Abstract
The present study characterizes the methanethiosulfonate (MTS) inhibition profiles of 26 consecutive cysteine-substituted mutants comprising transmembrane (TM) helix 6 of the human apical Na+-dependent bile acid transporter (hASBT, SLC10A2). TM6 is linked exofacially to TM7 via extracellular loop (EL) 3. TM7 was previously identified as lining part of the substrate permeation path (Hussainzada et al. Mol Pharmacol 70:1565, 2006). Most TM6 cysteine replacements were well-tolerated, except for five residues with either severely hampered (I229C, G249C) or abolished (P234C, G237C, G241C) activity. Disruption of protein synthesis or folding and stability may account for lack of activity for mutant P234C. Subsequent Pro234 amino acid replacement reveals its participation in both structural and functional aspects of the transport cycle. Application of polar MTS reagents (1 mM) significantly inhibited activity of six mutants (V235C, S239C, F242C, R246C, A248C, Y253C), for which rates of modification were almost fully reversed (except Y253C) upon inclusion of bile acid substrates or removal of Na+ from the MTS pre-incubation medium. Activity assessments at equilibrative [Na+] revealed numerous Na+ sensitive residues, suggesting their proximity in or around Na+ interaction sites. In silico modeling reveals the intimate and potentially cooperative orientation of MTS-accessible TM6 residues toward functionally important TM7 amino acids, substantiating TM6 participation during the transport cycle. We conclude a functional requirement for helical flexibility imparted by Pro234, Gly237 and Gly241, likely forming a “conformational switch” requisite for substrate turnover; meanwhile, MTS-accessible residues, which line a helical face spatially distinct from this switch, may participate during substrate permeation.
By coupling bile acid movement to the passive flow of Na+ ions down their concentration gradient, the human apical Na+-dependent bile acid transporter (ASBT; SLC10A2) concentrates bile acids within the cell interior. Physiologically, ASBT effectively conserves the body’s re-circulating bile acid pool (Trauner and Boyer, 2003) in tandem with numerous active transporters expressed along the enterohepatic pathway. Since cholesterol provides the precursor molecule in FXR- and hepatic CYP7A-mediated bile acid synthesis (Chiang et al., 2001; Pauli-Magnus et al., 2005), ASBT also constitutes a key modulator of cholesterol homeostasis. Numerous studies have recently underscored the exploitive potential of ASBT in cholesterol-lowering therapies (Huff et al., 2002; Izzat et al., 2000; Li et al., 2004; Oelkers et al., 1997), as well as emphasizing the utility of this high capacity, high affinity transporter in prodrug targeting (Balakrishnan and Polli, 2006; Geyer et al., 2006; Swaan et al., 1997). Consequently, ASBT’s unique pharmaceutical relevance coupled to the absence of a crystal structure has provided a strong impetus towards elucidation of its structure/function relationships.
Using cysteine mutagenesis and thiol modification (SCAM), our previous studies identified transmembrane (TM) domain 7 in forming part of the putative substrate permeation pathway (Hussainzada et al., 2006) with extracellular loop (EL) 3 containing Na+ and bile acid interaction sites (manuscript submitted). Presently, we continue SCAM analysis along TM6 based on the following rationale: (i) our previously published topology model predicts that TM6 lies adjacent to and may interact with TM7 in forming a putative translocation pathway (Zhang et al., 2004); (ii) EL3 amino acids link TM6 and TM7 membrane-spanning segments along the exofacial matrix; (iii) the highly conserved nature of TM6 amino acids corroborate a potential role during transport; (iv) presence of the charged, conserved Arg246 which could potentially participate in electrostatic interactions previously implicated during ligand binding (manuscript submitted); and finally, (v) presence of two conserved proline residues (Pro234, Pro251) which have been shown in other membrane-bound carriers to provide cation binding sites as well as enable formation of conformational switches essential for substrate translocation (Deber et al., 1990; Pajor and Randolph, 2005; Sansom and Weinstein, 2000). As in our previous studies, the C270A mutant provides the scaffold for subsequent cysteine introduction due to its insensitivity to methanethiosulfonate (MTS) reagents. Therefore, the present study assesses MTS sensitivity of 26 consecutive cysteine mutants introduced along TM6 of hASBT, thereby providing novel insight into the molecular workings of the ASBT translocation cycle. We demonstrate a functional prerequisite for TM6 helical flexibility in global conformational changes to protein structure leading to substrate turnover and the putative involvement of TM6 amino acids in lining portions of the permeation pathway.
MATERIALS AND METHODS
Materials
[3H]-Taurocholic acid (0.2 Ci/mmol) was purchased from American Radiolabeled Chemicals, Inc, (St. Louis, MO); Taurocholic acid (TCA) and glychodeoxycholic acid (GDCA) from Sigma (St. Louis, MO); (sulfosuccinimidyl-2 (biotinamido) ethyl-1,3-dithiopropionate (sulfo-NHS-SS-biotin) from Pierce Chemical Co. (Rockford, IL); MTS reagents: (2-Aminoethyl)-methanethiosulfonate (MTSEA), [2-(trimethylammonium) ethyl] methanethiosulfonate (MTSET), and methanethiosulfonate ethylsulfonate (MTSES) from Toronto Research Chemicals, Inc, (North York, ON, Canada). Cell culture media and supplies were obtained from Invitrogen (Rockville, MD). All other reagents and chemicals were of highest purity available commercially.
Cell Culture and Transient Transfections
COS-1 cells (ATCC CRL-1650) were maintained in Dulbecco’s modified Eagle’s Medium containing 10% fetal calf serum, 4.5g/L glucose, 100 units/ml penicillin and 100 µg/ ml streptomycin (Life Technologies, Inc, Rockville, MD) at 37°C in a humidified atmosphere with 5% CO2. Transient transfections were performed as previously described (Banerjee et al., 2005)
Site-directed Mutagenesis
Site-directed mutations were incorporated into hASBT cDNA using the QuikChange® Site-Directed mutagenesis kit from Stratagene (La Jolla, CA) and mutagenesis primers custom synthesized and purchased from Sigma Genosys (St. Louis, MO). Plasmid purifications were performed using a kit from Qiagen (Valencia, CA) and amino acid substitutions confirmed via DNA sequencing using an ABI 3700 DNA analyzer (Applied Biosystems, Foster City, CA) at the Plant-Microbe Genomics Facility of the Ohio State University.
Uptake Assay and Protein Membrane Expression
Initial rates of transport for each mutant were determined in transiently transfected COS-1 cells incubated in modified Hanks’ Balanced Salt Solution (MHBSS), pH 7.4 uptake buffer containing 5.0 µM [3H]-TCA at 37 °C for 12 min. We have demonstrated that this uptake period ensures linear steady-state kinetics in conjunction with an optimal signal-to-noise ratio for subsequent [3H]-TCA analysis via liquid scintillation counting (Banerjee et al., 2005; Banerjee and Swaan, 2006; Hussainzada et al., 2006). Uptake was halted by a series of washes with ice-cold Dulbecco’s phosphate buffered saline (DPBS), pH 7.4 containing 0.2% fatty acid free bovine serum albumin (BSA) and 0.5 mM TCA. Cells were lysed in 350µl of 1N NaOH and subjected to liquid scintillation counting using an LS6500 liquid scintillation counter (Beckmann Coulter, Inc., Fullerton, CA) and total protein quantification using the Bradford protein assay (Bio-Rad, Hercules, CA). Uptake activity was calculated as pmols of [3H]-TCA internalized per min per mg of protein.
