ABSTRACT
Paramyxoviruses, enveloped RNA viruses that include human parainfluenza virus type 3 (HPIV3), cause the majority of childhood viral pneumonia. HPIV3 infection starts when the viral receptor-binding protein engages sialic acid receptors in the lung and the viral envelope fuses with the target cell membrane. Fusion/entry requires interaction between two viral surface glycoproteins: tetrameric hemagglutinin-neuraminidase (HN) and fusion protein (F). In this report, we define structural correlates of the HN features that permit infection in vivo. We have shown that viruses with an HN-F that promotes growth in cultured immortalized cells are impaired in differentiated human airway epithelial cell cultures (HAE) and in vivo and evolve in HAE into viable viruses with less fusogenic HN-F. In this report, we identify specific structural features of the HN dimer interface that modulate HN-F interaction and fusion triggering and directly impact infection. Crystal structures of HN, which promotes viral growth in vivo, show a diminished interface in the HN dimer compared to the reference strain’s HN, consistent with biochemical and biological data indicating decreased dimerization and decreased interaction with F protein. The crystallographic data suggest a structural explanation for the HN’s altered ability to activate F and reveal properties that are critical for infection in vivo.
IMPORTANCE Human parainfluenza viruses cause the majority of childhood cases of croup, bronchiolitis, and pneumonia worldwide. Enveloped viruses must fuse their membranes with the target cell membranes in order to initiate infection. Parainfluenza fusion proceeds via a multistep reaction orchestrated by the two glycoproteins that make up its fusion machine. In vivo, viruses adapt for survival by evolving to acquire a set of fusion machinery features that provide key clues about requirements for infection in human beings. Infection of the lung by parainfluenzavirus is determined by specific interactions between the receptor binding molecule (hemagglutinin-neuraminidase [HN]) and the fusion protein (F). Here we identify specific structural features of the HN dimer interface that modulate HN-F interaction and fusion and directly impact infection. The crystallographic and biochemical data point to a structural explanation for the HN’s altered ability to activate F for fusion and reveal properties that are critical for infection by this important lung virus in vivo.
IMPORTANCE
Human parainfluenza viruses cause the majority of childhood cases of croup, bronchiolitis, and pneumonia worldwide. Enveloped viruses must fuse their membranes with the target cell membranes in order to initiate infection. Parainfluenza fusion proceeds via a multistep reaction orchestrated by the two glycoproteins that make up its fusion machine. In vivo, viruses adapt for survival by evolving to acquire a set of fusion machinery features that provide key clues about requirements for infection in human beings. Infection of the lung by parainfluenzavirus is determined by specific interactions between the receptor binding molecule (hemagglutinin-neuraminidase [HN]) and the fusion protein (F). Here we identify specific structural features of the HN dimer interface that modulate HN-F interaction and fusion and directly impact infection. The crystallographic and biochemical data point to a structural explanation for the HN’s altered ability to activate F for fusion and reveal properties that are critical for infection by this important lung virus in vivo.
Introduction
Infection by human parainfluenza viruses (HPIV) causes respiratory diseases, including croup, bronchiolitis, and pneumonia, which lead to illness or death in millions of infants and young children worldwide. Human parainfluenza virus type 3 (HPIV3), an enveloped virus, enters cells by fusing directly with the cell membrane. During entry, the viral surface glycoproteins HN (receptor-binding protein; hemagglutinin-neuraminidase) and F (fusion protein) cooperate to mediate fusion upon receptor binding. A series of coordinated interactions between HN and F glycoproteins are critical for infection. Interaction of HPIV3 HN with its sialic acid receptor is required in order for F to promote membrane fusion during infection (1, 2). Studies of other paramyxoviruses confirmed the paradigm that HN is essential to the F-mediated fusion process (3–5 reviewed in reference 6), with only a few exceptions (e.g., respiratory syncytial virus [RSV] [7, 8] and human metapneumovirus [hMPV] [9–11]). Before HPIV3 HN engages the receptor, HN stabilizes F in its prefusion state to prevent premature activation, and thereby inactivation, of F (12). Once engaged with its sialic acid receptor, HN activates the viral fusion protein (F) to undergo a series of structural rearrangements that ultimately lead to direct fusion of the viral envelope with the plasma membrane of the cell (13–17). Finally, HN plays another key role, when its sialic acid-cleaving (neuraminidase) activity facilitates release and spread of the virus. The four activities of HN, namely, receptor binding, receptor cleaving, F stabilization, and F activation, are regulated within a tetrameric type II membrane protein, consisting of a cytoplasmic domain, a membrane-spanning region, a stalk region, and a globular head that contains both the primary sialic acid binding/neuraminidase site and the putative secondary sialic acid binding site (18). The stalk domain, which confers specificity for the homologous fusion (F) protein in the fusion process (19–28), is responsible for mediating the activation of F, once it receives the signal from a sialic acid receptor-bound globular head (29, 30).
Once activated, F inserts its terminal fusion peptide into the target membrane and undergoes a series of conformational changes that lead to fusion. Receptor-engaged HPIV3 HN continues to act on F even after insertion of fusion peptide into the target membrane, promoting further structural rearrangement of the F protein that drives the viral membrane toward the cell-associated fusion peptide (29). This rearrangement brings the virus and cell membranes into proximity and facilitates membrane merger and viral entry.
A series of recent experiments showed that the communication between HN and F directly impacts infection in the natural host (31). An HPIV3 virus bearing an HN with Q552 at the putative secondary binding site (site II) at HN’s dimer interface (HNQ552), compared with the HPIV3 strain bearing a histidine at the same position (reference strain; H552), shows a greater level of fusion with cultured mammalian monolayer cells (2, 18, 32). The Q552 HN binds sialic acid receptor more avidly, dimerizes more efficiently, physically interacts more avidly with F, and produces more efficient F activation (32). However, the virus bearing HNQ552, while ideal in cultured monolayer cells, is unfit in differentiated human airway epithelium (HAE), a tissue system that mimics the natural host, and in vivo (cotton rat model for disease) compared to the reference strain containing HNH552 (31). These results suggest that increased F activation efficiency or increased physical interaction with F (32) may be disadvantageous in the natural host. The virus bearing the HN with enhanced F triggering (HNQ552) produces noninfectious viral particles in HAE (33), in which the F protein may have been prematurely activated before contacting target cells (31, 34).
The results obtained with HNQ552 present striking evidence for the role of the HN dimer interface-mediated function of F activation in modulating infection. This disadvantageous mutation for growth in vivo could be rescued during the virus adaptation to growth in natural host cells. During growth in HAE, the HPIV3 virus bearing HNQ552 adapted for survival by evolving to decrease F activation (33). Two sequential mutations in the F (G396D) and HN (Q559R) proteins decreased HN-receptor interaction and reduced F activation, impairing fusion/growth in immortalized monolayer cells but promoting successful growth in HAE. The doubly mutated HN (HNQ552/R559) responsible for conferring growth in HAE binds its receptor less avidly and promotes less fusion (33), revealing that these activities must attain a specific balance for growth in the natural host.
