Abstract
Gonadotropin-releasing hormone (GnRH) neurons are neuroendocrine cells, located in the hypothalamus, that play an essential role in mammalian reproduction. These neurons originate in the nasal placode and migrate during embryonic development, in association with olfactory/vomeronasal nerves, first in the nose, then through the cribriform plate to enter the forebrain, before settling in the hypothalamus. One of the molecules required for their early migration in the nose is the chemokine CXCL12, which is expressed in the embryonic nasal mesenchyme in an increasing ventral to dorsal gradient, presumably guiding GnRH neurons toward the forebrain. Mice lacking CXCR4, the receptor for CXCL12, exhibit defective GnRH cell movement and a significant reduction in their number, suggesting that CXCL12/CXCR4 signaling is important in the migration and survival of these neurons. Here, we investigated the role of the more recently identified second CXCL12 receptor, CXCR7, in GnRH neuron development. We demonstrate that CXCR7 is expressed along the migratory path of GnRH neurons in the nasal cavity and, although not expressed by GnRH neurons, it affects their migration as indicated by the ectopic accumulation of these cells in the nasal compartment in CXCR7−/− mice. Absence of CXCR7 caused abnormal accumulation of CXCL12-RFP at CXCR4-positive sites in the nasal area of CXCL12-RFP-transgenic mice and excessive CXCL12-dependent intracellular clustering of CXCR4 in GnRH neurons, suggesting internalization. These findings imply that CXCR7 regulates CXCL12 availability by acting as a scavenger along the migratory path of GnRH neurons and, thus, influences the migration of these cells in a noncell-autonomous manner.
Introduction
Gonadotropin-releasing hormone (GnRH) neurons are a small group of hypothalamic cells that control mammalian reproduction by secreting GnRH decapeptide into the portal vessels of the pituitary. There, GnRH stimulates the production of gonadotropins, which control gonadal function (Merchenthaler et al., 1984). During development, GnRH neurons originate in the nasal placode and migrate in association with olfactory/vomeronasal nerve axons to the hypothalamus (Schwanzel-Fukuda and Pfaff, 1989; Wray et al., 1989). In humans, failure of these neurons to reach the hypothalamus results in infertility (Hardelin, 2001). The mechanisms that guide the migration of GnRH neurons are complex, as they encounter different molecular environments during their migratory journey. Thus, their migration is affected by a wide range of factors (Cariboni et al., 2007b; Wray, 2010; Wierman et al., 2011; Giacobini and Prevot, 2013) including the chemokine CXCL12 (Schwarting et al., 2006; Toba et al., 2008; Casoni et al., 2012), which has an established role in directed cell migration (Tiveron and Cremer, 2008; Lewellis and Knaut, 2012). CXCL12 has been observed in the nasal mesenchyme (NM), whereas its receptor, CXCR4, has been localized in migrating GnRH neurons and olfactory/vomeronasal nerve axons. Further, CXCR4-deficient mice exhibit a loss of GnRH neurons and impaired migration, suggesting the importance of CXCL12/CXCR4 signaling in the development of this system (Schwarting et al., 2006; Toba et al., 2008).
Recent work established CXCR7 as the second CXCL12 receptor (Balabanian et al., 2005; Burns et al., 2006). Unlike CXCR4, however, CXCR7 fails to signal through Gαi proteins and does not facilitate cell migration in response to CXCL12, suggesting that it is an atypical chemokine receptor (Burns et al., 2006; Boldajipour et al., 2008; Rajagopal et al., 2010). CXCR7 has a 10-fold higher binding affinity to CXCL12 than CXCR4, and its property to rapidly internalize CXCL12 and to degrade large amounts of extracellular chemokine has led to the concept that it acts as a scavenger to control the levels of the chemokine (Luker et al., 2010; Naumann et al., 2010; Hoffmann et al., 2012). In this context, studies in zebrafish (Boldajipour et al., 2008) and mice (Sánchez-Alcaniz et al., 2011) have demonstrated that CXCR7 indeed plays a role in cell migration by regulating CXCL12 protein levels available to CXCR4-expressing cells in the process of migration.
Here we investigated the contribution of CXCR7 in CXCL12 signaling in GnRH neuron migration. We found that CXCR7 is expressed within the NM, especially in its ventral aspect, but not by GnRH neurons or their guiding axons. Mice deficient of CXCR7 exhibit impaired migration and ectopic GnRH neuron distribution in the nasal area, suggesting that this receptor affects their directed migration in an indirect manner, possibly by regulating CXCL12 availability. Indeed, a CXCL12/RFP fusion protein (CXCL12-RFP) accumulated at different sites in the nasal area of CXCL12-RFP-transgenic mice lacking functional CXCR7 than of CXCL12-RFP-transgenic controls. Furthermore, CXCR7-null mice exhibited severely reduced CXCR4 immunoreactivity and excessive intracellular clustering of CXCR4 in the entire nasal area including GnRH neurons, which suggests receptor hyperactivation due to CXCL12 overstimulation. In support of this hypothesis, CXCL12−/−;CXCR7−/− mice displayed lack of CXCR4 internalization and increased CXCR4 levels, pointing to a role for CXCR7 in controlling CXCL12 availability in this system.
Materials and Methods
Animals.
To establish mRNA expression patterns for CXCR7, CXCL12, and CXCR4 in the nasal area of wild-type mice, we used pregnant C57BL/6 mice purchased from Charles River. Embryos of transgenic CXCR7-eGFP and CXCR4-eGFP mice (The Gene Expression Nervous System Atlas project GENSAT; http://www.gensat.org/index.html), and transgenic mice expressing CXCL12-RFP under control of the CXCL12 promoter (Bhattacharyya et al., 2008), bred on CD1 background, were used for immunohistochemistry. CXCL12−/−, CXCR7−/−, and CXCR4−/− embryos were obtained by mating heterozygous mice (Nagasawa et al., 1996; Zou et al., 1998; Sierro et al., 2007), which were on C57BL/6 background. CXCR7-eGFP;CXCL12-RFP and CXCR7−/−;CXCL12-RFP mice were obtained by breeding CXCL12-RFP mice with CXCR7-eGFP and CXCR7+/− animals, respectively. Noon of the day after mating was defined as embryonic day 0.5 (E0.5). All embryos examined in this study were collected regardless of sex. All animal procedures were performed in accordance with UK Animals (Scientific Procedures) Act 1986 and German and EU guidelines and approved by the Thüringer Landesamt für Lebensmittelsicherheit und Verbraucherschutz (Reg.-Nr. 22-2684-04-02-015/11).
Riboprobes and in situ hybridization procedure.
The CXCL12 probe was transcribed from full-length mouse CXCL12α cDNA (Tashiro et al., 1993). Probes for mouse CXCR7 and mouse CXCR4 corresponded to the receptors' coding regions (Stumm et al., 2002; Sánchez-Alcaniz et al., 2011). All described cDNAs were subjected to DNA sequencing for control. Riboprobes were generated from the linearized vector constructs by in vitro transcription using digoxigenin-UTP (DIG; Roche) as label. In situ hybridization was performed as described previously (Faux et al., 2010). Briefly, embryonic heads were dissected in PBS, pH 7.4, fixed in 4% paraformaldehyde (PFA) made in PBS overnight, followed by cryoprotection in 30% sucrose, treated with diethyl pyrocarbonate (DEPC)/PBS, at 4°C for 1 d. Heads were frozen in Tissue-Tek OCT and sectioned coronally at 20 μm. Sections were dried at room temperature (RT) for 2 h, before overnight incubation at 65°C in hybridization buffer 1× DEPC-treated “salts” (200 mm NaCl, 5 mm EDTA, 10 mm Tris, pH 7.5, 5 mm NaH2PO4 · 2H2O, 5 mm Na2HPO4; Sigma-Aldrich), 50% deionized formamide (Ambion), 0.1 mg/ml RNase-free yeast tRNA (Invitrogen), 1× Denhardts (RNase/DNase free; Invitrogen), and 10% dextran sulfate (Sigma-Aldrich) containing 100–500 ng/ml DIG-labeled RNA probes. After hybridization, sections were washed three times in a solution containing 50% formamide 1× SSC (Ambion) and 0.1% Tween 20 (Sigma-Aldrich) at 65°C, and two times at RT in 1× MABT (20 mm maleic acid, 30 mm NaCl, 0.1% Tween 20; Sigma-Aldrich) before incubating in a solution containing 2% blocking reagent (Roche) and 10% normal goat serum (NGS) in MABT, followed by overnight incubation in alkaline phosphatase-conjugated anti-DIG antibody (1:1500; Roche). Nitrobluetetrazolium chloride (Roche)/5-bromo-4-chloro-3-indolyl phosphate (Roche) diluted 1:1000 in MABT with 5% polyvinyl alcohol (VWR International) was used for colorimetric detection for 6 h. Sections were mounted using Glycergel mounting medium (Dako). Specificity of the procedure was assessed with probes corresponding to the sense strand of CXCL12, CXCR4, and CXCR7. The sense probes produced only slight signals that were clearly distinct from the signals of the antisense probes.