Protein expression was determined by washing transiently transfected COS-1 cells in PBS followed by lysis in 0.2 ml lysis buffer B (25 mM Tris (pH 7.4), 300 mM NaCl, 1 mM CaCl2, 1% Triton X-100 and 0.5% Sigma Protease Inhibitor Cocktail). Cell lysates were separated on a 12.5% SDS-polyacrylamide gel and transferred onto an Immuno-Blot PVDF membrane (Bio-Rad, Hercules, CA). Blots were probed with rabbit anti-ASBT primary antibody (1:1000) and visualized using goat anti-rabbit IgG/HRP conjugated secondary antibody with chemiluminescent detection (ECL Plus Western Blot kit, Amersham Biosciences, Piscataway, NJ). Levels of cell surface protein expression were measured via biotin labeling, wherein transiently transfected COS-1 cells were incubated with sulfo-NHS-SS-biotin for 30 min at RT (Mitchell et al., 2004; Wong et al., 1995). After several washes with PBSCM (PBS containing 0.1mM CaCl2 and 1.0mM MgCl2), cells were disrupted with lysis buffer B at 4 °C for 20 minutes (Zhang et al., 2004) and biotinylated proteins recovered overnight at 4 °C using 100 µl of streptavidin agarose beads. Samples were eluted with SDS-PAGE buffer and immunoblotting performed as described above. Blots were probed for positive and negative controls, the plasma membrane marker α-integrin (150 kDa) and the endoplasmic reticulum membrane protein calnexin (90 kDa), respectively, to assess the integrity of the biotinylation procedure (calnexin data not shown). Relative hASBT membrane expression was standardized to integrin expression and quantified via densitometry as previously described (Hussainzada et al., 2006).
MTS Inhibition Studies
Sensitivity of mutants to charged, membrane-impermeant MTS reagents was determined by preincubation of transiently transfected COS-1 cells with either 1 mM MTSES, MTSET, or MTSEA for 10 min at room temperature. After MTS treatment, cells were washed twice in modified Hank’s Balanced Salt Solution (Sigma, St. Louis, MO) followed by [3H]-TCA uptake as described above. All MTS solutions were freshly prepared prior to each study due to the short aqueous half-life of these MTS reagents.
Cation and Substrate Protection Assays
To determine whether the presence or absence of Na+ and/or bile acid substrates alters MTSEA labeling, transiently transfected COS-1 cells were washed twice in 1X PBS, pH 7.4 followed by co-incubation with equal concentrations of MTSEA and GDCA (1 mM) prepared either in MHBSS or Na+-free buffer (MHBSS except choline chloride entirely substitutes NaCl) for 10 minutes at room temperature. After pre-incubation treatments, cells were washed twice in either MHBSS, pH 7.4 or Na+-free buffer and additionally equilibrated for 15 minutes at 37 °C in these buffers followed by determination of [3H]-TCA uptake as described above. All control wells were treated identically. For each mutant transporter, uptake values were determined by taking a ratio of mutant uptake at each experimental condition versus mutant uptake for its respective unmodified control. We normalize mutant ratios to C270A by expressing mutant ratios for each condition as a percentage of C270A ratios calculated in the same manner.
Sodium Activation
Measurement of [3H]-TCA uptake at equilibrative extracellular Na + concentrations (12 mM, i.e. at equilibrium with cytosolic [Na+]) was performed (uptake conducted as described above; choline chloride used as equimolar NaCl replacement) and expressed as a ratio of uptake at physiological (137 mM) Na+ concentrations to determine overall sensitivity of each mutant to the presence/absence of Na+. Theoretically, Na+ ratios equal to one imply little measurable difference in transporter activity despite the scarcity of Na+ ions, while fractions less than one indicate a greater necessity for physiological Na+ concentrations for proper transport function of a mutant transporter.
Data Analysis
For each mutant, data are represented as mean ± S.D. of at least three different experiments with triplicate measurements. Data analysis was performed with GraphPad Prism 4.0 (GraphPad Software, San Diego, CA) using analysis of variance (ANOVA) with Dunnett’s post-hoc test. Data were considered statistically significant at p ≤ 0.05.
RESULTS
Cysteine Scan of TM6
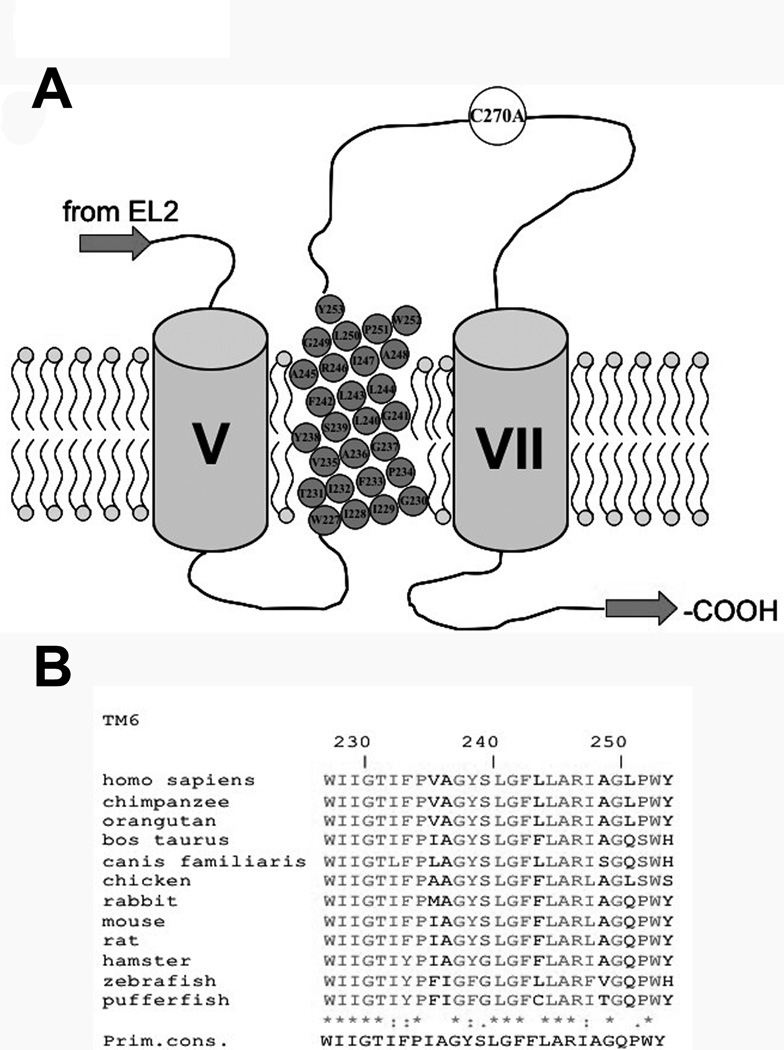
Based on our previously published topology model (Banerjee and Swaan, 2006; Zhang et al., 2004), residues spanning W227-Y253 are predicted to constitute TM6 of hASBT (Fig. 1A). High sequence homology is observed among various evolutionarily diverse species for this protein region (Fig. 1B), which lies in intimate proximity to critical protein regions previously described (Hallén et al., 2002; Hallén et al., 2000; Hussainzada et al., 2006; Kramer et al., 2001). Presently, systematic cysteine substitutions were incorporated along TM6 followed by structural and functional analysis of mutant transporters.
Figure 1. Multiple sequence alignment of TM6 amino acids.
[A] Secondary structure model of the last three transmembrane domains (TMD) of hASBT according to the 7TM model. Roman numerals indicate flanking TMD while TMD 6 amino acids are represented by gray circles inscribed with amino acid identity and position. Phospholipids of plasma membrane represented by circle (polar phosphate head group) with two tails (hydrophobic lipids). Top is exofacial, bottom cytosolic. [B] Sequence alignment of amino acids 227–253, putatively forming TM6, for all known ASBT paralogs. Sequences were retrieved from GeneBank and aligned via the MULTALIN routine with annotation performed via MPSA. Shaded regions denote complete amino acid conservation among all species. Amino acid positioning relative to human ASBT is indicated by numbering on top. Bottom line indicates primary consensus for the TM6 region.