In this study, we sought to define the structural correlates of the HN features that are advantageous for infection in vivo and to identify the impact of this structure on HN-F interaction. Crystal structures of HNQ552/R559 show a diminished interface in the HN dimer compared to HNH552/Q559 of the reference strain, consistent with biochemical and biological data also indicating a decrease in dimerization, and suggest a structural explanation for its altered fusion promotion activity. Thus, the region of HN that we identified by functional assays as a key operative site on HPIV3 HN—termed site II (18)—has a clear structural correlate and modulates viral growth in vivo. Single-residue alterations at this site lead to structural changes that affect the dimer interface, influence HN’s ability to activate F, and affect growth in the natural host. These three viruses allowed us to compare the biological effect of specific structural changes, since the three viruses differ only in the structure at the HN dimer interface and yet diverge greatly in their abilities to infect the airway. Comparisons between the three viruses revealed specific structural changes in the HN dimer interface that modulate HN-F interaction and fusion triggering, confirm that these functions directly impact viral infection, and show that the requirements for these functions differ when virus infects natural host tissues compared to a monolayer cell culture. The analysis of the biological effects of specific structural changes revealed properties that are critical for infection in vivo. Using the cotton rat model, we confirmed that the virus bearing the doubly mutated HN has acquired increased fitness in vivo, compared to both the parental strain containing HNQ552 and the reference strain of HPIV3, neither of which is fit in vivo.
RESULTS
Mutation Q559R in HN reduces the physical strength of HN-F interaction.
We previously showed that infection of HAE with HPIV3-HNQ552 yielded noninfectious viral particles (31, 33, 35). While infection with HPIV3-HNQ552 yielded zero titer up to 3 days postinfection, by 7–9 days postinfection of HAE, a population of small-plaque-forming virus (referred to as S18 in the previous publications) emerged. The viability of the emerging variants was associated with mutations in HN and F envelope glycoproteins, with respect to the parent virus, that decreased HN receptor avidity and F’s triggerability (33); the S18 virus carried the mutation G396D in F and the mutation Q559R in HN; the virus is called HPIV3-HNQ552/R559/FD396. The HNs with both mutations and HN with R559 alone were inherently less efficient at activating F than the parent with HNQ552 (33).
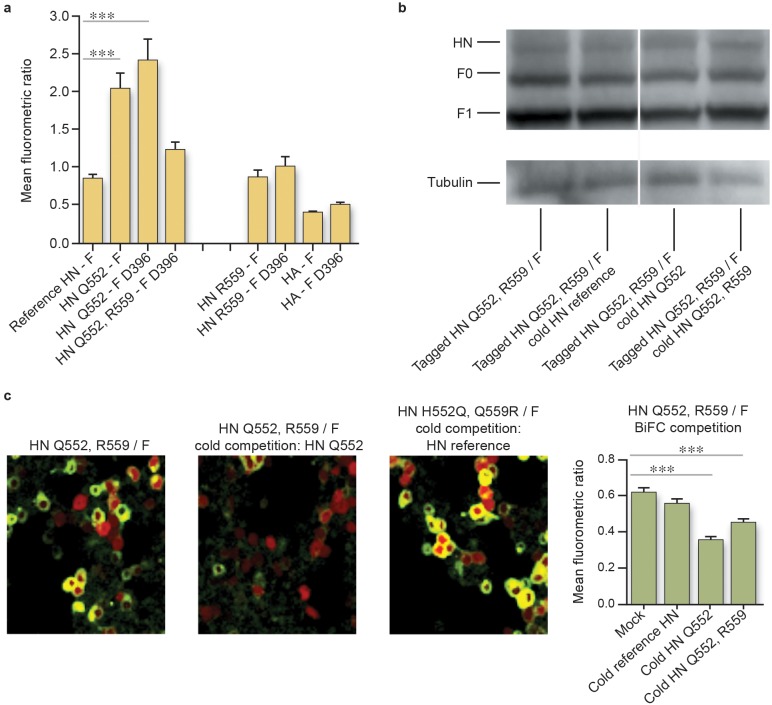
We previously described the use of a bimolecular fluorescence complementation (BiFC) technique to measure the strength of HN-F interaction (32). The BiFC strategy relies on the reconstitution of an active complex when fragments of fluorescent proteins, fused to putative interacting pairs, associate (36–41). We showed that this strategy can be effectively used to measure the relative strength of interaction between respective HN-F pairs by quantifying the relative mean fluorescence (mean fluorometric ratio [MFR]) produced upon HN-F interaction before receptor engagement (32). We now investigated the effect on HN-F interaction during the evolution of HPIV3-HNQ552 to HPIV3-HNQ552/R559 and HPIV3-HNQ552/R559/FD396. These experiments were performed first in the absence of sialic acid receptor engagement (using the small molecule zanamivir to prevent HN receptor engagement) in order to study the inherent strength of interaction between the two proteins.
HNQ552 exhibits increased interaction with parental F (MFR of 2.0 ± 0.2; values are means ± SE) or FD396 (MFR of 2.4 ± 0.3) compared to HN interaction with F (MFR of 0.8 ± 0.1) (Fig. 1A). The reference strain HN’s interaction with FD396 (MFR of 0.95 ± 0.2) (data not shown) is similar to its interaction with parental F. Thus, the G396D mutation in F has no measurable effect on its ability to interact with HN as assessed by BiFC. Neither parental F nor FD396 interacted significantly with uncleaved influenza HA as a negative control (Fig. 1a), with MFRs of 0.4 and 0.5, respectively.
FIG 1 .
The strength of HN-F interaction for the lung-fit virus HPIV3-HNQ552/R559/FD396 is reduced relative to that of the parental virus (HPIV3-HNQ552/FG396) in 293T cells. (a) Cells were cotransfected with constructs encoding the indicated F fused to C-CFP and HN fused to N-Venus (or HA fused to N-Venus) and treated overnight with 10 mM zanamivir. One hour prior to confocal microscopy analysis, fresh 10 mM zanamivir and cycloheximide were added. Fluorescence (the mean fluorometric ratio) resulting from HN-F interaction was quantified. Values are means and standard errors of the means (SEM) from 4 to 7 separate experiments. ***, P < 0.001 (one-way analysis of variance). (b) In a competition assay, HNQ552/R559 N-Venus and F C-CFP were cotransfected with pCAGGS (mock), HNH552 pCAGGS (cold HNH552), HNQ552 (cold HNQ552), or HNQ552/R559 (cold HNQ552/R559) and treated overnight with 10 mM zanamivir. The expression levels of HN and F (in the uncleaved precursor form [F0] and the proteolytically cleaved form [F1]) are similar for each HN-F pair, as shown by Western blotting using tubulin (55 kDa) as a loading control. (c) Fluorescence (the mean fluorometric ratio) resulting from HN-F interaction in the competition assay was quantified as described for panel a. Representative fluorescence images of cotransfected 293T cells 20 h after transfection show HN-F interaction on the cell surface. Cells were also transfected with mCherry (red).
