Abstract
Although the targets of most miRNAs have not been experimentally identified, microRNAs (miRNAs) have begun to be extensively characterized in physiological, developmental and disease-related contexts in recent years. Thus far, mainly computational approaches have been employed to predict potential targets for the large majority of miRNAs. Although miRNAs exert a major influence on the efficiency of translation of their targets in animals, most studies describing experimental identification of miRNA target genes are based on detection of altered mRNA levels.. miR-143 is a miRNA involved in tumorigenesis in multiple types of cancer, smooth muscle cell fate and adipocyte differentiation. Only a few miR-143 targets are experimentally verified, so we employed a SILAC-based quantitative proteomic strategy to systematically identify potential targets of miR-143. In total, we identified > 1,200 proteins from MiaPaCa2 pancreatic cancer cells, of which 93 proteins were downregulated > 2-fold in miR-143 mimic transfected cells as compared to controls. Validation of 34 of these candidate targets in luciferase assays showed that 10 of them were likely direct targets of miR-143. Importantly, we also carried out gene expression profiling of the same cells and observed that the majority of the candidate targets identified by proteomics did not show a concomitant decrease in mRNA levels confirming that miRNAs affect the expression of most targets through translational inhibition. Our study clearly demonstrates that quantitative proteomic approaches are important and necessary for identifying miRNA targets.
Introduction
MicroRNAs (miRNAs) are small, non-coding, single-stranded RNAs that control protein expression at the post-transcriptional level. miRNAs are initially transcribed by RNA Polymerase II and further processed by Drosha and Dicer, giving rise to ~22-nucleotide mature miRNAs. Mature miRNAs then bind to complementary sequences in 3’UTRs of target mRNA transcripts in the RNA-induced silencing complex (RISC). When a miRNA binds to its cognate mRNA with perfect complementarity, the mRNA will be degraded by RISC. However, most miRNAs in animals bind to their targets with imperfect complementarity. Six or seven nucleotides at the 5′ end of the miRNA are referred to as the ‘seed region’ for binding.1 The imperfect complementarity usually results in translational inhibition of the target mRNA with smaller effects at the level of mRNA degradation.2,3
It is critical to identify miRNA targets to understand functions of miRNAs under normal and/or diseased conditions. Thus far, only a relatively small subset of predicted miRNA targets have been experimentally validated,4 although a number of programs for predicting miRNA targets are available.5–7 High-throughput approaches based on gene expression microarrays have been used to experimentally identify miRNA targets.8 However, such approaches are likely to overlook targets regulated solely through translational repression. Proteomic approaches are promising to identify miRNA targets because protein abundance is used as the direct readout.9–11 In a previous study, we have successfully employed a quantitative proteomic approach using the iTAQ methodology to identify targets of miR-21 in breast cancer cells.11 Our previous results demonstrated that miR-21 affects the expression of many of its targets through translational inhibition instead of mRNA degradation.
In the present study, we sought to identify miR-143 targets at the protein level using a quantitative proteomic approach. miR-143 has been found to be strongly associated with tumorigenesis. For example, miR-143 is frequently observed to be downregulated in colorectal12 and gastric cancers,13 chronic lymphocytic leukemias and B-cell lymphomas.14 Overexpression of miR-143 in colorectal cancer cell lines reduces cell viability and increases sensitivity to 5-fluorouracil treatment.15 We also recently reported that miR-143 is frequently downregulated in pancreatic cancer cells.16 miR-143 has also been reported to be associated with smooth muscle cell fate. For example, Cordes et al. found that miR-143 was a transcriptional target of myocardin and other transcriptional factors involved in smooth muscle cell fate.17 It was downregulated in injured vessels containing less differentiated smooth muscle cells. miR-143 knockout mice are viable and do not display gross macroscopic abnormalities.18,19 However, neointima formation in miR-143 knockout mice was significantly blocked in response to vascular injury due to abnormalities of activity of serum response factor and actin dynamics regulated by miR-143.19 miR-143 has also been found to play a role in adipocyte differentiation.20–23 Esau et al reported that the expression of miR-143 was elevated in differentiating adipocytes and that inhibition of miR-143 could suppress differentiation of adipocytes.23 Ectopically expressed miR-143 in preadipocyte 3T3-L1 cells has been found to accelerate adipogenesis.20
Although miR-143 is clearly important in several biological processes, only a few targets of miR-143 have been described thus far. Mitogen-activated protein kinase 7 (MAPK7), also called extracellular signal-regulated kinase 5 (ERK5) was the first candidate target of miR-143 identified in colon cancer cells24 and malignant B-cells.14 Overexpression of miR-143 in both DLD-1 colon cancer cells and Raji B lymphoma cells resulted in significantly decreased expression of MAPK7 at the protein level, but not at the mRNA level.24 Cordes et al found that miR-143 repressed proliferation of vascular smooth muscle cells by targeting ELK1, member of EST oncogene family (ELK1) at the protein level, but not at the mRNA level.17
Stable isotope labeling with amino acids in cell culture (SILAC) (http://www.silac.org) is an accurate labeling strategy for mass spectrometry-based proteomics.25 It has been used to identify c-Src tyrosine kinase substrates in platelet-derived growth factor receptor signaling26 and to study effects of kinase inhibitors on signaling networks.27 Using a SILAC-based proteomic strategy in combination with strong cation exchange (SCX) chromatography, we carried out global proteomic profiling to identify targets of miR-143. In total, we have identified 94 putative targets of miR-143, including 93 downregulated and one upregulated protein following transfection of a miR-143 mimic. Using luciferase assays, we demonstrate that a subset of these putative targets represents direct targets of miR-143. Further studies for transcriptional profiling confirm that the many of the targets are regulated through translational inhibition without affecting the mRNA levels.
Results
The mechanisms by which miRNAs regulate their targets include both translational inhibition and mRNA degradation. However, most studies thus far have used strategies to detect mRNA changes instead of protein changes to identify miRNA targets. Here, we describe a SILAC-based quantitative proteomic strategy to identify candidate targets of miR-143.
