Abstract
A dual-specificity, paralogue-free Cdc14 phosphatase was located in the nuclei of Beauveria bassiana (filamentous entomopathogen) and functionally characterized. Inactivation of cdc14 caused defective cytokinesis due to multinucleate cells formed in Δcdc14 and 89% decrease of blastospore production, followed by slower growth and a loss of ≥ 96% conidial yield under normal conditions. These defects coincided well with drastic down-regulation of 25 genes required for mitosis and conidiation. Moreover, Δcdc14 became hypersensitive to oxidative, osmotic, and cell wall and mitosis perturbing stresses, and lost 41−70% of conidial thermotolerance, UV-B resistance and virulence, accompanied with transcriptional down-regualtion of various signaling factors and stress-responsive effectors and depressed phosphorylation signals of Hog1 and Slt2 in high-osmolarity glycerol and cell-wall integrity pathways. All changes were well restored by rescuing cdc14. Our findings indicate that Cdc14 vital for the fungal cytokinesis acts as a signaling hub in regulating not only asexual development but multi-stress responses and virulence.
Coordinating nuclear division with growth and cell cycle is vital for eukaryotic development1. Cdc14 is a key regulator of nuclear behavior in the family of dual-specificity phosphatases that dephosphosphorylate the residues of phosphotyrosine and phosphoserine/phosphothreonine2. This phosphatase is highly conserved in almost all eukaryotes and primarily involved in cell division3 as has been elucidated in yeasts4,5. In budding yeast, for instance, Cdc14 can inactivate cyclin dependent kinases (CDKs) at the end of mitosis for cell entry into G1 phase because Cdc14 inactivation may result in overexpressed CDC28-CLB kinase, elongated mitotic spindles and separated chromosomes6,7. Moreover, Cdc14 may act as a hub of five phosphatases and 23 kinases8, including mitosis-associated CDKs and mitogen-activated protein kinase (MAPK) cascades, which constitute the pathways of high-osmolarity glycerol (HOG), cell wall integrity (CWI), filamentous/invasive growth (FIG) and pheromone response (PR)9,10. Thus, Cdc14 is essential not only for cell cycle, checkpoint and metabolism but likely involved in multi-stress responses. In fact, Cdc14 orthologues take similar, but not always identical, parts in the regulation of cell division in some eukaryotes, such as human11 and nematode12. A Cdc14-like phosphatase Clp1 (also known as Flp1) in fission yeast is required for cell entry into mitosis rather than exit and for septation rather than cyclin B destruction2,6,13. Cdc14A and Cdc14B, two Cdc14 paralogues in human14, also function like yeast Cdc143. Deletion of cdc14p from Candida albicans affected late cell-cycle events and morphogenesis, such as large cell aggregates, decreased invasion into solid substrate and impaired hyphal growth15. Knockdown expression of Cdc14 orthologues may result in defective sporulation in Phytophthora1,16. However, most of the previous studies have focused primarily on Cdc14 contribution to nuclear events but paid little attention to possible effects on multi-stress responses and other biological aspects, such as virulence, although Cdc14 may interact with other phosphatases and MAPKs in budding yeast8. The functions of Cdc14 orthologues in filamentous fungi remain poorly understood.
Fungal virulence and multi-stress tolerance are determinant to the biocontrol potential of filamentous entomopathogens against arthropod pests because their cells, such as unicellular conidia, are excellent active ingredients of mycoinsecticides and mycoacaricides17. Genetic engineering of fungal candidate strains for improved virulence18 and stress tolerance19 often requires the understanding of involved molecular mechanisms. Genomic analysis of Beauveria bassiana20 has revealed the existence of a single Cdc14 in the fungal entomopathogen. This study sought to characterize B. bassiana Cdc14 by analyzing multiple phenotypes and transcriptional profiles of its mutants under various stresses. We found that Cdc14 controlled not only cytokinesis but also B. bassiana conidiation, virulence and responses to a wide range of nutritional, chemical and environmental stresses by governing the expressions of many stress-responsive effectors and signaling factors, such as phosphatases, protein kinases and cascaded MAPKs.
Results
Features of cdc14 and deduced protein in B. bassiana
The coding sequence of cdc14 amplified from the wild-type strain B. bassiana ARSEF 2860 (Bb2860 or WT herein) is 1962 bp long, encoding a protein of 642 amino acids (molecular weight: 71.51 kDa; isoelectric point: 8.97). The deduced Cdc14 is characteristic of a highly conserved signature motif typical for the superfamily of protein tyrosine phosphatases and four CDK consensus phosphorylation sites (S/TPXK/R), i.e., S43PRK46, T414RIR417, S534PMR537 and S599 PLR602. There are two to six similar sites in other fungal Cdc14 orthologues but none of them exists in Saccharomyces cerevisiae15. The deduced protein has no paralogue found in B. bassiana and shares 40−100% sequence identity with the fungal/yeast orthologues in NCBI database (Fig. S1A).
As a result of quantitative real-time PCR (qRT-PCR) analysis, the transcript level of cdc14 in Bb2860 was much higher during conidiation than during hyphal growth under normal conditions (Fig. S1B) and greatly elevated by different chemical stresses but less affected by heat shock (Fig. S1C).
A transformant expressing the fusion protein Cdc14::eGFP in Bb2860 was created to show intracellular location of Cdc14. As a result, green fluorescence was emitted from the nuclei of hyphal cells grown in Sabouraud dextrose broth (SDB) for 2 days at 25°C and the expressed Cdc14 was well stained with the nuclear stain DAPI (Fig. 1A). This confirmed the nuclear location of Cdc14 in B. bassiana.
