ABSTRACT
Bacteria in the biofilm mode of growth are protected against chemical and mechanical stresses. Biofilms are composed, for the most part, of extracellular polymeric substances (EPSs). The extracellular matrix is composed of different chemical constituents, such as proteins, polysaccharides, and extracellular DNA (eDNA). Here we aimed to identify the roles of different matrix constituents in the viscoelastic response of biofilms. Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus mutans, and Pseudomonas aeruginosa biofilms were grown under different conditions yielding distinct matrix chemistries. Next, biofilms were subjected to mechanical deformation and stress relaxation was monitored over time. A Maxwell model possessing an average of four elements for an individual biofilm was used to fit the data. Maxwell elements were defined by a relaxation time constant and their relative importance. Relaxation time constants varied widely over the 104 biofilms included and were divided into seven ranges (<1, 1 to 5, 5 to 10, 10 to 50, 50 to 100, 100 to 500, and >500 s). Principal-component analysis was carried out to eliminate related time constant ranges, yielding three principal components that could be related to the known matrix chemistries. The fastest relaxation component (<3 s) was due to the presence of water and soluble polysaccharides, combined with the absence of bacteria, i.e., the heaviest masses in a biofilm. An intermediate component (3 to 70 s) was related to other EPSs, while a distinguishable role was assigned to intact eDNA, which possesses a unique principal component with a time constant range (10 to 25 s) between those of EPS constituents. This implies that eDNA modulates its interaction with other matrix constituents to control its contribution to viscoelastic relaxation under mechanical stress.
IMPORTANCE
The protection offered by biofilms to organisms that inhabit it against chemical and mechanical stresses is due in part to its matrix of extracellular polymeric substances (EPSs) in which biofilm organisms embed themselves. Mechanical stresses lead to deformation and possible detachment of biofilm organisms, and hence, rearrangement processes occur in a biofilm to relieve it from these stresses. Maxwell analysis of stress relaxation allows the determination of characteristic relaxation time constants, but the biofilm components and matrix constituents associated with different stress relaxation processes have never been identified. Here we grew biofilms with different matrix constituents and used principal-component analysis to reveal that the presence of water and soluble polysaccharides, together with the absence of bacteria, is associated with the fastest relaxation, while other EPSs control a second, slower relaxation. Extracellular DNA, as a matrix constituent, had a distinguishable role with its own unique principal component in stress relaxation with a time constant range between those of other EPSs.
Introduction
Bacteria adhere to virtually all natural and artificial surfaces. Once they adhere to a surface, bacteria rapidly grow into a biofilm in which they are protected against chemical and mechanical stresses. The protection offered by the biofilm to organisms that inhabit it against chemical stresses, like antibiotic challenges, has been extensively studied (1–3) and is a huge problem in modern medicine, where biofilms account for 65% of all nosocomial infections in humans, costing over one billion dollars annually to treat in the United States alone (2, 4, 5). Few studies, however, have focused on how bacteria in a biofilm mode of growth cope with mechanical stresses. Oral biofilms on teeth are exposed to compressive stresses many times per day, especially when growing in fissures (6, 7). Also, intestinal biofilms are exposed to compressive stresses during peristaltic bowel movements. Compression of a biofilm leads to a more compact structure, which is undesirable from the perspective of nutrient penetration to deeper layers of a biofilm (8). In addition to compressive stresses, biofilms are subjected to tensile stresses. Tensile stresses develop in oral biofilms during tooth brushing and may eventually lead to detachment of biofilm organisms (9) and their subsequent death in the gastrointestinal tract. Also, tensile stresses on intestinal biofilms due to frictional forces arising from stool passage can cause detachment of biofilm organisms and their removal from their natural environment. Similar examples hold for other biofilms in the human body, as well as for biofilms in many natural and industrial environments (10).
The EPS (extracellular polymeric substance) matrix in which biofilm organisms embed themselves plays an important role in providing protection against chemical and mechanical stresses, which is required for their survival (11). This EPS consists of, among other components, different proteins, polysaccharides, and extracellular DNA (eDNA). Each constituent of the matrix has specific functions in maintaining overall biofilm health, including bacterial adhesion and cohesion, retention of water, formation of a protective barrier against chemical challenges, sorption of ions and compounds, and exportation of cell components (12). Different bacterial species have their own specific needs and thus require various proportions of matrix constituents in order to optimize their normal functioning. Pseudomonas aeruginosa biofilms contain copious amounts of eDNA (13) and extracellular polysaccharides, including alginates in cystic fibrosis pulmonary isolates (14, 15). Both Staphylococcus aureus and Staphylococcus epidermidis have extracellular proteins in their matrixes (16, 17). Interestingly, only S. aureus could be prevented from forming biofilms in the presence of DNase I (18). Streptococcus mutans uses extracellular glucans in the presence of sucrose to build its protective matrix (19, 20).