Histochemistry, antibodies, and immunohistochemistry.
Mouse embryos were fixed in Bouin/Hollande fixative for 16 h (E12.5–E14.5) or 24 h (E16.5) and subjected to routine paraffin embedding (Stumm et al., 2001). Sections were cut at 8 μm for immunohistochemistry, and labeling was performed as described previously (Stumm et al., 2007). Briefly, endogenous peroxidase was blocked with methanol/H2O2 after deparaffinization. For antigen retrieval, sections were bathed for 25 min in 0.01 m citrate buffer, pH 6.0, at 95°C. Sections were then blocked in 50 mm PBS, containing 5% bovine serum albumin (BSA), and incubated overnight with primary antibody in 50 mm PBS containing 1% BSA. The following primary antibodies were used: UMB-2 rabbit monoclonal anti-CXCR4 (1:1000; Fischer et al., 2008), rabbit anti-GnRH (1:200 for fluorescence; 1:1000 for DAB; Immunostar), chicken anti-GFP (1:500; Aves), rabbit anti-peripherin (1:500 for fluorescence; 1:1000 for diaminobenzidine (DAB); Millipore), mouse anti-βΙΙΙ-tubulin (1:500; Sigma-Aldrich), rabbit anti-cleaved caspace-3 (1:5000; Cell Signaling Technology), and rabbit anti-RFP (1:300; Abcam). For dual fluorescence, primary antibodies from different species were combined. TSA enhancement (Vector ABC kit; Vector Laboratories) was performed for UMB-2 before detection with AF555 and AF488 (Invitrogen). Double fluorescence immunohistochemistry for UMB-2 and GnRH (both rabbit) was performed by staining first for UMB-2 as aforementioned and, following washes in a series of ethanol (50% to 100% and back to 50%) and PBS, sections were blocked again in PBS/BSA for 1 h and incubated overnight in anti-GnRH antibody (1:100). Subsequent detection of UMB-2 was performed using streptavidin AF488 (1:200) and localization of GnRH with Alexa 568 (1:200, Invitrogen). Controls were used to ensure that secondary antibodies (Alexa 568) did not bind to UMB-2. Nuclei were counterstained with DAPI (Sigma-Aldrich). Immunofluorescence images were captured with a LSM510 confocal microscope (Zeiss).
For quantification of GnRH neurons, embryonic heads (E12.5–E16.5) were fixed in PFA, made in PBS, for 4–8 h at RT. After fixation, tissue was cryoprotected in 30% sucrose in PBS, embedded and frozen in Tissue-Tek OCT (Sakura Finetek), and sectioned sagittally at 20 μm using a cryostat (Bright Instruments). Sections were washed in PBS and incubated with H2O2 to quench endogenous peroxidase activity. They were then blocked in PBS, containing 5% NGS (Sigma-Aldrich) (v/v) and 0.1% Triton X-100 (v/v) (Sigma-Aldrich), at RT for 2 h. They were subsequently incubated in anti-GnRH (1:1000) antibody at RT overnight, followed by washes in PBS, and incubated in biotinylated anti-rabbit (1:200; Vector Laboratories) for 2 h. Immunoreactivity was visualized with the ABC kit (Vector Laboratories) followed by DAB (Sigma-Aldrich) development. For each genotype, we analyzed at least three embryos at each age, and all GnRH-positive cells were counted in every section of each head under a 40× microscope objective. For all experiments, data were expressed as mean ± SEM. To determine statistical significance, we used unpaired Student's t test, with p < 0.05 considered to be statistically significant.
CXCR4-immunoreactivity was quantified in UMB-2-stained coronal head sections of CXCR7−/−, CXCL12−/−;CXCR7−/−, and homozygous or heterozygous control littermates. For quantification, the nose region was photographed under identical imaging conditions with an Axio Imager A1 microscope (Zeiss) connected to a ProgRes C5 camera (Jenoptik). The sensory epithelium of the vomeronasal organ (VNO) and the olfactory epithelium (OE) were defined as areas of interest using ImageJ software. The proportion of the stained area at the region of interest was measured after setting a uniform threshold level. Measurements were expressed as percentage of the control group.
Results
CXCR7 is expressed in the nasal area
We first analyzed the spatiotemporal profile of CXCR7 expression in the developing nasal area from early to late embryonic life, and compared its pattern of expression to that of CXCR4 and their agonist, CXCL12. Analysis was performed in embryos from E12.5 to E16.5, a period that covers the early stage of migration (E12.5) of GnRH neurons, following their emergence from the nasal placode at E10.5 (Schwanzel-Fukuda and Pfaff, 1989), and the migration between the nasal compartment and the basal forebrain (E14.5–E16.5).
As we observed only minor differences in the overall patterns of the three genes between E12.5 and E14.5, we shall only provide details of their expression at E14.5 (Fig. 1). At this age, CXCR7 was strongly expressed in the VNO and the surrounding area (Fig. 1A,A′). Specifically, CXCR7 mRNA was detected in the sensory epithelium of the VNO (Fig. 1A′, arrow), the surrounding cartilaginous capsule, the cavernous body adjacent to the VNO, and the NM dorsomedial to the VNO (Fig. 1A′). CXCL12 expression was found to overlap with CXCR7 in the apical zone of the VNO (Fig. 1A′,B′). CXCR4 was present throughout the VNO with the most prominent signal being localized in the basal zone (Fig. 1C′). Cells in the nasal area that appeared to be migrating were also CXCR4 positive (Fig. 1C,C′, arrows) and resembled GnRH neurons as previously reported by Schwarting et al. (2006). The respiratory epithelium (RE), which is located lateral to the VNO in the ventral part of the nasal cavity, strongly expressed CXCR7 (Fig. 1A,A′), but contained only few CXCR4-positive cells (Fig. 1C,C′) and no CXCL12 signal (Fig. 1B,B′). More dorsally, in the OE, both CXCR7 and CXCR4 were detected (Fig. 1A″,C″). CXCL12 was absent in the OE except a small area in the dorsal-most part of the nasal cavity (Fig. 1B″).
Figure 1.
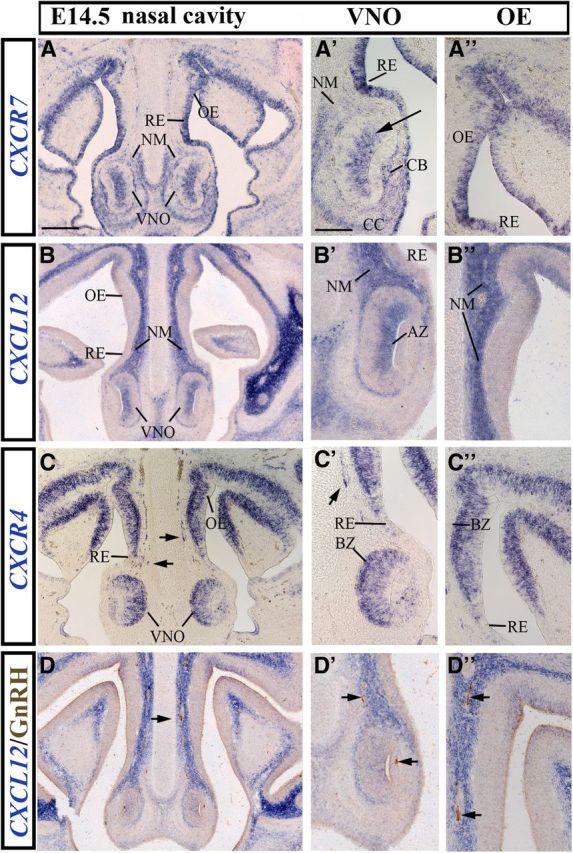
Expression of CXCL12 and its receptors, CXCR7 and CXCR4, in the nasal area of E14.5 mice as revealed by in situ hybridization. A, Low-magnification image of a coronal head section at the level of the nose reveals CXCR7 expression in the VNO, NM, RE, and OE. A′, A″, Higher magnification views show that CXCR7 is predominantly expressed in the apical zone of the sensory epithelium of VNO (arrow) and OE. B, CXCL12 is expressed in the VNO and NM. A′, B′, CXCL12 and CXCR7 overlap in the apical zone (AZ) of the VNO and the NM dorsomedial to the VNO. B″, Higher magnification of the OE area where CXCL12 is highly expressed in the NM and moderately expressed in the dorsal part of the sensory epithelium. Note that most of the OE is CXCL12 negative. C, CXCR4 is highly expressed in the sensory epithelium of VNO and OE and sparsely expressed in the RE. Presumed migrating GnRH neurons are CXCR4 positive (arrows). C′, C″, Higher magnification images illustrate predominant CXCR4 expression in the basal zone (BZ) of VNO (C′) and OE (C″). D, D′, In situ hybridization for CXCL12 and immunohistochemistry for GnRH illustrate migrating GnRH neurons (arrows) on CXCL12 expressing substrate in the NM. D′, D″, These neurons are shown in higher magnification in the area of the VNO (D′) and OE (D″). Scale bars: (in A) A–D, 200 μm; in (A′) A′–D″, 100 μm. CB, cavernous body; CC, cartilaginous capsule.
