrating concentration (12 mM), and assaying the transport of TCA by the TM5 cysteine mutants, it is possible to determine the involvement of the TM5 residues in sodium transport. Figure 3 shows the ratio of TCA transport for each mutant at 12 mM vs. 137 mM sodium concentrations in comparison to those of C270A. Only G201C shows a significant sensitivity to sodium in comparison to C270A.
Figure 3.
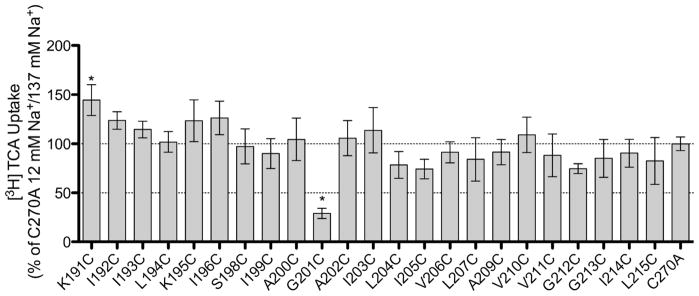
Sodium sensitivity of TM5 cysteine mutants. Uptake of [3H] TCA in pmol [3H] TCA transported per minute per milligram of protein. The graph represents a ratio of uptake at 12 mM/137 mM sodium concentrations (1:1.84 for C270A) and normalized as a percentage of the parental transporter, C270A. Bars represent the S.D. of at least three separate experiments performed in triplicate. A significant difference was determined by an ANOVA calculation with p < 0.05 represented with an asterisk (*).
Substituted Cysteine Solvent Accessibility of TM5 Mutants
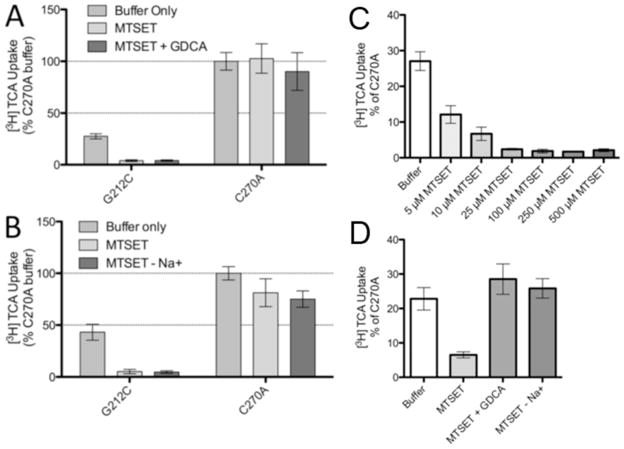
Sulfhydryl groups of cysteine residues are predominantly deprotonated in aqueous environments allowing for a facile reaction with MTSET, which reacts selectively with ionized sulfhydryl groups.20–22 Taking advantage of this chemistry, the MTSET reagent can be incubated with the cysteine mutants; then, the chemically-modified mutants can be assayed for transport function to identify residues that are in aqueous environments. In the TMs, these environments could signify a substrate pathway or in a binding pocket. The TM5 data demonstrate a highly significant decrease in transport activity after MTSET treatment for G212C only (Figure 4).
Figure 4.
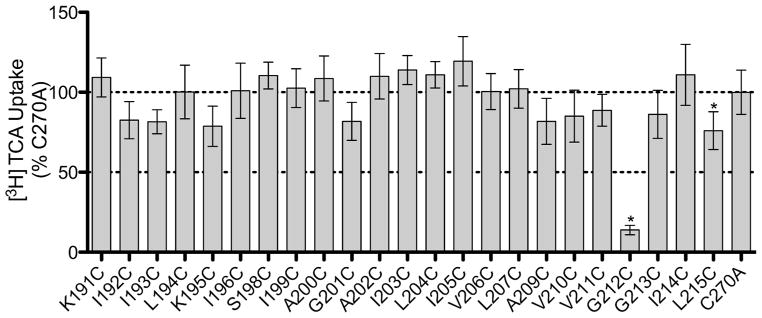
Identification of solvent accessible TM5 mutant residues by MTSET labeling. Prior to [3H] TCA uptake, COS-1 cells expressing C270A or cysteine mutants were incubated with 1 mM MTSET for 10 min at room temperature. Uptake in the absence of MTSET was compared to uptake in the presence of MTSET (MTSET/Initial) and represented as a percentage of C270A. Bars represent the S.D. of at least three separate experiments performed in triplicate. A significant difference was determined by an ANOVA calculation with p < 0.05 represented with an asterisk (*).