The introduction of R559 into the HNQ552 background decreases the interaction between HN and FD396 (MFR, 1.2 ± 0.1) (Fig. 1a). Thus, the introduction of the R559 alteration into this site not only reduced the receptor binding-to-cleavage ratio and the F-triggering ability of this HN (33) but also decreased the HN-F interaction. However, the effect of the R559 mutation in HN is significant only in the HNQ552 background. The MFRs for interaction between HNR559 and parental F and FD396 were 0.9 and 1.0, respectively (Fig. 1a).
To further compare the HN-F interactions mediated by HNQ552/R559 and HNH552, we carried out cold competition experiments (32, 38–41) that measured the ability of each HN to effectively compete for interaction with F (Fig. 1b and c). Cells were cotransfected with constructs encoding BiFC-tagged HNQ552/R559 and F along with one of the following constructs: empty vector (control) or untagged HNH552, HNQ552, or HNQ552/R559. Every sample thus contains two HNs, one BiFC tagged and one untagged, along with F. If the interaction of a given untagged (“cold”) HN with F is more avid than that of the BiFC-tagged HN, then the cold HN competes with tagged HN for interaction with F, and fluorescence decreases as a function of competitor. The presence of untagged HNQ552, but not of untagged HN from the reference strain with H552, decreases the BiFC of tagged HNQ552/R559 and F (Fig. 1c, which presents values collected from at least seven replicate experiments). Thus, HNQ552, but not HNH552, effectively competes with HNQ552/R559 for interaction with F, and together with the finding that untagged HNQ552/R559 does not compete with HNH552 for interaction with F (i.e., HNQ552/R559 and HNH552 do not displace each other), the data suggest that HNQ552/R559 and HNH552 interact similarly with F. A representative Western blot performed after BiFC analysis indicates that all proteins were expressed at similar levels for each HN-F pair (Fig. 1b).
Q552/R559 decreases HN-HN dimer association: biochemical and structural analyses.
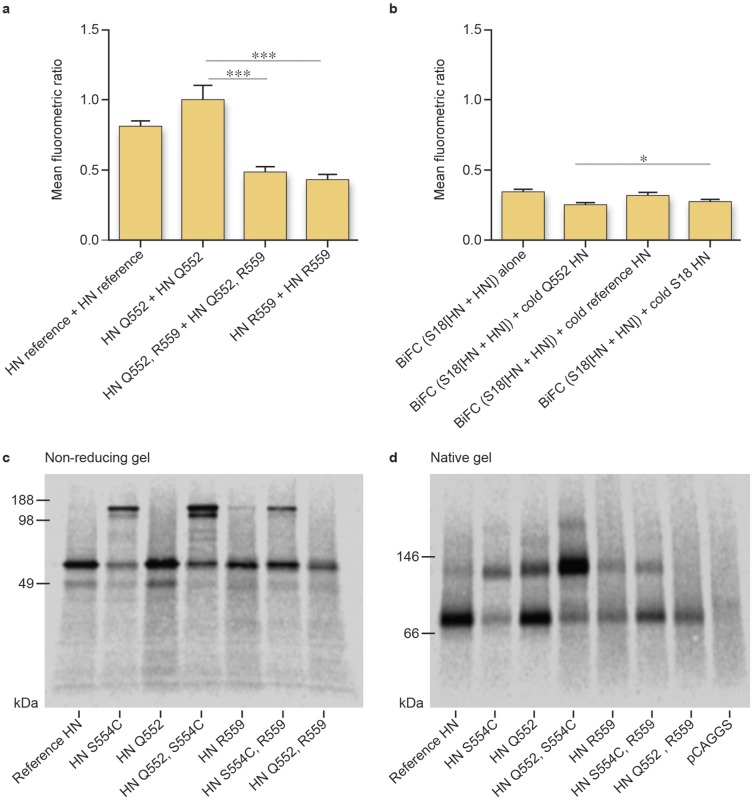
We have shown that the presence of a glutamine at position 552 at the dimer interface of the parental HPIV3 HN improves HN oligomerization and that oligomer alterations may modulate F triggering (32). We compare here the oligomerization tendency of HNQ552/R559 to that of the parental HNQ552. BiFC using tagged HNs in the absence of F protein suggests a decrease in homo-oligomerization for HNQ552/R559 and HNR559 compared to HNQ552 (Fig. 2a). The observation is further confirmed by cold competition experiments (Fig. 2b). Cells were cotransfected with BiFC-tagged HNQ552/R559 along with cold (untagged) HNQ552, cold HNH552, or empty vector (negative control). Cold HNQ552 outcompeted HNQ552/R559 for homo-oligomerization, decreasing BiFC fluorescence significantly more than by competition with cold HNH552 or cold HNQ552/R559 (Fig. 2b).
FIG 2 .
Hypofusion-triggering HN (Q552/R559) shows decreased HN dimerization. (a) Mean fluorometric ratios of BiFC resulting from the oligomerization of HN. Constructs containing R559 (HNQ552/R559 and HNR559) show lower mean fluorometric ratios and weaker association than the reference strain HN with H552 and HNQ552. Values are means and SEM of results from five experiments. ***, P < 0.001 (one-way analysis of variance). (b) 293T cells were cotransfected with HNQ552/R559 N-Venus, HNQ552/R559 C-Venus, and a nontagged (cold) HN construct. BiFC from the oligomerization of HNQ552/R559 N-Venus and HNQ552/R559 C-Venus can be disrupted by cold HN, which results in a decreased mean fluorometric ratio. Cold HNQ552 is more effective than cold HNH552 and cold HNQ552/R559 to compete with tagged HNQ552/R559 for homo-oligomerization. Values are means and SEM of results from five experiments. *, P < 0.05 (Student’s t test). (c) Nonreducing SDS-PAGE autoradiography of the indicated untagged HNs. (d) Blue native gel autoradiography of the indicated untagged HNs.
