Abstract
Congenital Nystagmus (CN) is a genetically heterogeneous ocular disease, which causes a significant proportion of childhood visual impairment. To identify the underlying genetic defect of a CN family, twenty-two members were recruited. Genotype analysis showed that affected individuals shared a common haplotype with markers flanking FRMD7 locus. Sequencing FRMD7 revealed a T > C transition in exon 8, causing a conservative substitution of Isoleucine to Tyrosine at codon 240. By protein structural modeling, we found the mutation may disrupt the hydrophobic core and destabilize the protein structure. We reviewed the literature and found that exons 2, 8, and 9 (11.4% of the sequence of FRMD7 mRNA) represent the majority (55.3%) of the reported FRMD7 mutations. In summary, we identified a novel mutation in FRMD7, showed its molecular consequence, and revealed the mutation-rich exons of the FRMD7 gene. Collectively, this provides molecular insights for future CN clinical genetic diagnosis and treatment.
Congenital nystagmus (CN) is a common oculomotor disorder (frequency of 1/1,500 live births) characterized by bilateral, involuntary, periodic, predominantly ocular oscillations, that may result in reduced or limited vision1. CN can be an idiopathic disease (idiopathic congenital nystagmus, ICN) or can be a feature of other ocular diseases, such as albinism (OCA1A, OMIN 203100), achromatopsia (ACHM3, OMIN 262300), and Leber congenital amaurosis (LCA1, OMIN 204000). Three different inheritance models have been reported: X-linked idiopathic congenital nystagmus (XLICN, OMIN 300628, 300589), autosomal recessive (OMIN 257400) and autosomal dominant (OMIN 164100, 608345, 193003)2. Studies showed that at least two disease loci of XLICN were mapped to Xq26–q27 and Xp11.4–Xp11.31,3, respectively. The corresponding gene for the first locus is FRMD74,5.
In the present study, to identify the gene which is responsible for causing CN in a five-generation family, twenty-two family members were recruited, informed consent was obtained from each, and each submitted to an ophthalmologic exam. To test for mutations in one gene reported to cause CN (FRMD7, NM_194277), recruited individuals were first genotyped with microsatellite markers flanking the FRMD7 locus. Mutations were identified by direct sequencing using gene specific primers. Combining sequence analysis, protein structural modeling, and a comprehensive literature review, we provide molecular insights for future CN genetic diagnosis and treatment.
Results
Clinical data
There were eleven affected individuals (seven males and four females) in this five-generation family. No male to male transmission was observed in this family. The proband (V:1) had nystagmus as the first symptom. Eye movement recordings revealed that nystagmus of the proband is predominantly a horizontal jerk waveform while pendular and vertical waveforms also exist. The ocular oscillation could be slowed down at a left gaze of 10° or at right gaze of 5°. The oscillations become stronger when the proband focused on observing or one eye was covered. No null zone or head nodding was observed in the proband, although some patients in this family showed head oscillations/nodding. Patients in this family were excluded from albinism and were confirmed as X-linked congenital nystagmus based on the detailed ophthalmologic examinations and family history.
Haplotype and linkage analysis
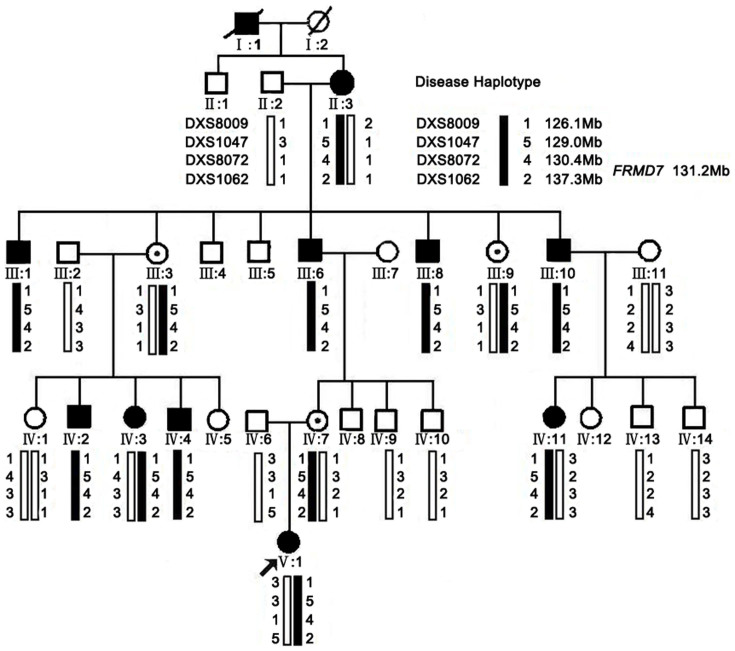
Haplotype analysis showed that the affected individuals in the family shared a common haplotype with markers DXS8009, DXS1047, DXS8072 and DXS1062 flanking FRMD7 (Figure 1). Linkage analysis generated a positive two-point LOD score (2.71 at recombination fraction 0) for the markers DXS1047, DXS8072, and DXS1062 (Supplementary data, Table S1), which further indicated the linkage of the disease in the family with the mutation in FRMD7. We observed the same LOD score with three STRs (DXS1047, DXS8072 and DXS1062), because similar heterozygosity exists in the family.