The chemical diversity and adaptability of the EPS matrix among different bacterial strains are important means through which a biofilm can protect itself against chemical challenges and mechanical stresses. The protection offered by the EPS matrix against chemical challenges has been well documented to result from reduced antimicrobial penetration of and adsorption to matrix constituents (2, 21–23). Biofilms are both viscous and elastic in nature, enabling bacteria in the biofilm mode of growth to survive mechanical stresses (24, 25), but it is unknown how the different biofilm components and matrix constituents contribute to the viscoelastic response of biofilms to mechanical stresses (26). One way to analyze the viscoelastic response of biofilms to mechanical stress is stress relaxation measurement. Stress relaxation measurement indicates how a biofilm relieves itself from external stresses, and by using Maxwell analyses (25), the different relaxation processes that occur in a biofilm under mechanical stress can be mathematically modeled. Maxwell analyses yield a spring constant and a characteristic time constant for each of the relaxation processes that occur, but interpretation has seldom gone beyond their mathematical background. Tentative interpretations have attributed the fastest relaxation element to the flow of water in mechanically stressed biofilms, as water has the lowest viscosity of all biofilm components. On the other hand, the organisms themselves represent the heaviest masses in a biofilm and their rearrangement can thus be expected to coincide with the slowest stress relaxation element. This leaves a wide array of stress relaxation elements with intermediate characteristic time constants that have been attributed to the flow of EPS. However, intuition has been the only underlying argument for these associations, while the roles of the different constituents of the EPS matrix in stress relaxation have remained obscure.
Here we have measured compressive stress relaxation of biofilms of different genera (see Table 1) and used a generalized Maxwell model with the aim to relate different biofilm components and matrix constituents to the different stress relaxation elements obtained, with a focus on the constituents of the EPS matrix. Biofilms were grown in which specific EPS constituents like polysaccharides, glucans, or eDNA were present, naturally absent, or chemically altered (see Table 1). The total range of characteristic relaxation time constants observed over 104 different biofilms was divided into seven time constant ranges and subjected to a principal-component analysis that yielded three new principal components that were subsequently related to the chemically derived matrix chemistries of the biofilms. A highly distinguishable role was assigned to intact eDNA as a matrix constituent that possesses its own unique principal component with a time constant range between those of other EPS constituents. This implies that intact eDNA, when present, may interact with other EPS constituents to form agglomerates with a unique response to the mechanical stresses imposed upon a biofilm.
TABLE 1 .
Matrix chemistries as chemically determined for biofilms of different species and resulting from different biofilm treatments, including the bacterial strains and growth mediums involved in this studya
| Treatment, matrix chemistryb | Bacterial strain | Growth mediumc | Avg soluble polysaccharide concn (μg/ml) ± SD | Avg eDNA concn (μg/ml) ± SD |
|---|---|---|---|---|
| No treatment, naturally occurring EPS (12) | P. aeruginosa SG81 | Nutrient broth | 134 ± 25 | 130.7 ± 5.7 |
| No treatment, no naturally occurring EPS (39) | P. aeruginosa SG81-R1 | Nutrient broth | 28 ± 0 | 49.2 ± 7.8 |
| MgCl2, naturally occurring EPS (13, 14) | P. aeruginosa SG81 | Nutrient broth | 76 ± 18 | 70.0 ± 3.4 |
| DNase I with MgCl2, EPS with less eDNA (18) | P. aeruginosa SG81 | Nutrient broth | 60 ± 17 | No intact DNA detectedd |
| Phosphate-buffered saline, naturally occurring EPS (13, 14) | P. aeruginosa SG81 | Nutrient broth | 103 ± 10 | 74.6 ± 1.8 |
| N-Acetyl-l-cysteine, EPS with less polysaccharides (40) | P. aeruginosa SG81 | Nutrient broth | 68 ± 12 | 68.5 ± 8.6 |
| 3.0% sucrose added to agar, glucan-rich EPS matrix (19, 20) | S. mutans ATCC 25175 | Trypticase soy broth | 33 ± 9 | 0.183 ± 0.216 |
| No treatment, naturally occurring EPS (41) | S. mutans ATCC 25175 | Trypticase soy broth | 4 ± 1 | 0.749 ± 0.105 |
| No treatment, naturally occurring EPS (42) | S. aureus ATCC 12600 | Trypticase soy broth | 10 ± 1 | 0.622 ± 0.593 |
| No treatment, no EPS matrix (43) | S. aureus 5298 | Trypticase soy broth | 10 ± 3 | 0.150 ± 0.068 |
| No treatment, naturally occurring EPS (44) | S. epidermidis HBH 45 | Trypticase soy broth | 12 ± 8 | 5.25 ± 2.90 |
| No treatment, no EPS matrix (45) | S. epidermidis ATCC 12228 | Trypticase soy broth | 9 ± 3 | 1.62 ± 0.93 |
Chemical determination was performed in triplicate on biofilms not subjected to deformation.
Chemical treatments were applied to fully grown biofilms, except for the addition of sucrose to the agar growth medium of S. mutans biofilms.
Bacteria were cultured on agar plates with growth medium appropriate for the specific strain (containing 12 g/liter agar).
See Fig. S2 in the supplemental material.
RESULTS
S. aureus, S. epidermidis, S. mutans, and P. aeruginosa (Table 1) were grown on filters, placed on agar plates, and subjected to chemical treatments to yield distinct EPS matrix chemistries, as chemically measured. DNase I treatment was used to break down the eDNA, and N-acetyl-l-cysteine was used to break down polysaccharides in the EPS matrix of P. aeruginosa. S. mutans biofilms were grown with extra sucrose in the growth medium to create a glucan-rich EPS matrix.