Identification of miR-143 targets using a quantitative proteomic approach
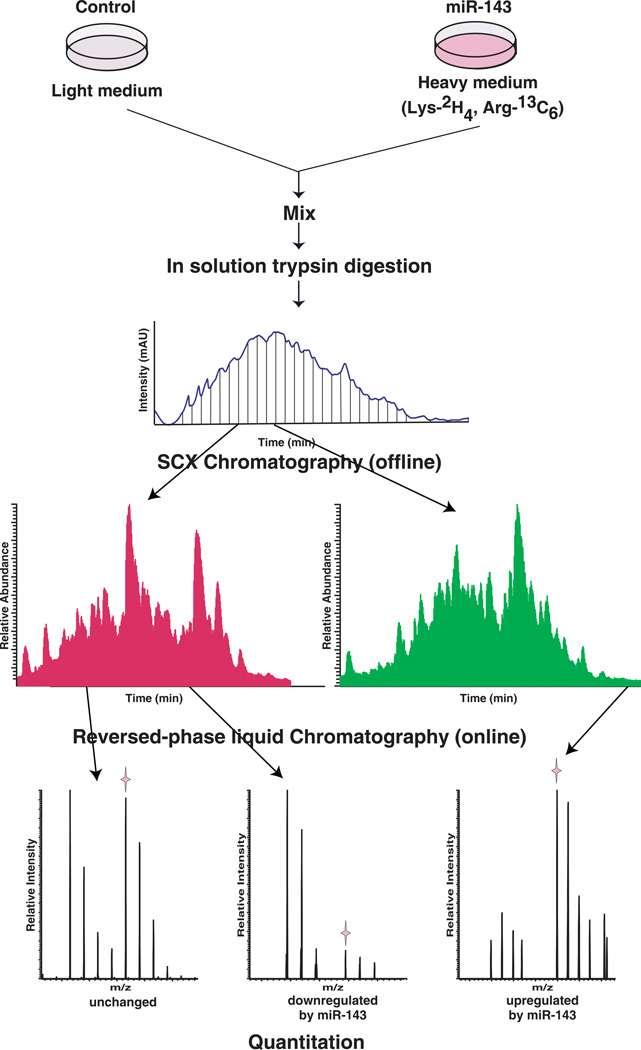
Because miR-143 is expressed at low levels in the pancreatic cancer cell line MiaPaCa2,16 we reasoned that overexpression of miR-143 would decrease the expression of its cognate targets in this setting. In order to overexpress miR-143, we transfected the cells with a miR-143 mimic oligo. An oligo without any specificity to mammalian miRNAs was used as a control. While miR-143 mimic was transfected to cells stably adapted in DMEM medium containing 2H4 lysine and 13C6 arginine (heavy state), control oligo was transfected to cells grown in normal DMEM medium (light state) as illustrated in Figure 1. Equals amount of total proteins were mixed from heavy cells with miR-143 mimic and light cells with control oligo, as illustrated in Figure 1. The mixture was digested with trypsin and fractionated into 25 fractions using SCX chromatography. LC-MS/MS analysis of the fractions from SCX was carried out using a high-resolution Fourier Transform mass spectrometer (LTQ-Orbitrap XL ETD).
Figure 1. Strategy for SILAC-based proteomic analysis.
miR-143 mimic was transfected in MiaPaCa2 pancreatic cancer cells stably adapted in DMEM medium containing 2H4 lysine and 13C6 arginine (heavy state) while a control oligo was transfected to the same cells grown in normal DMEM medium (light state). After 48 hours, total proteins were harvested and protein lysates were mixed and digested with trypsin. The mixed peptides were fractionated by strong cation exchange (SCX) chromatography. Twenty-five fractions were cleaned and analyzed by liquid chromatography based tandem mass spectrometry (LC-MS/MS). The fold changes were calculated from the ratio of intensity of peptide MS spectrum obtained from samples with miR-143 mimic to those with control oligo.