Transport Activity and Membrane Expression of Cysteine Mutants
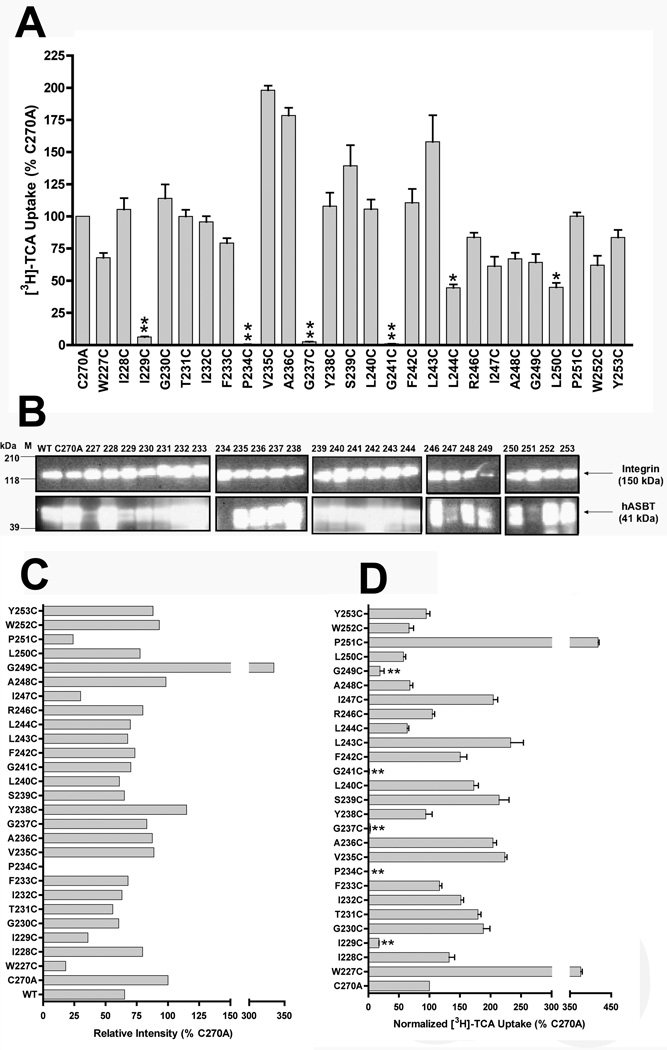
Due to its low background levels of bile acid transport (Hussainzada et al., 2006), the COS-1 cell line was used to transiently express all TM6 mutant transporters. Surface biotin labeling of membrane-expressed proteins was accomplished using the membrane-impermeant sulfo-NHS-SS-biotin and quantified via densitometry of protein bands (Fig. 2B). ASBT bands for each sample were standardized to an internal control (α-integrin) and expressed as a percentage of C270A intensity (Fig. 2C). Initial transport activities (Fig. 2A) were then normalized to relative membrane expression for each mutant transporter.
Figure 2. [3H]-TCA uptake activity and membrane expression of TM6 cysteine mutants.
[A] Uptake of [3H]-TCA was measured in COS-1 cells as described in “Experimental Procedures” and expressed as a percentage of the parental transporter, C270A. [B] Intact transfected COS-1 cells were treated with sulfo-NHS-SS-biotin as described in “Experimental Procedures” followed by Western Blot processing. Blots were probed with the anti-hASBT antibody (1:30,000 dilution) followed by horseradish peroxide-linked anti-rabbit immonuglobin (1:2000 dilution). Each blot was probed for the internal plasma membrane marker, α-integrin (150 kDa) and the absence of calnexin (90 kDa) (data not shown), an endoplasmic reticulum membrane protein representing the negative control in the biotinylated fractions. Marker lanes are shown on the left side of the individual blots. Mature glycosylated hASBT visualizes as the 41kDa band while the lower, 38 kDa band (not indicated) represents the unglycosylated species. [C] Densitometric analysis for cysteine mutants normalized to internal marker (α-integrin) and represented as a percentage of C270A parent. [D] [3H]-TCA uptake activity normalized to relative cell surface expression. Bars represent mean ± S.D. of three separate experiments with p ≤ *** 0.001, ** 0.01 and *0.05 respectively, using ANOVA with Dunnett’s post-hoc analysis.
After data normalization (Fig. 2D), most TM6 mutants retained appreciable levels of activity, except for five mutants either severely hampered (I229C, G249C) or inactivated (P234C, G237C, G241C) upon cysteine substitution. Only P234C lacked expression in biotinylated (Fig. 2B) and whole cell (data not shown) extracts. This may be due to disruptions in protein synthesis, but more likely, alterations in protein folding and stability occur that induce rapid protein degradation via ER-associated machinery. Interestingly, all five residues are conserved amongst known species of ASBT (Fig. 1B), suggesting primary roles in transport function that necessitate preservation of these amino acids. Due to their low activity levels, these mutants were excluded from further studies.
TM6 Mutants Demonstrate Substantial Na+ Sensitivity
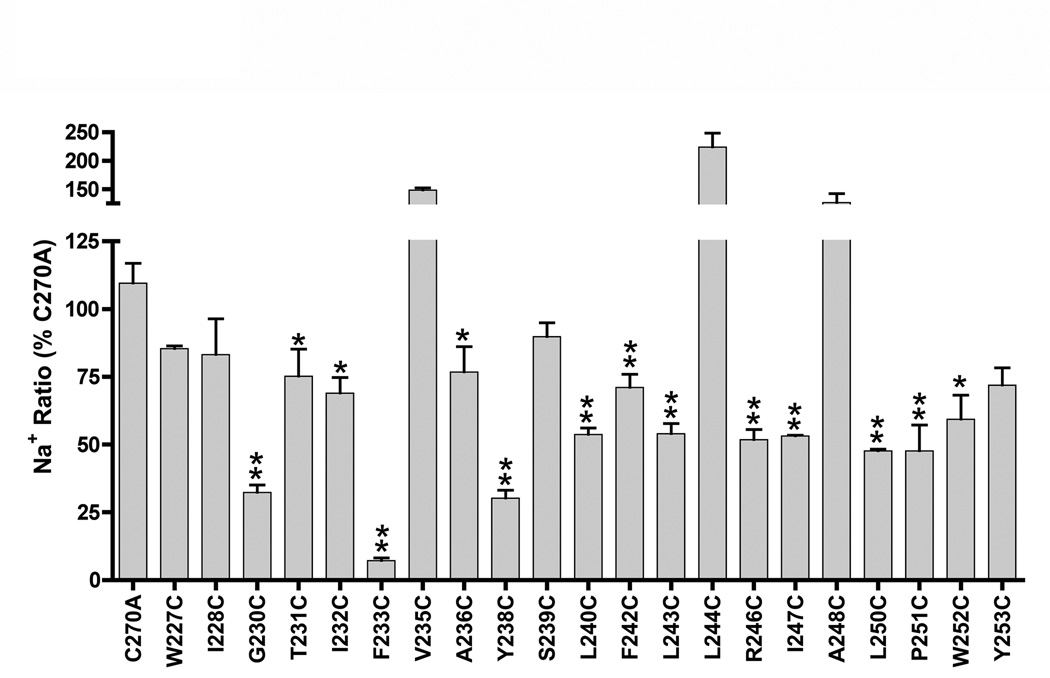
As a Na+ co-transporter, ASBT activity relies upon proper recognition, binding and translocation of two Na+ ions per one bile acid molecule (Weinman et al., 1998). Thus, we examined the consequences of equilibrative extracellular Na+ concentrations upon mutant activity. For each mutant, the ratio of transport rates at equilibrative (12 mM) versus physiological (137mM) Na+ concentrations was calculated and expressed as a percentage of the C270A Na+ ratio. This experimental scheme may uncover hidden functional defects in mutants otherwise unaffected by cysteine mutation. Accordingly, the C270A parental construct displays a Na+ ratio of 0.70 ± 0.04 (data not shown), indicating minimal consequences to transporter function upon alanine substitution at the native cysteine residue. In contrast, significant decreases in activity were observed for the majority (~ 64%) of TM6 cysteine mutants. Of 21 mutants assayed, 14 demonstrated hampered uptake rates at equilibrative [Na+] (Fig. 3). EL3 cysteine mutants from our earlier study also exhibited similarly extensive Na+ sensitivity (manuscript submitted), wherein uptake activities of 90% of assayed mutants were susceptible to equilibrative [Na+]. Since EL3 residues putatively form Na+ interaction sites (manuscript submitted), the widespread Na+ dependency observed for TM6 mutants implies their close proximity to such Na+ interaction sites and lends credence to TM6 participation during Na+ permeation.