Viscoelastic relaxation of compressed biofilms.
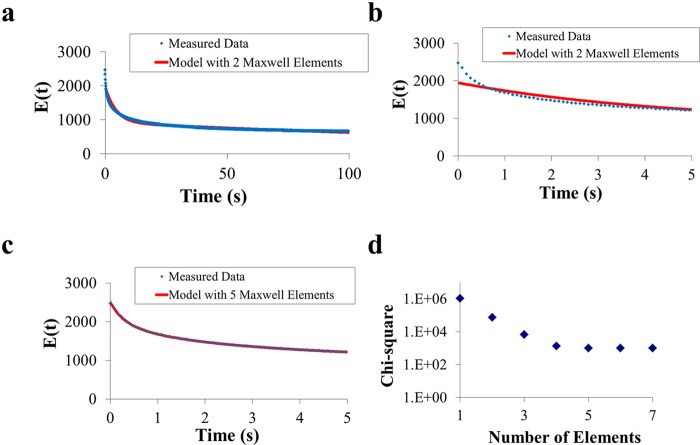
The biofilms were compressed to 80% of their original thickness and held in the deformed state for 100 s. The stress required to keep the biofilms in the deformed state decreased with time because of different rearrangement processes in the deformed biofilm (Fig. 1a to c), including flow of EPS matrix constituents and water. The decrease in stress for each biofilm was modeled by using a generalized Maxwell model with each element having a spring constant related to the elastic part of the biofilm and a characteristic relaxation time constant related to the ratio of the viscous and elastic parts of the biofilm. Initially, one Maxwell element was used to fit to the relaxation curve, after which additional elements were added. Four Maxwell elements generally sufficed to accurately model stress relaxation of the biofilms, and further addition of elements did not improve the quality of the fit (Fig. 1d). Biofilms containing an EPS matrix required more Maxwell elements (four or five) to describe the stress relaxation than biofilms without an EPS matrix (two or three).
FIG 1 .
Panels a to c represent the measured stress relaxation of a P. aeruginosa SG81 biofilm as a function of time, together with model fits to the data, obtained by using two (panels a and b) or five (panel c) Maxwell elements. Note that panel a extends over 100 s, while panels b and c refer only to the first 5 s of the relaxation process. Panel d represents the quality of the fit, indicated by chi-square values, as a function of the number of Maxwell elements used for the fit.
Principal-component analysis.
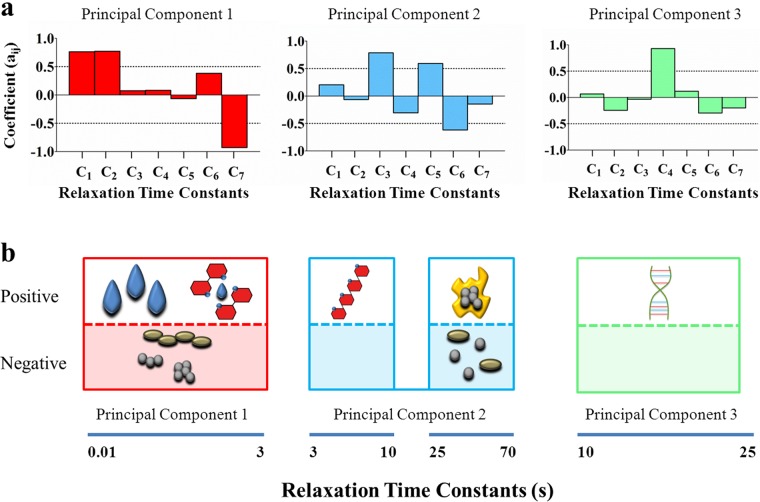
The relaxation time constants of all of the Maxwell elements of the 104 biofilms comprised in this study, taking replicate runs as a separate biofilm, were plotted as a function of the relative importance of their Maxwell elements (Fig. 2). Relaxation time constants spread over a wide time range, and hence, the total range of relaxation time constants observed over the 104 biofilms investigated was divided into seven relaxation time constant ranges on a semilog basis as follows: <1, 1 to 5, 5 to 10, 10 to 50, 50 to 100, 100 to 500, and >500 s. A principal-component analysis was carried out to determine possible interdependence among the different time constant ranges and to reduce the number of time constant ranges. However, on the basis of this division of the total range of relaxation time constants, it occurred 42 times in the total of 442 Maxwell elements measured that one biofilm possessed two data points in one relaxation time constant range. Since this impedes principal-component analysis, the semilog-based initial division was slightly adjusted to eliminate this redundancy. This led to a new division of relaxation time constant ranges (Ci) according to the relaxation time ranges <0.75, 0.75 to 3, 3 to 10, 10 to 25, 25 to 70, 70 to 460, and >460 s, which were subjected to a principal-component analysis. The principal-component analysis yielded three new principal components (PC1, PC2, and PC3) in terms of coefficients of the seven initial time ranges to describe the stress relaxation of the different biofilms (Fig. 3a), accounting, respectively, for 31, 22, and 15% of the variance observed. Incidentally, it was noted that no redundancy occurred when the total time constant range was divided into a higher number of subranges while yielding similar results for the resulting principal components.