Substrate Protection of G212C
Substrate protection of mutants in the presence of MTSET can occur in two conditions; the substrate can interact directly with the mutant residue and inhibit an interaction with MTSET, or it can interact at a site away from the residue, causing a change in the conformation of the protein and inhibiting the access of MTSET. In order to determine if ASBT substrates interact with G212C in the presence of MTSET, buffers containing GDCA or lacking Na+ were co-incubated with MTSET prior to the transport assay. In the case of both GDCA and Na+, the presence of substrate was not able to rescue the transport function of the mutant (Figs. 5A and B). These results indicate that G212C is solvent accessible in the presence or absence of substrate and can be modified by MTSET under both conditions. Therefore, it is likely that the interaction between G212C and MTSET alone is altering the activity of the mutant protein and that neither bile acid nor sodium is directly or indirectly interacting with G212C to alter its accessibility. Additionally, we validated the dose response of MTSET (5–500μM) to determine whether G212C is highly sensitive to inhibition by MTSET (Fig. 5C). At low [MTSET] (5–10μM) functional inhibition was reduced (apparent IC50 ~ 4.1μM) and subsequent experiments at 5μM MTSET (Fig. 5D) revealed that substrate (GDCA) and presence of Na+ were both able to rescue ASBT from MTSET inactivation. These data suggest that conformational changes may occur in the presence of bile acid and sodium that protects this residue from modification; this effect may be reversed by increasing [MTSET].
Figure 5.
Protection of G212C from MTSET modification and MTSET does response. COS-1 cells expressing C270A or G212C were incubated with buffer alone or 1 mM MTSET A. with or without GDCA or B. with or without sodium for 10 min at room temperature. After washing, cells were allowed to equilibrate to 37°C for 15 min prior to [3H] TCA uptake. Each uptake was represented as a percentage of C270A uptake in buffer alone. Bars represent the S.D. of at least three separate experiments performed in triplicate. A significant difference was determined by an ANOVA calculation with p < 0.05 represented with an asterisk (*). C. COS-1 cells expressing G212C were incubated with buffer alone or 5 to 500 μM MTSET for 10 min at room temperature. D. COS-1 cells expressing G212C were incubated with buffer alone or 5 μM MTSET with or without 200 μM GDCA or with or without 137 mM sodium for 10 min at room temperature. After washing, cells were allowed to equilibrate to 37 °C for 15 min prior to [3H] TCA uptake.
TCA and Sodium Kinetics of TM5 Mutants
Kinetic analysis was performed on select TM5 mutants in an attempt to further understand their roles. TCA affinity (KTCA) was not significantly altered in all five mutants tested (Table 1). However, Jmax was significantly lower in four of the five mutants tested. Additionally, a significant decrease in affinity for sodium (KNa) as well as a significant decrease in Jmax was also observed for G201C.
Table 1.
Substrate kinetics for select TM5 cysteine mutantsa
| Na+ Kinetics | TCA Kinetics | |||
|---|---|---|---|---|
| Jmax (pmol/min/mg protein) | KNa (mM) | Jmax (pmol/min/mg protein) | KTCA (μM) | |
| C270A | 249.1 ± 19.97 | 6.96 ± 2.36 | 1295 ± 131.6 | 9.60 ± 3.68 |
| K191C1 | 1216 ± 113.3 | 7.32 ± 3.01 | ||
| K195C1 | *360.7 ± 44.22 | 3.91 ± 2.77 | ||
| G201C1,2 | *30.25± 2.34 | *31.08 ± 6.84 | *140.4 ± 17.85 | 4.64 ± 3.14 |
| I205C1 | *950.0 ± 86.36 | 5.32 ± 2.42 | ||
| G212C1,3 | *199.4 ± 27.21 | 11.49 ± 5.86 | ||
decreased uptake upon cysteine mutation,
sodium sensitive,
MTSET accessible
Sodium concentrations ranged from 0–200 mM (5 μM TCA) and TCA concentrations ranged from 0–200 μM (137 mM Na+). Data, with standard error, represents at least three separate experiments performed in triplicate. Kinetics parameters were determined using GraphPad 5.0 software (San Diego, CA), with constants determined by nonlinear regression. Parameters determined to be significant compared to the C270A control ± standard deviation when analyzed by one-way ANOVA (TCA kinetics) or t-test (Na+ kinetics) are indicated,
p ≤ 0.05.