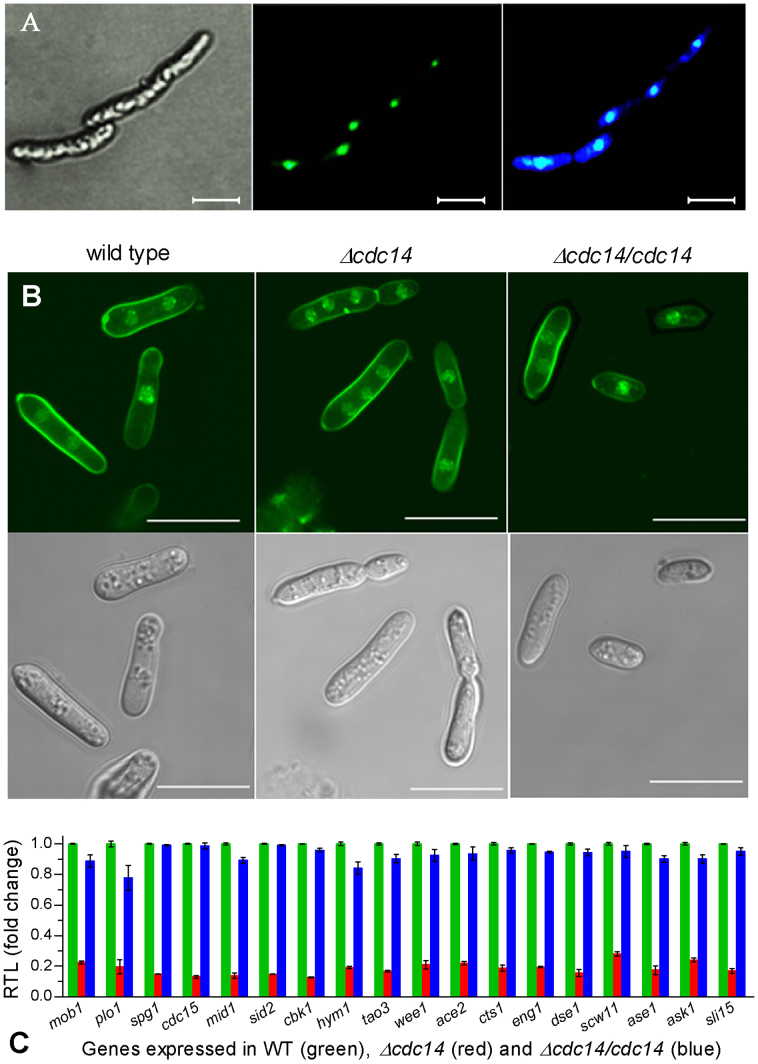
Figure 1. Disruption of cdc14 caused cytokinesis defect in B. bassiana.
(A) Microscopic images of transgenic hyphal cells expressing the fused protein eGFP::Cdc14 in SDB at 25°C. Left: bright image of the cells stained with DAPI. Middle: fluorescent image of the expressed eGFP in nuclei at the excitation/emission wavelengths of 488/507 nm. Right: fluorescent image of the DAPI-stained Cdc14 in nuclei at the excitation/emission wavelengths of 358/360 nm. (B) Bright (upper) and fluorescent (lower) images of hyphal cells stained with both DAPI and calcofluor white (a stain specific to cell wall). Note that cytokinesis was normal (with one or two nuclei per cell) in wild type but abnormal (with three or more nuclei per cell) in Δcdc14. Scale bars: 10 μm. (C) Relative transcript levels (RTL) of 18 cytokinesis-associated genes in Δcdc14 and Δcdc14/cdc14 versus wild type grown for 3 days in SDB at 25°C. Error bars: SD from three cDNA samples assessed via qRT-PCR with paired primers (Table S1).
Disruption of cdc14 caused defects in cytokinesis and asexual development
The disruption and complementation of cdc14 in Bb2860 were verified via PCR and Southern blotting analyses (Fig. S1D) with paired primers (Fig. S1E). Hyphal cells gained by shaking 106 conidia/ml SDB for 3 days at 25°C were stained with both DAPI and calcofluor white. In three batches of > 300 stained cells, Δcdc14 showed three or more nuclei at the mean (± SD) percentage of 13.3 ± 0.9 whereas only one or two nuclei were consistently present in two control strains (Fig. 1B). Strikingly, 18 genes involved in cytokinesis and cell division (Table S1) were all down-regulated by 72−87% in the SDB culture of Δcdc14 versus WT but their transcripts in Δcdc14/cdc14 were well restored to normal WT levels (Fig. 1C). These indicated that cytokinesis was defective in Δcdc14 due to the drastic down-regulation of those genes.
As a result of abnormal cytokinesis in Δcdc14, its blastospore yield was reduced by 89% in the SDB culture compared to the mean yield of 4.58 (± 0.17) × 107 blastospores/ml from two control strains (Fig. 2A). After 7-day growth at 25°C on nutrition-rich SDAY (Sabouraud dextrose agar plus 1% yeast extract), Δcdc14 colonies were 16% smaller than those (~7 cm2 each) of the control strains (F2,6 = 81.8, P < 0.0001). Conidial yields measured from the cultures during days 4−7 were reduced by ≥ 96% in Δcdc14 (Fig. 2B). Microscopic examination of colony samples revealed that, during the incubation, Δcdc14 failed to form zig-zag conidiophore clusters and spore balls typical for B. bassiana as observed in the control strains (Fig. S2). Eight genes essential for hyphal development and conidiation (Table S1) were assessed for their transcript levels in 3-day SDAY cultures via qRT-PCR. All of them except flbA were down-regulated by 24−94% in Δcdc14 versus the control strains (Fig. 2C), a possible cause of the severe defect of its conidiation. All the data indicated that Cdc14 was indispensable for both cytokinesis and asexual development of B. bassiana.
Figure 2. Disruption of cdc14 in B. bassiana caused severe defects in the production of blastospores and conidia.