The mature HN tetrameric complex is composed of a dimer of dimers. While our competition experiments showed a decrease in fluorescence in the presence of untagged HNQ552, the experiment does not distinguish whether the alteration was due to a decrease in dimer versus tetramer formation. Q559 is at the HN-HN dimer interface in the HN HPIV3 crystal structure (42), and we proposed that Q559R mutation decreases the stability of the HN-HN dimer (33). We used blue native polyacrylamide gel electrophoresis (BN-PAGE) to determine whether the decreased oligomerization of HNQ552/R559 was due to disruption of the dimer-dimer interface in the tetramer, or the HN-HN dimer interface. We previously applied this approach to investigate the oligomeric state of HNQ552 compared to HNH552 (32), as others have done for PIV5 and measles viruses (43, 44). We also took advantage of a mutation in HPIV3 HN (S554C) that leads to disulfide linkage across the HN-HN dimer interface, which provides a reference for the stable dimer state (45). Cells were transfected either with HNH552, with HNS554C, with HNR559, or with the double mutant HNR559/S554C, radiolabeled for 3 h, synchronized by adding cycloheximide, and treated with neuraminidase in order to observe HN oligomerization in the absence of receptor interaction (32). Cell lysates were immunoprecipitated as described in Materials and Methods and subjected to nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2c) and BN-PAGE (Fig. 2d) to examine the migration patterns of the four different HN molecules. In the native gel, HN constructs carrying the S554C mutation have a larger percentage of the protein sample migrating as a dimer of HN (Fig. 2d). These HN-HN dimers are covalently linked by the introduced cysteines (Fig. 2c). The results obtained with the S554C mutation were expected and serve as proof of concept for the experimental design. HNQ552 is more enriched in the dimer fraction than HNH552 on BN-PAGE (Fig. 2d), through noncovalent protein-protein interactions (Fig. 2c). HNQ552/R559 is present in monomeric form, in contrast to HNQ552 and HNH552, consistent with the HN-Q552/R559 dimer being the least stable (Fig. 2d). Even for the mutant HNS554C, the introduction of R559 somewhat reduces dimerization, with the fraction of the molecule in dimer form being lower than that in monomer form (Fig. 2c); however, the functional alterations conferred by the R559 mutation in the context of the double mutant are more impressive than its simple effect on dimerization (see below) (33).
Structural analysis of HNQ552/R559 reveals the basis of the diminished tendency toward HN-HN dimerization. Recombinant HNH552 and HNQ552/R559 were expressed and purified for crystallization and structural determination to examine the impact of the mutations on the HN dimer interface. The purified recombinant HN ectodomain is monomeric in solution but forms noncrystallographic dimers in the crystal structure. HNH552 (reference strain) and HNQ552/R559 were crystallized under similar conditions and share the same crystal packing. These two mutants diffracted to 1.65- and 1.80-Å resolutions, respectively (Table 1). The HN globular head, but not the stalk region, is well ordered in the structures. The overall structure of the HN reference strain protomer is very similar to a previously published HN structure from a clinical strain (46), with a RMSD (root mean square deviation) of 0.18 Å for 395 out of 431 Cα atoms in the head domain.
TABLE 1 .
Data collection and refinement statistics (molecular replacement)a
| Parameter | HN | HNQ552/R559 |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 83.8, 94.8, 105.5 | 84.0, 96.6, 105.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–1.65 (1.71–1.65)a | 30–1.80 (1.86–1.80) |
| Rsym or Rmerge | 4.8 (69.2) | 6.2 (73.9) |
| I/σ(I) | 23.1 (2.5) | 29.2 (3.0) |
| Completeness (%) | 99.7 (99.7) | 99.9 (99.9) |
| Redundancy | 5.5 (5.5) | 6.9 (6.3) |
| Refinement | ||
| Resolution (Å) | 47.4–1.65 | 28.9–1.8 |
| No. of reflections | 100,646 | 79,882 |
| Rwork/Rfree | 16.8/19.8 | 15.8/18.7 |
| No. of atoms | ||
| Protein | 6,841 | 6,842 |
| Ligand/ion | 288 | 361 |
| Water | 550 | 638 |
| B factors | ||
| Protein | 29.5 | 30.2 |
| Ligand/ion | 57.2 | 59.3 |
| Water | 36.0 | 35.8 |
| RMSD | ||
| Bond lengths (Å) | 0.007 | 0.007 |
| Bond angles (°) | 1.25 | 1.25 |
Data collected from one crystal were used for each structure. Values in parentheses are for the highest-resolution shell.
The introduction of residues Q552 and R559 induces significant local conformational changes, but not to the overall dimer arrangement (Fig. 3A). One monomer within the dimer shifted slightly about 1 Å when the other protomer was superimposed between the two structures. Other than that, most of the structural differences are around the 550 loop at the dimer interface, which harbors residues 552 and 559 (Fig. 3B). In HNH552 (from the reference strain), the two 550 loops within the dimer are adjacent to each other, with extensive interactions. Together, the 550 loops contribute to a 468-Å2 buried interface area for the dimer formation, out of a 3,701-Å2 total buried area at the HN-HN dimer interface. The imidazole functional groups from the His552 residues are about 4 Å apart, positioned for potential π-π interaction. Gln559 side chains point toward the neighboring HN subunit and are well accommodated in the cavity at the dimer interface. In the HNQ552/R559 structure, the two Arg559 residues are oriented away from each other, parallel to the dimer interface, which likely reflects charge repulsion between these two positively charged residues within the cavity. These residues further induce a rigid body rotation of the 550 loop, moving it away from the dimer interface (Fig. 3B). These conformational changes at the 550 loop are propagated to nearby loops at the dimer interface and significantly reduce dimer association, as measured by buried interface areas. HNH552 (reference) buries 3,701 Å2 at the dimer interface while, in HNQ552/R559, 2,867 Å2 of protein surface is contributed to dimer association (Fig. 4A and B).
FIG 3 .
Introduction of a Q559R mutation weakens the HN-HN dimer interaction. (A) Structural comparison between HPIV3-HNH552 (reference strain) and HNQ552/R559. The HPIV3 H552 (reference strain) is shown in gray, and the two HNQ552/R559 protomers that form a noncrystallographic dimer are superimposed on the reference strain and are in green and cyan, respectively. The mutations induce local conformational changes on the dimer interface. Most notably, HNQ552/R559 displays a slightly wider separation across the dimer interface near the 550 loop region. (B) Dimer interface around the 550 loop in HNH552 and HNQ552/R559. The two HNQ552/R559 protomers in the noncrystallographic dimer are in green and cyan, respectively. H552 (from the reference strain) is shown in gray for comparison.
FIG 4 .
Footprints of dimer interface in HNH552 (reference strain) (A) and HNQ552/R559 (B). HN residues in contact in the dimer interface are shown in red. Introduction of the double mutation results in the loss of about 800 of 3,700 Å2 buried interface area in the dimer, mostly near the 550 loops.
The HN Q559R and F G396D mutations in HPIV3-HNQ552/R559/FD396 confer enhanced fitness in vivo over the parental HNQ552-containing virus.
To assess whether and how specific residues in HN-F affect viral fitness in vivo, viral fitness coinfection competition experiments (47–50) were performed. The goal of these experiments was to determine whether one virus would dominate growth in HAE (31, 33, 51), a setting that is relevant to human lungs. The competition between HPIV3-H552 (reference strain) and HPIV3-HNQ552/R559/FD396 was assessed. Coinfections with the HPIV3 reference strain and HPIV3-HNQ552/R559/FD396 were performed with percent ratios of 90:10, 50:50, or 10:90 HPIV3 reference strain to HPIV3-HNQ552/R559/FD396. Production of infectious HPIV3-HNQ552/R559/FD396 surpassed that of HPIV3 reference strain (Fig. 5a; viral titers are on the left and percent viral genome is on the right). Even when HAE cultures were infected with 90% HPIV3 reference strain (Fig. 5a, red bars) and 10% HPIV3-HNQ552/R559/FD396 (yellow bars), at 3 days postinfection, 90% of the viral population consisted of HPIV3-HNQ552/R559/FD396. In the same experiment, 58% of the viral genomes released at 2 days postinfection and 78% at 3 days carried the Q559R mutation in HN (corresponding to HPIV3-HNQ552/R559/FD396). A similarly dominant HPIV3-HNQ552/R559/FD396 growth was observed with infection ratios of 50:50 and 10:90 HPIV3 reference strain to HPIV3-HNQ552/R559/FD396 (Fig. 5A). The growth of HPIV3-HNQ552/R559/FD396 appears to outcompete the growth of the reference HPIV3 strain, revealing its enhanced fitness for growth in the human airway. As previously described, HPIV3-HNQ552/R559/FD396 also produced more infectious particles in HAE than the HPIV3 reference strain (33).