Select Conservative Mutations of TM5
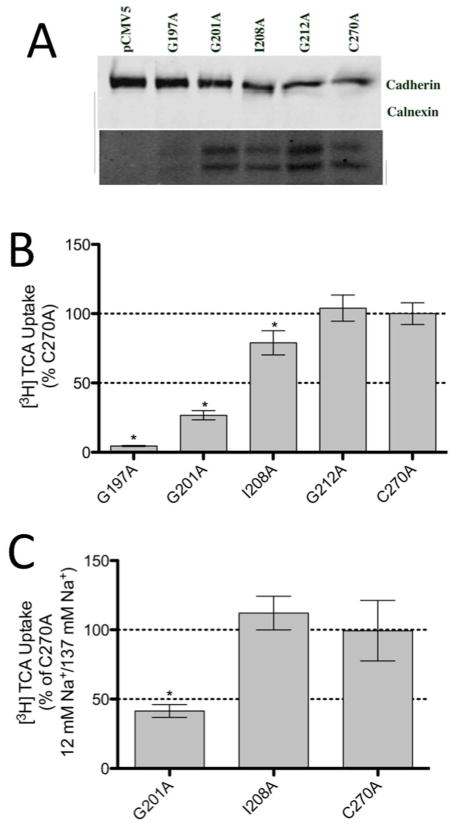
The above results, suggesting that neither TCA nor sodium is being directly transported by the residues in TM5, prompted a further investigation into the structural relevance of the residues exhibiting a decreased activity. Residues displaying a complete or nearly complete loss of activity due to cysteine mutation (G197C, G201C, I208C and G212C) were mutated to the more conservative alanine in the C270A background. Alanine mutants were assessed for function by a [3H] TCA uptake assay (Figure 6). While G197A remained inactive and the activity of G201A remained relatively similar to that of G201C, the activities of I208A and G212A increased significantly from their cysteine counterparts. The sodium sensitivity of G201A and I208A were also determined (Figure 6b). While G201A remains sodium sensitive, I208A was found to be insensitive to sodium. These results support earlier data suggesting that, other than G201, TM5 does not play a role in sodium transport and that G201 may be indirectly involved.
Figure 6.
[3H] TCA uptake and sodium sensitivity of select TM5 alanine mutants. A. Western Blot of TM5 alanine mutants expressed on the cell surface of COS-1 cells. ASBT is seen a two bands. Cadherin (140 kDa), expressed at the cell surface, was used as a positive loading control while Calnexin (90 kDa), expressed in the ER, was used as a negative control. B. Uptake of [3H] TCA expressed as a percentage of the parental transporter, C270A, in pmol [3H] TCA transported per minute per milligram of protein. C. Uptake of [3H] TCA in pmol [3H] TCA transported per minute per milligram of protein. The graph represents a ratio of uptake at 12 mM/137 mM sodium concentrations and normalized as a percentage of the parental transporter, C270A. Bars represent the S.D. of at least three separate experiments performed in triplicate. A significant difference was determined by an ANOVA calculation with p < 0.05 represented with an asterisk (*).
DISCUSSION
hASBT, primarily expressed in the distal ileum, is involved in the recycling of bile acids and is critical for cholesterol homeostasis. Given its biological importance and its potential role in drug transport, characterization at the functional and structural level would enable rational drug design for ASBT as a pharmacological target. Previous studies in our lab have elucidated roles for several TMs and extracellular loops in ASBT.12–17, 19, 23 For example, TM3 and TM4 have been shown to play a role in TCA translocation on the cytosolic half to the protein.15, 16 In the present study, the structure-function relationship of the TM5 residues involved in transport has been revealed. While TM5 residues may not play a direct role in the transport of either bile acids or sodium, several appear to play a critical role in alpha helical stability.