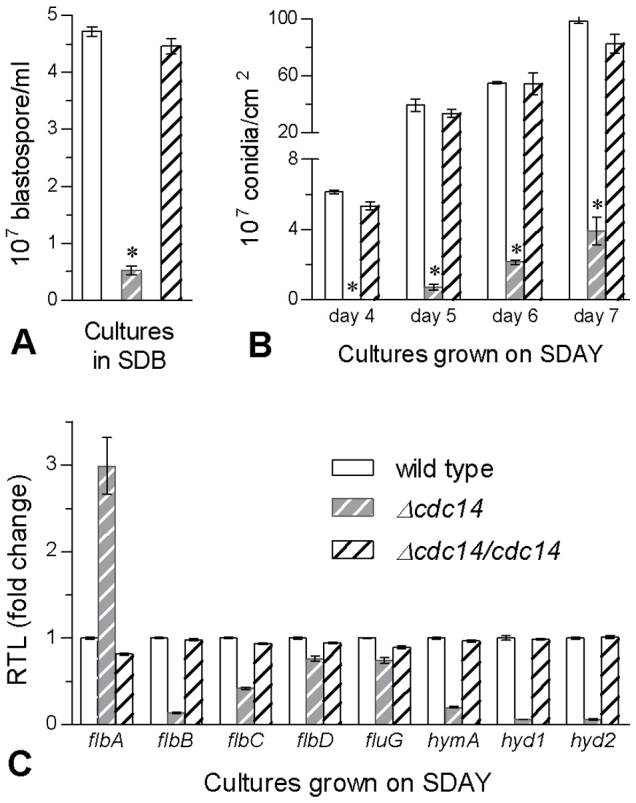
(A) Blastospore yields in liquid SDB cultures initiated with 106 conidia/ml and shaken for 3 days at 25°C. (B) Conidial yields over the period of incubation on SDAY plates at 25°C. The asterisked bar in each group differs significantly from those unmarked (Tukey's HSD, P < 0.05). (C) Relative transcript levels (RTL) of eight conidiation-associated genes in 3-day SDAY cultures at 25°C based on qRT-PCR with paired primers (Table S1). Error bars: SD from three replicates.
Disruption of cdc14 reduced B. bassiana adaptation to environment and host
During 8-day incubation at 25°C on minimal CZA (Czapek agar) and eight CZA-derived media, Δcdc14 grew significantly slower than the control strains (P < 0.01 in F tests). The final colony size of the disruptant was reduced by 46−58% on the mere carbon source of sucrose, glucose, galactose, acetate or glycerol and 35−45% on the mere nitrogen source of NO3−, NO2− or NH4+, and more affected by the starvation of carbon than nitrogen (Fig. 3A).
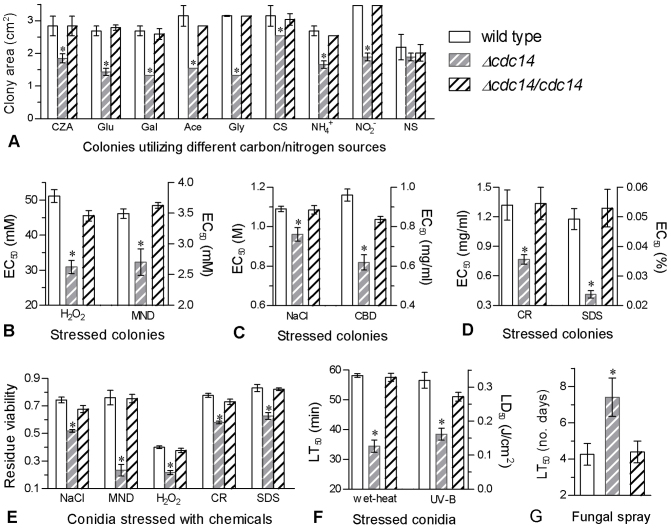
Figure 3. Multi-phenotypic defects observed in Δcdc14 versus two control strains.
(A) Colony sizes after 6-day growth on minimal CZA (3% sucrose as mere carbon and 0.3% NaNO3 as mere nitrogen) and CZA-derived media [altered carbon: glucose (Glu), galactose (Gal), glycerol (Gly), acetate (Ace) or carbon starvation (CS); altered nitrogen: NaNO2, NH4Cl or nitrogen starvation (NS)]. (B −D) Effective concentrations (EC50s) of stressful chemicals required for the suppression of 50% colony growth after 6-day incubation at 25°C on the plates of 1/4 SDAY supplemented with the gradients of H2O2, menadione (MND), NaCl, carbendazim (CBD), Congo red (CR) and SDS respectively. (E) Residue viability of conidia after 24 h incubation at 25°C on germination medium supplemented with NaCl (1.2 M), MND (0.2 mM), H2O2 (4 mM), CR (0.5 mg/ml) or SDS (0.04%). (F) Median lethal time (LT50) for conidial tolerance to wet-heat stress at 45°C and median lethal dose (LD50) for conidial UV-B resistance. (G) LT50 (no. days) for conidial virulence to S. litura second-instar larvae under a uniform spray. The asterisked bar in each group differs significantly from those unmarked (Tukey's HSD, P < 0.05). Error bars: SD from three repeated assays.
Modeling analyses of relative growth trends over the gradients of stressful chemicals added to 1/4 SDAY for 6-day incubation at 25°C generated effective concentration (EC50) of each inhibiting 50% colony growth. Compared to the control strains, Δcdc14 became most sensitive to Congo red and SDS, followed by H2O2, carbendazim, menadione and NaCl in order (Fig. 3B−D).
A sensitive concentration was used to assess the effects of NaCl, H2O2, menadione, Congo red and SDS on conidial germination. Based on residual viability (germination), Δcdc14 was 24−69% less tolerant to the stressful chemicals (Fig. 3E). Conidial thermotolerance and UV-B resistance were reduced by 41% and 50% in Δcdc14, as indicated by lowered LT50 and LD50 (Fig. 3F) respectively. Under a uniform spray of 107 conidia/ml suspension, the LT50 of Δcdc14 against Spodoptera litura second-instar larvae was 3-day longer than those (~4.3 days) from the control strains (Fig. 3G).
All the data indicated that Cdc14 acted as a positive regulator of B. bassiana virulence and responses to nutritional, oxidative, hyperosmotic, cell wall disturbing, fungicidal, thermal and UV-B stresses and contributed greatly to the fungal biocontrol potential.
Disruption of cdc14 altered transcriptional profiles of stress-responsive genes and phosphorylaiton levels of CWI and HOG pathways
Up to 61 stress-responsive genes were assessed for their transcript levels in the 3-day cultures of 1/4 SDAY supplemented with menadione, Congo red, NaCl and carbendazim via qRT-PCR with paired primers (Table S1), including those involved in various signal transduction pathways (STP), such as protein kinases (pkA, pkC and Snf1), Ras GTPases (Ras1 and Ras2), and all MAPK cascades. Under each chemical stress, all effector and/or SIP genes were transcribed at similar levels in two control strains, as those shown for cytokinesis (Fig. 1C) and conidiation (Fig. 2C).