In agreement with a previous report (Schwarting et al., 2006), CXCL12 was prominently expressed in the NM (Fig. 1B–B″). Dual labeling for CXCL12 and GnRH showed that the CXCL12-positive NM corresponds to the migration corridor of GnRH neurons (Fig. 1D–D″, arrows). Interestingly, CXCR7 was also detected in the NM (Fig. 1A,A′) but, unlike CXCL12, it was concentrated ventrally (dorsomedial to the VNO) and expressed weakly in the dorsal NM.
CXCR7 is not expressed in migrating GnRH neurons and their guiding axons
It has been established that CXCR7 expressed in non-migrating cells along CXCL12-positive migration paths can modulate CXCL12/CXCR4-guided cell migration (Boldajipour et al., 2008). Recent reports suggested that CXCR7 expressed in migrating neurons can directly influence the migration process (Sánchez-Alcaniz et al., 2011; Wang et al., 2011). We thus wondered if CXCR7 modulates GnRH neuron migration in a direct or indirect manner and sought to determine the identity of CXCR7- and CXCR4-expressing cells in the nasal area. In these experiments, we used BAC transgenic mice carrying eGFP under the control of CXCR7 and CXCR4 promoters in double immunofluorescence for GnRH/eGFP. We found that the overall expression patterns of CXCR7- and CXCR4-eGFP (Fig. 2A,D–G) matched the distribution of the respective mRNAs (Fig. 1A,C). However, in the NM, strong CXCR7-eGFP signal was present along the dorsoventral extent (Fig. 2A), whereas strong CXCR7 mRNA signal was restricted to the ventral mesenchyme surrounding the VNO (Fig. 1A). This difference could be due to stability of eGFP, which permits accumulation of the reporter protein despite low gene expression. Nevertheless, GnRH neurons showed no colocalization of eGFP in CXCR7-eGFP mice at the three ages examined (E12.5, E14.5, and E16.5; Fig. 2C–C″), indicating that GnRH neurons do not express CXCR7 during their migration. Since olfactory/vomeronasal axons serve as guides to migrating GnRH neurons, we tested whether CXCR7 is present in this axonal scaffold by staining sections through the nasal compartment of mice at E12.5–E16.5 for peripherin. These experiments indicated that peripherin-labeled axons were also negative for CXCR7-eGFP (Fig. 2B, arrowheads). In similar experiments, using CXCR4-eGFP mice, we confirmed that, in contrast to CXCR7, the majority of GnRH neurons (Schwarting et al., 2006; Fig. 2I–I″) and their guiding axons (Toba et al., 2008; Fig. 2H, arrowheads) express CXCR4 at all ages examined.
Figure 2.
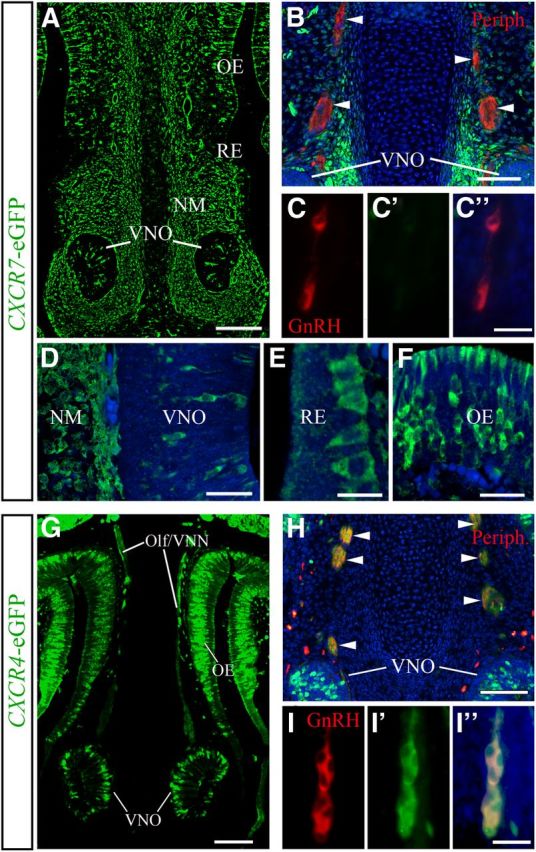
GnRH neurons and their axonal scaffold do not express CXCR7, but do express CXCR4. A, G, Coronal head sections of E14.5 transgenic CXCR7- and CXCR4-eGFP mice reveal the relative expression patterns of CXCR7 and CXCR4 in the nasal area. B, H, Olf/VNN axons (arrowheads) used by migrating GnRH neurons and stained for peripherin (red) are CXCR7 negative (B), but CXCR4 positive (H). C, I, Dual immunofluorescence for GnRH and eGFP demonstrates that GnRH neurons are eGFP negative in CXCR7-eGFP mice (C–C″) and eGFP positive in CXCR4-eGFP mice (I–I″) indicating that GnRH neurons express CXCR4, but not CXCR7. D–F, High-magnification images reveal CXCR7-eGFP in the VNO area (D), RE (E), and OE (F) of E14.5 mice. Specimens shown in B–F, H, and I were counterstained with DAPI. A–I, All images were acquired by confocal microscopy. Scale bars: A, 140 μm; B, 50 μm; (in C″) C–C″, 50 μm; D, 30 μm; E, 14 μm; F, 30 μm; G, 100 μm; H, 100 μm; (in I″) I–I″, 50 μm. Olf/VNN, olfactory/vomeronasal nerve; Periph, peripherin.
CXCR7 regulates CXCL12 availability and CXCR4 activation in the nasal area
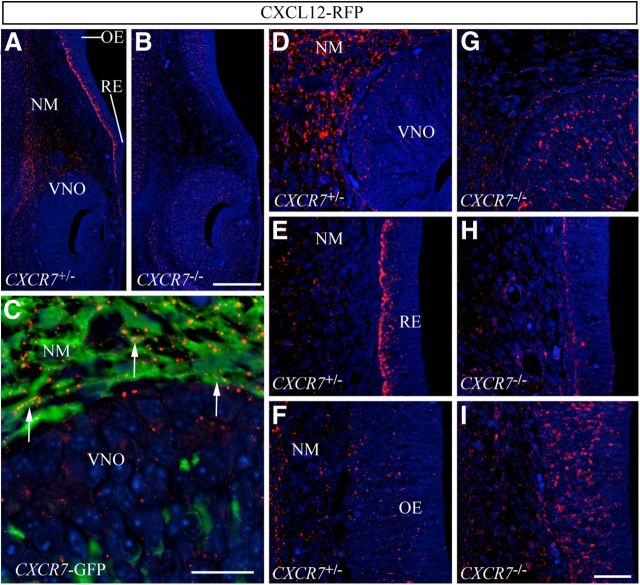
If CXCR7 is not present in GnRH neurons and their scaffold, could it exert an indirect effect on their CXCL12/CXCR4-guided migration? To approach this question, we sought to establish whether CXCR7 influences CXCL12 availability and CXCR4 activation in the nasal area. Since chemokine receptors internalize and accumulate their bound ligands in intracellular vesicles, identification of RFP-labeled cells in transgenic mice expressing RFP-labeled chemokine proved useful to reveal sites of chemokine/chemokine receptor interaction (Jung et al., 2009). We examined the nasal area of BAC-transgenic mice expressing an RFP-fused version of CXCL12 (CXCL12-RFP) under the control of the CXCL12 promoter (Bhattacharyya et al., 2008), and found that RFP transcripts (data not shown) exactly recapitulated the pattern of CXCL12 mRNA in wild-types (Fig. 1B–B″), whereas CXCL12-RFP protein (Fig. 3A) did not. Specifically, the NM dorsomedial to the VNO showed matching strong CXCL12 mRNA and robust RFP expression (Figs. 1B′, 3A,D). The VNO, however, which showed moderate expression of CXCL12 mRNA (Fig. 1B′), contained low RFP signal (Fig. 3D). The most striking mismatch between CXCL12 expression and RFP was observed in the RE, which showed very strong RFP signal (Fig. 3A,E) despite being CXCL12 mRNA negative (Fig. 1B). Most of the OE was sparsely RFP positive (Fig. 3A,F) and CXCL12 mRNA negative (Fig. 1B,B″; a small dorsal area moderately expressed CXCL12 and contained sparse RFP). In CXCR7-eGFP;CXCL12-RFP bitransgenic mice, we observed that the strongly RFP-positive NM and the RE (data not shown) were eGFP positive. High-power confocal analysis confirmed that RFP-signal was often contained within CXCR7-eGFP-positive cells (Fig. 3C, arrows), suggesting that the signal could, at least in part, result from CXCR7-mediated CXCL12-RFP accumulation.
Figure 3.