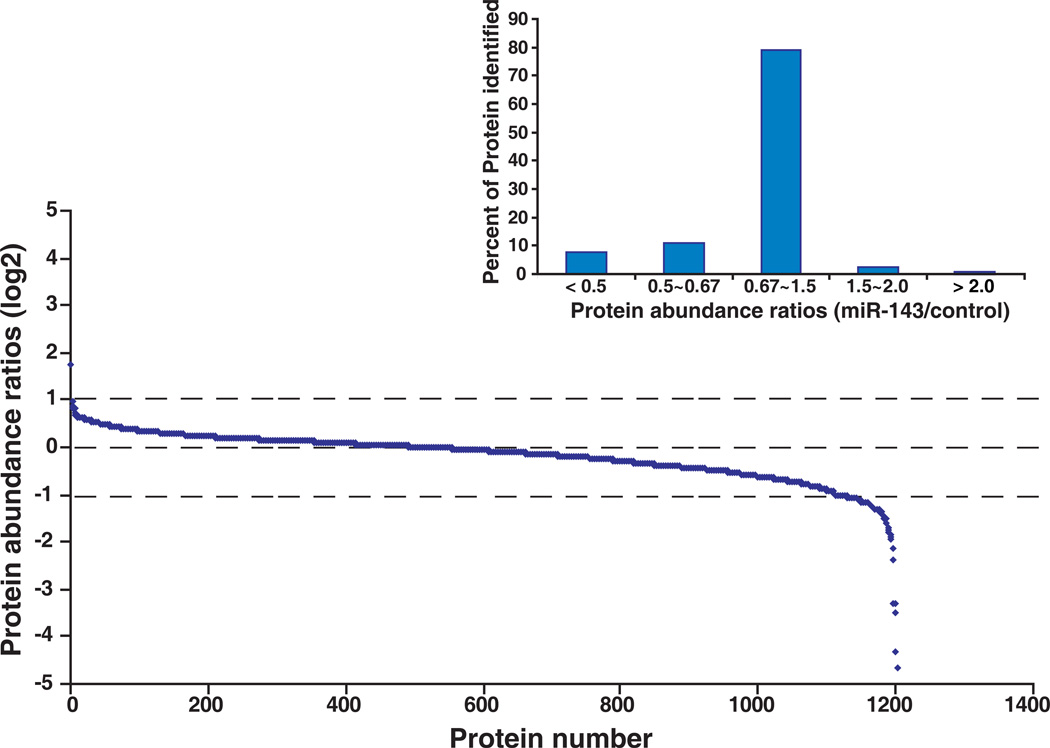
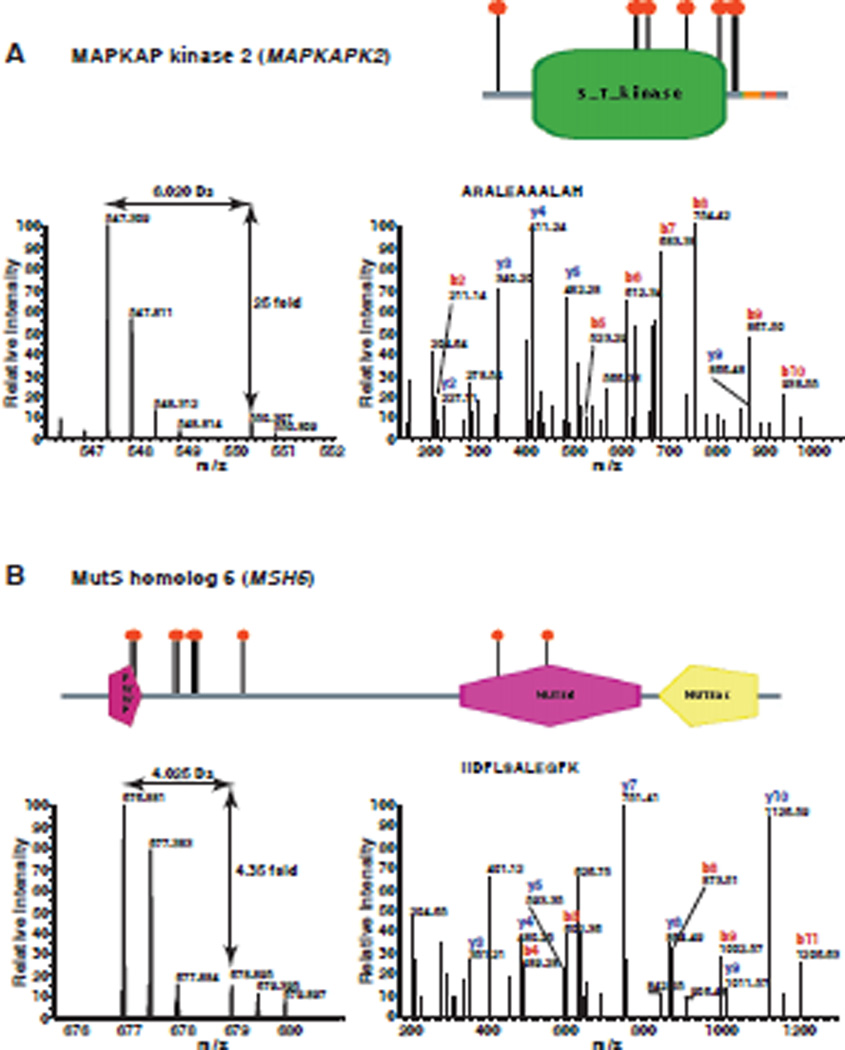
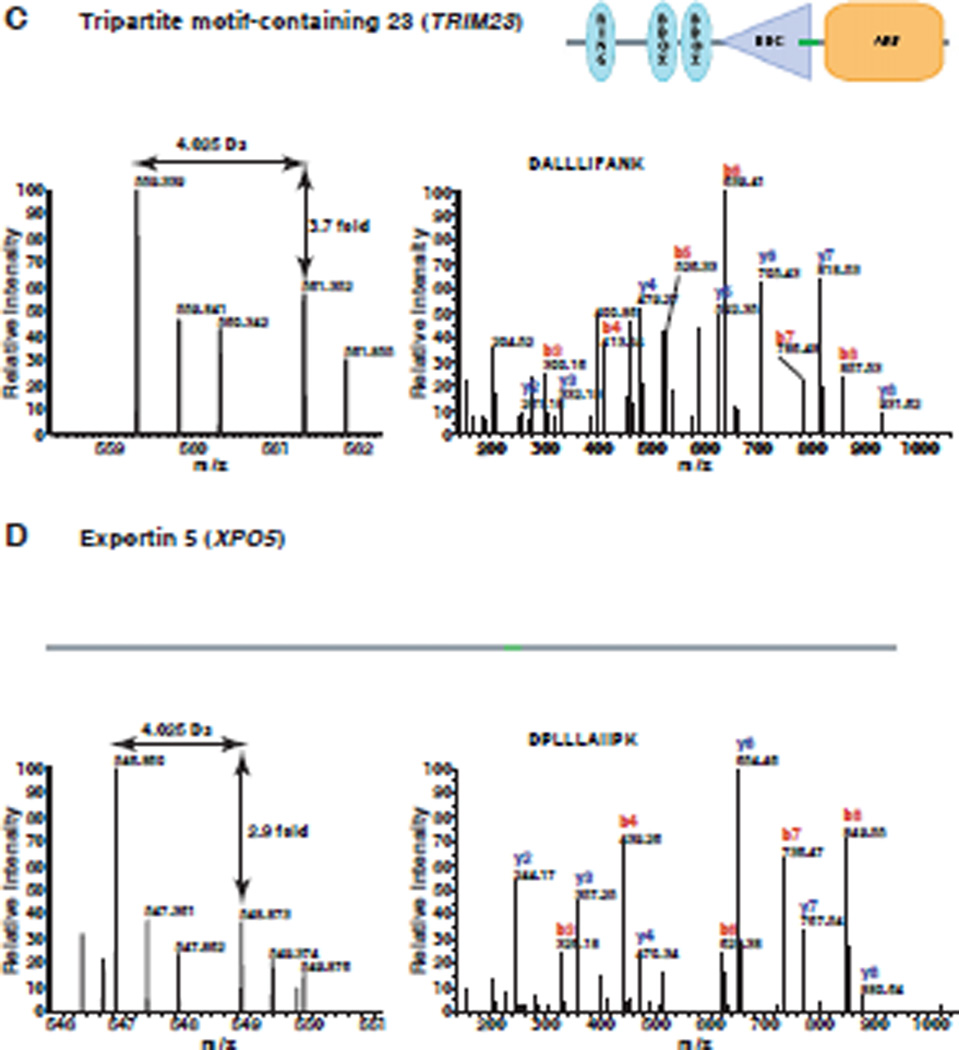
A total of 1,202 proteins were confidently identified and quantitated (Supplementary table 1). Relative changes in the levels of identified proteins were determined by the ratio of intensity from miR-143 transfected samples to control oligo transfected samples. We observed a number of proteins with > 2-fold change after overexpression of miR-143. With this cut-off, we found 93 (7.7 %) downregulated proteins and only one upregulated protein following transfection of the miR-143 mimic. The distribution of ratios of all quantitated proteins is shown in Figure 2. Many studies have shown that miRNAs downregulate their targets and the preponderance of downregulated proteins in our data also confirms this. Representative MS and MS/MS spectra of 4 putative targets, MAPKAP kinase 2, MutS homolog 6, tripartite motif-containing 23 and exportin 5 are shown in Figures 3 and 4. All of the four targets are firstly identified as putative targets of miR-143.
Figure 2. Ratio distribution of all quantitated proteins.
In total, 1,202 proteins were identified and quantitated using the SILAC-based proteomic approach. Relative changes of identified proteins are demonstrated by the ratio of intensity from miR-143 transfected sample to control oligo transfected sample. The distribution of the protein ratio is illustrated and the right insert shows percent of proteins with different ratio ranges.