Figure 3. Sodium sensitivity of cysteine mutants.
COS-1 cells expressing mutant transporters were incubated in uptake medium (5 µM [3H]-TCA) containing low (12 mM) or physiological (137 mM) Na+ concentrations as described under “Experimental Procedures”. Sodium ratios were calculated for each mutant as the quotient of activity at 12 mM versus 137 mM [Na+] and expressed as a percentage of C270A. Bars represent mean activity ± S.D. (n=3). Asterisk (*) denotes p ≤ 0.05.
Substrate and Cation Binding Modulate Accessibility of Cysteine Mutants to MTS Modification
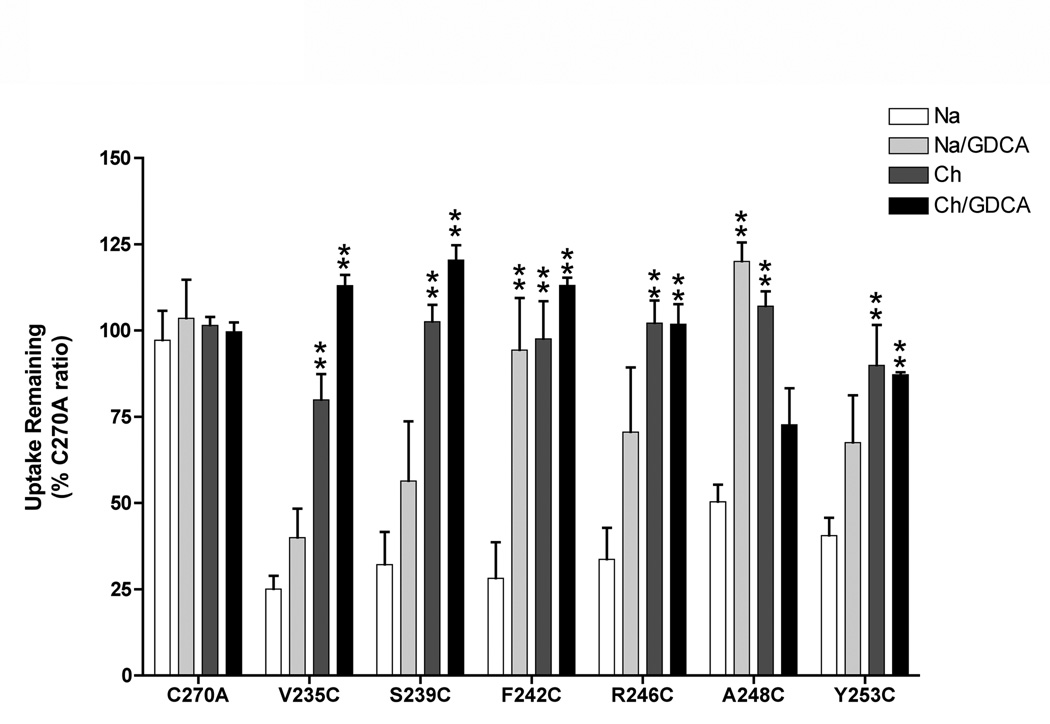
Both positively and negatively charged MTS reagents were used to probe the solvent accessibility of 21 cysteine mutants demonstrating measurable uptake activity. Intact monolayers of COS-1 cells expressing mutant transporters were pre-incubated with either 1.0 mM MTSES, MTSET or MTSEA followed by uptake assessments. MTSET (109 Å3) and MTSES (90 Å3) exhibited similar inhibition profiles, in which activities of only mutants V235C, S239C, F242C and R246C were significantly reduced (data not shown), suggesting minimal electrostatic effects in accessibility at those sites. The remainder of TM6 mutants assayed were either inaccessible to these MTS reagents or modification was functionally silent. Incubation with the relatively smaller MTSEA (69 Å3) inhibited uptake at sites accessed by the larger MTS reagents (i.e. V235C, S239C, F242C, R246C; Fig. 4), as well as two additional sites (A248C, Y253C; Fig. 4).
Figure 4. Cation and substrate protection of TM6 cysteine mutants.
Transiently transfected COS-1 cells expressing TM6 cysteine mutants were pre-incubated in buffer, pH 7.4 containing 1 mM MTSEA and either 137 mM NaCl (white bar); 137 mM NaCl and 1 mM GDCA (grey bar); 137 mM choline chloride (dark grey bar); or 137 mM choline chloride and 1 mM GDCA (black bar) and followed by [3H]-TCA uptake as described in “Experimental Procedures”. Choline chloride does not activate the transporter and provides equimolar replacement for NaCl. All control wells were treated identically. Bars represent mean ± S.D. of at least three separate measurements. Data expressed as percentage of C270A values for each condition as described under “Materials and Methods”. Student’s t-test analysis performed with *p < 0.05, **p < 0.01.
Since MTSEA (1 mM) application produced the most pronounced inhibition of the three MTS reagents used (similar to our previous study with TM7 (Hussainzada et al., 2006)), the effects of Na+ and bile acid substrate on MTSEA accessibility of mutants V235C, S239C, F242C, R246C, A248C and Y253C was evaluated. Our previous studies with EL3 and TM7 have shown that co-incubation of MTS reagents with bile acids and/or removal of Na+ from the pre-incubation buffer caused a reversal of the inhibitory effect observed with MTS incubation alone (manuscript submitted; Hussainzada et al., 2006). Similarly in the present study, all TM6 mutants inhibited by MTSEA (1 mM) demonstrated significant uptake recovery when MTSEA incubation was performed in the absence of Na+ and/or presence of 1.0 mM glycodeoxycholic acid (GDCA) (Fig. 4). Specifically, removal of Na+ from the MTSEA pre-incubation medium significantly restored transport activity for mutants V235C, S239C, F242C, R246C and A248C, while co-incubation with GDCA (Km = 2.0 ± 0.4 µM) significantly protected mutants F242C and A248C. Concurrent removal of Na+ and addition of the high affinity substrate GDCA (1 mM) resulted in significant MTSEA protection for mutants V235C, S239C, F242C and R246C. In all cases, mutant activities were restored to control (C270A) levels (within standard deviation; Fig. 4).