CXCR7 regulates spatial distribution of CXCL12-RFP along the migration route of GnRH neurons. A–I, Immunostaining for RFP in coronal sections of the nasal area of E14.5 transgenic mice expressing CXCL12-RFP fusion protein. A, B, Low-magnification confocal images reveal strongly decreased RFP signal in the NM and RE and increased RFP signal in the VNO and OE of a CXCL12-RFP;CXCR7−/− embryo (B) as compared with a CXCL12-RFP;CXCR7+/− littermate (A). D–I, High-magnification confocal views of VNO and adjacent NM (D, G), RE (E, H), and OE (F, I). C, High-magnification confocal view of a dual immunofluorescence for RFP and eGFP in a CXCL12-RFP;CXCR7-eGFP bitransgenic mouse shows RFP signal in eGFP-positive mesenchymal cells (arrows). Scale bars: (in B) A, B, 50 μm; C, 14 μm; (in I) D–I, 14 μm.
We then generated CXCL12-RFP-transgenic CXCR7-deficient (CXCL12-RFP;CXCR7−/−) mice to test this assumption, and found that the pattern of CXCL12-RFP was severely altered in the absence of CXCR7. The most obvious difference was noted in the RE (Fig. 3E,H), which normally expresses CXCR7 and contains strong RFP signal. In CXCL12-RFP;CXCR7−/− animals, this structure was virtually RFP negative (Fig. 3H). Since the RE does not express CXCL12 and contains only few CXCR4-positive cells, this observation suggests that CXCR7 normally accumulates/sequesters RFP in this tissue. Also in the NM dorsomedial to the VNO, the RFP signal was severely reduced in the absence of CXCR7 (Fig. 3D,G). The residual RFP signal most likely reflects the strong CXCL12-RFP expression. Interestingly, VNO (Fig. 3D,G) and OE (Fig. 3F,I) showed increased RFP labeling in CXCL12-RFP/CXCR7−/− mice. Given that both structures highly express CXCR4 (Fig. 1C–C″), the increased RFP labeling might be due to enhanced CXCL12-RFP/CXCR4 interactions. Collectively, our analysis indicates that CXCL12-RFP preferentially accumulates at sites expressing CXCR7. In the absence of CXCR7, CXCL12-RFP accumulates in CXCR4-expressing structures.
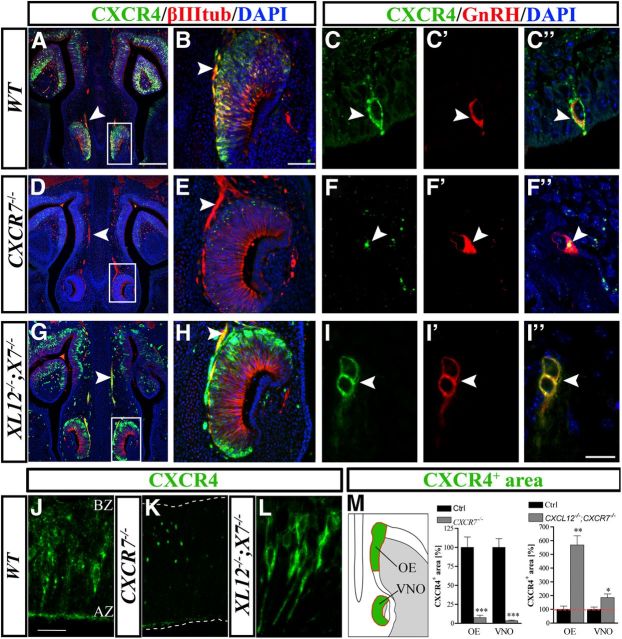
If absence of CXCR7 does lead to increased availability of CXCL12, it would increase CXCL12/CXCR4 interaction, promote CXCR4 internalization, and, thus, alter the subcellular distribution of CXCR4. Careful analysis of the CXCR4 staining in the VNO (Fig. 4D,E) and OE (Fig. 4K) in CXCR7−/− mice revealed the intracellular accumulation of reaction product (dots), suggesting increased internalization or allocation of the receptor in the cytoplasm of cells. In wild-types, most of CXCR4 was present at the surface and only a fraction appeared internalized in cells of the VNO (Fig. 4B) and OE (Fig. 4J). Quantitative analysis of the CXCR4-immunoreactive area in OE and VNO confirmed that CXCR4 staining was severely reduced in CXCR7−/− mice (Fig. 4M, middle). Double immunohistochemistry with CXCR4 and βIII-tubulin, which labels both neuronal somata and processes, revealed absence of CXCR4 from the olfactory/vomeronasal axons in CXCR7−/− mice (Fig. 4D,E, arrowheads). Having shown that lack of CXCR7 affects the spatial distribution of CXCL12-RFP and CXCR4 throughout the nasal area, we turned to GnRH neurons and found that CXCR4 immunoreactivity was also reduced or even absent when compared with controls (Fig. 4C–C″,F–F″). The possibility, that the altered pattern of CXCR4 in the nasal area is due to altered CXCR4 mRNA expression, was ruled out by in situ hybridization for CXCR4, which gave similar results in CXCR7−/− mice and control littermates (data not shown).
Figure 4.
Subcellular redistribution and loss of CXCR4 in the nasal area of CXCR7−/− mice. A–M, Immunostained frontal sections of E14.5 mice. A, B, Double labeling for CXCR4 (green) and βIII tubulin (red) shows that in a WT animal, CXCR4 is localized in axons (arrowheads) and in cells in the VNO (B). D, E, CXCR4 is not detectable in axons (arrowheads) and shows a punctate/internalized pattern in the VNO of a CXCR7−/− animal (E). G, H, The loss of CXCR4 observed after CXCR7 ablation (D, E) is reversed in a CXCL12−/−;CXCR7−/− animal (XL12−/−;X7−/−). C, F, I, Similar changes in CXCR4 localization were noted in GnRH neurons. For example, in one of these neurons of a WT animal (C–C″, arrowhead), CXCR4 is clearly expressed on the cell membrane. However, in a GnRH neuron of CXCR7−/− mutant, the receptor is localized in the cytoplasm and not detected on the plasma membrane (F–F″, arrowhead). In the absence of CXCL12, CXCR4 is upregulated on the cell membrane and intracellular clusters of CXCR4 are not observed in CXCR7-deficient GnRH neurons (I–I″, arrowhead). J–L, High-magnification views demonstrate altered subcellular distribution of CXCR4 in the OE of a CXCR7−/− mutant (K) and a CXCL12−/−;CXCR7−/− double mutant (L) as compared with a WT control (J). A–L, All images were acquired by confocal microscopy. M, Quantification of the CXCR4-immunoreactive area in the OE and VNO of CXCR7−/− and CXCL12−/−;CXCR7−/− mice and their control littermates (n = 4). *p < 0.05, **p < 0.01,***p < 0.001, ANOVA. Green areas in the schematic represent the regions of interest used for quantification. The nasal septum is to the left and the nasal cavity (shaded) to the right. Scale bars: (in A) A, D, G, 200 μm; (in B) B, E, H, 50 μm; (in I″) C, F, I, 20 μm; (in J) J–L, 20 μm. WT, wild-type.
To assess whether altered subcellular distribution of CXCR4 and reduced CXCR4 staining in the nasal area of CXCR7−/− mice depends on CXCL12, we generated CXCL12−/−;CXCR7−/− mice. We found that absence of CXCL12 resulted in upregulation of CXCR4 on the plasma membrane and increased CXCR4 immunoreactivity in cells of the VNO and OE (Fig. 4G,H,L,M, right) as compared with controls. Double immunofluorescence for CXCR4/βIII-tubulin and CXCR4/GnRH revealed high levels of CXCR4 in vomeronasal axons (Fig. 4G,H, arrowheads) and targeting of CXCR4 to the plasma membrane of GnRH neurons (Fig. 4I–I″). Thus, loss of CXCR4 from the surface of cells in the nasal area observed in CXCR7-deficient mice, a typical response to high levels of CXCL12, was reversed in an experiment that combined removal of both CXCL12 and CXCR7.