FIG 5 .
HPIV3-HNQ552/R559/FD396 has enhanced fitness for growth in HAE compared to HPIV3-HNH552 (reference strain) and HPIV3-HNQ552. HAE cultures were coinfected with 4,000 PFU of the HPIV3 reference strain and HPIV3-HNQ552/R559/FD396 (a) or of HPIV3-HNQ552 and HPIV3-HNQ552/R559/FD396 (b) at the ratios indicated. Viral titers (left) were determined at 1, 2, and 3 days postinfection. The ratios of viral genomes released (right) containing the codon CAA (glutamine) or CGA (arginine) corresponding to residue 559 on HN were quantified by qRT-PCR by SNP genotyping. The HPIV3 reference strain and HPIV3-HNQ552 genomes carried Q559, while HPIV3 small plaques (HPIV3-HNQ552/R559/FD396) carried R559. Titers on days 1 to 3 after HAE infection were determined by plaque assay in CV1 cells based on plaque size. Viral genome was quantified using SNP genotyping specific to Q or R at position 559 in HN. All data points are means and standard deviations (SD) from triplicate experiments.
The competition between HPIV3-HNQ552/R559/FD396 and its parental HPIV3 strain bearing HNQ552 was also examined. Coinfections with HPIV3-HNQ552 and HPIV3-HNQ552/R559/FD396 were performed with percent ratios of 90:10, 50:50, or 10:90 HPIV3-HNQ552 to HPIV3-HNQ552/R559/FD396. HPIV3-HNQ552/R559/FD396’s production of infectious virus surpassed that of the parental HPIV3 (HNQ552) regardless of the coinfection ratio used (Fig. 5B). At 2 days after infection, over 90% of the viral genome released carried HNR559, corresponding to HPIV3-HNQ552/R559/FD396.
Coinfection competition experiments were also performed in CV1 monolayer cell cultures using the virus combinations and ratios in Fig. 5. The HPIV3 reference strain and the parental HPIV3 (HPIV-HNQ552) both outcompete HPIV3-HNQ552/R559/FD396 in these cultures (data not shown). In cells infected with HPIV3-HNQ552/R559/FD396 alone, new plaque morphology was observed, and sequencing analysis revealed that HPIV3-HNQ552/R559/FD396 acquired new mutations only in the F protein. These are the subjects of ongoing study.
HPIV3-HNQ552/R559/FD396 is fit in the cotton rat model.
To assess the growth of HPIV3-HNQ552/R559/FD396 in vivo, we used the well-characterized cotton rat model for HPIV3 infection (31, 35). Cotton rats (five per group) were infected with the HPIV3 reference strain bearing HNH552, HPIV3-HNQ552, or HPIV3-HNQ552/R559/FD396. The animals were sacrificed 3 days after infection, and the lungs were used for viral titration (Fig. 6). As previously observed, growth of the virus bearing HPIV3-HNQ552 was attenuated relative to that of the reference HPIV3 (HNH552) (31, 35). HPIV3-HNQ552/R559/FD396 grew to titers 0.5 log higher than the HPIV3 reference strain and 2 to 3 logs higher than the parental virus, HPIV3-HNQ552, revealing enhanced fitness for growth in vivo.
FIG 6 .
HPIV3-HNQ552/R559/FD396 is fit in vivo. Cotton rats (5 per group) were infected with HPIV3-HNH552 (reference strain), HPIV3-HNQ552, or HPIV3-HNQ552/R559/FD396. At 3 days postinfection, the viral titer (PFU/g lung tissue) was determined by plaque assay.
DISCUSSION
Infection and growth of HPIV3 in the lung constitute a positive selection process in which the strength of binding to host cells and the strength of fusion activation strike a suitable balance. In the experiments presented here, adaptation of the HPIV3 HN-F fusion machinery for growth in the natural host highlights the critical nature of the balance between activation and stabilization of the fusion complex and identifies specific sites on HN that regulate HN-F interaction. A mutation at the HN dimer interface emerged that promotes the ability of the virus to infect lung tissue in explant models and in vivo. Structural analysis of these adaptively mutated HNs reveals that the mutation alters the dimer interface and weakens HN dimer association. Biological and biochemical data confirm the impact of the adaptive mutation on HN dimerization and HN-F interaction. Both HN dimerization and HN-F interaction affect HN’s ability to activate F and to mediate infection in the lung. The correlation between the structural consequences of mutations in HN and F that emerged during adaptation to lung growth, their impact on the function of HN and F, and their effect on viral infection in vivo indicate that the dimer interface (and its modulation of HN-F interaction) is critical to infection in the host.
HN is the driving force for fusion initiation and then for sustaining F’s role in mediating viral entry (13, 29, 52). In a variety of in vitro experimental situations, F can fuse alone (12, 29, 30, 53), or a “headless” HN may be sufficient to mediate F activation (54). However, this work shows that the function of specific residues in the globular head of HN is essential for infection in the host. Even a subtle change at the dimer interface of the globular domain can affect HN dimer association, impact the HN-F fusion machine, and markedly alter host infection.
We have identified critical features of the HN-F complex that correlate with infection in the host. Infection in natural host tissue with virus bearing HNQ552—an HN with high receptor avidity and active F-triggering capacity—was ineffective, although this HN confers ideal properties for growth in monolayer tissue culture. Sequential mutations that led to optimal growth in natural host tissue (and in vivo) led to a reduction in efficiency of the fusion machinery. While the adaptation of this virus may not reflect evolution of viruses in the population, since we started with a tissue culture-adapted strain, the requirements for infection in vivo were revealed. In the progression from fitness in monolayer culture toward fitness in host tissue, the virus underwent a decrease in receptor avidity, decrease in neuraminidase activity, decrease in fusion triggering capacity (33), decrease in HN homo-oligomerization, and decrease in the strength of interaction between the HN and F. The combination of these characteristics led to a diminished HN-F activation capacity and decreased fusion. The serial changes to the combined functions of HN and F coincide with optimal growth in host tissue. For optimal growth in the lung, we propose that viruses with lower HN-F interaction, a lower binding/neuraminidase ratio, and slower HN-F activation kinetics—in sum, a less active HN-F fusion machinery—will be at a selective advantage. In contrast, viruses that grow well in monolayer cell cultures have more avid HN-F interaction, a higher binding/neuraminidase ratio, faster HN-F activation kinetics and more fusogenic HN-F fusion machineries. It will be of interest to determine whether HPIV clinical isolates share the combination of HN-F complex features predicted by our results.