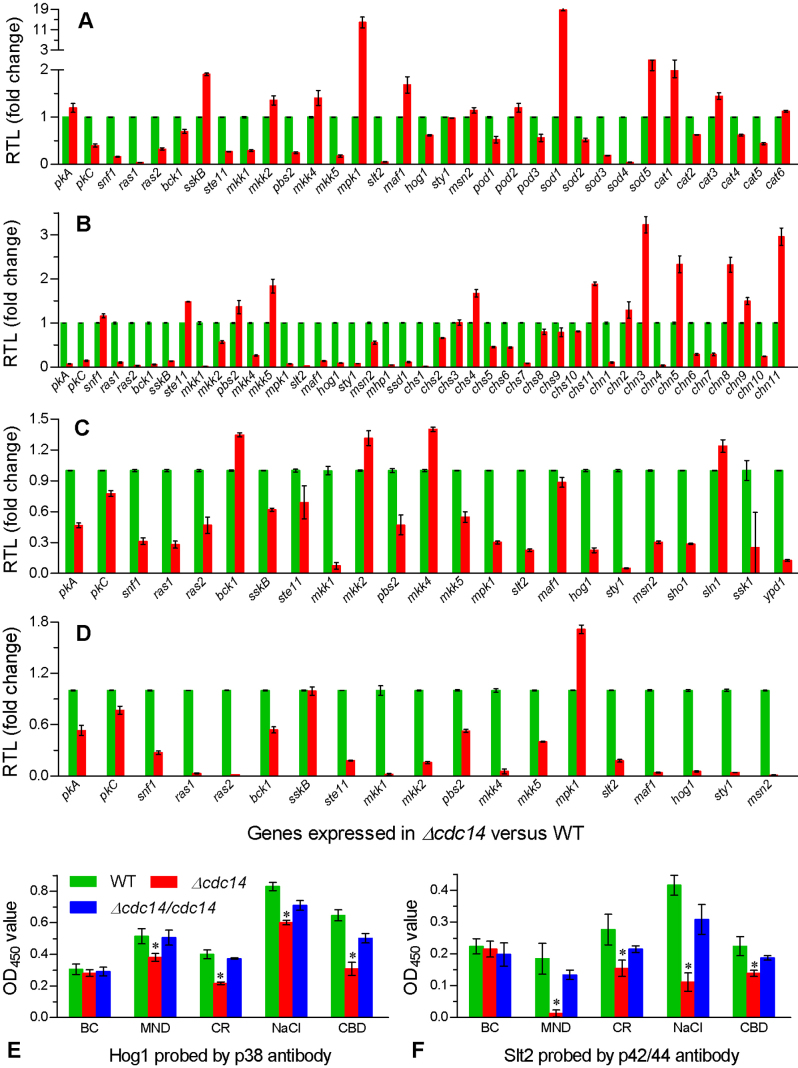
In contrast, many of the examined genes were down-regulated in Δcdc14 versus WT. Transcript levels of 11 STP genes were lowered by 30−96% under oxidative stress (Fig. 4A), including ras1/2, pkC, snf1, mkk5 and the full cascades of HOG (ste11, pbs2 and hog1) and CWI (bck1, mkk1 and slt2). Also, three catalases (Cat2/4/5), two peroxidases (Pod1/3) and three superoxide dismutases (SODs) (Sod2−4) were transcriptionally down-regualted by 32−99% under the same stress. Cell wall disturbance by Congo red resulted in 43−98% reductions of all the STP gene transcripts except those of snf1, ste11, pbs2 and mkk5 (Fig. 4B), accompanied with 20−97% transcriptional down-regulation of nine chitin synthases (Chs1/2/5−10), two cell wall biosynthesis proteins (Mhp1 and Ssd1) and five chitinases (Chn1/4/6/7/10). Under osmotic stress, 13 STP and four osmosensitive genes were down-regulated by 31−95% and 74−94% respectively (Fig. 4C). Inclusion of carbendazim in the culture also down-regulated all the STP genes except sskB and mpk1 (Fig. 4D). Notably, the STP genes repressed by Congo red, NaCl and carbendazim were involved in almost all the MAPK cascades while Msn2, an important transcription factor with multiple stress-responsive elements, was highly responsive to all the chemical cues except menadione.
Figure 4. Disruption of cdc14 altered the transcripts of stress-responsive genes and the phosphorylation of Hog1 and Slt2.
(A−D) Relative transcript levels (RTL) of stress-responsive genes in the 1/4 SDAY cultures of Δcdc14 versus wild type supplemented with MND (menadione 0.2 mM), CR (Congo red 0.5 mg/ml), NaCl (0.8 M) and CBD (carbendazim 0.5 μg/ml) for 3-day growth at 25°C respectively (assessed via qRT-PCR with paired primers in Table S1). (E and F) OD450 values for the respective phosphorylation levels of Hog1 and Slt2 in the protein extracts of fungal cultures grown under the same stresses (BC: unstressed blank control). The extracts were probed with phospho-p38 and phospho-p42/44 antibodies. The asterisked bar in each group differs significantly from those unmarked (Tukey's HSD, P < 0.05). Error bars: SD from three samples.
In ELISA experiments with available phospho-antibodies, the phosphorylation levels of Hog1 and Slt2 as hallmarks of the HOG and CWI pathways of Δcdc14 were significantly lowered by 26−52% (Fig. 4E) and 38−93% (Fig. 4F) under all the chemical stresses respectively. In contrast, their phosphorylation levels did not differ significantly among the tested strains under normal culture conditions (P ≥ 0.58 in F tests) and were restored in the complemented mutant under the chemical stresses.
All the data indicated that Cdc14 was a core phosphatase to activate the expressions of various STP and phenotype-related effector genes under the chemical stresses of different types.
Discussion
As a dual-specificity phosphatase located in the nuclei of B. bassiana, Cdc14 has proved to play a crucial role in cytokinesis, a key step of cell cycle well known in eukaryotes. The nuclear behavior of Cdc14 exerted profound effects not only on asexual development but on virulence and multi-stress tolerances of B. bassiana, thereby contributing greatly to the fungal biocontrol potential. All phenotypic and transcriptional events altered by Cdc14 inactivation are discussed below.