CXCR7-deficient mice exhibit impaired GnRH neuron migration
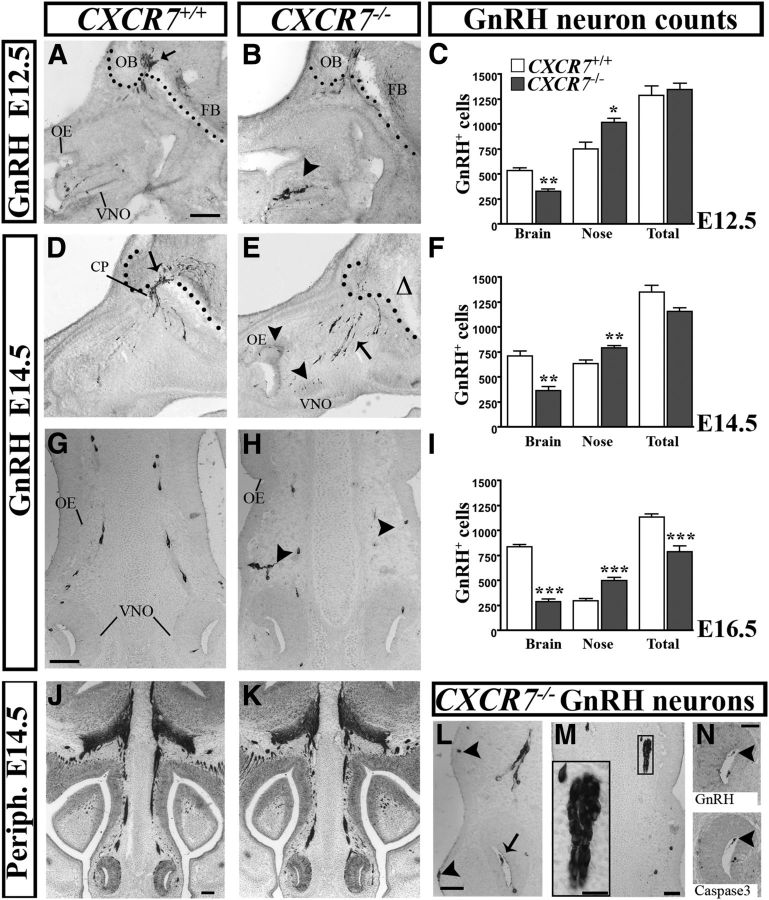
Given our findings suggest that CXCR7 controls CXCL12 availability in the nasal area and CXCL12/CXCR4 interactions in GnRH neurons, we investigated whether its loss might impact on GnRH neuron migration. For this purpose, the number and distribution of GnRH neurons in sagittal sections through the nose and forebrain of CXCR7−/− mice were compared with control littermates at three developmental stages (E12.5, E14.5, and E16.5). Regional analysis revealed an increase in the number of GnRH neurons in the nasal compartment and a concomitant decrease in the forebrain of the CXCR7−/− mice compared with controls at all ages examined (Fig. 5C,F,I). In detail, at E12.5, there was no significant difference between the total GnRH neuron population of CXCR7−/− mice and controls (knock-out (KO): 1346 ± 64 as compared with control: 1289 ± 92, p = 0.64; n = 3; Fig. 5C), implying that the production of these neurons is not affected by the absence of the receptor. Nonetheless, CXCR7−/− mice exhibited a significant increase in GnRH cell numbers in the nasal compartment (with cells accumulating around the VNO; Fig. 5B, arrowhead) as compared with controls (Fig. 5A, arrow) (KO: 1017 ± 42, control: 754 ± 68, *p = 0.031 < 0.05; Fig. 5C) and a decrease in the forebrain (Fig. 5B) (KO: 329 ± 23, control: 535 ± 29, **p = 0.0052 < 0.01; Fig. 5C). This finding suggests a role for CXCR7 in the early steps of GnRH neuron migration, when these cells exit the VNO and start migrating toward the forebrain. At E14.5, when >50% of GnRH neurons in control animals have already passed the cribriform plate and are directed toward the forebrain (Fig. 5D), the equivalent proportion of neurons in CXCR7-deficient mice is 34% (KO: 368 ± 37 as compared with control: 714 ± 50, **p = 0.001, n = 6; Fig. 5F) with many GnRH cells remaining in the nasal compartment (Fig. 5E, arrow) (KO: 793 ± 24, control: 637 ± 33, **p = 0.009 < 0.01; Fig. 5F). Strikingly, GnRH neurons in CXCR7-deficient mice often formed large multicellular clusters, similar to those previously reported in GNR23 mice carrying a deletion in Epha5 gene (Gamble et al., 2005; Fig. 5M), whereas they migrated in chain-like formations in wild-types (Fig. 5G). Further, many cells were found abnormally in the nasal epithelium (Fig. 5H,L, arrowheads) and a small number appeared in the VNO lumen (Fig. 5L, arrow), where they eventually die (Fig. 5N, arrowheads), indicating misguided migration. Still at this stage (E14.5), the total GnRH cell number did not differ significantly between the two groups of animals, although there was a slight reduction in the mutants (KO: 1161 ± 32, control: 1351 ± 69, p = 0.069 > 0.05; Fig. 5F). To determine the fate of the cells that accumulate in the nose, we analyzed CXCR7-null mice of E16.5. By this age, the majority of GnRH neurons in control mice have migrated to the brain, whereas in CXCR7-null animals most GnRH neurons remained in the nasal area (KO: 500 ± 34, control: 298 ± 22, ***p = 0.0001 < 0.001; n = 3; Fig. 5I) and only a small percentage (∼37%), similar to that observed at E14.5, had passed the cribriform plate (KO: 289 ± 26 as compared with control: 837 ± 21, ***p = 3.21E-09 < 0.001; Fig. 5I). Interestingly, the mean total number of GnRH neurons in CXCR7−/− mice of this later age appeared significantly reduced when compared with control animals (KO: 789 ± 57, control: 1135 ± 30, ***p = 0.0004 < 0.001; Fig. 5I), implying that abnormally accumulating GnRH neurons in the nose could have been eliminated by programmed cell death.
Figure 5.
Migration of GnRH neurons is altered in CXCR7-null mice. Sagittal sections (A, B) through the nose at E12.5, stained for GnRH, show that cells tend to remain in the area of the VNO (B, arrowhead) in CXCR7−/− mice whereas, in the corresponding controls, they appear to cross the cribriform plate (arrow) and migrate toward the forebrain (FB). Black dots separate the nasal area from the brain. D, E, A similar picture is evident at 14.5, with most GnRH neurons still migrating in the nasal area in CXCR7-null mice (E, arrow). Additionally, some cells are still present in the VNO and, abnormally, in the OE (E, arrowheads). The relative absence of GnRH neurons in the forebrain is indicated with a triangle. G, H, Coronal sections stained for GnRH reveal that, in CXCR7−/− mice, migrating cells deviate from their normal chain-like formation (G) and, instead, appear in the OE and RE (arrowheads in H and L). Further, they form abnormal multicellular clusters in the NM (rectangle in M and at higher magnification in the inset in M) and, sometimes, in the lumen of the VNO (L, arrow). N, GnRH cells in the lumen of the VNO (arrowhead) show cleaved caspace-3 in a contiguous section, suggesting they are destined to die. J, K, However, guiding axons stained for peripherin appear similar in both genotypes. C, F, I, Compartmental quantification of GnRH cells in the three ages examined showed significant increase of neurons in the nose and a concomitant decrease in the brain of CXCR7−/− mice compared with WT. Moreover, while the total number of cells was similar in the two groups at E12.5 and E14.5, it declined in the KO mice at E16.5 (I). *p < 0.05, **p < 0.01, ***p < 0.001; Scale bars: (in A) A, B, 150 μm; D, E, G, H, L, M, 50 μm; (in J) J, K, 500 μm; N, 30 μm. CP, cribriform plate; OB, olfactory bulb.
We next examined the patterning of olfactory/vomeronasal axons stained for peripherin and confirmed that, in CXCR7-null mice, the axonal substrates used by migrating GnRH neurons were normally fasciculated and properly positioned (Fig. 5J,K), suggesting that CXCR7 is not involved in the patterning/targeting of these axons.
Comparative analysis of CXCL12-, CXCR4-, and CXCR7-null mice
Previous studies have demonstrated that CXCR4-deficient mice display decreased GnRH cell number and impaired migration (Schwarting et al., 2006; Toba et al., 2008). However, comparable analysis of mice lacking the ligand, CXCL12, has not been performed to date. After CXCR7 was identified as a second CXCL12 receptor (Balabanian et al., 2005), we reasoned that examination of CXCL12-null mice in comparison with animals lacking CXCR4 or CXCR7 would shed light on the role of each of these molecules in the development of the GnRH neuron system and enhance our knowledge of the mechanisms that regulate this receptor system.
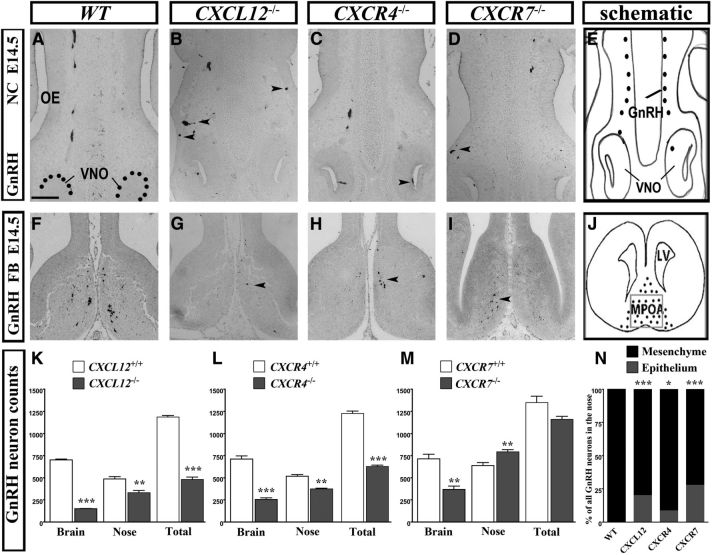
Thus, we analyzed CXCL12 defective mice at E14.5 and found a gross decrease in the total number of GnRH neurons when compared with control littermates (KO: 480 ± 25, control: 1187 ± 16; ***p = 1.98E-05 < 0.0001; n = 3; Fig. 6K). We then re-examined CXCR4−/− mice at the same age and, in agreement with previous studies (Schwarting et al., 2006), found reduced total GnRH cell number (KO: 628 ± 12, control: 1227 ± 25; ***p = 2.96E-05; n = 3; Fig. 6L).