Comparison of the homodimerization tendency of different HN molecules—HNQ552 and HNQ552/R559—reveals that the Q559R mutation decreases HN-HN dimer association. Even when a dimer is “artificially” supported by an S554C mutation, the perturbation caused by Q559R affects HN function; the dimerization occurring in the presence of S554C does not rescue functional properties in the presence of Q559R (data not shown). These results, supported by the structural study and combined with the biological data, indicate that HN oligomerization is likely correlated with F activation and fusion. The H552 and Q559 residues are located at the HN dimer interface observed in the crystal structure. While the H552Q mutation confers higher avidity for receptor and increased F triggering relative to HNH552, the introduction of R559 into the HN protein has the opposite effect. Introducing R559 into the background of HNQ552 decreased HN’s avidity for receptor and its ability to activate the F protein. The introduction of R559 was the final step in the progression from a virus unfit for growth in lung to a stable HPIV3 variant that is highly infectious in HAE and in cotton rats. We proposed that the larger arginine at this position might disrupt the dimer due to charge repulsion and side chain clashes with other amino acids (33), and the structural analysis presented here shows that the introduced charge repulsion leads to a decreased buried surface area at the dimer interface.
Viral competition experiments reveal that HPIV3’s adaptation to changes in the host environment favors a homogenous population. We have previously shown that the constellation of functional properties of the viral surface proteins required for growth in vivo are different from those required in monolayer cell culture (33). The progression of an HPIV3 strain toward optimal growth in the environment of HAE and cotton rat lungs now reveals several of the mechanisms that account for this difference, and their structural correlates. We speculate that the density of receptors in the lung tissue may be a factor that impairs the fitness of the parent HPIV3-HNQ552 virus in HAE. HNQ552, avid for receptor and efficient at F activation, would promote excessive binding to sialic acid-containing moieties in the lung and premature activation of F in released virions and noninfectious particle production (31, 34). Mucus, rich in sialic acid-containing molecules, could provide abundant triggers for HN to activate F prematurely, thus producing noninfectious particles, since once F has been activated distant from the target, the virus is no longer viable. The virus that emerged after selective pressure for growth in HAE (HPIV3-HNQ552/R559/FD396 carries an HN with lower avidity for receptor and less efficient activation of F and is infectious in vivo. Ongoing studies will determine whether these precise slow-fusion characteristics are shared by HPIV3 clinical isolates.
Understanding the molecular attributes of HN and F as well as the timing, kinetics, and interaction between HN and F as a complex in lung tissue will improve our understanding of HPIV entry. We now consider the HN-F fusion machinery a highly interactive biological complex with its various components in balance—in contrast to the conventional notion, which focuses on specific isolated domains or residues. Our approach also enables us to design antiviral strategies that target specific biological roles of the fusion machinery and to test these strategies in a model that reflects the requirements for growth in vivo.
MATERIALS AND METHODS
Ethics statement.
All animal work with parainfluenza virus was performed in the laboratory animal facility at the Ohio State University (OSU). OSU is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All research involving animals was conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals (55). The animal use protocol was approved by the Institutional Animal Care and Use Committee at OSU (Animal Use Protocol 2009A0183; approved 20 October 2009).
Human airway epithelial cultures.
The EpiAirway AIR-100 system (MatTek Corporation) consists of normal, human-derived tracheobronchial epithelial cells that have been cultured to form a pseudostratified, highly differentiated mucociliary epithelium closely resembling that of epithelial tissue in vivo. Upon receipt from the manufacturer, HAE cultures were handled as previously described (31, 35). HAE cultures were infected with 4,000 PFU of the HPIV3 reference strain or variant as previously described (31, 35).
Monolayer cells.
CV-1 (African green monkey kidney) cells and 293T (human kidney epithelial) were grown in Dulbecco’s modified Eagle’s medium (Cellgro; Mediatech) supplemented with 10% fetal bovine serum and antibiotics at 37°C in 5% CO2.
Viruses.
Viral titers were determined as previously described (33). Virus isolation was performed as previously described (33).
Viral genome analysis.
RNA isolation and sequence analysis of HN and F genes were performed as previously described (33). To measure the amount of viral genome released, quantitative real-time PCR (qRT-PCR) was used. Thirty microliters of fluid containing released virus was processed for RNA extraction; then the genome was quantified using the real-time one-step qRT-PCR pathogen detection kit specific for HPIV3 (PrimerDesign) per the manufacturer’s instructions. To detect changes in the viral genome, we employed a custom TaqMan single-nucleotide polymorphism (SNP) genotyping assay (Applied Biosystems) as previously described (33).
Transient expression of HN and F genes.
Constructs used were as previously described (32, 33). Transfections were performed according to the Lipofectamine Plus or Lipofectamine 2000 manufacturer’s protocols (Invitrogen).
Bimolecular fluorescence complementation microscopy (BiFC) assay.
BiFC was performed as previously described (32). 293T cells were transiently transfected on biocoated Delta TPG dishes (Fisher Scientific) with the cDNA combinations indicated in Fig. 1 and 10% red fluorescence protein (RFP) using Lipofectamine 2000 according to the manufacturer’s instructions. The transfection ratio of the N-Venus/C-Venus–cyan fluorescent protein (CFP) constructs was 2:1. After 4 h of incubation at 37°C, the transfection mixture was replaced with complete medium (Dulbecco’s modified Eagle medium [DMEM]; 10% fetal bovine serum [FBS]–1% penicillin-streptomycin) supplemented with 10 mM zanamivir. Fifteen hours later, the medium was replaced with Optimem supplemented with 10 mM zanamivir and 100 ng/ml cycloheximide (Sigma) for 1 h at 37°C. Fluorescent images and the mean fluorometric ratio (calculated as the fluorescence intensity produced by BiFC divided by that of the RFP) were acquired using a confocal laser scanning microscope (Nikon TE-2000U Digital Eclipse C1si equipped with a spectral detector) using EZ-C1 acquisition and analysis software and a 60×, 1.4-numerical aperture (NA) oil objective. Venus, hybrid fluorescent complex, and RFP were excited at 488 nm (emission, 529 nm), 488 nm (emission, 513 nm) and 561 nm (emission, 610 nm), respectively. The same laser power and gain settings were used for all samples and for all replicate experiments.
Immunoprecipitation (IP) and capture of native HN complexes.