Figure 3. MS/MS spectra of representative targets identified by proteomics.
Panels A–D show the MS and MS/MS spectra of representative peptides from MAPKAP kinase 2 (MAPKAPK2), MutS homolog 6 (MSH6), Tripartite motif-containing 23 (TRIM23) and exportin 5 (XPO5), respectively. The left panel in each case shows the MS spectrum for quantitation of the peptide. The right panel in each case shows the MS/MS spectrum for identification of the peptide. The domain structure of each protein is taken from Human Protein Reference Database (www.hprd.org)43.
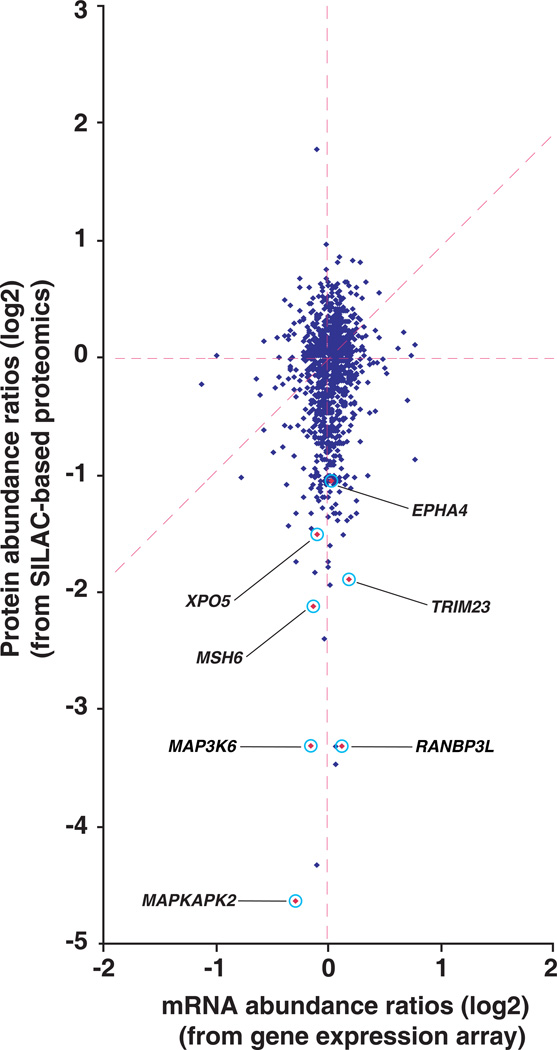
Figure 4. Comparison of proteome and transcriptome.
The dataset containing 1,117 genes, which have quantitative data at both protein and mRNA levels was used to generate a scatter plot of the protein ratios (miR-143 mimic/control oligo) versus mRNA ratios (miR-143 mimic/control oligo) are shown. Select interesting molecules MAPKAP kinase 2 (MAPKAPK2), mitogen-activated protein kinase kinase kinase 6 (MAP3K6), RAN binding protein 3-like (RANBP3L), MutS homolog 6 (MSH6), Tripartite motif-containing 23 (TRIM23), exportin 5 (XPO5) and ephrin receptor EphA4 (EPHA4) are highlighted in the figure.
Sequence analysis of candidate targets identified through proteomics
In order to find miR-143 seed matches in transcripts of candidate targets identified by proteomics, we searched nucleotide sequence databases for the transcripts of all genes identified by proteomics and obtained their 3’UTR, 5’UTR and coding sequences. Using mirMatcher program, these sequences were searched for miR-143 seed matches. The distribution of perfect 6-mer seed matches (complementary to nucleotides 2–7 at the 5’end of miR-143) is shown in Table 2. Noticeably, the 6-mer seed matches exist in 31% of 3’UTRs of downregulated genes (ratio ≤ 0.5). Only 17 % of 3’UTRs of unchanged genes contains the 6-mer seed matches. The difference between downregulated (ratio ≤ 0.5) and unchanged (ratio = 0.5~2) reaches significance (p < 0.01) when evaluated by chi-square test. These results indicate that the 6-mer seed matches are significantly enriched in 3’UTRs of genes with > 2-fold changes at protein levels.
Table 2.
The distribution of 6-mer seed matches of miR-143 in transcripts of identified genes by proteomics
| Fold-change | 3'UTR | 5'UTR | CDS |
|---|---|---|---|
| ≤ 0.5 | 29 (31.2%) | 2 (2.2%) | 32 (34.4%) |
| 0.5 ~ 2 | 193 (17.4) | 28 (2.5) | 332 (30.0%) |
| ≥ 2 | 0 | 0 | 1 (100%) |
We also used PicTar, TargetScan and miRanda programs to predict potential targets of miR-143 and compared the predictions with our proteomic data. Only a limited number of putative targets identified by our proteomic analysis were predicted by these three prediction methods (Table 3). For example, only 2, 3 and 4 genes were predicted to be miR-143 targets among 93 downregulated genes by PicTar, TargetScan and miRanda, respectively. In contrast, 3’UTRs of 29 genes containing 6-mer seed matches of miR-143 were among the downregulated genes. Because the computational prediction systems have relatively high false-positives and negatives, additional experimental data for different miRNAs are needed for increasing the accuracy of prediction programs.
Table 3.
Predicted miR-143 targets by PicTar, TargetScan and miRanda in identified genes by proteomics
| Fold-change | PicTar | TargetScan | Miranda |
|---|---|---|---|
| ≤ 0.5 | 2 (2.2%) | 3 (3.2%) | 4 (4.3%) |
| 0.5~2.0 | 6 (0.4%) | 10 (0.9%) | 42 (3.8%) |
| ≥ 2.0 | 0 (0%) | 0 (0%) | 0 (0%) |
Transcript changes induced by miR-143
We also carried out gene expression profiling to detect mRNA changes following miR-143 overexpression using Agilent gene expression arrays, which contain 41,000 unique probes for transcripts including 26,705 genes. We observed that overexpression of miR-143 in MiaPaCa2 only caused a mild change at the mRNA level. Using 2-fold as a cut-off, only 9 genes exhibited a significantly decreased mRNA levels following miR-143 overexpression (p < 0.05). Similarly, 15 genes showed >2-fold increase (p < 0.05) after miR-143 overexpression. Because of lack of both mRNA and protein data for many genes, we constructed a dataset where we had obtained quantitative data at both mRNA and protein levels. In this set of 1,117 genes, 84 genes showed downregulation while one gene was upregulated ≥2 fold at the protein level. Almost all of the genes with downregulated protein levels demonstrated very mild changes at the mRNA level as shown in Figure 4. Our data again underscores the fact the regulation of targets occurs at the protein level. Some interesting molecules such as RANKBP3L, MAP3K6 and MAPKAPK2 are highlighted in Figure 4, whose protein expression was inhibited more than 2-fold.