Although mechanistic details of the ASBT translocation cycle are as yet unresolved, ordered binding of ligands followed by translocation likely occurs, similar to many other Na+-coupled transporters (Jung, 2001; Pajor and Randolph, 2005; Quick and Jung, 1997; Zhang and Rudnick, 2005). In this scenario, the ordered binding of Na+ and bile acids would trigger the protein to assume various discrete structural conformations eventually leading to carrier reorientation within membrane leaflets and substrate turnover. Within TM6, the lack of Na+ binding events (simulated by substitution of Na+ with choline+ in pre-incubation buffers) significantly decreased MTSEA modification rates for all sites (Fig. 4), suggesting that protein conformational states assumed before the binding of Na+ occlude these thiol groups from subsequent modification. Furthermore, the binding of bile acid substrate (GDCA), either in the presence or absence of Na+, also triggers protein conformations that significantly decrease MTSEA access to all sites (Fig. 4). Due to the close association between TM6 and protein regions previously implicated during ligand binding and translocation (i.e. TM7 and EL3), the observed substrate protection may result from; (i) occlusion of these sites via conformational changes; (ii) the physical presence of substrates preventing access; or (iii) a combination of both scenarios. The alternating accessibility of TM6 sites to thiol modification suggests the first scenario, while the restoration of mutant activity to control levels via substrate protection infers the second scenario. Likely, a union of both situations prevails, in which TM6 residues may line portions of the permeation pore and also transduce conformational changes resulting from ligand interactions at adjacent protein regions (TM7, EL3), although further studies are needed to unequivocally conclude origin of substrate protection. However, it is noteworthy that our data highlight a trend toward protection from MTS modification in the absence of Na+, which is entirely plausible given that EL3 residues contain putative Na+ interaction sites (manuscript submitted). It may be that lack of Na+ binding at EL3 regions prevents downstream conformational changes that “open” TM6 residues to MTS modification. Overall, we conclude that TM6 amino acids likely line portions of the substrate permeation route due to their spatial proximity with EL3 and TM7 residues and their solvent-accessibility profile.
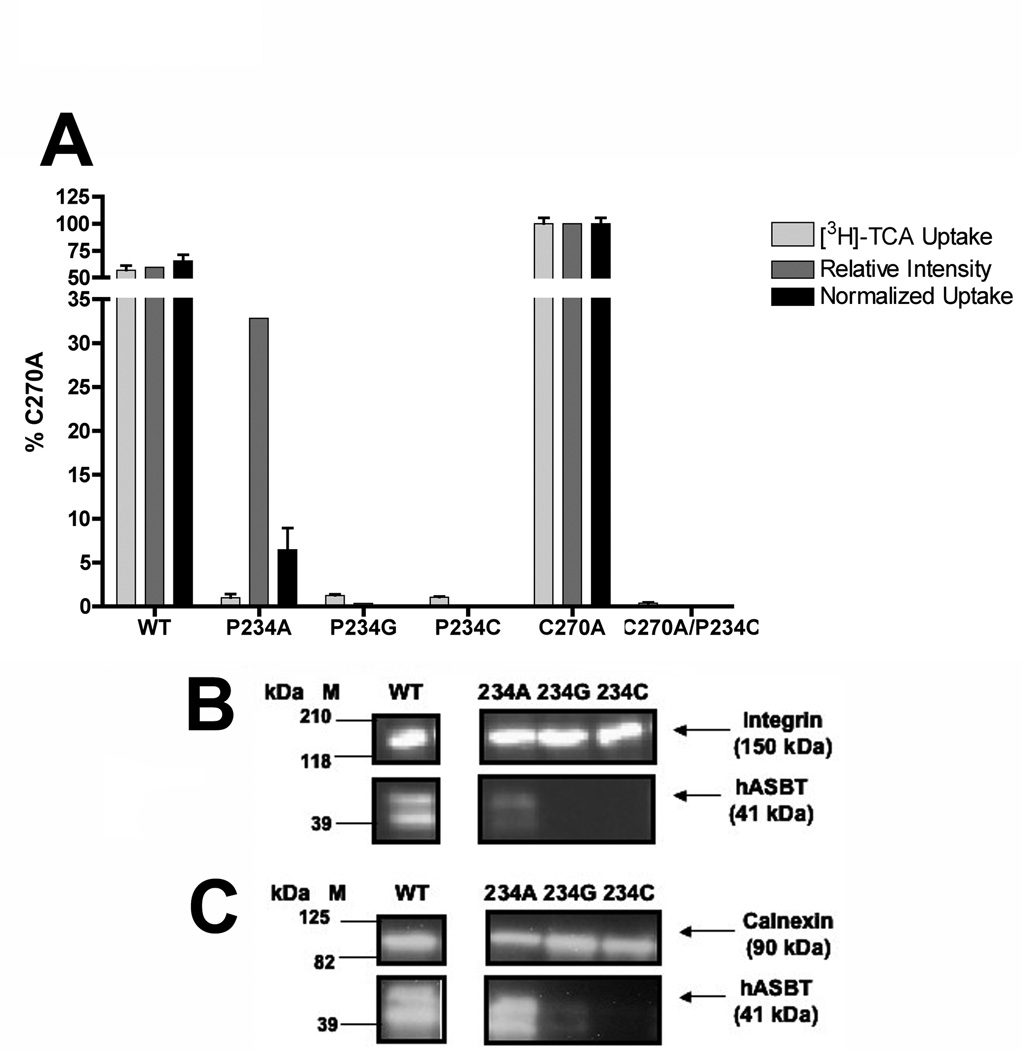
Mutation of Pro234 Affects Both Transporter Expression and Function
Since the P234C double mutant (C270A/P234C) lacked expression both at the plasma membrane (Fig. 2B) and in whole cell extracts (data not shown), additional replacements were made at this site to determine if Pro234 makes functional contributions during the hASBT transport cycle. Using the wild-type (WT) species as the scaffold, alanine, glycine and cysteine replacements were incorporated and analyzed with respect to uptake activity and membrane expression. As expected from our results using the kinetically similar C270A template (Banerjee et al., 2005), the P234C mutant constructed against the WT background lacked expression both in membrane (Fig. 5B) and whole cell (Fig. 5C) extracts, confirming that cysteine replacement at this position affects transporter expression levels irrespective of the mutational template used. Glycine replacement (P234G) also appears to disrupt protein expression, resulting in minimal transporter expression in membrane (Fig. 5B) and whole cell (Fig. 5C) extracts. In contrast, the alanine mutant (P234A) displayed plasma membrane expression, albeit at reduced levels (30 % of C270A levels; Fig. 5A). After normalization to cell surface expression levels, uptake function of the P234A mutant remained severely inhibited (Fig. 5A), suggesting functional as well as structural impairments to transporter function. Defective trafficking to the plasma membrane may account for the lowered membrane expression of the P234A mutant, since the ratio of its plasma membrane expression to whole cell expression is approximately half of the similar ratio for the WT species (i.e. 0.452 vs. 0.814), however further studies are needed for unequivocal evidence. We conclude that Pro234 participates in both protein expression and transporter function, confirming the overall importance of this atypical imino acid.
Figure 5. [3H]-TCA uptake and membrane expression of Pro234 mutants.
[A] The initial [3H]-TCA uptake (grey bars), relative intensity of immunoblotting (dark grey bars), and normalized uptake activities (black bars) for Pro234 single and double mutants and their parental templates are depicted. Bars represent mean ± S.D. of at least three separate measurements. Data expressed as percentage of C270A values for each condition. Student’s t-test analysis performed with *p < 0.05, **p < 0.01. Activities of the Pro234 constructs are statistically different that both WT and C270A (p < 0.01), however asterisks (**) have been omitted for visual clarity. [B] Intact transfected COS-1 cells were treated with sulfo-NHS-SS-biotin as described in “Materials and Methods” followed by Western Blot processing. Marker lanes are shown on the left side of the individual blots. Mature glycosylated hASBT visualizes as the 41kDa band while the lower, 38 kDa band (not indicated) represents the unglycosylated species; [C] Immunoblotting of whole cell extracts from transfected COS-1 cells as described in “Materials and Methods” showing glycosylated (41 kDa) and unglycosylated (38 kDa) hASBT species.