FIG 2 .
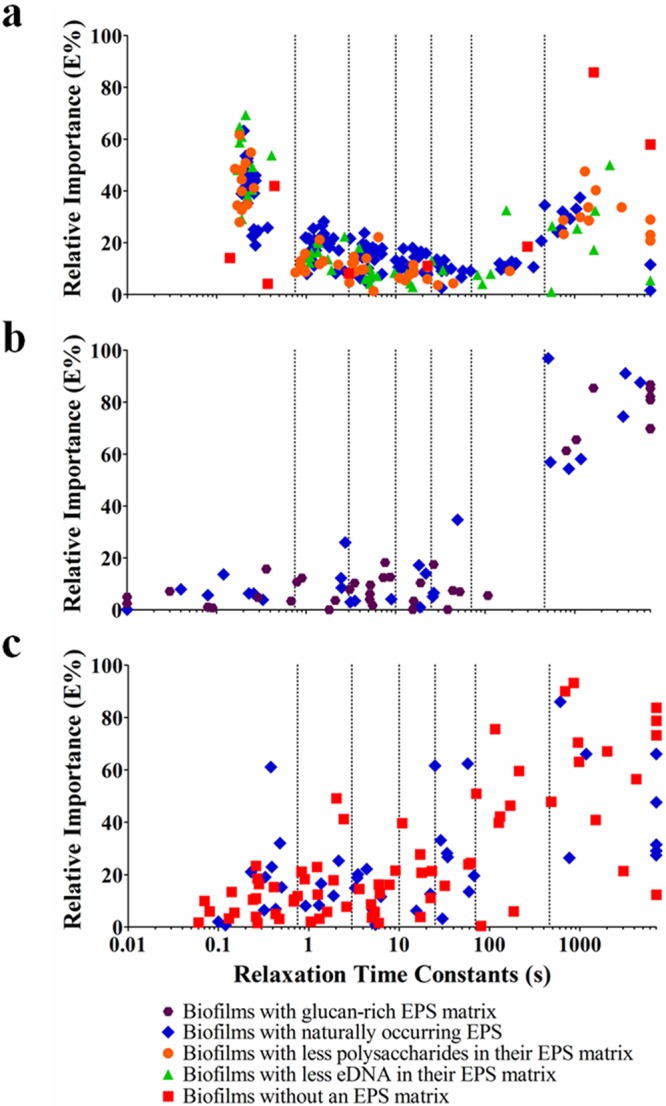
Relative importance of the individual Maxwell elements of different biofilms as a function of their characteristic relaxation time constants in relation to the different matrix chemistries according to Table 1. Each data point represents one Maxwell element with its time constant plotted against its relative importance. Each individual biofilm possessed an average of four or five Maxwell elements. Similar biofilms were grown and investigated minimally three times with separate initial bacterial cultures. Maxwell elements with 0% relative importance have no accompanying time constant and are not plotted, while characteristic time constants exceeding 7,000 s have been assigned a value of 7,000 s. Vertical lines indicate divisions of relaxation time constant ranges (Ci). Panels: a, P. aeruginosa biofilms; b, S. mutans biofilms; c, S. aureus and S. epidermidis biofilms.
FIG 3 .
Identification of matrix chemistries and biofilm components responsible for stress relaxation in biofilms.
Prominent properties of the biofilms significantly represented in each of the principal components were identified by statistical comparison (Mann-Whitney U test, P < 0.05; see Fig. S1 in the supplemental material), as related to the strain-specific biofilm properties listed in Table 1. The principal component comprising the two fastest initial elements (PC1) is negatively impacted by the slowest initial element (Fig. 3a). Rearrangement of bacteria within a deformed biofilm can be considered the slowest process, as bacteria constitute the heaviest masses. At the same time, water, with dissolved components, has the lowest viscosity in a biofilm and its flow will form the basis of fast relaxation. Therewith, the process assignment to this principal component becomes quite logical, since the presence of water implies the absence of bacterial cells. The second principal component (PC2) encompasses intermediate time constant ranges (Fig. 3a) of 3 to 10 and 25 to 70 s that are associated by statistical comparison with EPS (see also Fig. S1). This too is quite logical, as EPS is a more viscous material than water. Interestingly, the third principal component (PC3) contains only one initial time constant range that was uniquely associated with the absence or presence of intact eDNA as an extracellular matrix constituent (Fig. 3a).
DISCUSSION
The viscoelasticity of biofilms reflects their structure and composition and serves, among other functions, to protect a biofilm against mechanical and chemical challenges. Little is known, however, about the biofilm components and matrix constituents that are responsible for the stress relaxation processes within a biofilm as a response to mechanical deformation. In this study, we analyzed the stress relaxation of >100 different biofilms and chemically determined their matrix chemistries. Stress relaxation obeyed a generalized Maxwell model generally comprising four or five Maxwell elements. Using principal-component analysis, we are the first to establish that three components suffice to describe the viscoelastic relaxation of mechanically deformed biofilms.