Figure 6.
GnRH neuron migration in CXCL12−/−, CXCR4−/−, and CXCR7−/− mice at E14.5. A–D, In the nose, GnRH neurons migrate in a chain-like formation in WT animals but, in KOs, they form clusters (B, arrowheads), and are often found abnormally in the lumen of the VNO (C, arrowhead) or in the epithelium (D, arrowhead). F–I, GnRH staining of coronal sections at the level of the MPOA shows the distribution of these cells in WT animals (F) and the relative reduction in numbers in the CXCL12−/− (G), CXCR4−/− (H), and CXCR7−/− (I) mice. K–M, Compartmental quantification of GnRH neurons shows an overall decrease in CXCL12−/− and CXCR4−/−, but not in CXCR7−/− animals, and a significant reduction in both nose and brain when compared with control animals. N, Spread/distribution analysis of GnRH neurons in the nasal area, divided in two compartments, epithelium and NM, show that 22% of all cells in the nasal area are abnormally localized in the epithelium of CXCL12−/− animals. The respective percentage is 28% in CXCR7−/− and 8.5% in CXCR4−/− mice. No GnRH neurons are found in the epithelium in control animals. E, J, Schematics show the nasal compartment (NC) and the medial preoptic area (MPOA) in the forebrain (FB) as photographed in A–D and F–I, respectively. K–N, *p < 0.05, **p < 0.01,***p < 0.001. Scale bars: (in A) A–D, F–I, 100 μm. FB, forebrain; LV, lateral ventricle.
As CXCR7 expression was shown to be absent from GnRH neurons, at least within the time frame of our analysis (E12.5–16.5; KO mice die in late gestation), we considered CXCR4 to be the only receptor mediating CXCL12 signaling on GnRH neurons. The similarity between the two phenotypes (significant decrease in the total number of GnRH neurons in CXCR4−/− and CXCL12−/− mice), together with our finding that CXCR7−/− mice do not display such defect (Fig. 6M), provide credence to this hypothesis.
In addition to the defect in total cell number in both CXCR4 and CXCL12-null mice, the regional distribution of GnRH neurons differed from that of control animals. Specifically, similar to embryos lacking CXCR7 (Fig. 6D,I,M), GnRH neurons in CXCL12-deficient mice were found accumulated in the nasal compartment (area between the VNO and nasal/forebrain junction) and not migrating to the brain (Fig. 6B,G). Thus, at E14.5, there were twice as many neurons in the nose compared with the brain when, in control mice, most of GnRH neurons had already crossed the nasal/forebrain junction and were directed toward the hypothalamus. Counts of cells residing in the forebrain revealed that control animals had five times as many GnRH neurons as CXCL12−/− mice (KO: 148 ± 3, control: 701 ± 9; ***p < 0.0001; Fig. 6F,G,K). Similarly, in CXCR4-lacking mice, the majority (60%) of the cells were still present in the nasal compartment as compared with 42% in control littermates. Concomitantly, the number of GnRH neurons in the brain differed significantly from control mice (KO: 254 ± 17, control: 710 ± 36; ***p = 0.0003; Fig. 6C,H,L).
To gain greater insight on why GnRH neurons remain in the nasal compartment, we carefully examined their appearance and spatial distribution in this area. As depicted in Fig. 6N, considerably more cells in these animals were found abnormally in the OE (Fig. 6B,D, arrowheads). Specifically, when we analyzed the positions of GnRH neurons in the three groups (CXCL12, CXCR4, and CXCR7) of KO mice, we observed that in CXCL12- and CXCR7-deficient mice, 22 and 28% of the total GnRH cell population present in the nose was aberrantly located in the epithelium; in the CXCR4−/− mutant, that percentage was 8.5%. In comparison, hardly any (0–2%) cells were identified in the OE in all control littermates. These observations strongly suggest that this chemokine/receptor system keeps GnRH neurons to their migratory path. We should add that we found no apparent defect in the peripherin-stained vomeronasal axons of CXCL12−/− and CXCR4−/− mice (data not shown).
Discussion
The migration of GnRH neurons represents a unique example of neuronal migration as these cells are born outside the brain and follow a long and tortuous route to their destinations in the forebrain. Chemokines expressed along migration corridors have been reported to serve as guidance cues for migrating neurons (Borrell and Marín, 2006; Stumm et al., 2007). Accordingly, CXCL12 has been found to be expressed in the NM and to guide migrating GnRH neurons toward the forebrain (Schwarting et al., 2006; Toba et al., 2008).
CXCL12 exerts its functions by binding to CXCR4. In the developing brain, CXCL12 and its receptor CXCR4 are highly expressed in the neocortex, cerebellum, and hippocampus, and regulate cell migration and axonal pathfinding (Ma et al., 1998; Chalasani et al., 2003; Tran and Miller, 2003). Although the CXCR4 gene is downregulated in most brain regions postnatally, persistent expression in the hypothalamus and CXCL12/CXCR4-mediated modulation of hypothalamo-pituitary systems points to a role of this receptor in neuroendocrine regulation (Callewaere et al., 2006; Li and Ransohoff, 2008; Rostène et al., 2011).
GnRH neurons express CXCR4 early in their development, as they leave the VNO to begin their migration to the brain. CXCR4-deficient mice exhibit impaired GnRH neuron migration and a significant decrease in their total number (Schwarting et al., 2006; Toba et al., 2008), pointing to the importance of CXCL12 signaling in this system. CXCR4 was long considered the only receptor for CXCL12, until it was shown recently that it binds with higher affinity to an additional receptor, CXCR7 (Balabanian et al., 2005). While signaling properties of CXCR7 are presently controversial (Thelen and Thelen, 2008; Luker et al., 2009; Rajagopal et al., 2010; Hoffmann et al., 2012; Kumar et al., 2012; Odemis et al., 2012), it appears to act as a CXCL12 scavenger controlling chemokine levels in the extracellular environment by ligand sequestration (Boldajipour et al., 2008; Naumann et al., 2010; Hoffmann et al., 2012). In this context, loss of CXCR7 leads to extracellular accumulation and perturbed gradients of the chemokine (Boldajipour et al., 2008; Sánchez-Alcaniz et al., 2011). Similar to CXCL12 and CXCR4, CXCR7 is expressed in the developing hematopoietic system, cardiovascular system, and brain, and CXCR7-deficient mice die perinatally with cardiovascular defects (Sierro et al., 2007).
Here, we provide evidence that CXCR7 is important in the early steps of GnRH neuron migration, as lack of this receptor results in aberrant accumulation of these cells in the nasal cavity. The defects are associated with severe reduction and perturbed subcellular distribution of CXCR4 protein in cells throughout the nasal area, including GnRH neurons.
CXCR7 expression in the nasal area
The first indication that CXCR7 may be involved in GnRH neuron migration came from its expression at the start and along the well defined migratory path of these cells in the nose. Specifically, we identified CXCR7 expression within and around the VNO, where GnRH neurons emerge. Its expression in the more dorsal parts of the NM was significantly reduced up to the cribriform plate, where GnRH neurons enter the brain. Given that CXCR7 sequesters CXCL12 from the extracellular environment (Boldajipour et al., 2008; Naumann et al., 2010; Hoffmann et al., 2012), we may suggest that its pattern of expression may help form a ventrodorsally increasing chemokine gradient. An additional mechanism that could form a CXCL12 gradient is the ventrodorsally increasing CXCL12 expression within the nasal area (Schwarting et al., 2006). Our observation that GnRH neurons as well as the supporting axons of the olfactory/vomeronasal nerve do not express CXCR7, is consistent with the established role of this receptor in shaping CXCL12 gradients. Interestingly, CXCR7 was also expressed in the RE and OE, tissues that are normally avoided by migrating GnRH neurons. Although the RE does not express CXCL12 mRNA, it contained strong RFP signal in transgenic mice expressing CXCL12-RFP fusion protein under the CXCL12 promoter. The observation that the RE in CXCL12-RFP;CXCR7−/− mice was devoid of RFP lends support to the concept that CXCR7-expressing cells sequester CXCL12 in the nasal area.
Impaired migration and abnormal distribution of GnRH neurons in CXCR7-null mice
If shaping the CXCL12 gradient by CXCR7 is important for GnRH neuron migration, then this process would be affected in mice lacking the receptor. Indeed, our analysis of E12.5 and E14.5 CXCR7-deficient mice revealed a defect in GnRH neuron migration when, at the same time, the total cell number was unaffected. This suggests that CXCR7 is not involved in the early generation of GnRH neurons. In these null mice, half of the GnRH neuron population failed to migrate to the hypothalamus and, instead, distributed randomly in the nasal area, formed clusters, and positioned themselves ectopically in the OE, suggesting that CXCR7 plays an important role in their directed migration toward the forebrain. Interestingly, although the overall GnRH cell number did not differ from control littermates at early developmental stages (E12.5–E14.5), it did decline significantly at E16.5. It is likely that ectopic GnRH neurons that failed to migrate to the brain eventually die or cease to express the GnRH gene. The partial phenotype observed here may be attributed to the reported heterogeneous makeup of the GnRH neuron population (Giacobini et al., 2008; Jasoni et al., 2009; Forni et al., 2011), and abundant evidence clearly indicates that no single genetic mutation may prevent all GnRH neurons from reaching their destinations (Cariboni et al., 2007a, 2011; Schwarting et al., 2007; Giacobini et al., 2008; Messina et al., 2011). The impact of reduced GnRH neurons in reproduction has been assessed in lines of transgenic mice that survive to adulthood. These studies have invariably shown abnormal gonadal development and disrupted fertility (Cariboni et al., 2005, 2007a; Messina et al., 2011).