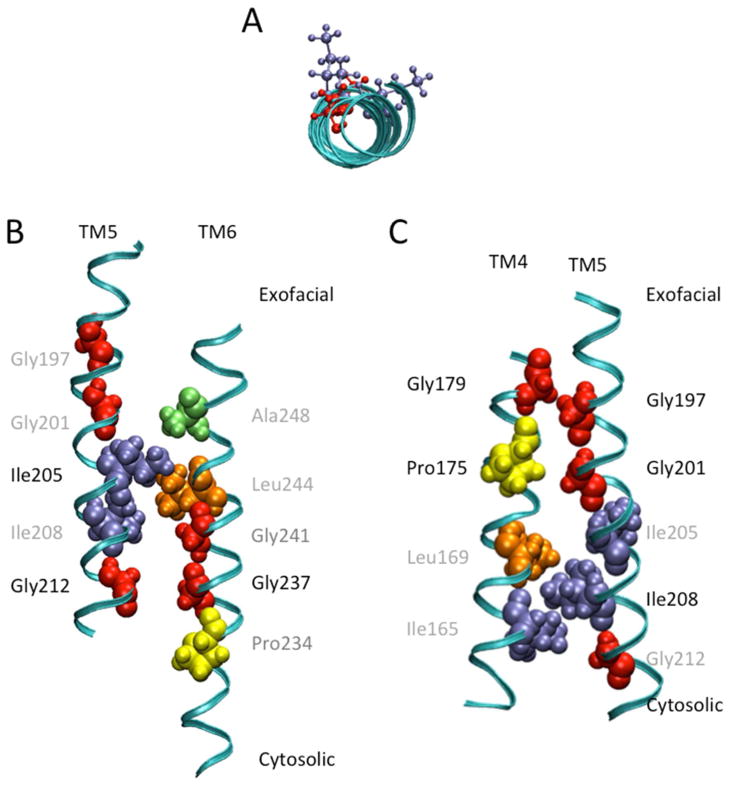
The alignment of TM5 residues from humans to those of nine eukaryotic orthologs demonstrates a highly conserved sequence with 22 of the 25 residues being either identical or highly similar (Figure 1). As expected, cysteine-scanning mutagenesis of these residues indicated that the majority of residues, 96%, were sensitive to cysteine mutation (Figure 2). However, most mutants maintained more than 35% activity. Of the remaining evolutionarily conserved, low activity residues, the activity of G201C (5.6%) and G212C (8.8%) was severely hampered by cysteine substitution while residues G197C and I208C had a total loss of function. Interestingly, all four residues reside on the same face of the helix (Figure 7a).12 Upon alanine mutation, G197A remained inactive; however, the activity of the other residues was restored partially (G201A; 27%) or completely (I208A, G212A) (Figure 6). While it is possible that the mutation of these glycine residues alone could disrupt the helical structure in TM5, the high concentration of functionally relevant glycines on one face of the helix warranted additional investigation.
Figure 7.
In silico representation of TM5 residues relevant for ASBT function. A. A top down view of TM5 from the exofacial side of the membrane highlighting relevant amino acids residing on the same face of the helix. Ball and stick representations in red denote glycine residues while purple denote isoleucine residues. B. In silico prediction of TM5 and TM6 functional group orientation. The GxxxG motif of TM6 (Gly237 and Gly241) potentially associates with Gly212 of TM5 at the backbone level to stabilize the helices. Ile205 may participate to constrain the crossing angle of the two helices. C. In silico prediction of TM4 and TM5 functional group orientation. The GxxxG motif in TM5 (Gly197 and Gly201) potentially interacts with Gly179 and Pro175, respectively, to allow for a strong and close association of the two helices. Ile208 interacts with Ile165 and Leu169 to anchor and constrain the crossing angles of the helices. Images were generated with VMD 1.8.6 software.
To determine the TM5 mutants’ sensitivity to sodium, the effect of TCA transport was determined at equilibrative sodium concentrations and compared to those at physiological concentrations. One residue, G201C, was found to be sensitive to sodium concentrations (Figure 3). The sensitivity to sodium was reproduced in the G201A mutant (Figure 6b). Kinetic studies of G201C revealed a significant decrease in sodium affinity (Table 1). These results indicate a role for G201 in sodium transport, but it is unlikely to be a direct interaction as other TM5 residues are not involved (vide infra).
The identification of solvent accessible residues can reveal a substrate translocation pathway. We applied the substituted cysteine accessibility method (SCAM) by incubating each mutant, expressed in COS-1 cells, with the membrane-impermeable MTSET reagent prior to uptake studies (Figure 4). This study pointed to only one solvent accessible residue, G212C. Additionally, TCA kinetic data on this residue indicated a slight decrease in substrate affinity in comparison to the C270A control (Table 1). In an attempt to modify accessibility of this residue to MTSET, experiments were conducted either in the presence of GDCA or the absence of sodium (Figure 5). As observed previously with wild-type (WT) and other TM mutants, substrate protection can partially or fully recover protein activity to levels seen when uptake is conducted in buffer alone. Co-incubation of MTSET with GDCA or in the absence of sodium did not alter the solvent accessibility, suggesting that neither substrate is directly interacting with G212 and that conformational changes are not occurring that would inhibit MTSET from accessing the residue. So, while G212C is solvent accessible, it does not appear to be in the substrate or co-substrate pathway.