Figure 1. XLICN pedigree, haplotype analysis.
The squares and circles symbolize males and females, respectively. Dots in the middle of the circle denote that the female is a carrier. Black and white denotes affected and unaffected status, respectively. An individual is identified (ID) by generation number and the aforementioned symbols. Pedigree and haplotype analysis of the XLICN family showing segregation of four microsatellite markers, listed in descending order from the centromeric end.
Mutation analysis and protein structure modeling
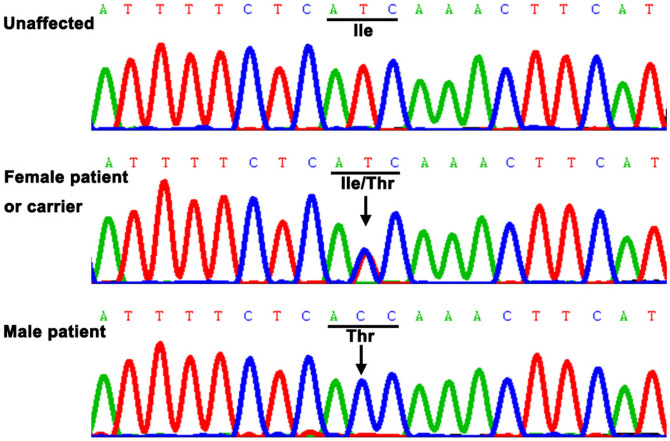
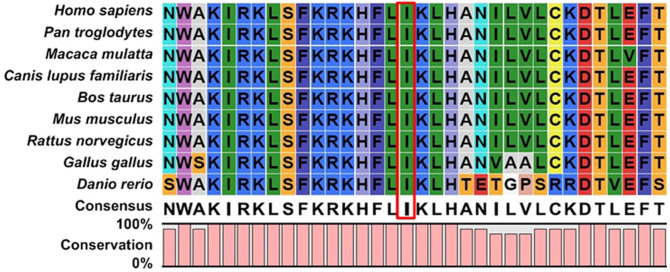
Sequencing FRMD7 revealed a T > C transition (c.719T > C) in exon 8 (Figure 2), which causes a conservative substitution of isoleucine to tyrosine at codon 240 (p.I240T). This mutation co-segregated with all affected individuals and was present in the obligate, non-penetrant female carriers. However, the mutation was not observed in unaffected familial males or 200 control males. We searched the SNPs database and found it doesn't present in any database. In addition, the FRMD7 p.I240T mutant was predicted with high confidence to be “possibly damaging” and “damaging” by Polyphen-2 (score = 0.992) and SIFT (score = 0), respectively. Furthermore, multiple sequence alignments were performed and we found that codon 240, where the mutation (p.I240T) occurred, was located within a phylogenetically conserved region (Figure 3).
Figure 2. DNA sequence chromatograms.
DNA sequence chromatograms of the unaffected members, carriers, and affected members in an XLICN is shown. There is a single base T > C transition in exon 8 of FRMD7 that causes a conservative substitution of isoleucine to tyrosine at codon 240 (p.I240T).
Figure 3. Multiple-sequence alignment of FRMD7 from different species.
Multiple-sequence alignment in FRMD7 from different species revealed that codon 240, where the mutation (p.I240T) occurred, was located within a highly conserved region.
The p.I240T substitution predicted replacement of the hydrocarbon side chain of isoleucine by the hydroxy side-chain of tyrosine, which results in a nonpolar residue change to polar one.
Considering all lines of evidence, the conclusion could be drawn that the p.I240T substitution was a causative mutation rather than a benign polymorphism in linkage disequilibrium with the disease.