As the GnRH system has never been investigated in mice lacking CXCL12, we analyzed the number and distribution of GnRH neurons in these mice and compared their phenotype to that of CXCR4- and CXCR7-deficient animals. We found that the phenotype in CXCL12−/− mice closely resembled that of CXCR4−/− mice, both in terms of migration and overall decrease in cell number, strongly suggesting that CXCL12 signals through CXCR4 in this system to control GnRH neuron survival and migration. Moreover, the aberrant distribution of GnRH neurons in the nasal area of CXCL12, CXCR4, and CXCR7-null mice points to a direct role for CXCL12 in guiding the migration of these cells, especially since the supporting axonal scaffold was unaffected in the three KO lines.
CXCR7 controls CXCL12 availability
Previous studies have shown that sustained excess of CXCL12 leads to desensitization and degradation of CXCR4 (Marchese and Benovic, 2001). In addition, CXCR7 has been shown to regulate CXCR4 function. This can be done either in a cell-autonomous manner, when it is coexpressed with CXCR4 in the same cell (Sánchez-Alcaniz et al., 2011), or in a noncell-autonomous manner, when it is expressed in the tissue surrounding CXCR4-positive cells (Boldajipour et al., 2008; Haege et al., 2012). Our present data support for the first time the latter hypothesis for migrating neurons and thus, are clearly distinct from earlier findings proposing a cell-autonomous function for CXCR7 in neuronal migration (Tiveron et al., 2010; Sánchez-Alcaniz et al., 2011; Wang et al., 2011).
GnRH neurons express CXCR4, but not CXCR7, during their migration through the nasal compartment, and are attracted by progressively higher levels of CXCL12 as they target the cribriform plate (Schwarting et al., 2006). In the case of compromised CXCR7 activity, we found clustering of CXCR4 in the intracellular compartment suggesting internalization of the receptor and, in some cases, loss of CXCR4 protein in GnRH neurons and in olfactory/vomeronasal cells and their axons. This finding, the function of CXCR7 as a scavenger of extracellular CXCL12 (Naumann et al., 2010; Cruz-Orengo et al., 2011) and our observation that, in absence of CXCR7, more CXCL12-RFP accumulates at CXCR4 sites, prompted us to suggest that CXCR4 re-allocation from the plasma membrane to the cytoplasm of neurons in the nasal compartment is an indicator for excessive CXCL12-promoted CXCR4 activation. Thus, if increased levels of CXCL12 in CXCR7-null mice result in CXCR4 internalization, then concomitant removal of the ligand (and CXCR7) would prevent excessive internalization and loss of CXCR4. Indeed, the apparent intracellular clustering of CXCR4 in CXCR7-deficient mice was reversed in CXCL12−/−;CXCR7−/− animals, strongly suggesting that the function of CXCR7 in this system is to control CXCL12 levels. It has been proposed that GnRH neurons switch off the CXCR4 pathway once they reach the forebrain (Toba et al., 2008). Here, we may suggest a rapid gene regulation-independent mechanism by which this may be achieved: GnRH neurons, while in the nasal compartment, migrate toward higher levels of CXCL12, retaining CXCR4 activity through CXCR7 function. When they reach the cribriform plate, which is rich in CXCL12, but almost devoid of CXCR7, CXCR4 starts to become highly internalized and degraded as a result of hyperactivation, rendering GnRH neurons no longer responsive to the effects of CXCL12. Once GnRH neurons enter the forebrain, they are subjected to the influences of other molecules as they negotiate their migration toward the hypothalamus.
In summary, our studies have shown that CXCR7 is required for GnRH neuron migration. Our data support a mechanism, whereby CXCR7 controls CXCR4 activity in migrating GnRH neurons by regulating spatial availability and possibly levels of CXCL12. It is pertinent to note that recent studies in zebrafish have implicated β-arrestin in this mechanism (Mahabaleshwar et al., 2012). It would be interesting to assess whether this process is conserved in mice, and whether mice with defects in β-arrestin/CXCR7 interaction exhibit a similar phenotype to CXCR7-null animals.
Footnotes
This work was supported by a Wellcome Trust Programme Grant (089775) and a Deutsche Forschungsgemeinschaft Grant (STU 295/7-1). We thank C. Anders, H. Bechmann, S. Bechmann, and H. Stadler for excellent technical assistance. We also thank Mathilda Mommersteeg, Bill Andrews, and Mason Yeh for technical advice and fruitful discussions.
References
- Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M, Bachelerie F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya BJ, Banisadr G, Jung H, Ren D, Cronshaw DG, Zou Y, Miller RJ. The chemokine stromal cell-derived factor-1 regulates GABAergic inputs to neural progenitors in the postnatal dentate gyrus. J Neurosci. 2008;28:6720–6730. doi: 10.1523/JNEUROSCI.1677-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B, Mahabaleshwar H, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Borrell V, Marín O. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat Neurosci. 2006;9:1284–1293. doi: 10.1038/nn1764. [DOI] [PubMed] [Google Scholar]
- Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ, Wei K, McMaster BE, Wright K, Howard MC, Schall TJ. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaere C, Banisadr G, Desarménien MG, Mechighel P, Kitabgi P, Rostène WH, Mélik Parsadaniantz S. The chemokine SDF-1/CXCL12 modulates the firing pattern of vasopressin neurons and counteracts induced vasopressin release through CXCR4. Proc Natl Acad Sci U S A. 2006;103:8221–8226. doi: 10.1073/pnas.0602620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariboni A, Rakic S, Liapi A, Maggi R, Goffinet A, Parnavelas JG. Reelin provides an inhibitory signal in the migration of gonadotropin-releasing hormone neurons. Development. 2005;132:4709–4718. doi: 10.1242/dev.02033. [DOI] [PubMed] [Google Scholar]
- Cariboni A, Hickok J, Rakic S, Andrews W, Maggi R, Tischkau S, Parnavelas JG. Neuropilins and their ligands are important in the migration of gonadotropin-releasing hormone neurons. J Neurosci. 2007a;27:2387–2395. doi: 10.1523/JNEUROSCI.5075-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariboni A, Maggi R, Parnavelas JG. From nose to fertility: the long migratory journey of gonadotropin-releasing hormone neurons. Trends Neurosci. 2007b;30:638–644. doi: 10.1016/j.tins.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Cariboni A, Davidson K, Rakic S, Maggi R, Parnavelas JG, Ruhrberg C. Defective gonadotropin-releasing hormone neuron migration in mice lacking SEMA3A signalling through NRP1 and NRP2; implications in the aetiology of hypogonadotropic hypogonadism. Hum Mol Genet. 2011;20:336–344. doi: 10.1093/hmg/ddq468. [DOI] [PubMed] [Google Scholar]
- Casoni F, Hutchins BI, Donohue D, Fornaro M, Condie BG, Wray S. SDF and GABA interact to regulate axophilic migration of GnRH neurons. J Cell Sci. 2012;125:5015–5025. doi: 10.1242/jcs.101675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani SH, Baribaud F, Coughlan CM, Sunshine MJ, Lee VM, Doms RW, Littman DR, Raper JA. The chemokine stromal cell-derived factor-1 promotes the survival of embryonic retinal ganglion cells. J Neurosci. 2003;23:4601–4612. doi: 10.1523/JNEUROSCI.23-11-04601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Orengo L, Holman DW, Dorsey D, Zhou L, Zhang P, Wright M, McCandless EE, Patel JR, Luker GD, Littman DR, Russell JH, Klein RS. CXCR7 influences leukocyte entry into the CNS parenchyma by controlling abluminal CXCL12 abundance during autoimmunity. J Exp Med. 2011;208:327–339. doi: 10.1084/jem.20102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux C, Rakic S, Andrews W, Yanagawa Y, Obata K, Parnavelas JG. Differential gene expression in migrating cortical interneurons during mouse forebrain development. J Comp Neurol. 2010;518:1232–1248. doi: 10.1002/cne.22271. [DOI] [PubMed] [Google Scholar]