A new dimer model (manuscript submitted) shows that G212 lies near the proposed, solvent accessible, dimer interface.24 The interface is formed between TM1-TM7-TM6 regions. TM5 is close to TM6 of the same monomer and next to TM1 of the other monomer in the hASBT dimer. Because TM5 is in close proximity to the water accessible interface, it is possible that G212 is effected by MTSET though this pathway.
Glycine and proline residues are known to destabilize and impart flexibility upon α-helices.25 This increase in flexibility is important in transmembrane proteins which do not usually possess much inherent flexibility within the membrane but require conformational changes during turnover.26 Alternatively, these glycine residues may be involved in the stabilization of the helix. The loss of flexibility due to the mutation of the glycine residues in TM5 to cysteines or even to alanines could, in part, explain the decrease or loss in activity observed. The GxxxG motif is commonly found in alpha-helices of transmembrane proteins,25, 27–33 and has been found to be important in the association of two adjacent TMs. The small size of the glycine residues on the same face of the helix allows for a close proximity of the two interacting helices so that the backbone structure of the helix can interact, either through hydrogen bonding or van der Waals forces, with the backbone of the adjacent helix.27, 28, 31
According to our validated structural model of hASBT, Gly212, on the cytosolic side of the membrane, is facing TM6 residue Gly237 that is part of the Pro234/Gly237/Gly241 conformational switch (Figure 7b).13 The presence of the GxxxG motif in the conformational switch and its interaction with Gly212 could explain why the mutation of Gly212 to cysteine resulted in decreased function. It is possible that the mutation of Gly212 to alanine resulted in a fully functional transporter because the small size of the alanine could still allow for the required interactions between helices. Another possibility is that Gly213 was able to moderately compensate for the loss of Gly212. On the exofacial side of the membrane, Gly197 and Gly201 of TM5 are facing TM4 residues Gly179 and Pro175, respectively (Figure 7c). Like Gly197, Gly179 also has a total loss of function when mutated to cysteine.16 Additionally, both G201 and P175 are sodium sensitive. In the case of Gly197, it is possible that the alanine mutation was sufficient to disrupt the packing of the helices that resulted in a total loss of function. However, it seems clear that these helix-helix associations are likely responsible for the stability of the helices and thus the GxxxG motifs and their interacting residues are required for function. Further, while it is possible that sodium could interact with the backbone residues of G201, it is more likely that G201 plays a role in the helix-helix interaction and that its role in sodium transport is indirect. The sodium sensitivity of G201 mutants may be attributed to conformational changes due to the disruption of the helical interactions.
Recently, a crystal structure of an ASBT ortholog from Neisseria meningitis (AsbtNM) was reported,34 with 26% sequence identity to hASBT. AsbtNM was found to contain 10 TM domains34 differing from the verified 7 TM topology model of hASBT.11, 12 TM7 of AsbtNM coincides with TM5 in hASBT and has 12% identity when aligned with the 10 eukaryotic species seen in Figure 1. TM7 of AsbtNM transects the membrane from the cytosolic side of the membrane to the exofacial side while TM5 of hASBT transects the membrane in the opposite direction. Additionally, the crystal structure indicates that the residues of TM7 in AsbtNM, corresponding to the functionally significant residues of TM5 in hASBT, interact with the nonconserved TM1. In hASBT, Asn10 in the N-terminal segment is glycosylated and, therefore, the N-terminal segment does not reside within the membrane as it does in AsbtNM. The many differences in topology and sequence identity could be due to many factors, including the evolutionary distance and the differences in membrane composition. Also, as the native substrate for AsbtNM has not been identified, substrate affinity differences could contribute to the diversity of topology and sequence identity. At this point, it remains unclear as to whether the structure of AsbtNM has any correlation with or relevance to that of hASBT but further investigation into the structure-function relationship between residues in AsbtNM may provide more insight.