The first principal component comprises the fastest two initial time ranges (<0.75 and 0.75 to 3 s). The fastest time range was associated with the flow of water on the basis of its low viscosity and incompressibility. Similar stress relaxation times have been found for the cytoplasm of a macrophage (27) under creep and flow of aqueous solutions through micrometer-size channels (25). With water having been associated with the fastest relaxation time constant range, the next fastest initial time range could be associated with soluble polysaccharides (Fig. 3b). The second principal component comprises two initial intermediate time ranges (3 to 10 and 25 to 70 s) that could be associated with other EPS polymers, including glucans (Fig. 3b), with a noticeable separation.
The third principal component is the most interesting one, as it comprises a single, relatively narrow relaxation time range (10 to 25 s) that could be uniquely associated with the presence of intact eDNA as a matrix constituent (Fig. 3b). eDNA as a matrix constituent originates from chromosomal DNA and is thought to be produced through active processes such as autolysis or vesicular secretion (12, 28, 29). Several recent reports have shown that eDNA is involved in different stages of biofilm formation, including initial bacterial adhesion, aggregation (12, 30), biofilm architecture (31), and mechanical stabilization of biofilms (29). eDNA performs its role as a pivotal matrix constituent through acid-base interactions with bacterial cell surfaces and polysaccharides (30, 32). The ability of eDNA to interact with polysaccharides coincides with the position of the third principal component, which was found to be uniquely due to intact eDNA, between two initial time ranges associated with the presence of other EPS polymers. The filamentous structure of eDNA (12) allows it to form agglomerates with smaller polysaccharides and globular proteins found in the EPS matrix by means of acid-base interactions. The current highly distinguishable role of intact eDNA in the stress relaxation of deformed biofilms suggests that these agglomerates are well-defined structures, otherwise they could not form a single principal component with a narrow time constant range. The narrow range of the relaxation time constant associated with the presence of intact eDNA in a biofilm is likely controlled by the length of the DNA strands. Shorter fragments of eDNA strands are more quickly adsorbed onto bacterial cells, while longer fragments tend to have more functional responsibilities (33) and are thus more readily involved in the formation of agglomerates controlling stress relaxation.
Conclusions.
Mechanical stresses lead to deformation and possible detachment of biofilm organisms, and hence, rearrangement processes occur in a biofilm to relieve it from these stresses and maintain its integrity. Maxwell analysis of the stress relaxation of biofilms allows the determination of characteristic relaxation time constants, but hitherto, biofilm components and matrix constituents associated with stress relaxation have never been identified. Using specific bacterial pairs with distinct EPS chemistries as chemically determined, we have, for the first time, related purely mathematical Maxwell elements describing stress relaxation to biofilm components and matrix constituents on the basis of the characteristic relaxation time constants of the Maxwell elements in terms of three principal components. The presence of water or the absence of bacteria, for that matter, was associated with the fastest relaxation process, while the rearrangement of other EPS polymers controls a second, slower relaxation process. Interestingly, intact eDNA as a matrix constituent had a distinguishable role in stress relaxation, with its own unique principal component. Although several functions of eDNA as a matrix constituent have been demonstrated in recent years, such a distinguishable role in stress relaxation is new and adds to the importance of eDNA in biofilm structure and function.
MATERIALS AND METHODS
Biofilm growth and application of chemical treatments.
Bacterial strains (Table 1) were stored at −80°C in 7% dimethyl sulfoxide, grown on sheep blood agar plates, and cultured in 10 ml of growth medium (37°C, 17 h). Bacteria were sonicated (10 W, 10 s, 0°C) to disrupt possible aggregates and enumerated in a Bürker Türk counting chamber. Sterile demineralized water (100 ml) and 1 × 108 bacteria were deposited on a membrane filter (0.4-µm pore size, 4.6-cm diameter, HTTP; Millipore, Tullagreen, Carrigtwohill, Ireland) under negative pressure and washed in demineralized water (50 ml) for an additional 30 s. Subsequently, the filter with the appropriate growth medium for each bacterial strain (Table 1) was moved onto agar plates containing 12 g/liter Bacto agar (BD, Le Pont de Claix, France) with the bacterial side up and incubated at 37°C for 48 h. All growth media were purchased from Oxoid (Basingstoke, United Kingdom), while chemicals were purchased from Sigma (St. Louis, MO) or Merck (Darmstadt, Germany).
Two P. aeruginosa strains were used, SG81 and SG81-R1; SG81-R1 is an isogenic mutant deficient in matrix production. In order to increase or decrease the prevalence of different constituents in the EPS matrix of P. aeruginosa SG81, biofilms were subjected to treatment (2 h, 37°C) with phosphate-buffered saline (PBS; 5 mM K2HPO4, 5 mM KH2PO4, 150 mM NaCl, pH 7.0), PBS supplemented with 10 mM MgCl2 and DNase I (0.25 U/ml; Fermentas Life Sciences, Roosendaal, The Netherlands), PBS supplemented with 10 mM MgCl2, or PBS supplemented with 2 mg/ml N-acetyl-l-cysteine. S. mutans biofilms were grown in the absence or presence of sucrose to vary the amount of glucans in the matrix. S. aureus ATCC 12600 and strain 5298 and S. epidermidis HBH45 and ATCC 12228 were selected as representatives of the genus Staphylococcus for their known ability to produce biofilms with or without an EPS matrix, respectively. Table 1 summarizes the chemical characteristics of the EPS matrixes of the different biofilms grown, as chemically derived in this study.