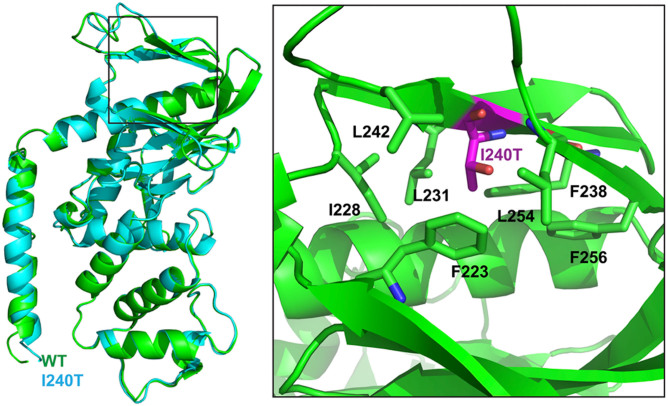
Using Phyre2 program, we predicted that the secondary structure of the wild-type FRMD7 protein, modeled the 3D structure of the wild type FRMD7 protein (1–336), and mapped out I240 in the structure (Figure 4). Mutation of I240T (Purple) introduces a hydrophobic amino acid into a hydrophobic pocket, which may disrupt this hydrophobic core, and cause structural instability.
Figure 4. Structural model of the FRMD7 protein.
The structural analysis of the point mutation I240T. Left, Structure modeling of both WT and I240T, and structure alignment shows that WT (green) and I240T (cyan) share similar overall structure. Right, the hydrophobic pocket to which I240 localizes.
Discussion
Genetically, at least four loci have been proposed for familial idiopathic congenital nystagmus, of which two have been identified for XLICN. From the pedigree described in Figure 1, it is obvious that the inherited trait is X-linked because there was no male-to-male transmission, but there was frequent female-to-male transmission and so far, only one disease-causing gene (FRMD7) was identified. To identify the underlying genetic defect, we first selected four STRs flanking the FRMD7 gene to perform genotyping to confirm that the disease gene is FRMD7. The results of genotyping showed that the affected individuals shared a common haplotype with markers DXS8009, DXS1047, DXS8072 and DXS1042 at Xq25–26.3. We sequenced the coding and flanking intron sequences of FRMD7, which results in identification of a novel mutation in FRMD7.
The human FRMD7 gene comprises 12 exons and encodes a 714-residue polypeptide. Mutations in FRMD7 were first reported in 20064. Since then, many additional mutations have been reported in different populations4,5,6,7,8,9,10,11,12,13,14,15, including two novel mutations from our previous studies6,7. We reviewed the literature and found that 50% and 47% of the XLICN pedigrees have yielded FRMD7 mutations in the Western and Chinese, one of Eastern populations, respectively, which indicates that it is very meaningful to screen for FRMD7 mutations in XLICN families6.
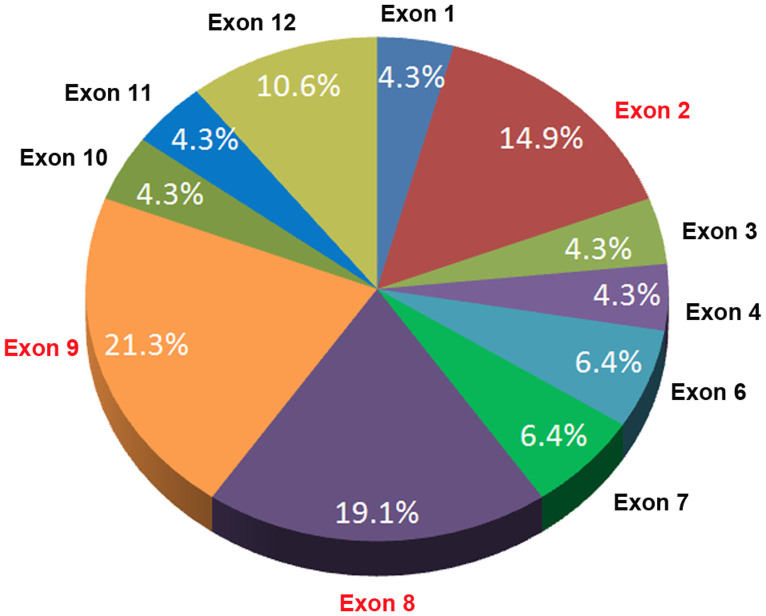
It is still an open question whether there is a mutation-rich exon(s) in the FRMD7. To address it, we reviewed the literature4,5,6,7,8,9,10,11,12,13,14,15, and found forty six mutations of FRMD7 reported (including the present one). Charting these mutations indicates that three of the twelve exons (exons 2, 8, 9) of FRMD7 represent a mutation-rich region (55.3%) for the reported mutations (Figure 5). The 361 bp comprising these three exons represents 11.4% of the 3157 bp of the entire gene. Thus, 55.3% of known mutations have been mapped to a region spanning 11.4% of the sequence of FRMD7 (361/3157). This indicates that more attention should be paid to these three exons when screening for mutations causing XLICN. Meanwhile, the protein sequence encoded by these exons is likely to be the key functional region of FRMD7, which is consistent with a recent review19 and our previous studies16.