Overall, data suggests that residues along one face of TM5 do not play a direct role in substrate transport but are critical for ASBT function, most likely through helical stability. The GxxxG motifs and their interacting residues along TM4, TM5 and TM6 potentially allow for a close association of the helices at various positions, by serving as scaffolds for the other portions of the helices to interact with substrate. Additionally, I205 and I208 may constrain the crossing angles of TM5 and the other helices and may direct the change in interaction of TM5 from TM4 to TM6. This further insight into the structure-function relationship of ASBT may aid in future studies involving drug or prodrug design for the treatment of various diseases.
Acknowledgments
This research was supported by a grant from the Nation Institutes of Health, National Institute for Diabetes and Digestive and Kidney Diseases DK061425 to Dr. Peter W. Swaan.
The authors would like to thank Dr. Tatiana Claro da Silva for her technical guidance and support. We would also like to thank Dr. Sairam Mallajosyula for assistance with modeling and the VMD software.
ABBREVIATIONS
- GDCA
glycodeoxycholic acid
- hASBT
human apical sodium-dependent bile acid transporter
- MTSET
[2-(Trimethylammonium) ethyl] methanethiosulfonate
- SCAM
substituted cysteine accessibility method
- TCA
taurocholic acid
- TM
transmembrane domain
References
- 1.Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res. 2009;50:2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinman SA, Carruth MW, Dawson PA. Bile acid uptake via the human apical sodium-bile acid cotransporter is electrogenic. J Biol Chem. 1998;273:34691–34695. doi: 10.1074/jbc.273.52.34691. [DOI] [PubMed] [Google Scholar]
- 3.Izzat NN, Deshazer ME, Loose-Mitchell DS. New molecular targets for cholesterol-lowering therapy. J Pharmacol Exp Ther. 2000;293:315–320. [PubMed] [Google Scholar]
- 4.Kitayama K, Nakai D, Kono K, van der Hoop AG, Kurata H, de Wit EC, Cohen LH, Inaba T, Kohama T. Novel non-systemic inhibitor of ileal apical Na+-dependent bile acid transporter reduces serum cholesterol levels in hamsters and monkeys. Eur J Pharmacol. 2006;539:89–98. doi: 10.1016/j.ejphar.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Kramer W, Glombik H. Bile acid reabsorption inhibitors (BARI): novel hypolipidemic drugs. Curr Med Chem. 2006;13:997–1016. doi: 10.2174/092986706776361003. [DOI] [PubMed] [Google Scholar]
- 6.Balakrishnan A, Polli JE. Apical sodium dependent bile acid transporter (ASBT, SLC10A2): a potential prodrug target. Mol Pharm. 2006;3:223–230. doi: 10.1021/mp060022d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balakrishnan A, Wring SA, Polli JE. Interaction of native bile acids with human apical sodium-dependent bile acid transporter (hASBT): influence of steroidal hydroxylation pattern and C-24 conjugation. Pharm Research. 2006;23:1451–1459. doi: 10.1007/s11095-006-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagedahl M, Swaan PW, Redemann CT, Tang M, Craik CS, Szoka FC, Jr, Oie S. Use of the intestinal bile acid transporter for the uptake of cholic acid conjugates with HIV-1 protease inhibitory activity. Pharm Research. 1997;14:176–180. doi: 10.1023/a:1012044526054. [DOI] [PubMed] [Google Scholar]
- 9.Swaan PW, Hillgren KM, Szoka FC, Jr, Øie S. Enhanced transepithelial transport of peptides by conjugation to cholic acid. Bioconjug Chem. 1997;8:520–525. doi: 10.1021/bc970076t. [DOI] [PubMed] [Google Scholar]
- 10.Tolle-Sander S, Lentz KA, Maeda DY, Coop A, Polli JE. Increased acyclovir oral bioavailability via a bile acid conjugate. Mol Pharmaceutics. 2004;1:40–48. doi: 10.1021/mp034010t. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee A, Swaan PW. Membrane topology of human ASBT (SLC10A2) determined by dual label epitope insertion scanning mutagenesis. New evidence for seven transmembrane domains. Biochemistry. 2006;45:943–953. doi: 10.1021/bi052202j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang EY, Phelps MA, Banerjee A, Khantwal CM, Chang C, Helsper F, Swaan PW. Topology scanning and putative three-dimensional structure of the extracellular binding domains of the apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2004;43:11380–11392. doi: 10.1021/bi049270a. [DOI] [PubMed] [Google Scholar]