First of all, Cdc14 is required for the cytokinesis and asexual development of B. bassiana because cytokinesis was abnormal in Δcdc14 (Fig. 1B). The hypersensitivity of Δcdc14 cells to the mitosis inhibitor carbendazim (Fig. 3C) implicates that the defective cytokinesis could result from abnormal mitosis, as was evidenced with 13% multinucleate cells formed in our Δcdc14 and reported previously from fission yeast13,21. The abnormal nuclear behavior in Δcdc14 is well supported by the drastic downregulation of 18 genes (Fig. 1C), such as sli15, ask1, fin1 and ase1 involved in the stabilization and extension of anaphase spindle22,23,24, ace2, cts1, eng1, dse1 and scw11 associated with cell wall hydrolysis25,26,27,28,29, and cdc15 required for septum formation21.
Accompanied with the abnormal cytokinesis, Δcdc14 became defective in hyphal growth and much worse in the formation of blastospores and conidia (Fig. 2). Its defective growth could be attributed to less efficient use of carbon and nitrogen sources (Fig. 3A). The severe conidiation defect is consistent with those caused by knockdown cdc14 expression in two oomycetes1,16 and likely attributable to the repressed expression of hymA required for conidiophore development30, hyd1/2 essential for hydrophobin biosynthesis and spore coat rodlet layer assembly during conidiation31, and flbB−D as transcriptional factors required for morphogenesis and asexual development32,33,34,35. The exceptional up-regulation of flbA might contribute to brlA activation for the premature initiation of development36 but its effect on brlA could be shaded by the repressed flbB−D and fluG, another brlA activator37.
Moreover, B. bassiana Cdc14 may act as a hub of cellular signaling network in fungal response to various stresses. The fact that Δcdc14 was hypersensitive to oxidative, osmotic, and cell wall and mitosis perturbing agents (Fig. 3B−E) confirmed for the first time that Cdc14 is a powerful, positive regulator of multi-stress responses in filamentous fungi. The responses of our Δcdc14 to the stresses of different types are well in agreement with increased sensitivity of yeast Cdc14 mutants to both cell wall and mitosis disturbance8,38 but reverse to the decreased osmosensitivity in the yeast mutants8. We found that many of the examined effector genes were down-regulated in Δcdc14 under the chemical stresses of different types (Fig. 4A−D). These effector genes could lower the activities of antioxidant and osmotolerant enzymes/proteins and impaire cell wall composition, thus increasing Δcdc14 sensitivity to each of the stressful chemicals. More importantly, all cascaded kinases in the CWI and HOG pathways of Δcdc14 were transcriptionally down-regulated by menadione and even more by NaCl, Congo red and carbendazim. With available phospho-antibodies, we confirmed that the phosphorylation levels of both Hog1 and Slt2 in Δcdc14 were significantly lowered by all the chemical stresses. Taking together, Cdc14 is a core phosphatase in the signaling network of B. bassiana as shown in budding yeast8. Thus, we propose that the positive regulation of multi-stress responses by Cdc14 is likely achieved by its interplay with multiple STP components because cellular multi-stress responses are well controlled by the pathways of HOG39, CWI40, Ras GTPases41 and pkA/pkC42. However, we could not find more commercial phospho-antibodies to detect phosphorylation signal changes in other MAPK pathways.
Finally, Cdc14 contributes greatly to the biocontrol potential of B. bassiana against arthropod pests. Conidial virulence, thermotolerance and UV-B resistance determinant to the fungal biocontrol potential were greatly decreased by the cdc14 disruption (Fig. 3). Previously, cellular antioxidant capability and cell wall integrity were linearly correlated with either UV-B resistance or virulence of B. bassiana41,43. In this study, Δcdc14 lost large parts of conidial tolerances to both oxidative and cell wall perturbing stresses, an interpretation for the decreased UV-B resistance and virulence. Moreover, Cat2, a single thermosensitive member in the catalase family of B. bassiana44, was transcirptionally down-regulated in Δcdc14 under oxidative stress, partially interpreting the deceased thermotolerance. Therefore, Cdc14 is also a positive regulator of the fungal biocontrol potential.
Methods
Microbial strains and culture conditions
The wild-type strain Bb2860 was used as a recipient of target gene manipulation and cultivated in SDAY (4% glucose, 1% peptone and 1.5% agar plus 1% yeast extract) at 25°C and 12:12 h (light:dark cycle). CZA (3% sucrose, 0.3% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4 and 0.001% FeSO4 plus 1.5% agar) and 1/4 SDAY (SDAY nutrients diluted to 1/4) were used in phenotypic assays of fungal mutants. Escherichia coli DH5α from Invitrogen (Shanghai, China) grown in Luria-Bertani medium at 37°C was used for plasmid propagation. Agrobacterium tumefaciens AGL-1 cultivated at 28°C in YEB medium45 was used as a T-DNA donor for fungal transformation.
Cloning and analysis of cdc14 in B. bassiana
The Cdc14 gene was blasted from the genome of Bb2860 under the NCBI accession ADAH0000000020. Its coding region (Tag code: BBA_07962) was amplified from Bb2860 via PCR and verified by sequencing at Invitrogen. The deduced Cdc14 sequence was compared with the Cdc14 homologues of other fungal and yeast species in NCBI protein database via online structural and phylogenetic analyses.
Cellular localization of Cdc14
To localize Cdc14 in wild-type cells, a cassette with multiple restriction enzyme sites of 5′-PmeI-SpeI-EcoRV-EcoRI-BamHI-3′ (x) was constructed and introduced to the C-terminus of PtrpC promoter via PCR with designed primers using the template of p0380-bar43. The resultant fragment was digested with NotI/BamHI and ligated into pAN52-1N46 to remove original PgpdA, forming pAN52-x. The bar marker cut from p0380-bar was digested with XbaI/HindIII and ligated into the same enzyme sites of pAN52-x, yielding pAN52-x-bar. The coding sequence of cdc14 was amplified from WT cDNA with paired primers (5′-AAAAAACTAGTATGGCTCCCGCCTCCCGCC-3′/5′-AAAAAGAATTCTGCGCCCGACACCTTGCGGG-3′). The reporter gene of enhanced green fluorescent protein (eGFP) was amplified from pAN52-eGFP with designed primers47. The fragments of cdc14 and eGFP were digested with SpeI/EcoRI and EcoRI/BamHI and introduced to the respective enzyme sites in pAN52-x-bar. The resultant plasmid pAN52-cdc14-eGFP-bar was linearized with NotI and then integrated into Bb2860 using the method of blastospore transformation47.