Figure 5. FRMD7 mutations and their relative frequencies in XLICN.
About 14.9%, 19.1%, and 21.3% of reported mutations are located in exons 2, 8, and 9 of FRMD7, respectively, indicating that these exons represent a mutation-rich exons (55.3%) for the reported mutations.
In the present study, a novel mutation, p.I240T, in FRMD7 was identified. To dissect the molecular mechanism at the protein structural level, we predicted the secondary structure of human wild-type and mutant FRMD7 proteins using I-TASSER online software. We rationalize that the point mutation (p.I240T) may potentially disrupt FRMD7 function through one of the following mechanisms. First, Mutation of I240T introduces a hydrophobic amino acid into a hydrophobic pocket, which may disrupt this hydrophobic core, and cause structural instability. Secondly, T260 is predicted to be a phosphorylation site, and the p.I240T mutation will introduce a new phosphorylation event, thus altering the regulation of FRMD7 via phosphyorylation17.
In summary, this study adds a novel mutation (p.I240T) to the existing spectrum of FRMD7 mutations with XLICN. Furthermore, we showed the molecular consequence of the p.I240T mutation by protein structural modeling. To our knowledge, this is the first report that it showed the mutation-rich exons in FRMD7, which is a very meaningful improvement for clinical genetic diagnosis. While these data represent significant advances, further functional studies are needed to provide new insights to this inherited ocular disease.
Methods
Clinical evaluations and DNA specimens
This study followed the tenets of the Declaration of Helsinki, and the protocol was approved by the ethics committee at eye hospital, Wenzhou Medical University. Written informed consent was obtained from each participant. If the participants are younger than 18 years old, written informed consents were signed by the parents on behalf of the children.
A five-generation family with non-syndromic congenital nystagmus was recruited. Twenty-two individuals participated in this study, including 10 affected and 12 unaffected individuals (Figure 1). Routine ophthalmologic examinations were performed on all 22 family members.
Genotyping and linkage analysis
Four STRs (Short tandem repeat, DXS8009, DXS1047, DXS8072 and DXS1062) flanking the FRMD7 gene were genotyped in available family members as described previously18 to confirm that mutations in FRMD7 were the causative agent of CN in this family. Pedigree and haplotype data were managed using Cyrillic (version 2.1) software. Linkage analysis was performed with the LINKAGE (version 5.1) package using an X-linked dominant genetic model18 and an assumed gene frequency of 0.0001. The marker order and distances between the markers were taken from the Ensemble Genome browser (http://asia.ensembl.org/index.html).
Mutational analysis
The entire coding exons and splice junctions of the human FRMD7 gene were amplified by PCR using PCR primers and conditions, which have been described previously7. PCR products were purified and directly sequenced on an ABI A3730 Automated Sequencer (Applied Biosystems). When a suspected mutation was found in the proband, it was further confirmed in all other available family members as well as in 200 control males from the same ethnic background. Mutation descriptions follow the nomenclature recommended by the Human Genomic Variation Society.
Amino acid sequences of FRMD7 in various species were retrieved from the NCBI website. Multiple sequence alignments were performed using CLC Sequence Viewer 6 (CLC bio A/S, www.clcbio.com). The possible impact of amino acid substitution on the function of FRMD7 protein was predicted using online tools including PolyPhen-2 (Polymorphism Phenotyping v2, http://genetics.bwh.harvard.edu/pph2)19 and SIFT (Sorting Intolerant From Tolerant, http://sift.jcvi.org)20.
Protein structural modeling
The secondary structure of human wild-type and mutant FRMD7 were predicted using I-TASSER and the results saved in PDB file format. The PDB files obtained for the two samples were then used by Pymol to visualize the structures of these proteins.