- 13.Hussainzada N, Khandewal A, Swaan PW. Conformational flexibility of helix VI is essential for substrate permeation of the human apical sodium-dependent bile acid transporter. Mol Pharmacol. 2008;73:305–313. doi: 10.1124/mol.107.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussainzada N, Banerjee A, Swaan PW. Transmembrane domain VII of the human apical sodium-dependent bile acid transporter ASBT (SLC10A2) lines the substrate translocation pathway. Mol Pharmacol. 2006;70:1565–1574. doi: 10.1124/mol.106.028647. [DOI] [PubMed] [Google Scholar]
- 15.Hussainzada N, Claro Da Silva T, Swaan PW. The cytosolic half of helix III forms the substrate exit route during permeation events of the sodium/bile acid cotransporter ASBT. Biochemistry. 2009;48:8528–8539. doi: 10.1021/bi900616w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khantwal CM, Swaan PW. Cytosolic half of transmembrane domain IV of the human bile acid transporter hASBT (SLC10A2) forms part of the substrate translocation pathway. Biochemistry. 2008;47:3606–3614. doi: 10.1021/bi702498w. [DOI] [PubMed] [Google Scholar]
- 17.Claro da Silva T, Hussainzada N, Khantwal CM, Polli JE, Swaan PW. Transmembrane helix 1 contributes to substrate translocation and protein stability of bile acid transporter SLC10A2. J Biol Chem. 2011;286:27322–27332. doi: 10.1074/jbc.M110.217802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee A, Ray A, Chang C, Swaan PW. Site-directed mutagenesis and use of bile acid-MTS conjugates to probe the role of cysteines in the human apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2005;44:8908–8917. doi: 10.1021/bi050553s. [DOI] [PubMed] [Google Scholar]
- 19.Hussainzada N, Da Silva TC, Zhang EY, Swaan PW. Conserved aspartic acid residues lining the extracellular loop 1 of sodium-coupled bile acid transporter ASBT Interact with Na+ and 7α-OH moieties on the ligand cholestane skeleton. J Biol Chem. 2008;283:20653–20663. doi: 10.1074/jbc.M802885200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Absalom NL, Schofield PR, Lewis TM. Pore structure of the Cys-loop ligand-gated ion channels. Neurochem Res. 2009;34:1805–1815. doi: 10.1007/s11064-009-9971-2. [DOI] [PubMed] [Google Scholar]
- 21.Karlin A, Akabas MH. Substituted-cysteine accessibility method. Meth Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- 22.Ray A, Banerjee A, Chang C, Khantwal CM, Swaan PW. Design of novel synthetic MTS conjugates of bile acids for site-directed sulfhydryl labeling of cysteine residues in bile acid binding and transporting proteins. Bioorg Med Chem Lett. 2006;16:1473–1476. doi: 10.1016/j.bmcl.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee A, Hussainzada N, Khandelwal A, Swaan PW. Electrostatic and potential cation-pi forces may guide the interaction of extracellular loop III with Na+ and bile acids for human apical Na+-dependent bile acid transporter. Biochem J. 2008;410:391–400. doi: 10.1042/BJ20071300. [DOI] [PubMed] [Google Scholar]
- 24.Mallajosyula SS, Sabit H, Swaan PW, MacKerell J, Alexander D. Human Bile Acid Transporter (hASBT) Functions as a Dimer. 2013. Under Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaback HR, Wu J. From membrane to molecule to the third amino acid from the left with a membrane transport protein. Quart Rev Biophys. 1997;30:333–364. doi: 10.1017/s0033583597003387. [DOI] [PubMed] [Google Scholar]
- 27.Kleiger G, Eisenberg D. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through C(alpha)-H.. O hydrogen bonds and van der waals interactions. J Mol Biol. 2002;323:69–76. doi: 10.1016/s0022-2836(02)00885-9. [DOI] [PubMed] [Google Scholar]
- 28.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 29.Schneider D, Engelman DM. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J Mol Biol. 2004;343:799–804. doi: 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- 30.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Eilers M, Patel AB, Liu W, Smith SO. Comparison of helix interactions in membrane and soluble alpha-bundle proteins. Biophys J. 2002;82:2720–2736. doi: 10.1016/S0006-3495(02)75613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 33.Mackenzie KR. Folding and stability of alpha-helical integral membrane proteins. Chem Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 34.Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011;478:408–411. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]