DISCUSSION
The present manuscript details the solvent accessible sites along TM6 of hASBT as revealed by cysteine-scanning mutagenesis and thiol modification (SCAM). This follows as a logical extension of our previous studies using this methodology that revealed putative Na+ and substrate interaction sites along EL3 (manuscript submitted) and demonstrated TM7 participation during substrate permeation (Hussainzada et al., 2006). Here, the key findings include: (i) identification of a TM6 helical face demonstrating significant MTS accessibility; (ii) modulation of thiol modification rates along this helical face via Na+ and bile acid binding events; (iii) the spatially intimate association of MTS-accessible TM6 amino acids with functionally important TM7 residues; and finally, (iv) identification of a potential “conformational switch” necessary for substrate permeation that lines a helical face spatially distinct from the MTS-accessible face of TM6.
Of the 26 amino acids constituting TM6, the majority proved tolerant to cysteine substitution, whereas five were either functionally inactive (P234C, G237C, G241C) or significantly hampered (I229C, G249C) upon mutation (Fig. 2D). With the exception of I229, these residues en bloc display an approximate α-helical periodicity (Fig. 6B). Interestingly, cysteine substitution at Pro234 decreased protein expression levels, as evidenced by lack of expression during immunoblotting of membrane (Fig. 2B) and whole cell (data not shown) protein fractions. To determine if Pro234 contributes in a functional capacity during ASBT transport, additional amino acid substitutions were incorporated using the native (WT) protein template (which kinetically behaves very similarly to the C270A construct required for thiol modification studies). Again cysteine, as well as glycine, replacement severely decreased whole cell protein expression levels (Fig. 5C), suggesting disruptions during either protein synthesis, or more likely, protein folding and stability that lead to rapid degradation. Only alanine substitution (P234A) at this site resulted in significant membrane expression, however transport function remained abrogated (Fig. 5). Thus, the highly conserved Pro234 functions in a dual capacity, likely essential for both proper protein expression and function. This two-pronged contribution of prolines, especially those situated in membrane segments, has been previously documented for many other important solute carriers and exchangers as well (Joshi and Pajor, 2006; Labro et al., 2003; Lin et al., 2000; Lu et al., 2001; Shelden et al., 2001; Slepkov et al., 2004). As observed by Kaback and colleagues for Lac permease, a paradigm of secondary active transporters, relatively few amino acids are essential for transport, unless directly involved in forming linkages with substrate; instead, such carriers must be “highly flexible proteins capable of widespread conformational changes during turnover” (Kaback and Wu, 1997). Since membrane-bound carriers are tightly woven through the lipid bilayer, usually via multiple transmembrane segments, such flexibility is not inherent, and must instead be imparted by specific residues situated along the protein. Proline residues, specifically, can impart this flexibility, since they cyclize back onto their backbone nitrogen atom and prevent hydrogen bonding between protons of the ith residue to (i – 4)th carbonyl oxygens, a requirement for maintaining classical a-helical structure (Woolfson and Williams, 1990), and ultimately destabilize the helix and increase overall flexibility (Bright et al., 2002; Cordes et al., 2002). Such perturbations of backbone dihedral angles of α-helices that lead to increased flexibility are also observed with glycine residues, albeit to a lesser extent (Cordes et al., 2002; Javadpour et al., 1999). Compellingly, a total of four glycine (G230, G237, G241, G249) and two proline (P234, P251) residues line the TM6 helix (Fig. 1B), of which all, except Pro251, are conserved among all known species of ASBT (Fig. 1B). Accordingly, with the exception of Gly230 and Pro251, these residues were found to be essential for transport function of ASBT (Fig. 2). Thus, glycines and particularly prolines, may impart the necessary flexibility required to assume discrete protein conformational states during substrate turnover. Interestingly, numerous studies (Gibbs et al., 1997; Javadpour et al., 1999; Tieleman et al., 1999) have noted the preferred pairing of proline and glycine residues, particularly at positions lining a shared helical face, which may reflect possible modulation of proline-induced distortions by the presence of similarly “flexible” glycine residues. We observe this trend for inactive mutants P234A, G237C, G241C, and G249C, which approximately line a single helical face (Fig. 6B) that could facilitate potential glycine modulation of P234-induced helical distortions. Finally, in the case of G230C and P251C, substantial loss of activity occurs in equilibrative Na+ concentrations, suggesting auxiliary roles for these residues during the translocation cycle, possibly in transduction of conformational signals.
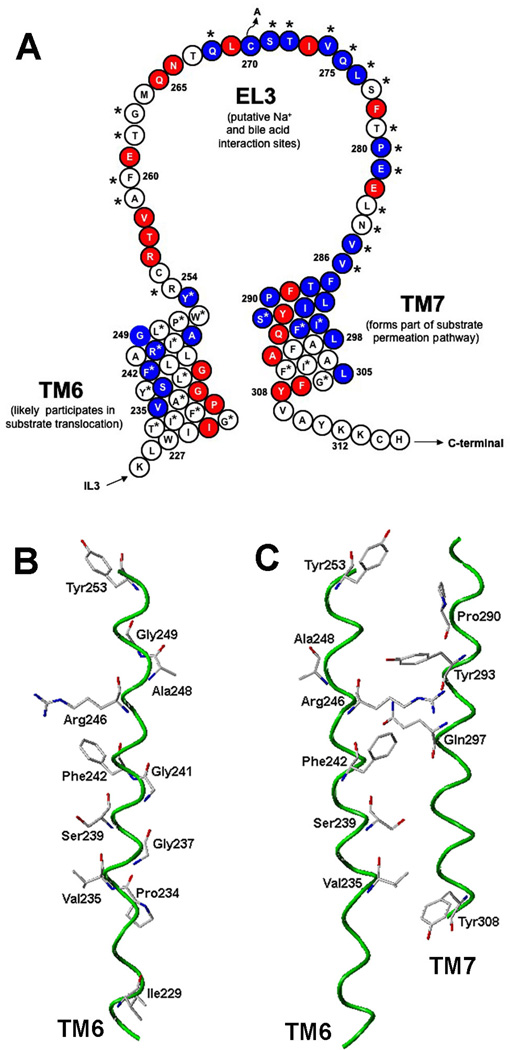
Figure 6. ASBT MTS inhibition profile and TM6 in silico model.
[A] Secondary structural model of hASBT regions submitted to SCAM analysis thus far. Red shading indicates residues inactivated by cysteine substitution while blue shading indicates significant (p < 0.05) MTS inhibition. Asterisks (*) indicate significant (p < 0.05) Na+ sensitivity. Taken en masse, our MTS inhibition data reflect the widespread solvent accessibility of these protein regions, corroborating their roles in ligand interaction and translocation. The prevalence of mutation-inactivated sites also confirms the essential nature of amino acids lining these protein segments. As noted, our earlier studies putatively assign ligand interaction sites along EL3 and TM7 with the substrate permeation path comprising TM7 (Hussainzada et al., 2006) and likely TM6 amino acids. [B] In silico prediction of functional group orientation of TM6 sites inhibited by 1 mM MTSEA and protected by GDCA addition (1 mM) or Na+ removal from MTS preincubation medium (i.e. V235, S239, F242, R246, A248, Y253). Also shown are the spatially distinct mutation-inactivated sites (I229, P234, G237, G241, G249). Occurrence of proline and mutiple glycine residues along inactivated helical face indicates functional requirement for helical flexibility crucial for transporter activity; [C] In silico prediction of spatially intimate association of TM6 amino acids demonstrating MTSEA inhibition and substrate protection (i.e. V235, S239, F242, R246, Y253) and TM7 residues previously shown as either highly MTS accessible (P290, S294) or inactivated by cysteine substitution (Y293, Q297, Y308) (Hussainzada et al., 2006). In silico model spans amino acids W227-Y253 (TM6) and F286-Y308 (TM7) and was generated using THREADER with visualization by SYBYL 6.9 (3). Top exofacial; bottom cytosolic.