Author Contributions
F.G. and J.H.Y. formulated the idea of the paper and supervised the research. Y.H.Z., J.F.Z., X.L.G. and J.H.Y. collected the samples and performed the experiments. X.L.G., X.Z., Z.W. performed data analyses, X.Z. and J.S. performed structural modeling. F.G. wrote the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Supplementary Table
Acknowledgments
The authors thank the family and all subjects for taking part in this study. This work was supported by grants from Natural Science Foundation of China (81201181/H1818, F.G.; 81270999/H1204, Y.H.Z.), Natural Science Foundation of Fujian Province (2010J06010, J.H.Y.), Program for New Century Excellent Talents in Fujian Province University (JA10127, J.H.Y.) and Professor Academic Development Fund of Fujian Medical University (JS12003, J.H.Y.), Zhejiang provincial & Ministry of Health research fund for medical sciences (201339279, F.G.) and Wenzhou Medical University starting grant (QTJ 12011, F.G.).
References
- Kerrison J. B., Vagefi M. R., Barmada M. M. & Maumenee I. H. Congenital motor nystagmus linked to Xq26–q27. Am J Hum Genet. 64, 600–607 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrison J. B. et al. A gene for autosomal dominant congenital nystagmus localizes to 6p12. Genomics. 33, 523–526 (1996). [DOI] [PubMed] [Google Scholar]
- Cabot A. J. et al. A gene for X-linked idiopathic congenital nystagmus (NYS1) maps to chromosome Xp11.4–p11.3. Am J Hum Genet. 64, 1141–1146 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey P. et al. Mutations in FRMD7, a newly identified member of the FERM family, cause X-linked idiopathic congenital nystagmus. Nat Genet. 38, 1242–1244 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna U. et al. Novel homozygous, heterozygous and hemizygous FRMD7 gene mutations segregated in the same consanguineous family with congenital X-linked nystagmus. Eur J Hum Genet. 20, 1032–1036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. et al. A novel frameshift mutation in FRMD7 causing X-linked idiopathic congenital nystagmus. Genet Test. 12, 607–613 (2008). [DOI] [PubMed] [Google Scholar]
- He X. et al. A novel mutation in FRMD7 causing X-linked idiopathic congenital nystagmus in a large family. Mol Vis. 14, 56–60 (2008). [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. Novel mutations of the FRMD7 gene in X-linked congenital motor nystagmus. Mol Vis. 13, 1674–1679 (2007). [PubMed] [Google Scholar]
- Zhang Q., Xiao X., Li S. & Guo X. FRMD7 mutations in Chinese families with X-linked congenital motor nystagmus. Mol Vis. 13, 1375–1378 (2007). [PubMed] [Google Scholar]
- Li N. et al. Five novel mutations of the FRMD7 gene in Chinese families with X-linked infantile nystagmus. Mol Vis. 14, 733–738 (2008). [PMC free article] [PubMed] [Google Scholar]
- Thomas S. et al. Phenotypical characteristics of idiopathic infantile nystagmus with and without mutations in FRMD7. Brain. 131, 1259–1267 (2008). [DOI] [PubMed] [Google Scholar]
- Fingert J. H. et al. Novel intragenic FRMD7 deletion in a pedigree with congenital X-linked nystagmus. Ophthalmic Genet. 31, 77–80 (2010). [DOI] [PubMed] [Google Scholar]
- Du W. et al. A novel frame-shift mutation in FRMD7 causes X-linked idiopathic congenital nystagmus in a Chinese family. Mol Vis. 17, 2765–2768 (2011). [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Identification of a novel FRMD7 splice variant and functional analysis of two FRMD7 transcripts during human NT2 cell differentiation. Mol Vis. 17, 2986–2996 (2011). [PMC free article] [PubMed] [Google Scholar]
- Hu Y. et al. A novel splicing mutation of the FRMD7 gene in a Chinese family with X-linked congenital nystagmus. Mol Vis. 18, 87–91 (2012). [PMC free article] [PubMed] [Google Scholar]
- Watkins R. J. et al. The Role of FRMD7 in Idiopathic Infantile Nystagmus. J. Ophthalmol. 2012, 460956 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts-Henderson J. et al. The nystagmus-associated FRMD7 gene regulates neuronal outgrowth and development. Hum Mol Genet. 19, 342–351 (2010). [DOI] [PubMed] [Google Scholar]
- Gu F. et al. A novel mutation in AlphaA-crystallin (CRYAA) caused autosomal dominant congenital cataract in a large Chinese family. Hum Mutat. 29, 769 (2008). [DOI] [PubMed] [Google Scholar]
- Adzhubei I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods. 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Henikoff S. & Ng P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 4, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table