A positive transformant expressing the fusion BbCdc14::eGFP was grown in SDB (i.e., agar-free SDAY) containing106 conidia/ml. After 2-day shaking at 25°C, mycelia were collected, rinsed with PBS buffer (pH 7.0), stained with the nuclear stain DAPI (4′,6′-diamidine-2′-phenylindole dihydrochloride) (Sigma) and visualized for their microscopic images of bright field and fluorescence at the excitation/emission wavelengths of 358/460 nm and 488/507 nm for the location of DAPI-stained Cdc14 and expressed eGFP in cells respectively.
Disruption and complementation of cdc14
The 5′ and 3′ ends (1500 and 1500 bp) of cdc14 were amplified from Bb2860 via PCR with paired primers (Fig. S1E) and LaTaq DNA polymerase (Promega, Madison, MI, USA) and inserted into the respective sites of EcoRI/BamHI and BglII/SpeI in p0380-bar, forming disruption plasmid p0380-bar-cdc14D, which vectors phosphinothricin (PPT) resistant bar gene as marker 1. To rescue the target gene, p0380-sur-gateway43 was used as a backbone. The full-length cdc14 sequence with flanking regions (6450 bp in total) was cloned from Bb2860 with rCdc14-F/R (Fig. S1E) and ligated into the backbone to replace the gateway fragment, yielding the plasmid p0380-sur-cdc14C, which vectors chlorimuron ethyl resistance gene sur as marker 2.
The two plasmids were transformed into Bb2860 and Δcdc14 mutant respectively through Agrobacterium-mediated transformation42. Putative mutant colonies grown on selective medium were screened in terms of their bar and sur resistance to PPT (200 μg/ml) and chlorimuron ethyl (10 μg/ml) and identified sequentially through PCR, qRT-PCR and Southern blotting with paired primers and amplified probe (Fig. S1E). Positive Δcdc14 mutant was evaluated together with WT and Δcdc14/cdc14 (control strains) in triplicate experiments below.
Cytokinesis assay
For WT and mutant strains, 50 ml cultures were initiated by suspending 106 conidia in 1 ml SDB. After 3-day shaking at 25°C, hyphal cells were harvested, washed twice and resuspensed in 50 ml dd-H2O. Three 1 ml aliquots of the suspension were stained with 1 μg DAPI and 200 μl calcofluor white (Sigma), a stain specific to cell wall. The stained cells were microscopically examined for the images of bright field and fluorescence as above to reveal cytokinesis change in Δcdc14 versus control strains and their nuclei were counted.
Measurement of growth and conidiation parameters
Aliquots (1 μl) of 106 conidia/ml suspension were spotted centrally onto the plates (9 cm diameter) of nutrition-rich SDAY, minimal CZA and eight CZA-derived media. The derived media were prepared by deleting 3% sucrose (carbon starvation) or 0.3% NaNO3 (nitrogen starvation) from standard CZA, replacing 3% sucrose with 3% of glucose, galactose, glycerol or acetate (NaAc) as sole carbon source, and replacing 0.3% NaNO3 with 0.3% of NaNO2 or NH4Cl as sole nitrogen source respectively. During 8-day incubation at 25°C and 12:12 h, colony diameters were daily cross-measured to calculate colony area as an index of growth rate.
To assess sporulation capacity of each strain, 100 μl aliquots of 107 conidia/ml suspension (the same below unless specified) were evenly spread on SDAY plates, followed by 7-day incubation at 25°C and 12:12 h. From day 3 onwards, colony discs (5 mm diameter) were daily cut off from the plates and conidia on each disc were washed into 1 ml of 0.02% Tween 80 by vibration. After removal of hyphal debris by filtration, the conidial concentration was determined with microscopic counts and converted to the number of conidia per cm2 colony. During the incubation, fungal mass samples taken from the colonies were examined under microscope to reveal possible morphological changes in their conidiophores and conidia.
Assaying cellular responses to chemical and environmental stresses
SDAY plates overlaid with cellophane were spread with 100 μl aliquots of conidial suspension to produce uniform cultures after 3-day incubation at 25°C. Fungal mass discs (5 mm diameter) cut from the culture of each strain were placed centrally onto the plates (90 mm diameter) of 1/4 SDAY supplemented with the gradients of NaCl (0.4−2 M), menadione (2−8 mM), H2O2 (20−80 mM), Congo red (0.5−3 mg/ml), SDS (0.02−0.12%) and carbendazim (0.4–2 μg/ml) respectively. After 6-day stressful incubation at 25°C and 12:12 h, diameters of all colonies in the control (free of any chemical stress) and stress treatments were cross-measured to compute their net area increases.
Conidial tolerances to oxidation, hyperosmolarity and cell wall disturbance were assayed by spreading 100 μl aliquots of conidial suspension onto the plates of germination medium (GM: 2% sucrose and 0.5% peptone plus 1.5% agar) supplemented with menadione (0.2 mM), H2O2 (4 mM), NaCl (1.2 M), Congo red (1 mg/ml) and SDS (0.04%) respectively. After 24 h incubation at 25°C, conidial germination on each of the plates stressed or not (control) was assessed using three counts of germinated and ungerminated conidia under microscope. Residue viability was calculated as the ratio of percent germination under each stress over that in the control. Conidial tolerances to wet-heat stress of 15−120 min at 45°C and UV-B irradiation (weighted 312 nm) of 0.1–0.8 J/cm2 in Bio-Sun++ chamber (Vilber Lourmat, Marne-la-Vallée, France) were assayed using our previous protocols48,49. After exposure to a given intensity of heat or UV-B stress, conidia were incubated for 24 h at 25°C under saturated humidity and percent germination was determined using microscopic counts.
Bioassay of fungal virulence
A standardized cabbage leaf disc system19 was used to bioassay the virulence of each strain to the second-instar larvae of S. litura. Briefly, batches of 30−40 larvae on a cabbage leaf disc were separately exposed to a spray of 1 ml conidial suspension (treatment) or 0.02% Tween 80 (control) in automatic Potter Spray Tower (Burkard Scientific Ltd, Uxbridge, UK) at uniform working pressure. After spray, all larvae were reared in Petri dishes (with leaf discs changed daily for their feeding) for 8 days at 25°C and 12:12 h and monitored daily for mortality records.