Verification of miR-143 targets using luciferase assays
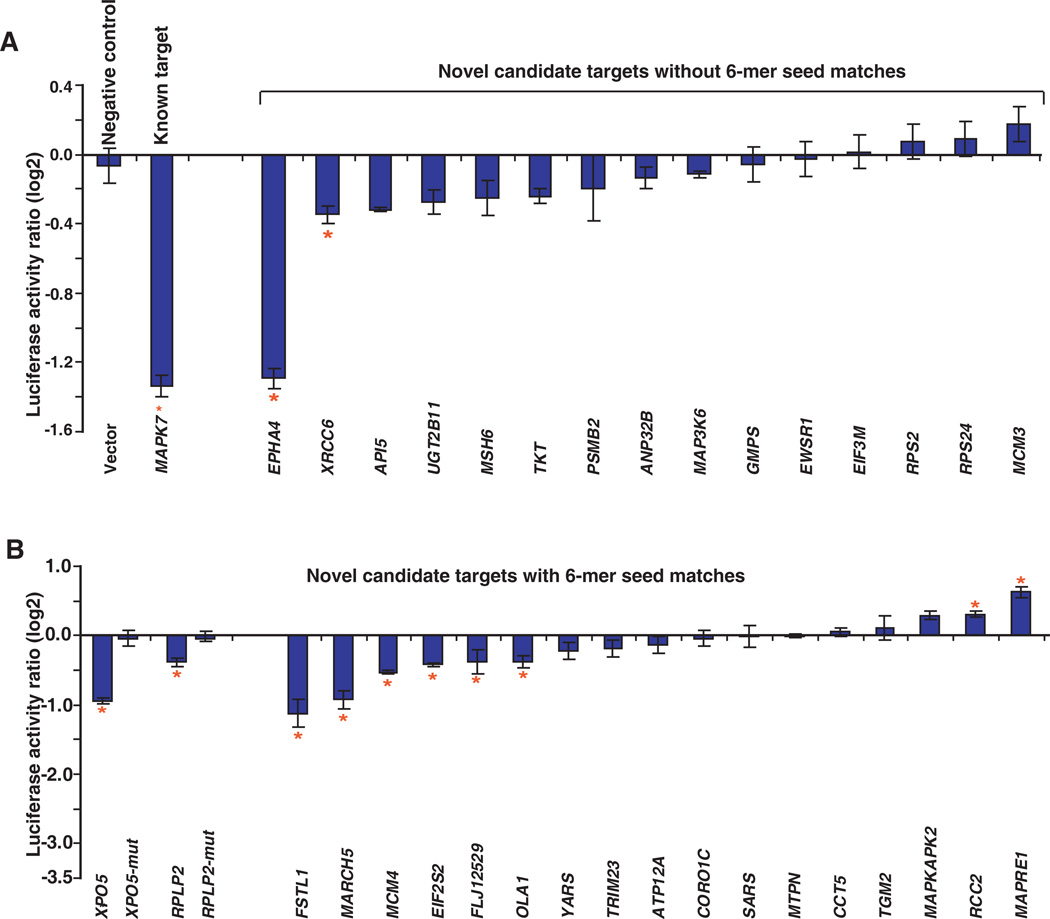
We performed luciferase assays to determine if miR-143 directly regulates some or all of the putative targets that we identified using our proteomic approach. Nineteen candidate targets with 6-mer seed matches in the 3’UTR regions and 15 candidate targets without 6-mer seed matches in the 3’UTR regions were randomly chosen for luciferase assays. The 3’UTR regions of these targets were cloned downstream of the luciferase gene. The constructs were co-transfected with miR-143 mimic or control oligos into MiaPaCa2 cells. Empty vector (pGL3-control) without any 3’UTR fragment was used as a negative control and one known target, mitogen-activated protein kinase 7 (MAPK7), was used as a positive control. As expected, while the known target MAPK7 showed a significant reduction in luciferase activity induced by miR-143, there was no observed change in the luciferase activity for vector only as shown in Figure 5A. Only 2 out of 15 candidate targets without 6-mer seed matches in the 3’UTR regions had 20% reduction (p<0.05) as illustrated in Figure 5A. In contrast, 8 out of 19 candidate targets containing 6-mer seed matches in the 3’UTR regions showed > 20% reduction (p < 0.05) in luciferase activity (Figure 5B). The results indicate that 6-mer seed matches of miR-143 are enriched in downregulated genes with decreased luciferase activity. We also mutated 6-mer seed matches of miR-143 in 3’UTR regions of 2 targets to confirm the specificity of the binding sites. As shown in Figure 5B, mutation of 6-mer seed matches in 3’UTR regions of XPO5 and RPLP2 transcripts rescued reduced luciferase activity by miR-143.
Figure 5. Verification of miR-143 targets using luciferase assays.
Luciferase assays were performed to verify miR-143 candidate targets identified by our proteomics. Vector control and one known target Mitogen-activated protein kinase 7 (MAPK7) were used as negative and positive controls, respectively. Shown are relative luciferase activity normalized to corresponding transfections with control oligo. Data are shown as the mean ± SD (log2) of 3 replicates and are representative of 3 independent experiments. *p<0.05 using two-tailed t-test (miR-143 mimic vs control oligo). A: Fifteen candidate targets without 6-mer seed matches (complementary to nucleotides 2 – 7 at the 5’end of miR-143). B: Nineteen candidate targets with 6-mer seed matches. XPO5-mut and RPLP2-mut indicate constructs where a mutation was introduced within the seed match region located in the 3’UTRs of XPO5 or RPLP2 transcripts, respectively.
Discussion
Our understanding of the molecular mechanisms by which miRNAs modulate cellular signaling pathways will remain incomplete until the full set of miRNA targets is identified and validated. Strategies based on detecting mRNA expression changes, such as microarray analysis, have been used to identify miRNA targets. Because a miRNA may regulate the expression of its targets only through translational inhibition, such mRNA approaches may be inherently limited. Here, we demonstrate the utility of quantitative proteomic approaches to discover targets of miRNAs in an unbiased fashion. Our mRNA based gene expression array data clearly show that miR-143 regulates many of its targets without affecting mRNA abundance (Figure 4). However, Baek et al observed that mRNA destabilization played a major role in the repression of miRNA targets.10 The reason for the discrepancy may be that different miRNAs in different cells have different mechanisms to regulate their targets.