For radiolabeling, 293T cells were transiently transfected using Lipofectamine 2000 according to the manufacturer’s instructions with untagged HN cDNAs. The cells were incubated overnight in 30 mU exogenous neuraminidase (Sigma). The cells were starved for 2 h using DMEM without l-methionine or l-cystine (GIBCO) with 30 mU neuraminidase. The cells were washed and then incubated in medium containing 55 µCi of 35S-labeled methionine/cystine (PerkinElmer) and 30 mU neuraminidase for 2 h. Cycloheximide (100 ng/ml) was added to each well for 1 h. Cells were then lysed and immunoprecipitated using the Pierce cross-link IP kit (ThermoScientific) and a custom-made cDNA polyclonal HN antibody as described in the manufacturer’s instructions. For nonreducing gel analysis, immunoprecipitated samples were resuspended and boiled in 2× Novex Tris-glycine SDS sample buffer (Invitrogen), resolved by 4-to-20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto a polyvinyl difluoride membrane by electroblotting. Membranes were dried and then exposed to a storage phosphor screen for 5 h. The screen was scanned with a Typhoon imager (GE Healthcare). For native gel analysis, immunoprecipitated samples were resuspended in 4× native PAGE sample buffer (Invitrogen), resolved by 4-to-16% native PAGE on Novex bis-Tris gels (Invitrogen), and transferred onto a polyvinyl difluoride membrane by electroblotting. Membranes were dried and exposed to a storage phosphor screen for 5 h. The screen was scanned with a Typhoon imager (GE Healthcare).
Animals, infection, virus titration, and histology of animals.
Inbred cotton rats were obtained from Harlan (Indianapolis, IN). Female animals, 6 to 10 weeks of age, were infected as previously described (31, 35).
Cloning, expression, and purification of recombinant HN.
The gene corresponding to the ectodomain of hemagglutinin-neuraminidase (HN) (strain Wash/47885/57) (residues 54 to 572) was inserted into a baculovirus transfer vector, pFastbacHT-A (Invitrogen), with an N-terminal gp67 signal peptide, a His6 tag, and a thrombin cleavage site between the HN ectodomain and the His6 tag. The mutations H552Q and Q559R in HN were introduced by the polymerase incomplete primer extension cloning method (56). HN proteins were produced by infecting suspension cultures of Hi5 cells with recombinant baculovirus and incubated at 28°C with shaking at 110 rpm. After 72 h, Hi5 cells were removed by centrifugation. Supernatants containing secreted, soluble HNs were concentrated and buffer exchanged into 20 mM Tris (pH 8.0), 150 mM NaCl, 2.5 mM CaCl2, 10 mM imidazole. The HNs were recovered from the cell supernatants by metal affinity chromatography using nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen). The stem region of the HN sample was degraded during storage at 4°C, probably from contaminating protease activity. The remaining HN receptor-binding domain was further purified with gel filtration columns and concentrated before crystallization.
Crystallization and structural determination of HN.
HN proteins of the reference strain (3 mg/ml) and the HNQ552/R559 mutant (11 mg/ml) were crystallized by mixing with an equal volume (0.5 µl) of precipitant solution using the sitting-drop vapor diffusion method at 22.5°C. For reference strain HN, the precipitant in the reservoir contained 26% polyethylene glycol (PEG) 1000, 0.1 M phosphate citrate (pH 4.2), and 0.1 M Li2SO4. The HNQ552/R559 mutant was crystallized under similar conditions but with a lower PEG 1000 concentration (20%). Crystals were soaked in reservoir solution containing 20% PEG 1000 plus 15% ethylene glycol and flash cooled in liquid nitrogen. For the reference strain HN, diffraction data were collected at 100K on beamline 12-2 (wavelength, 0.97950 Å) at the Stanford Synchrotron Radiation Lightsource and processed with HKL2000 (57). For HNQ552/R559, diffraction data were collected at 100K on beamline 11-1 (wavelength, 0.97945 Å) at the Stanford Synchrotron Radiation Lightsource and processed with HKL2000 (57). The structures were solved by molecular replacement by Phaser (58) using the coordinates of PIV3 HN (PDB no. 1V3B). The structures were then adjusted using COOT (59) and refined with PHENIX (60). Statistics for data collection and structure refinement are presented in Table 1. In the final model of reference strain HN, 95.6% of the residues were in favored regions of the Ramachandran plot, with 4.3% in additional allowed regions. In the final model of HNQ552/R559, 94.7% of the residues were in favored regions of the Ramachandran plot, with 5.1% in additional allowed regions.
Protein data accession number.
The atomic coordinates and structure factor have been deposited in the Protein Data Bank under accession numbers 4MZA and 4MZE (http://www.rcsb.org).
ACKNOWLEDGMENTS
We thank Tim McGraw and Carl Nathan for helpful scientific discussions and advice and Christine C. Yokoyama for critical reading of the manuscript.
We are grateful to Ashton Kutcher and Jonathan Ledecky for their support, without which the microscopy critical to this work would not have been possible, to Dan and Nancy Paduano for their essential support of innovative research projects, and to the Friedman Family Foundation for our laboratories at Weill Cornell Medical College. This work was supported by NIH grant R01 AI31971 to A.M., NIH grant 3R01 AI031971-19S1 (Research Supplement to Promote Diversity in Health-Related Research Program) to A.M., and NIH grant R56 AI099275 to I.A.W. M.P. is a Friedman Family Research Scholar in Pediatric Infectious Diseases.
R.X., S.G.P., M.P., L.M.P., I.A.W., and A.M. conceived and designed the experiments. R.X., S.P., M.P., L.M.P., and S.N. performed the experiments. R.X., S.G.P., M.P., L.M.P., S.N., I.A.W., and A.M. analyzed the data. R.X., S.G.P., M.P., L.M.P., L.F.C., S.N., and A.M. contributed reagents, materials, and/or analysis tools. R.X., S.G.P., M.P., S.N., I.A.W., and A.M. wrote the paper.
Footnotes
Citation Xu R, Palmer SG, Porotto M, Palermo LM, Niewiesk S, Wilson IA, Moscona A. 2013. Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo. mBio 4(5):e00803-13. doi: 10.1128/mBio.00803-13.