Soluble-polysaccharide determination.
Forty-eight-hour biofilms were submerged in 5 ml of PBS and vortexed for 1 min, and the resulting fluid was centrifuged (BHG HEKA no. 29380 centrifuge, setting 4, 10 min). One milliliter of the supernatant was mixed with 2 ml of anthrone (1 mg/ml in concentrated H2SO4). The samples were allowed to react for 10 min, and the absorbance at 630 nm was read (Spectronic GENESYS 20) and compared against glucose standards. Final sugar concentrations were reported in equivalent glucose units (34, 35). Polysaccharide determination was performed in triplicate (Table 1).
eDNA determination.
Forty-eight-hour biofilms were submerged in 2 ml of eDNA extraction buffer (10 mM EDTA, 0.9% NaCl) and vortexed for 1 min. Additional buffer was added to P. aeruginosa biofilms to help pellet the matrix material during centrifugation (BHG HEKA no. 29380 centrifuge, setting 4, 10 min). Dilutions were made for the supernatant in extraction buffer, and 1 ml of solution was combined with 500 µl of phenol (8.3 g/ml) and 500 µl of chloroform. After centrifugation (2,700 × g, 5 min, 10°C), the aqueous layer was collected and an additional 500 µl of chloroform was added before centrifugation (2,700 × g, 5 min, 10°C). Two 700-µl aliquots of the aqueous layer were mixed with 140 µl of 3 M sodium acetate and 460 µl of isopropyl alcohol. The aliquots were centrifuged (15,300 × g, 20 min, 10°C), 690 µl of the aqueous layer was removed, and 500 µl of 100% ethanol was added. The samples were centrifuged (15,300 × g, 15 min, 10°C), and the liquid phase was removed, leaving 50 to 100 µl to evaporate overnight. Samples were reconstituted with 45 µl of extraction buffer (4 h, 20°C). The two aliquots were recombined, forming one sample, and treated with 4 µl of RNase A (20 mg/ml, 30 min, 37°C) (36). Samples were analyzed with the CYQuant kit (Molecular Probes) for fluorescence intensity against DNA standards (480 and 520 nm; FLUOstar OPTIMA plate reader). eDNA determination was performed in triplicate (Table 1).
In order to verify whether the analysis was pertinent to intact eDNA, biofilms of P. aeruginosa SG81 prior to and after treatment with MgCl2 and DNase I were run on a 1% agarose gel for 90 min at 65 W (see Fig. S2 in the supplemental material).
Biofilm compression and analysis of viscoelastic relaxation.
Biofilms were deformed with a low-load compression tester (37). Briefly, a stainless steel plunger (diameter, 0.25 cm) was lowered toward a sample stage and the position of the stage was recorded. Next the plunger was lowered toward the top of the biofilm until a touch load of 0.01 g was achieved and its position was recorded again. The difference between plunger positions determined the thickness of the biofilms. Next, the biofilms were deformed 20% (strain 0.2) in 1 s and the deformation was subsequently held constant for 100 s while stress development was monitored over time (38). Stress relaxation as a function of time, E(t), was fitted by using a generalized Maxwell model according to the equation
where E(t) is the total stress divided by the induced strain expressed as the sum of i Maxwell elements with a spring constant Ei and characteristic relaxation time τi. Model fitting was performed with the Microsoft Excel 2007 Solver module without imposing any restrictions on the value of Ei or τi, except that the value had to be positive to maintain its physical relevance and τi had to be >0.01 s. Initially, one Maxwell element was used to fit to the stress relaxation data and then additional elements were added until no further decrease in chi-squared values was observed (Fig. 1). For each biofilm, a relative importance was assigned to each element on the basis of the value of its spring constant, Ei, and expressed as its spring constant’s percentage of the sum of all of the elements’ spring constants at t = 0 according to the equation
Identification of biofilm components and matrix constituents that influence the viscoelastic deformation of compressed biofilms.
The first step in the identification of biofilm components and matrix constituents that influence the viscoelastic relaxation of compressed biofilms was to divide the total range of relaxation time constants observed over the 104 biofilms investigated into seven initial relaxation time constant ranges on a semilog basis as follows: <1, 1 to 5, 5 to 10, 10 to 50, 50 to 100, 100 to 500, and >500 s. On the basis of the relative importance, Ēi, of the data in each time range, a principal-component analysis (SPSS v. 16.0 for Windows, data reduction factor analysis, principal-component analysis with a maximum of 25 iterations) was carried out to identify which combinations of relaxation time constant ranges could explain the variance in the data set best. The resulting new principal components (PCj) comprised coefficients from the seven initial relaxation time ranges according to the equation
for j = 1 to 3 and in which Ēi is the relative importance of the spring constants in each initial time constant range i and aij, the corresponding coefficients. The value for each principal component was calculated by using the equation above.
Next, median values of the principal components for each stress relaxation experiment were calculated according to the equation above. Results for bacterial pairs with known differences in their matrix chemistry were compared by using a Mann-Whitney U test. Whenever median values for biofilms with a specific known matrix chemistry were significantly higher than data for biofilm lacking that specific chemistry at a level of P < 0.05, the chemistry was related to a specific principal component, PCj (see also Fig. S1 in the supplemental material).