In total, we identified 93 potential targets of miR-143, which showed ≥ 2-fold downregulation by miR-143 mimic. For example, MAPKAP kinase 2 (MAPKAPK2), also called MK2, is a serine/threonine kinase. It is a major substrate of p38 MAPK and has been shown to play a pivotal role in inflammatory processes.32 MK2 knockout mice were shown to develop significantly fewer skin tumors compared with wild-type mice.33 Overexpression of miR-143 resulted in 25-fold decreased expression of MK2, which indicates that miR-143 may inhibit tumor growth through inhibiting MK2 expression. MutS homolog 6 (MSH6) protein is similar to MutS protein in E. coli, which helps in the recognition of mismatched nucleotides in DNA mismatch repair.34 Mutations in the MSH6 gene have been identified in individuals with hereditary nonpolyposis colon cancer (HNPCC) and endometrial cancer.35 MSH6 protein showed > 4-fold downregulation by miR-143, which suggests that miR-143 might modulate mismatch repair. Tripartite motif-containing 23 (TRIM23) is a member of the tripartite motif (TRIM) family. TRIM23 is also a member of the ADP ribosylation factor family, which plays a role in the formation of intracellular transport vesicles. Recently, TRIM23 was identified as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus.36 TRIM23 protein was observed to be downregulated >3-fold by miR-143. Our results also uncover an unexpected link between miR-143 and miRNA transport. Exportin 5 (XPO5) mediates the export of pre-miRNA from the nucleus to cytoplasm, an essential step in miRNA biogenesis. XPO5 protein not only acts as an export factor but also protects pre-miRNAs from digestion by nucleases.37,38 XPO5 protein showed almost 3-fold downregulation upon miR-143 overexpression. This result raises the intriguing possibility that miR-143 may regulate miRNA biogenesis in some settings.
Although miRNAs are largely known to downregulate gene expression, a few studies have recently demonstrated that miRNAs can also upregulate their targets under certain circumstances.39–42 For example, Vasudevan et al have shown that miRNA let-7 and synthetic miRNA miRcxcr4 could increase translation of their targets upon cell cycle arrest.42 In another study, Tsai at al found that miR-346 activated activity of receptor-interacting protein 14 by increasing its protein expression.39 In our study, we observed 28 proteins to be upregulated > 1.5-fold and one protein to be elevated > 2-fold by miR-143 - these upregulated molecules could also represent potential targets of miR-143. It is also possible that the upregulation of those molecules may due to indirect effects of miR-143.
Surprisingly, there are two genes showing a change in the opposite direction in luciferase activity compared with protein levels. Both regulator of chromosome condensation 2 (RCC2) and microtubule-associated protein, RP/EB family, member 1 (MAPRE1) are downregulated by miR-143 at protein level, However, both genes showed upregulation in luciferase activity when co-transfected with miR-143 mimic (Figure 5B). This could perhaps be due to some secondary effect of miR-143.
In conclusion, we have identified 93 genes as candidate targets of miR-143 using a global quantitative proteomic approach. Ten of 34 tested target genes are likely to be direct targets of miR-143 as shown by luciferase assays - 8 of these contain perfect 6-mer miR-143 seed matches. These data coupled with results from gene expression arrays, clearly show that miR-143 regulates many of its targets at the translational level without affecting mRNA abundance.
Experimental section
Cell culture, miRNA transfection and SILAC labeling
A pancreatic cancer cell line, MiaPaCa2, was cultured in Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen, Carlsbad, CA) supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. MiaPaCa2 cells were seeded in 150 mm dishes and transfected with 50 nM of miR-143 mimic oligonucleotides or control oligo (Dharmacon, Lafayette, CO) using oligofectamine (Invitrogen). The SILAC labeling procedure for MiaPaCa2 cells was carried out according to the protocol described earlier.25 While miR-143 mimic was transfected to cells stably adapted in DMEM medium containing 2H4 lysine and 13C6 arginine (heavy state), control oligo was transfected to cells grown in normal DMEM medium (light state).
SCX fractionation
The cells were harvested after rinsing the cells with ice cold PBS 6 times 48 hours post-transfection. The cells were lysed in 0.5% SDS by sonication (Duty cycle 30%, output control at 3, on Sonifier 250, Branson). Equal amount of clear protein lysates from control and miR143 transfected cells were combined, reduced (5 mM DTT), alkylated (20 mM iodoacetaminde) and digested using trypsin (1:20, Promega, Madison, WI) overnight.25 The digested peptides were acidified using 1M phopshoric acid and equilibrated with 10 mM potassium phosphate buffer containing 25% acetonitrile, pH 2.85 (Solvent A). The peptides were fractionated using strong cation exchange chromatography on a Polysulfoethyl A column (PolyLC, Columbia, MD) (300Å, 5µm, 100 × 2.1mm) using an Agilent 1100 HPLC system containing a binary pump, UV detector and a fraction collector.28 The peptides were eluted using a linear salt gradient (0 to 100%) between solvent A and solvent B (10 mM potassium phosphate buffer containing 25% acetonitrile, 350 mM KCl, pH 2.85). The fractions were completely dried and reconstituted in 40µl of 0.2% formic acid prior to reversed-phase (RP) liquid chromatography based tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS analysis
Tandem mass spectrometry analysis of SILAC labeled peptides was carried out on a Fourier Transform LTQ Orbitrap XL ETD mass spectrometer using an Agilent 1100 system LC system interfaced with the mass spectrometer. The RP-LC system consisted of a desalting column (75 µm × 3 cm, C18 material 5–10 µm, 120 Å) and an analytical column (75 µm × 10 cm, C18 material 5 µm, 120 Å) with a nanoflow solvent delivery. Electrospray source is fitted with an emitter tip 8µm (New Objective, Woburn, MA) and maintained at 2000 V ion spray voltage. Peptide samples were loaded onto a trap column in 0.1% formic acid, 5% acetonitrile for 15 min and LC-MS/MS data were acquired during peptide elution using gradient of acetonitrile in 0.1% formic acid (5–40%) for 60 min with a flow rate of 300 nl/min. The MS spectra were acquired at a resolution of 60,000 at m/z 400 with high resolution CID MS/MS detection in Orbitrap (resolution 7500). The precursors were isolated using m/z 3.5 window, 35% normalized collision energy and 30 millisec activation time. Monoisotopic precursor mass selection and peptides containing 2+ and above charges were enabled for MS/MS analysis. For each cycle of data dependent analysis 4 most abundant peptides were selected for MS/MS. The data from the LC-MS/MS analyses was processed together and search results were merged.