- Fischer T, Nagel F, Jacobs S, Stumm R, Schulz S. Reassessment of CXCR4 chemokine receptor expression in human normal and neoplastic tissues using the novel rabbit monoclonal antibody UMB-2. PLoS One. 2008;3:e4069. doi: 10.1371/journal.pone.0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni PE, Taylor-Burds C, Melvin VS, Williams T, Wray S. Neural crest and ectodermal cells intermix in the nasal placode to give rise to GnRH-1 neurons, sensory neurons, and olfactory ensheathing cells. J Neurosci. 2011;31:6915–6927. doi: 10.1523/JNEUROSCI.6087-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble JA, Karunadasa DK, Pape JR, Skynner MJ, Todman MG, Bicknell RJ, Allen JP, Herbison AE. Disruption of ephrin signaling associates with disordered axophilic migration of the gonadotropin-releasing hormone neurons. J Neurosci. 2005;25:3142–3150. doi: 10.1523/JNEUROSCI.4759-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacobini P, Prevot V. Semaphorins in the development, homeostasis and disease of hormone systems. Semin Cell Dev Biol. 2013;24:190–198. doi: 10.1016/j.semcdb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Giacobini P, Messina A, Morello F, Ferraris N, Corso S, Penachioni J, Giordano S, Tamagnone L, Fasolo A. Semaphorin 4D regulates gonadotropin-releasing hormone-1 neuronal migration through PlexinB1-Met complex. J Cell Biol. 2008;183:555–566. doi: 10.1083/jcb.200806160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haege S, Einer C, Thiele S, Mueller W, Nietzsche S, Lupp A, Mackay F, Schulz S, Stumm R. CXC chemokine receptor 7 (CXCR7) regulates CXCR4 protein expression and capillary tuft development in mouse kidney. PLoS One. 2012;7:e42814. doi: 10.1371/journal.pone.0042814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardelin JP. Kallmann syndrome: towards molecular pathogenesis. Mol Cell Endocrinol. 2001;179:75–81. doi: 10.1016/S0303-7207(01)00462-2. [DOI] [PubMed] [Google Scholar]
- Hoffmann F, Müller W, Schütz D, Penfold ME, Wong YH, Schulz S, Stumm R. Rapid uptake and degradation of CXCL12 depend on CXCR7 carboxyl-terminal serine/threonine residues. J Biol Chem. 2012;287:28362–28377. doi: 10.1074/jbc.M111.335679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasoni CL, Porteous RW, Herbison AE. Anatomical location of mature GnRH neurons corresponds with their birthdate in the developing mouse. Dev Dyn. 2009;238:524–531. doi: 10.1002/dvdy.21869. [DOI] [PubMed] [Google Scholar]
- Jung H, Bhangoo S, Banisadr G, Freitag C, Ren D, White FA, Miller RJ. Visualization of chemokine receptor activation in transgenic mice reveals peripheral activation of CCR2 receptors in states of neuropathic pain. J Neurosci. 2009;29:8051–8062. doi: 10.1523/JNEUROSCI.0485-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Tripathi V, Ahmad M, Nath N, Mir RA, Chauhan SS, Luthra K. CXCR7 mediated Gialpha independent activation of ERK and Akt promotes cell survival and chemotaxis in T cells. Cell Immunol. 2012;272:230–241. doi: 10.1016/j.cellimm.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Lewellis SW, Knaut H. Attractive guidance: how the chemokine SDF1/CXCL12 guides different cells to different locations. Semin Cell Dev Biol. 2012;23:333–340. doi: 10.1016/j.semcdb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ransohoff RM. Multiple roles of chemokine CXCL12 in the central nervous system: a migration from immunology to neurobiology. Prog Neurobiol. 2008;84:116–131. doi: 10.1016/j.pneurobio.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Gupta M, Steele JM, Foerster BR, Luker GD. Imaging ligand-dependent activation of CXCR7. Neoplasia. 2009;11:1022–1035. doi: 10.1593/neo.09724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Steele JM, Mihalko LA, Ray P, Luker GD. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599–4610. doi: 10.1038/onc.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci U S A. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahabaleshwar H, Tarbashevich K, Nowak M, Brand M, Raz E. beta-arrestin control of late endosomal sorting facilitates decoy receptor function and chemokine gradient formation. Development. 2012;139:2897–2902. doi: 10.1242/dev.080408. [DOI] [PubMed] [Google Scholar]
- Marchese A, Benovic JL. Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J Biol Chem. 2001;276:45509–45512. doi: 10.1074/jbc.C100527200. [DOI] [PubMed] [Google Scholar]
- Merchenthaler I, Görcs T, Sétáló G, Petrusz P, Flerkó B. Gonadotropin-releasing hormone (GnRH) neurons and pathways in the rat brain. Cell Tissue Res. 1984;237:15–29. doi: 10.1007/BF00229195. [DOI] [PubMed] [Google Scholar]
- Messina A, Ferraris N, Wray S, Cagnoni G, Donohue DE, Casoni F, Kramer PR, Derijck AA, Adolfs Y, Fasolo A, Pasterkamp RJ, Giacobini P. Dysregulation of Semaphorin 7A/β1-integrin signaling leads to defective GnRH-1 cell migration, abnormal gonadal development and altered fertility. Hum Mol Genet. 2011;20:4759–4774. doi: 10.1093/hmg/ddr403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- Naumann U, Cameroni E, Pruenster M, Mahabaleshwar H, Raz E, Zerwes HG, Rot A, Thelen M. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odemis V, Lipfert J, Kraft R, Hajek P, Abraham G, Hattermann K, Mentlein R, Engele J. The presumed atypical chemokine receptor CXCR7 signals through G(i/o) proteins in primary rodent astrocytes and human glioma cells. Glia. 2012;60:372–381. doi: 10.1002/glia.22271. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Kim J, Ahn S, Craig S, Lam CM, Gerard NP, Gerard C, Lefkowitz RJ. Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc Natl Acad Sci U S A. 2010;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostène W, Guyon A, Kular L, Godefroy D, Barbieri F, Bajetto A, Banisadr G, Callewaere C, Conductier G, Rovère C, Mélik-Parsadaniantz S, Florio T. Chemokines and chemokine receptors: new actors in neuroendocrine regulations. Front Neuroendocrinol. 2011;32:10–24. doi: 10.1016/j.yfrne.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Sánchez-Alcaniz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, López-Bendito G, Stumm R, Marín O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- Schwarting GA, Henion TR, Nugent JD, Caplan B, Tobet S. Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain. J Neurosci. 2006;26:6834–6840. doi: 10.1523/JNEUROSCI.1728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarting GA, Wierman ME, Tobet SA. Gonadotropin-releasing hormone neuronal migration. Semin Reprod Med. 2007;25:305–312. doi: 10.1055/s-2007-984736. [DOI] [PubMed] [Google Scholar]
- Sierro F, Biben C, Martínez-Muñoz L, Mellado M, Ransohoff RM, Li M, Woehl B, Leung H, Groom J, Batten M, Harvey RP, Martínez AC, Mackay CR, Mackay F. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm RK, Rummel J, Junker V, Culmsee C, Pfeiffer M, Krieglstein J, Höllt V, Schulz S. A dual role for the SDF-1/CXCR4 chemokine receptor system in adult brain: isoform-selective regulation of SDF-1 expression modulates CXCR4-dependent neuronal plasticity and cerebral leukocyte recruitment after focal ischemia. J Neurosci. 2002;22:5865–5878. doi: 10.1523/JNEUROSCI.22-14-05865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm R, Culmsee C, Schafer MK, Krieglstein J, Weihe E. Adaptive plasticity in tachykinin and tachykinin receptor expression after focal cerebral ischemia is differentially linked to gabaergic and glutamatergic cerebrocortical circuits and cerebrovenular endothelium. J Neurosci. 2001;21:798–811. doi: 10.1523/JNEUROSCI.21-03-00798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm R, Kolodziej A, Schulz S, Kohtz JD, Höllt V. Patterns of SDF-1alpha and SDF-1gamma mRNAs, migration pathways, and phenotypes of CXCR4-expressing neurons in the developing rat telencephalon. J Comp Neurol. 2007;502:382–399. doi: 10.1002/cne.21336. [DOI] [PubMed] [Google Scholar]
- Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- Thelen M, Thelen S. CXCR7, CXCR4 and CXCL12: an eccentric trio? J Neuroimmunol. 2008;198:9–13. doi: 10.1016/j.jneuroim.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Cremer H. CXCL12/CXCR4 signalling in neuronal cell migration. Curr Opin Neurobiol. 2008;18:237–244. doi: 10.1016/j.conb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Boutin C, Daou P, Moepps B, Cremer H. Expression and function of CXCR7 in the mouse forebrain. J Neuroimmunol. 2010;224:72–79. doi: 10.1016/j.jneuroim.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Toba Y, Tiong JD, Ma Q, Wray S. CXCR4/SDF-1 system modulates development of GnRH-1 neurons and the olfactory system. Dev Neurobiol. 2008;68:487–503. doi: 10.1002/dneu.20594. [DOI] [PubMed] [Google Scholar]
- Tran PB, Miller RJ. Chemokine receptors in the brain: a developing story. J Comp Neurol. 2003;457:1–6. doi: 10.1002/cne.10546. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JL. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierman ME, Kiseljak-Vassiliades K, Tobet S. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front Neuroendocrinol. 2011;32:43–52. doi: 10.1016/j.yfrne.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J Neuroendocrinol. 2010;22:743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci U S A. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]