SUPPLEMENTAL MATERIAL
Principal-component (PCj) values were determined for each biofilm and statistically compared for specific bacterial pairs possessing distinctly different matrix chemistries (Mann-Whitney U test, P < 0.05 [*]). Matrix compositions (Table 1; for colors, see Fig. 2) were used to isolate matrix constituents involved and assigned to a principal component in case that constituent was expressed significantly more by strains having a higher value of the principal component. Download
One percent agarose gels containing eDNA from the matrices of differently treated P. aeruginosa SG81 biofilms. (a) Molecular weight ladder consisting of genomic DNAs of different strand lengths (1 to 10 kb). (b) Untreated biofilm. High-molecular-weight DNA is indicated by the arrow. (c) Biofilm after MgCl2 treatment. High-molecular-weight DNA is indicated by the arrow. (d) Biofilm after treatment with MgCl2 and DNase I and containing smears of degraded genomic DNA and a unique region of increased DNA density consisting of smaller strands (<1 kb), indicated by the white rectangle, as also demonstrated by Lee et al. (CI Lee, SH Leong, AE Png, KW Choo, C Syn, DT Lim, HY Law, and OL Kon, Nat. Protoc. 1:2185–2194, 2006). Download
Footnotes
Citation Peterson BW, van der Mei HC, Sjollema J, Busscher HJ, Sharma PK. 2013. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. mBio 4(5):e00497-13. doi:10.1128/mBio.00497-13.
REFERENCES
- 1. Goward CR, Nicholls DJ. 1994. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 3:1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mah TF, O’Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 3. Schmid T, Burkhard J, Yeo BS, Zhang W, Zenobi R. 2008. Towards chemical analysis of nanostructures in biofilms I: imaging of biological nanostructures. Anal. Bioanal. Chem. 391:1899–1905 [DOI] [PubMed] [Google Scholar]
- 4. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 5. Archibald LK, Gaynes RP. 1997. Hospital-acquired infections in the United States. The importance of interhospital comparisons. Infect. Dis. Clin. North Am. 11:245–255 [DOI] [PubMed] [Google Scholar]
- 6. Bowden GH, Hamilton IR. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54–85 [DOI] [PubMed] [Google Scholar]
- 7. Alexandridis C, Thanos CE, Caputo AA. 1981. Distribution of stress patterns in the human zygomatic arch and bone. J. Oral Rehabil. 8:495–505 [DOI] [PubMed] [Google Scholar]
- 8. Sjollema J, Rustema-Abbing M, Van der Mei HC, Busscher HJ. 2011. Generalized relationship between numbers of bacteria and their viability in biofilms. Appl. Environ. Microbiol. 77:5027–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Busscher HJ, Jager D, Finger G, Schaefer N, Van der Mei HC. 2010. Energy transfer, volumetric expansion, and removal of oral biofilms by non-contact brushing. Eur. J. Oral Sci. 118:177–182 [DOI] [PubMed] [Google Scholar]
- 10. Stoodley P, Cargo R, Rupp CJ, Wilson S, Klapper I. 2002. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J. Ind. Microbiol. Biotechnol. 29:361–367 [DOI] [PubMed] [Google Scholar]
- 11. Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229–1238 [DOI] [PubMed] [Google Scholar]
- 12. Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 13. Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 14. Orgad O, Oren Y, Walker SL, Herzberg M. 2011. The role of alginate in Pseudomonas aeruginosa EPS adherence, viscoelastic properties and cell attachment. Biofouling 27:787–798 [DOI] [PubMed] [Google Scholar]
- 15. Coulon C, Vinogradov E, Filloux A, Sadovskaya I. 2010. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS One 5:e14220. 10.1371/journal.pone.0014220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gustafsson E, Jacobsson G, Nilsson P, Enroth H, Beronius MK, Andersson R, Arvidson S. 2009. Invasive Staphylococcus aureus strains are highly variable in PFGE patterns, agr group and exoprotein production. Scand. J. Infect. Dis. 41:577–583 [DOI] [PubMed] [Google Scholar]
- 17. Hussain M, Herrmann M, Von Eiff C, Perdreau-Remington F, Peters G. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65:519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duarte S, Klein MI, Aires CP, Cury JA, Bowen WH, Koo H. 2008. Influences of starch and sucrose on Streptococcus mutans biofilms. Oral Microbiol. Immunol. 23:206–212 [DOI] [PubMed] [Google Scholar]
- 20. Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AH, Hamaker BR, Lemos JA, Koo H. 2010. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS One 5:e13478. 10.1371/journal.pone.0013478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fux CA, Costerton JW, Stewart PS, Stoodley P. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34–40 [DOI] [PubMed] [Google Scholar]
- 22. Stewart PS. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107–113 [DOI] [PubMed] [Google Scholar]
- 23. Szomolay B, Klapper I, Dockery J, Stewart PS. 2005. Adaptive responses to antimicrobial agents in biofilms. Environ. Microbiol. 7:1186–1191 [DOI] [PubMed] [Google Scholar]
- 24. Shaw T, Winston M, Rupp CJ, Klapper I, Stoodley P. 2004. Commonality of elastic relaxation times in biofilms. Phys. Rev. Lett. 93:098102 [DOI] [PubMed] [Google Scholar]