Data Analysis
The mass spectrometry data was processed using Proteome Discoverer (Version 1.1) software (Thermo Scientific) workflow which combines data from Mascot and Sequest search engines and performs the quantitation. Proteome Discoverer software performs automated statistical analysis of the results and uses unique peptides to calculate accurate relative protein quantitation. Normalization of peptide concentration corrects for experimental errors. The mass spectrometry data were searched against NCBI RefSeq-35 human protein database containing 38,114 sequences. The peptide and protein data were extracted using high peptide score threshold and top peptide rank filters. False discovery rate was calculated by enabling the peptide sequence analysis using a decoy database. We used 1% FDR as a cut-off to export results from the analysis. In addition, we carried out a manual interpretation of MS/MS spectra with score more than 25 when using Mascot. The average ratio and standard deviation were calculated for protein quantitation wherever multiple peptides were identified for a protein The standard deviation values were obtained for single hit peptides using MSQuant by calculating the ratios of heavy and light peptide intensities measured at different time points of LC peaks. Quantitation data from MSQuant and Proteome Discoverer were merged together and peptide ratios were manually inspected.
Gene expression microarray analysis
Total RNA from transfected cells was isolated with miRNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. The RNAs were used for two-color gene expression microarray analysis as previously described.29 Briefly 400 ng of total RNAs from miR-143 mimic and control oligo transfected samples was amplified and labeled using Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies, Santa Clara, CA). Entire amount of each samples labeled with Cy3 or Cy5 are mixed, fragmented and hybridized. Agilent whole human genome microarrays (G4112F) were used, which contain 41,000 unique probes for transcripts including 26,705 genes with RefSeq or UniGene Accession Number. Microarrays were scanned using an Agilent G2505B Scanner controlled by Agilent Scan Control 7.0 software. Data were extracted with Agilent Feature Extraction 9.1 software. Data normalization and analysis were performed using GeneSpring GX 10.0.1 following software developer’s recommendation (Agilent).
Plasmid constructs and luciferase assays
3’UTR fragments of selected genes were PCR amplified from human cDNA and cloned downstream of the luciferase open reading frame in pGL3-control vector (Promega). Mutagenesis of 6-mer seed match in 3’UTR regions of each target was generated from corresponding wild type constructs using QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) according to manufacturer’s instructions. Twenty-four hours prior to transfection, 3 × 104 cells were plated per well in a 48-well plate. pGL3 constructs (100 ng) plus 10 ng of the Renilla luciferase plasmid phRL-SV40 (Promega) were co-transfected with miR-143 mimic or control oligo using Lipofectamine 2000 (Invitrogen) to MiaPaCa2 cells. Luciferase assays were performed using the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well. For each experimental trial, cells were transfected in triplicate. For each construct,the values from miR-143 mimic are normalized to control oligo. A p-value was calculated with the two-tailed t-test to compare relative luciferase activities of miR-143 mimic transfection with control oligo transfection.
Bioinformatics analysis
For determining of the frequency of miR-143 seed regions, genes identified in the present study with annotated 3' UTR, 5' UTR and coding sequence were selected. The corresponding mRNA sequences were downloaded from GenBank30 (August 2009 build). mirMatcher, an in-house Python program (Python 2.4) was developed to find perfect seed matches in 3' UTR, 5' UTR and CDS region and was reported. The enrichment of motifs complementary to 6-mer (2 to 7 nucleotide) of the 5' end of miR-143 was evaluated by chi square test. Pictar (http://pictar.bio.nyu.edu/)6, Targetscan4.1 (http://www.targetscan.org/)5,31 and miRanda (http://www.microrna.org/microrna/home.do)7 were used to computationally predict targets of miR-143.
Table 1.
A partial list of candidate targets of miR-143 identified by proteomics
| Gene Symbol |
Protein Name | Protein Ratio ± SD |
mRNA Ratio ± SD |
Luciferase Ratio ± SD |
|---|---|---|---|---|
| MAPKAPK2 | MAPKAP kinase 2 | 0.04 ± 0.01 | 0.82 ± 0.12 | 1.22 ± 0.19 |
| MAP3K6 | mitogen-activated protein kinase kinase kinase 6 | 0.10 ± 0.01 | 0.91 ± 0.07 | 0.92 ± 0.01 |
| UGT2B11 | UDP glucuronosyltransferase 2 family, polypeptide B11 | 0.10 ± 0.06 | 1.05 ± 0.12 | 0.83 ± 0.04 |
| MSH6 | mutS homolog 6 | 0.23 ± 0.02 | 0.91 ± 0.01 | 0.84 ± 0.06 |
| TRIM23 | tripartite motif-containing 23 | 0.27 ± 0.24 | 1.14 ± 0.09 | 0.88 ± 0.01 |
| RPS24 | ribosomal protein S24 isoform c | 0.29 ± 0.08 | 1.00 ± 0.03 | 1.07 ± 0.07 |
| MARCH5 | membrane-associated ring finger (C3HC4) 5 | 0.30 ± 0.08 | 0.82 ± 0.03 | 0.53 ± 0.05 |
| XPO5 | exportin 5 | 0.35 ± 0.18 | 0.94 ± 0.03 | 0.14 ± 0.03 |
| RCC2 | regulator of chromosome condensation 2 | 0.37 ± 0.13 | 0.78 ± 0.05 | 1.24 ± 0.04 |
| EWSR1 | Ewing sarcoma breakpoint region 1 isoform EWS | 0.39 ± 0.05 | 1.15 ± 0.15 | 0.98 ± 0.07 |
| RPS2 | ribosomal protein S2 | 0.39 ± 0.16 | 1.00 ± 0.21 | 1.05 ± 0.02 |
| SARS | seryl-tRNA synthetase | 0.40 ± 0.05 | 0.90 ± 0.04 | 0.99 ± 0.11 |
| EIF3M | eukaryotic translation initiation factor 3, subunit M | 0.40 ± 0.12 | 1.05 ± 0.07 | 1.01 ± 0.06 |
| MCM4 | minichromosome maintenance complex component 4 | 0.40 ± 0.17 | 0.82 ± 0.04 | 0.69 ± 0.01 |
| MAPRE1 | microtubule-associated protein, RP/EB family, member 1 | 0.40 ± 0.21 | 1.05 ± 0.18 | 1.55 ± 0.09 |
| PSMB2 | proteasome beta 2 subunit | 0.42 ± 0.16 | 1.04 ± 0.09 | 0.87 ± 0.17 |
| OLA1 | GTP-binding protein PTD004 isoform 1 | 0.44 ± 0.08 | 0.97 ± 0.13 | 0.77 ± 0.04 |