Transcriptional analysis of phenotype-associated genes
Aliquots of 100 μl conidial suspension were spread onto cellophane-overlaid plates of 1/4 SDAY alone (control) or supplemented with NaCl (0.8 M), menadione (0.2 mM), carbendazim (0.5 μg/ml), and Congo red (0.5 mg/ml) respectively, followed by 3-day incubation at 25°C and 12:12 h. Total RNAs were extracted from the stressed and unstressed (control) cultures with RNAiso™ Plus Reagent (TaKaRa, Dalian, China) and reversely transcribed into cDNAs with PrimeScript® RT reagent Kit (TaKaRa). Each cDNA (10 × dilution) was used as template to assess the transcript levels of 90 phenotype-associated genes via qRT-PCR with paired primers (Table S1) under the action of SYBR® Premix Ex TaqTM (TaKaRa). The transcript level of each gene in cDNA was assessed using the 2−ΔΔCT method50 and the fungal 18 S rRNA as internal standard. The relative transcript level of each gene under a given stress was calculated as the ratio of its transcript in each mutant over that in WT.
Assaying the phosphorylation levels of Hog1 and Slt2
The method of enzyme-linked immunosorbent assay (ELISA)51 was adopted to assess the phosphorylation levels of Hog1 and Slt2 in the HOG and CWI pathways of Δcdc14 and control strains using their commercial phospho-antibodies respectively. Protein extracts from the 4-day cultures grown on 1/4 SDAY plates under normal conditions and chemical stresses were suspended in 1 ml PBS (pH 7.5). After 10 min homogenization on ice, the mixture was centrifuged at 10,000 g for 10 min at 4°C and the supernatant was centrifuged for another 10 min. The protein concentration in each extract was assessed with the BCA Protein Assay Kit (KeyGen Biotech, Nanjing, China). For ELISA, 100 μl aliquots of each supernatant (5 μg/ml 0.05 M carbonate buffer, pH 9.6) were uploaded onto 96-well ELISA plate (Jet Biofil, Guangzhou, China) for overnight blocking at 4°C. Subsequently, the plate was repeatedly washed with PBS and added with 100 μl confining liquid [1% bovine serum albumin (BSA) in 0.05 M carbonate buffer] for 1 h incubation at 37°C. Washed again, the plate reacted with the rabbit phospho-p38 antibody (200 × dilution) against Hog1 or phospho-p44/42 antibody (500 × dilution) against Slt2 (Cell Signaling Technology, Boston, MA, USA) for 1 h at 37°C and then with the second antibody goat anti-rabbit IgG-HRP (Sigma) for 1 h at 37°C, followed by rinsing. Finally, 100 μl aliquots of TMB/H2O2 substrate (Amresco, Solon, OH, USA) were added to the wells on the plate for 20 min incubation at 37°C and the reaction was terminated with 2 M H2SO4 (100 μl per well). The phosphorylation level of Hog1 or Slt2 in each supernatant sample was measured as an OD reading at 450 nm (i.e., OD450). Each assay included three samples as replicates.
Data analyses
The ratio of colony size or conidial germination rate under a given stress over that in the control was defined as relative viability (VR). For each of the tested strains, the VR trends over the concentrations (C) of each chemical, the time lengths (T) of wet-heat stress, and the doses (D) of UV-B irradiation were fitted to the equation VR = 1/[1 + exp(a + bx)], where x is C, D or T, a and b are parameters to be estimated. When VR = 0.5, the fitted equations gave solutions (−a/b) to effective concentration (EC50) of each stressful chemical required to suppress 50% colony growth and median lethal responses of conidia to heat (LT50, min) and UV-B (LD50, J/cm2) stresses. Time-mortality trends from the bioassay were subjected to probit analysis, yielding LT50 (no. days) of each strain against the larvae. All the solutions or observations from three repeated assays were differentiated among the tested strains by one-way analysis of variance.
Author Contributions
M.G.F. and J.W. designed the research. J.W. and M.G.F. analyzed the data. J.W., J.L., Y.H. and S.H.Y. performed the experiments. M.G.F. and J.W. wrote the paper. All authors reviewed the manuscript.
Supplementary Material
Figure S1-2 and Table S1
Acknowledgments
This work was financially supported by the Ministry of Science and Technology of China (Grant No.: 2011AA10A204) and the Natural Science Foundation of China (Grant Nos.: 31270537 and 31021003).
References
- Ah Fong A. M. & Judelson H. S. Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus-like oomycete Phytophthora infestans. Mol Microbiol 50, 487–494 (2003). [DOI] [PubMed] [Google Scholar]
- Mocciaro A. & Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci 123, 2867–2876 (2010). [DOI] [PubMed] [Google Scholar]
- Cho H. P. et al. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol Cell Biol 25, 4541–4551 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D. & Amon A. At the interface between signaling and executing anaphase -Cdc14 and the FEAR network. Genes Dev 18, 2581–2595 (2004). [DOI] [PubMed] [Google Scholar]
- Krapp A., Gulli M. P. & Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol 14, R722–R730 (2004). [DOI] [PubMed] [Google Scholar]
- Cueille N. et al. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J Cell Sci 114, 2649–2664 (2001). [DOI] [PubMed] [Google Scholar]
- Yuste-Rojas M. & Cross F. R. Mutations in CDC14 result in high sensitivity to cyclin gene dosage in Saaccharomyces cerecisiae. Mol Gen Genet 263, 60–72 (2000). [DOI] [PubMed] [Google Scholar]
- Breitkreutz A. et al. A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H. & Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem 136, 267–272 (2004). [DOI] [PubMed] [Google Scholar]
- Rispail N. et al. Comparative genomics of MAP kinase and calcium–calcineurin signaling components in plant and human pathogenic fungi. Fungal Genet Biol 46, 287–298 (2009). [DOI] [PubMed] [Google Scholar]
- Kaiser B. K., Zimmerman Z. A., Charbonneau H. & Jackson P. K. Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol Biol Cell 13, 2289–2300 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Glotzer M., Gartner A. & Nigg E. A. The CeCDC-14 phosphatase is required for cytokinesis in the Caenorhabditis elegans embryo. J Cell Biol 158, 901–914 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S. et al. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol 11, 931–940 (2001). [DOI] [PubMed] [Google Scholar]
- Li L. et al. A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J Biol Chem 272, 29403–29406 (1997). [DOI] [PubMed] [Google Scholar]
- Clemente-Blanco A. et al. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J Cell Sci 119, 1130–1143 (2006). [DOI] [PubMed] [Google Scholar]