- 25. Cense AW, Peeters EA, Gottenbos B, Baaijens FP, Nuijs AM, Van Dongen ME. 2006. Mechanical properties and failure of Streptococcus mutans biofilms, studied using a microindentation device. J. Microbiol. Methods 67:463–472 [DOI] [PubMed] [Google Scholar]
- 26. Gautieri A, Vesentini S, Redaelli A, Buehler MJ. 2012. Viscoelastic properties of model segments of collagen molecules. Matrix Biol. 31:141–149 [DOI] [PubMed] [Google Scholar]
- 27. Bausch AR, Möller W, Sackmann E. 1999. Measurement of local viscoelasticity and forces in living cells by magnetic tweezers. Biophys. J. 76:573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas VC, Thurlow LR, Boyle D, Hancock LE. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190:5690–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barnes AM, Ballering KS, Leibman RS, Wells CL, Dunny GM. 2012. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio 3:e00193-12. 10.1128/mBio.00193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Das T, Sharma PK, Busscher HJ, Van der Mei HC, Krom BP. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76:3405–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu W, Li L, Sharma S, Wang J, McHardy I, Lux R, Yang Z, He X, Gimzewski JK, Li Y, Shi W. 2012. DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS One 7:e51905. 10.1371/journal.pone.0051905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Das T, Sharma PK, Krom BP, Van der Mei HC, Busscher HJ. 2011. Role of eDNA on the adhesion forces between Streptococcus mutans and substratum surfaces: influence of ionic strength and substratum hydrophobicity. Langmuir 27:10113–10118 [DOI] [PubMed] [Google Scholar]
- 33. Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6:37–53 [DOI] [PubMed] [Google Scholar]
- 34. Helbert JR, Brown KD. 1956. Color reaction of hexuronic acids with anthrone. J. Anal. Chem. 28:1098–1100 [Google Scholar]
- 35. Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350–356 [Google Scholar]
- 36. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 37. Körstgens V, Flemming HC, Wingender J, Borchard W. 2001. Uniaxial compression measurement device for investigation of the mechanical stability of biofilms. J. Microbiol. Methods 46:9–17 [DOI] [PubMed] [Google Scholar]
- 38. Peterson BW, Busscher HJ, Sharma PK, Van der Mei HC. 2012. Environmental and centrifugal factors influencing the visco-elastic properties of oral biofilms in vitro. Biofouling 28:913–920 [DOI] [PubMed] [Google Scholar]
- 39. Gómez-Suárez C, Pasma J, Van der Borden AJ, Wingender J, Flemming HC, Busscher HJ, Van der Mei HC. 2002. Influence of extracellular polymeric substances on deposition and redeposition of Pseudomonas aeruginosa to surfaces. Microbiology 148:1161–1169 [DOI] [PubMed] [Google Scholar]
- 40. Marchese A, Bozzolasco M, Gualco L, Debbia EA, Schito GC, Schito AM. 2003. Effect of fosfomycin alone and in combination with N-acetylcysteine on E. coli biofilms. Int. J. Antimicrob. Agents 22(Suppl 2):95–100 [DOI] [PubMed] [Google Scholar]
- 41. Kreth J, Zhu L, Merritt J, Shi W, Qi F. 2008. Role of sucrose in the fitness of Streptococcus mutans. Oral Microbiol. Immunol. 23:213–219 [DOI] [PubMed] [Google Scholar]
- 42. Møretrø T, Hermansen L, Holck AL, Sidhu MS, Rudi K, Langsrud S. 2003. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 69:5648–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rasyid HN, Van der Mei HC, Frijlink HW, Soegijoko S, Van Horn JR, Busscher HJ, Neut D. 2009. Concepts for increasing gentamicin release from handmade bone cement beads. Acta Orthop. 80:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Van der Mei HC, van de Belt-Gritter B, Reid G, Bialkowska-Hobrzanska H, Busscher HJ. 1997. Adhesion of coagulase-negative staphylococci grouped according to physico-chemical surface properties. Microbiology 143:3861–3870 [DOI] [PubMed] [Google Scholar]
- 45. Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577–1593 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal-component (PCj) values were determined for each biofilm and statistically compared for specific bacterial pairs possessing distinctly different matrix chemistries (Mann-Whitney U test, P < 0.05 [*]). Matrix compositions (Table 1; for colors, see Fig. 2) were used to isolate matrix constituents involved and assigned to a principal component in case that constituent was expressed significantly more by strains having a higher value of the principal component. Download
One percent agarose gels containing eDNA from the matrices of differently treated P. aeruginosa SG81 biofilms. (a) Molecular weight ladder consisting of genomic DNAs of different strand lengths (1 to 10 kb). (b) Untreated biofilm. High-molecular-weight DNA is indicated by the arrow. (c) Biofilm after MgCl2 treatment. High-molecular-weight DNA is indicated by the arrow. (d) Biofilm after treatment with MgCl2 and DNase I and containing smears of degraded genomic DNA and a unique region of increased DNA density consisting of smaller strands (<1 kb), indicated by the white rectangle, as also demonstrated by Lee et al. (CI Lee, SH Leong, AE Png, KW Choo, C Syn, DT Lim, HY Law, and OL Kon, Nat. Protoc. 1:2185–2194, 2006). Download