| ATP12A | ATPase, H+/K+ transporting, nongastric, alpha polypeptide | 0.44 ± 0.17 | 0.93 ± 0.06 | 0.91 ± 0.08 |
| EPHA4 | ephrin receptor EphA4 | 0.46 ± 0.13 | 1.15 ± 0.10 | 0.41 ± 0.02 |
| YARS | tyrosyl-tRNA synthetase | 0.46 ± 0.21 | 0.90 ± 0.14 | 0.86 ± 0.07 |
| RPLP2 | ribosomal protein P2 | 0.47 ± 0.09 | 0.96 ± 0.10 | 0.77 ± 0.03 |
| ANP32B | acidic (leucine-rich) nuclear phosphoprotein 32 family, member B | 0.47 ± 0.12 | 0.99 ± 0.03 | 0.91 ± 0.04 |
| MCM3 | minichromosome maintenance complex component 3 | 0.47 ± 0.15 | 0.91 ± 0.08 | 1.13 ± 0.08 |
| MTPN | myotrophin | 0.47 ± 0.29 | 1.04 ± 0.09 | 1.00 ± 0.02 |
| XRCC6 | ATP-dependent DNA helicase II, 70 kDa subunit | 0.48 ± 0.09 | 1.01 ± 0.04 | 0.79 ± 0.03 |
| GMPS | guanine monophosphate synthetase | 0.48 ± 0.10 | 1.03 ± 0.08 | 0.96 ± 0.05 |
| CCT5 | chaperonin containing TCP1, subunit 5 (epsilon) | 0.49 ± 0.02 | 0.99 ± 0.10 | 1.04 ± 0.11 |
| API5 | apoptosis inhibitor 5 | 0.49 ± 0.06 | 1.18 ± 0.12 | 0.80 ± 0.05 |
| TKT | transketolase | 0.49 ± 0.16 | 1.11 ± 0.10 | 0.85 ± 0.02 |
| FSTL1 | follistatin-like 1 precursor | 0.50 ± 0.01 | 1.12 ± 0.01 | 0.46 ± 0.07 |
| FLJ12529 | pre-mRNA cleavage factor I, 59 kDa subunit | 0.50 ± 0.04 | 1.01 ± 0.18 | 0.77 ± 0.10 |
| TGM2 | transglutaminase 2 isoform a | 0.50 ± 0.07 | 0.94 ± 0.03 | 1.08 ± 0.17 |
Acknowledgement
This study was supported by Department of Defense Era of Hope Scholar award (W81XWH-06-1-0428) and NIH roadmap grant (U54RR020839) for “Technology Centers for Networks and Pathways”. JTM is a Howard Hughes Medical Institute Early Career Scientist. We thank Dr. Wayne Yu for his expert assistance in gene expression array analysis.
Abbreviations
- SILAC
stable isotope labeling with amino acid in cell culture
- RISC
RNA-induced silencing complex
- SCX
strong cation exchange chromatography
- LC-MS/MS
liquid chromatography based tandem mass spectrometry
Footnotes
Supplementary Table 1: The entire list of proteins identified in this study
References
- 1.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 3.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. Nat. Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 4.Sethupathy P, Corda B, Hatzigeorgiou AG. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 6.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 9.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 10.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A. Proteomics. 2009;9:1374–1384. doi: 10.1002/pmic.200800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li JJ, Rocken C, Ebert MP, Kwok TT, Sung JJ. Br. J. Cancer. 2009;101:699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 14.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CM. Febs. J. 2009;276:6689–6700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- 16.Kent OA, Mullendore M, Wentzel EA, Lopez-Romero P, Tan AC, Alvarez H, West K, Ochs MF, Hidalgo M, Arking DE, Maitra A, Mendell JT. Cancer Biol. Ther. 2009;8:2013–2024. doi: 10.4161/cbt.8.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie H, Lim B, Lodish HF. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. J. Cell Physiol. 2009;218:444–449. doi: 10.1002/jcp.21621. [DOI] [PubMed] [Google Scholar]
- 22.Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H, Kita T, Satoh N, Shimatsu A, Hasegawa K. Biochem. Biophys. Res. Commun. 2008;376:728–732. doi: 10.1016/j.bbrc.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 23.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 24.Akao Y, Nakagawa Y, Naoe T. Oncol. Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 25.Harsha HC, Molina H, Pandey A. Nat. Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 26.Amanchy R, Zhong J, Hong R, Kim JH, Gucek M, Cole RN, Molina H, Pandey A. Mol. Oncol. 2009;3:439–450. doi: 10.1016/j.molonc.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan C, Olsen JV, Daub H, Mann M. Mol. Cell Proteomics. 2009;8:2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaerkady R, Kerr CL, Marimuthu A, Kelkar DS, Kashyap MK, Gucek M, Gearhart JD, Pandey A. J. Proteome Res. 2009;8:1315–1326. doi: 10.1021/pr8006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashyap MK, Marimuthu A, Kishore CJ, Peri S, Keerthikumar S, Prasad TS, Mahmood R, Rao S, Ranganathan P, Sanjeeviah RC, Vijayakumar M, Kumar KV, Montgomery EA, Kumar RV, Pandey A. Cancer Biol. Ther. 2009;8:36–46. doi: 10.4161/cbt.8.1.7090. [DOI] [PubMed] [Google Scholar]
- 30.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. Nucleic Acids Res. 38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansen C, Funding AT, Otkjaer K, Kragballe K, Jensen UB, Madsen M, Binderup L, Skak-Nielsen T, Fjording MS, Iversen L. J. Immunol. 2006;176:1431–1438. doi: 10.4049/jimmunol.176.3.1431. [DOI] [PubMed] [Google Scholar]
- 33.Johansen C, Vestergaard C, Kragballe K, Kollias G, Gaestel M, Iversen L. Carcinogenesis. 2009;30:2100–2108. doi: 10.1093/carcin/bgp238. [DOI] [PubMed] [Google Scholar]
- 34.Hargreaves VV, Shell SS, Mazur DJ, Hess MT, Kolodner RD. J. Biol. Chem. 2010 doi: 10.1074/jbc.M109.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsoekh D, Wagner A, van Leerdam ME, Dooijes D, Tops CM, Steyerberg EW, Kuipers EJ. Hered. Cancer Clin. Pract. 2009;7:17. doi: 10.1186/1897-4287-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole E, Groves I, MacDonald A, Pang Y, Alcami A, Sinclair J. J. Virol. 2009;83:3581–3590. doi: 10.1128/JVI.02072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 38.Yi R, Qin Y, Macara IG, Cullen BR. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai NP, Lin YL, Wei LN. Biochem. J. 2009;424:411–418. doi: 10.1042/BJ20090915. [DOI] [PubMed] [Google Scholar]
- 40.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. Embo. J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orom UA, Nielsen FC, Lund AH. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Vasudevan S, Tong Y, Steitz JA. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 43.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]