Superimposed upon a secondary structural model of ASBT, our en masse MTS inhibition profiles generated thus far (Fig. 6A) strongly implicate the participation of TM6 residues during substrate translocation as well. Using physicochemically diverse thiol modifiers, four sites were significantly inhibited by pre-incubation with either 1.0 mM MTSES or MTSET (V235, S239, F242, R246; data not shown); while MTSEA application (1.0 mM) inhibited six sites total (V235, S239, F242, R246, A248, Y253; Fig. 4). Based on its similar MTS inhibition profiles, electrostatic and steric effects minimally influence TM6 modification rates. Using our in silico three-dimensional model (Banerjee and Swaan, 2006; Zhang et al., 2004) four MTS-modified sites (V235, S239, F242, R246) exhibiting an α-helical periodicity were found to line a TM6 face spatially distinct from the proline and glycine “conformational switch” discussed above (Fig. 6B). Next, we modeled the functional group orientation of these four TM6 residues with respect to TM7 residues previously implicated in lining the substrate permeation path (Hussainzada et al., 2006) to determine the spatial feasibility of TM6 participation during translocation (Fig. 6C). Strikingly, the functional groups of the four MTS-accessible TM6 sites (V235, S239, F242, R246) are situated in line with TM7 sites previously shown to be either highly MTS accessible (P290, S294) or inactivated by mutation (Y293, Q297, Y308). Thus, our in silico model may delineate the shared solvent-accessible molecular spaces between TM7 and TM6 residues. It is tempting to speculate that such communal volumes of molecular space could form a translocation route to enable transfer of charged substrates across the forbiddingly hydrophobic environment of the lipid bilayer. The presence of polar (S239, S294, Q297) and charged (R246) residues along the collective TM7-TM6 “pathway” strengthens this hypothesis, as they may form hydrophilic surfaces promoting Na+ and bile acid movement into the cell interior (Fig. 6C). Further validation of the in silico hypotheses can be inferred from our experimental data, in particular substrate protection studies of the six MTSEA-accessible TM6 mutants. Removal of Na+ or addition of the high affinity substrate, GDCA (1.0 mM), to the MTSEA (1.0 mM) pre-incubation medium significantly decreased MTSEA modification rates for all inhibited sites, such that mutant activities were restored to C270A control levels (Fig. 4). Thus, our experimental results suggest the physical presence of substrates in or around these sites previously predicted in silico as forming translocation paths with TM7. Integrating our in silico model, MTS inhibition profiles and mutagenesis results, we can speculate that the TM6 “conformational switch” (i.e. P234, G237, G241, G249C) functions to promote helical flexibility necessary for widespread protein conformational changes triggered after ligand binding, while the spatially distinct solvent-accessible face (i.e. V235, S239, P242, R246) forms putative permeation routes with TM7 (and likely other protein regions) during substrate turnover. Loss of TM6 helical flexibility, as occurs upon replacement of the highly helix-distorting Pro234 with the more rigid alanine (P234A), renders the transporter incapable of the changes in protein structure that are a necessary prerequisite for function.
Finally, the ubiquitous Na+ sensitivity of TM6 residues is unsurprising, since it suggests proximity of this protein region to putative Na+ interaction sites. Since EL3 residues link TM6 and TM7 helical segments and our earlier study demonstrates Na+ interaction sites along EL3 (manuscript submitted), amino acid mutation along TM6 likely perturbs efficient transduction of conformational changes triggered after Na+ binding along EL3 contact points. Although these consequences are masked at physiological [Na+], equilibrative Na+ concentrations expose the functional defects resulting from mutagenesis of this protein region. Of 22 mutants assayed, ~64% exhibited significant decreases in activity at equilibrative Na+ concentrations; furthermore, of those mutants affected, approximately 71% demonstrated less than half of their original activity (Fig. 3). As mentioned earlier, G230C and P251C are included in this category, reinforcing the contribution of helical flexibility to ASBT function. Overall, the widespread and severe response of TM6 cysteine mutants to reduced levels of extracellular Na+ infers the proximity of this region to Na+ binding sites while supporting a functional role for TM6 amino acids during ASBT transport.
To conclude, analysis of data generated via cysteine-scanning mutagenesis of TM6 in the context of our previous SCAM studies and homology model has provided novel insight into the molecular workings of the ASBT translocation cycle. Per our earlier work, putative ligand binding domains and portions of the substrate permeation pathway were identified (manuscript submitted; Hussainzada et al., 2006); in the present manuscript, we demonstrate a functional prerequisite for TM6 helical flexibility in global conformational changes to protein structure leading to substrate turnover and the putative involvement of TM6 amino acids in lining portions of the permeation pathway.
Acknowledgments
This work was supported, in part, by the National Institutes of Health under grant DK61425 (to P. W. S.).
Abbreviations
- EL
Extracellular loop
- GDCA
glychodeoxycholic acid
- hASBT
human apical sodium-dependent bile acid transporter
- IL
intracellular loop
- MTS
methanethiosulfonate
- MTSES
methanethiosulfonate ethylsulfonate
- MTSET
[2-(trimethylammonium)ethyl]-methane-thiosulfonate bromide
- MTSEA
[(2-aminoethyl)-methanethiosulfonate
- SCAM
substituted cysteine accessibility method
- TM
transmembrane
REFERENCES
- Balakrishnan A, Polli JE. Apical sodium dependent bile acid transporter (ASBT, SLC10A2): a potential prodrug target. Mol Pharm. 2006;3(3):223–230. doi: 10.1021/mp060022d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Ray A, Chang C, Swaan PW. Site-directed mutagenesis and use of bile acid-MTS conjugates to probe the role of cysteines in the human apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2005;44(24):8908–8917. doi: 10.1021/bi050553s. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Swaan PW. Membrane topology of human ASBT (SLC10A2) determined by dual label epitope insertion scanning mutagenesis. New evidence for seven transmembrane domains. Biochemistry. 2006;45(3):943–953. doi: 10.1021/bi052202j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright JN, Shrivastava IH, Cordes FS, Sansom MS. Conformational dynamics of helix S6 from Shaker potassium channel: simulation studies. Biopolymers. 2002;64(6):303–313. doi: 10.1002/bip.10197. [DOI] [PubMed] [Google Scholar]
- Chiang JY, Kimmel R, Stroup D. Regulation of cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRalpha) Gene. 2001;262(1–2):257–265. doi: 10.1016/s0378-1119(00)00518-7. [DOI] [PubMed] [Google Scholar]
- Cordes FS, Bright JN, Sansom MS. Proline-induced distortions of transmembrane helices. J Mol Biol. 2002;323(5):951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- Deber CM, Glibowicka M, Woolley GA. Conformations of proline residues in membrane environments. Biopolymers. 1990;29(1):149–157. doi: 10.1002/bip.360290120. [DOI] [PubMed] [Google Scholar]