REFERENCES
- 1. Moscona A, Peluso RW. 1991. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J. Virol. 65:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moscona A, Peluso RW. 1993. Relative affinity of the human parainfluenza virus type 3 hemagglutinin-neuraminidase for sialic acid correlates with virus-induced fusion activity. J. Virol. 67:6463–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagai S, Lamb RA. 1995. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between Simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J. Virol. 69:6712–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. 1992. Biological activity of paramyxovirus fusion proteins: Factors influencing formation of syncytia. J. Virol. 66:4564–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu XL, Ray R, Compans RW. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lamb RA. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1–11 [DOI] [PubMed] [Google Scholar]
- 7. Chaiwatpongsakorn S, Epand RF, Collins PL, Epand RM, Peeples ME. 2011. Soluble respiratory syncytial virus fusion protein in the fully cleaved, pretriggered state is triggered by exposure to low-molarity buffer. J. Virol. 85:3968–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. San-Juan-Vergara H, Sampayo-Escobar V, Reyes N, Cha B, Pacheco-Lugo L, Wong T, Peeples ME, Collins PL, Castaño ME, Mohapatra SS. 2012. Cholesterol-rich microdomains as docking platforms for respiratory syncytial virus in normal human bronchial epithelial cells. J. Virol. 86:1832–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang A, Masante C, Buchholz UJ, Dutch RE. 2012. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J. Virol. 86:3230–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schildgen V, van den Hoogen B, Fouchier R, Tripp RA, Alvarez R, Manoha C, Williams J, Schildgen O. 2011. Human metapneumovirus: lessons learned over the first decade. Clin. Microbiol. Rev. 24:734–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. 2009. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. 83:1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Porotto M, Salah ZW, Gui L, Devito I, Jurgens EM, Lu H, Yokoyama CC, Palermo LM, Lee KK, Moscona A. 2012. Regulation of paramyxovirus fusion activation: the hemagglutinin-neuraminidase protein stabilizes the fusion protein in a pre-triggered state. J. Virol. 86:12838–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4:613–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lamb RA, Paterson RG, Jardetzky TS. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee B, Ataman ZA. 2011. Modes of paramyxovirus fusion: a henipavirus perspective. Trends Microbiol. 19:389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moscona A. 2005. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J. Clin. Invest. 115:1688–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith EC, Popa A, Chang A, Masante C, Dutch RE. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J 276:7217–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Porotto M, Fornabaio M, Kellogg GE, Moscona A. 2007. A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J. Virol. 81:3216–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J. Virol. 85:12855–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng R, Wang Z, Mirza AM, Iorio RM. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457–469 [DOI] [PubMed] [Google Scholar]
- 21. Melanson VR, Iorio RM. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Porotto M, Murrell M, Greengard O, Doctor L, Moscona A. 2005. Influence of the human parainfluenza virus 3 attachment protein’s neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 79:2383–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Porotto M, Murrell M, Greengard O, Moscona A. 2003. Triggering of human parainfluenza virus 3 fusion protein(F) by the hemagglutinin-neuraminidase (HN): an HN mutation diminishing the rate of F activation and fusion. J. Virol. 77:3647–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sergel T, McGinnes LW, Peeples ME, Morrison TG. 1993. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 193:717–726 [DOI] [PubMed] [Google Scholar]
- 25. Stone-Hulslander J, Morrison TG. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73:3630–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanabayashi K, Compans RW. 1996. Functional interactions of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2011. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. U. S. A. 108:14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuasa T, Kawano M, Tabata N, Nishio M, Kusagawa S, Komada H, Matsumura H, Ito Y, Tsurudome M. 1995. A cell fusion-inhibiting monoclonal antibody binds to the presumed stalk domain of the human parainfluenza type 2 virus hemagglutinin-neuraminidase protein. Virology 206:1117–1125 [DOI] [PubMed] [Google Scholar]
- 29. Porotto M, Devito I, Palmer SG, Jurgens EM, Yee JL, Yokoyama CC, Pessi A, Moscona A. 2011. Spring-loaded model revisited: paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J. Virol. 85:12867–12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Porotto M, Salah Z, Devito I, Talekar A, Palmer SG, Xu R, Wilson IA, Moscona A. 2012. The second receptor binding site of the globular head of the Newcastle disease virus (NDV) hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J. Virol. 86:5730–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palermo LM, Porotto M, Yokoyama CC, Palmer SG, Mungall BA, Greengard O, Niewiesk S, Moscona A. 2009. Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J. Virol. 83:6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Porotto M, Palmer SG, Palermo LM, Moscona A. 2012. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J. Biol. Chem. 287:778–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palmer SG, Porotto M, Palermo LM, Cunha LF, Greengard O, Moscona A. 2012. Adaptation of human parainfluenza virus to airway epithelium reveals fusion properties required for growth in host tissue. mBio 3:e00137-12. 10.1128/mBio.00137-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farzan SF, Palermo LM, Yokoyama CC, Orefice G, Fornabaio M, Sarkar A, Kellogg GE, Greengard O, Porotto M, Moscona A. 2011. Premature activation of the paramyxovirus fusion protein before target cell attachment with corruption of the viral fusion machinery. J. Biol. Chem. 286:37945–37954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moscona A, Porotto M, Palmer S, Tai C, Aschenbrenner L, Triana-Baltzer G, Li QX, Wurtman D, Niewiesk S, Fang F. 2010. A recombinant sialidase fusion protein effectively inhibits human parainfluenza viral infection in vitro and in vivo. J. Infect. Dis. 202:234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hernandez FP, Sandri-Goldin RM. 2010. Head-to-tail intramolecular interaction of herpes simplex virus type 1 regulatory protein ICP27 is important for its interaction with cellular mRNA export receptor TAP /NXF1. mBio 1:e00268-10. 10.1128/mBio.00268-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hernandez FP, Sandri-Goldin RM. 2010. Herpes simplex virus 1 regulatory protein ICP27 undergoes a head-to-tail intramolecular interaction. J. Virol. 84:4124–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789–798 [DOI] [PubMed] [Google Scholar]
- 39. Hu CD, Kerppola TK. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kerppola TK. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin HP, Vincenz C, Eliceiri KW, Kerppola TK, Ogle BM. 2010. Bimolecular fluorescence complementation analysis of eukaryotic fusion products. Biol. Cell 102:525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Porotto M, Murrell M, Greengard O, Lawrence MC, McKimm-Breschkin JL, Moscona A. 2004. Inhibition of parainfluenza type 3 and Newcastle disease virus hemagglutinin-neuraminidase receptor binding: Effect of receptor avidity and steric hindrance at the inhibitor binding sites. J. Virol. 78:13911–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brindley MA, Plemper RK. 2010. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J. Virol. 84:12174–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robach JG, Lamb RA. 2010. Analysis of parainfluenza virus-5 hemagglutinin-neuraminidase protein mutants that are blocked in internalization and degradation. Virology 406:189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mahon PJ, Mirza AM, Musich TA, Iorio RM. 2008. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J. Virol. 82:10386–10396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335:1343–1357 [DOI] [PubMed] [Google Scholar]
- 47. Batschelet E, Domingo E, Weissmann C. 1976. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene 1:27–32 [DOI] [PubMed] [Google Scholar]
- 48. Chao L. 1990. Fitness of RNA virus decreased by Muller’s ratchet. Nature 348:454–455 [DOI] [PubMed] [Google Scholar]
- 49. Holland JJ, de la Torre JC, Clarke DK, Duarte E. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martínez MA, Carrillo C, González-Candelas F, Moya A, Domingo E, Sobrino F. 1991. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. J. Virol. 65:3954–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang L, Bukreyev A, Thompson CI, Watson B, Peeples ME, Collins PL, Pickles RJ. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79:1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steffen DL, Xu K, Nikolov DB, Broder CC. 2012. Henipavirus mediated membrane fusion, virus entry and targeted therapeutics. Viruses 4:280–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Russell CJ, Jardetzky TS, Lamb RA. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc. Natl. Acad. Sci. U. S. A. 109:E2625–E2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 56. Klock HE, Lesley SA. 2009. The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 498:91–103 [DOI] [PubMed] [Google Scholar]
- 57. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 58. McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. D61:458–464 [DOI] [PubMed] [Google Scholar]
- 59. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. D60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 60. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58:1948–1954 [DOI] [PubMed] [Google Scholar]