- Zhao W. et al. Transient silencing mediated by in vitro synthesized double-stranded RNA indicates that PsCdc14 is required for sporangial development in a soybean root rot pathogen. Sci China Life Sci 54, 1143–1150 (2011). [DOI] [PubMed] [Google Scholar]
- de Faria M. R. & Wraight S. P. Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol Control 43, 237–256 (2007). [Google Scholar]
- Qin Y. et al. Integration of insecticidal protein vip3Aa1 into Beauveria bassiana enhances fungal virulence to Spodoptera Litura larvae by cuticle and per os infection. Appl Environ Microbiol. 76, 4611–4618 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. Q., Li F., Ying S. H. & Feng M. G. Additive contributions of two manganese-cored superoxide dismutases(MnSODs) to antioxidation, UV tolerance and virulence of Beauveria bassiana. PloS One 7, e30298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G. H. et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep 2, 483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C. & Simanis V. The Schizosaccharomyces pombe cdc14 gene is required for septum formation and can also inhibit nuclear division. Mol Biol Cell 4, 531–539 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G. & Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302, 2120–2124 (2003). [DOI] [PubMed] [Google Scholar]
- Higuchi T. & Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433, 171–176 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J. & Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol 177, 981–993 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury E. L. & Morgan D. O. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol 9, 106–112 (2007). [DOI] [PubMed] [Google Scholar]
- Baladrón V. et al. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot Cell 1, 774–786 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A., Chin T. E. & Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107, 739–750 (2001). [DOI] [PubMed] [Google Scholar]
- Doolin M. T., Johnson A. L., Johnston L. H. & Butler G. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol Microbiol 40, 422–432(2001). [DOI] [PubMed] [Google Scholar]
- Nelson B. et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell 14, 3782–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karos M. & Fischer R. Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol Gen Genet 260, 510–521 (1999). [DOI] [PubMed] [Google Scholar]
- Zhang S. Z., Xia Y. X., Kim B. & Keyhani N. O. Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Mol Microbiol 80, 811–826 (2011). [DOI] [PubMed] [Google Scholar]
- Arratia-Quijada J., Sánchez O., Cazzocchio C. & Aguirre J. FlbD, a Myb transcription factor of Aspergillus nidulans is uniquely involved in both asexual and sexual differentiation. Eukaryot Cell 11, 1132–1142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxebeste O. et al. The bZIP-type transcription factor FlbB regulates distinct morphogenetic stages of colony formation in Aspergillus nidulans. Mol Microbiol 73, 775–789 (2009). [DOI] [PubMed] [Google Scholar]
- Kwon N. J. et al. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol Microbiol 77, 1203–1219 (2010). [DOI] [PubMed] [Google Scholar]
- Xiao P., Shin K. S., Wang T. & Yu J. H. Aspergillus fumigatus flbB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production. Eukaryot Cell 9, 1711–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. N. & Adams T. H. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol Microbiol 114, 323–34 (1994). [DOI] [PubMed] [Google Scholar]
- Seo J. A., Guan Y. & Yu J. H. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172, 1535–1544 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., Rajagopalan S. & McCollum D. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev Cell 7, 755–762 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang Y. J. et al. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl Environ Microbiol 75, 3787–3795 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. D. et al. The MAP kinase Bbslt2 controls growth, conidiation, cell wall integrity, and virulence in the insect pathogenic fungus Beauveria bassiana. Fungal Genet Biol 49, 544–555 (2012). [DOI] [PubMed] [Google Scholar]
- Xie X. Q., Guan Y., Ying S. H. & Feng M. G. Differentiated functions of Ras1 and Ras2 proteins in regulating the germination, growth, conidiation, multi-stress tolerance and virulence of Beauveria bassiana. Environ Microbiol 15, 447–462 (2013). [DOI] [PubMed] [Google Scholar]
- Fang W. G., Pava-ripoll M., Wang S. B. & St. Leger R. J. Protein kinase A regulates production of virulence determinants by the entomopathogenic fungus, Metarhizium anisopliae. Fungal Genet Biol 46, 277–285 (2009). [DOI] [PubMed] [Google Scholar]
- Xie X. Q. et al. A new manganese superoxide dismutase identified from Beauveria bassiana enhances virulence and stress tolerance when overexpressed in the fungal pathogen. Appl Microbiol. Biotechnol 86, 1543–1553 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Z. L., Zhang L. B., Ying S. H. & Feng M. G. Catalases play differentiated roles in the adaptation of a fungal entomopathogen to environmental stresses. Environ Microbiol 15, 409–418 (2013). [DOI] [PubMed] [Google Scholar]
- Fang W. G. et al. Agrobacterium tumefaciens- mediated transformation of Beauveria bassiana using an herbicide resistance gene as a selection marker. J Invertebr Pathol 85, 18–24 (2004). [DOI] [PubMed] [Google Scholar]
- Punt P. J. et al. Transformation of Aspergillus based on the hygromycin B resistance marker form Escherichia coli. Gene 56, 17–124 (1987). [DOI] [PubMed] [Google Scholar]
- Ying S. H. & Feng M. G. Novel blastospote-based transformation system for integration of phosphinothricin resistance and green fluorescence protein genes into Beauveria bassiana. Appl Microbiol Biotechnol 72, 206–210 (2006). [DOI] [PubMed] [Google Scholar]
- Huang B. F. & Feng M. G. Comparative tolerances of various Beauveria bassiana isolates to UV-B irradiation with a description of a modeling method to assess lethal dose. Mycopathologia 168, 145–152 (2009). [DOI] [PubMed] [Google Scholar]
- Li J. & Feng M. G. Intraspecific tolerance of Metarhizium anisopliae conidia to the upper thermal limits of summer with a description of a quantitative assay system. Mycol Res 113, 93–99 (2009). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Kashyap R. S. et al. Diagnosis of tuberculosis in an Indian population by an indirect ELISA protocol based on detection of Antigen 85 complex: a prospective cohort study. BMC Infect Dis 7, 74 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1-2 and Table S1