- Geyer J, Wilke T, Petzinger E. The solute carrier family SLC10: more than a family of bile acid transporters regarding function and phylogenetic relationships. Naunyn Schmiedebergs Arch Pharmacol. 2006;372(6):413–431. doi: 10.1007/s00210-006-0043-8. [DOI] [PubMed] [Google Scholar]
- Gibbs N, Sessions RB, Williams PB, Dempsey CE. Helix bending in alamethicin: molecular dynamics simulations and amide hydrogen exchange in methanol. Biophys J. 1997;72(6):2490–2495. doi: 10.1016/S0006-3495(97)78893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallén S, Bjorquist A, Ostlund-Lindqvist AM, Sachs G. Identification of a region of the ileal-type sodium/bile acid cotransporter interacting with a competitive bile acid transport inhibitor. Biochemistry. 2002;41(50):14916–14924. doi: 10.1021/bi0205404. [DOI] [PubMed] [Google Scholar]
- Hallén S, Fryklund J, Sachs G. Inhibition of the Human Sodium/ Bile Acid Cotransporters by Side Specific Methanethiosulfonate Sulphydryl Reagents: Substrate Controlled Accessibility of Se of Activation. Biochemistry. 2000;39:6743–6570. doi: 10.1021/bi000577t. [DOI] [PubMed] [Google Scholar]
- Huff MW, Telford DE, Edwards JY, Burnett JR, Barrett PH, Rapp SR, Napawan N, Keller BT. Inhibition of the apical sodium-dependent bile acid transporter reduces LDL cholesterol and apoB by enhanced plasma clearance of LDL apoB. Arterioscler Thromb Vasc Biol. 2002;22(11):1884–1891. doi: 10.1161/01.atv.0000035390.87288.26. [DOI] [PubMed] [Google Scholar]
- Hussainzada N, Banerjee A, Swaan PW. Transmembrane domain VII of the human apical sodium-dependent bile acid transporter ASBT (SLC10A2) lines the substrate translocation pathway. Mol Pharmacol. 2006;70(5):1565–1574. doi: 10.1124/mol.106.028647. [DOI] [PubMed] [Google Scholar]
- Izzat NN, Deshazer ME, Loose-Mitchell DS. New molecular targets for cholesterol-lowering therapy. J Pharmacol Exp Ther. 2000;293(2):315–320. [PubMed] [Google Scholar]
- Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophysical journal. 1999;77(3):1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AD, Pajor AM. Role of conserved prolines in the structure and function of the Na+/dicarboxylate cotransporter 1, NaDC1. Biochemistry. 2006;45(13):4231–4239. doi: 10.1021/bi052064y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H. Towards the molecular mechanism of Na(+)/solute symport in prokaryotes. Biochim Biophys Acta. 2001;1505(1):131–143. doi: 10.1016/s0005-2728(00)00283-8. [DOI] [PubMed] [Google Scholar]
- Kaback HR, Wu J. From membrane to molecule to the third amino acid from the left with a membrane transport protein. Q Rev Biophys. 1997;30(4):333–364. doi: 10.1017/s0033583597003387. [DOI] [PubMed] [Google Scholar]
- Kramer W, Girbig F, Glombik H, Corsiero D, Stengelin S, Weyland C. Identification of a ligand-binding site in the Na+/bile acid cotransporting protein from rabbit ileum. J Biol Chem. 2001;276(38):36020–36027. doi: 10.1074/jbc.M104665200. [DOI] [PubMed] [Google Scholar]
- Labro AJ, Raes AL, Bellens I, Ottschytsch N, Snyders DJ. Gating of shaker-type channels requires the flexibility of S6 caused by prolines. J Biol Chem. 2003;278(50):50724–50731. doi: 10.1074/jbc.M306097200. [DOI] [PubMed] [Google Scholar]
- Li H, Xu G, Shang Q, Pan L, Shefer S, Batta AK, Bollineni J, Tint GS, Keller BT, Salen G. Inhibition of ileal bile acid transport lowers plasma cholesterol levels by inactivating hepatic farnesoid X receptor and stimulating cholesterol 7 alpha-hydroxylase. Metabolism. 2004;53(7):927–932. doi: 10.1016/j.metabol.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Lin Z, Itokawa M, Uhl GR. Dopamine transporter proline mutations influence dopamine uptake, cocaine analog recognition, and expression. Faseb J. 2000;14(5):715–728. doi: 10.1096/fasebj.14.5.715. [DOI] [PubMed] [Google Scholar]
- Lu H, Marti T, Booth PJ. Proline residues in transmembrane alpha helices affect the folding of bacteriorhodopsin. J Mol Biol. 2001;308(2):437–446. doi: 10.1006/jmbi.2001.4605. [DOI] [PubMed] [Google Scholar]
- Mitchell SM, Lee E, Garcia ML, Stephan MM. Structure and function of extracellular loop 4 of the serotonin transporter as revealed by cysteine-scanning mutagenesis. J Biol Chem. 2004;279(23):24089–24099. doi: 10.1074/jbc.M311173200. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Kirby LC, Heubi JE, Dawson PA. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2) J Clin Invest. 1997;99(8):1880–1887. doi: 10.1172/JCI119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajor AM, Randolph KM. Conformationally sensitive residues in extracellular loop 5 of the Na+/dicarboxylate co-transporter. J Biol Chem. 2005;280(19):18728–18735. doi: 10.1074/jbc.M501265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli-Magnus C, Stieger B, Meier Y, Kullak-Ublick GA, Meier PJ. Enterohepatic transport of bile salts and genetics of cholestasis. J Hepatol. 2005;43(2):342–357. doi: 10.1016/j.jhep.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Quick M, Jung H. Aspartate 55 in the Na+/proline permease of Escherichia coli is essential for Na+-coupled proline uptake. Biochemistry. 1997;36(15):4631–4636. doi: 10.1021/bi963063w. [DOI] [PubMed] [Google Scholar]
- Sansom MS, Weinstein H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol Sci. 2000;21(11):445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- Shelden MC, Loughlin P, Tierney ML, Howitt SM. Proline residues in two tightly coupled helices of the sulphate transporter, SHST1, are important for sulphate transport. Biochem J. 2001;356(Pt 2):589–594. doi: 10.1042/0264-6021:3560589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepkov ER, Chow S, Lemieux MJ, Fliegel L. Proline residues in transmembrane segment IV are critical for activity, expression and targeting of the Na+/H+ exchanger isoform 1. Biochem J. 2004;379(Pt 1):31–38. doi: 10.1042/BJ20030884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaan PW, Hillgren KM, Szoka FC, Jr, Oie S. Enhanced transepithelial transport of peptides by conjugation to cholic acid. Bioconjug Chem. 1997;8(4):520–525. doi: 10.1021/bc970076t. [DOI] [PubMed] [Google Scholar]
- Tieleman DP, Sansom MS, Berendsen HJ. Alamethicin helices in a bilayer and in solution: molecular dynamics simulations. Biophys J. 1999;76(1 Pt 1):40–49. doi: 10.1016/S0006-3495(99)77176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83(2):633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- Weinman SA, Carruth MW, Dawson PA. Bile acid uptake via the human apical sodium-bile acid cotransporter is electrogenic. J Biol Chem. 1998;273(52):34691–34695. doi: 10.1074/jbc.273.52.34691. [DOI] [PubMed] [Google Scholar]
- Wong MH, Oelkers P, Dawson PA. Identification of a mutation in the ileal sodium-dependent bile acid transporter gene that abolishes transport activity. J Biol Chem. 1995;270(45):27228–27234. doi: 10.1074/jbc.270.45.27228. [DOI] [PubMed] [Google Scholar]
- Woolfson DN, Williams DH. The influence of proline residues on alpha-helical structure. FEBS Lett. 1990;277(1–2):185–188. doi: 10.1016/0014-5793(90)80839-b. [DOI] [PubMed] [Google Scholar]
- Zhang EY, Phelps MA, Banerjee A, Khantwal CM, Chang C, Helsper F, Swaan PW. Topology scanning and putative three-dimensional structure of the extracellular binding domains of the apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2004;43(36):11380–11392. doi: 10.1021/bi049270a. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Rudnick G. Cysteine-scanning Mutagenesis of Serotonin Transporter Intracellular Loop 2 Suggests an α-Helical Conformation. J Biol Chem. 2005;280(35):30807–30813. doi: 10.1074/jbc.M504087200. [DOI] [PubMed] [Google Scholar]