Summary
The brain of a patient with Alzheimer’s disease (AD) undergoes changes starting many years before the development of the first clinical symptoms. The recent availability of large prospective datasets makes it possible to create sophisticated brain models of healthy subjects and patients with AD, showing pathophysiological changes occurring over time. However, these models are still inadequate; representations are mainly single-scale and they do not account for the complexity and interdependence of brain changes. Brain changes in AD patients occur at different levels and for different reasons: at the molecular level, changes are due to amyloid deposition; at cellular level, to loss of neuron synapses, and at tissue level, to connectivity disruption. All cause extensive atrophy of the whole brain organ. Initiatives aiming to model the whole human brain have been launched in Europe and the US with the goal of reducing the burden of brain diseases. In this work, we describe a new approach to earlier diagnosis based on a multimodal and multiscale brain concept, built upon existing and well-characterized single modalities.
Keywords: Alzheimer’s disease , hypermodel , multimodal integration , multiscale approach
Introduction
The diagnosis of neurodegenerative diseases such as Alzheimer’s disease (AD) is changing dramatically. For the first time in 27 years, experts have proposed a major change in the criteria, making it possible to diagnose and treat AD earlier. The new guidelines ( McKhann et al., 2011) state that new instrumental technologies can be used to detect the disease even before evident memory problems or other symptoms arise.
For the first time, diagnosis aims at identifying the disease as it is developing, using results from multiple biomarker tests, such as fluorodeoxyglucose (FDG) brain scans, magnetic resonance imaging (MRI) scans and spinal taps, able to reveal indicative signs of brain degeneration. These biomarkers have been developed and tested only recently ( Sperling and Johnson, 2013; Johansson et al., 2011; Petersen, 2012), which explains why none had been previously approved for AD diagnosis. One of the newest, the positron emission tomography (PET) scan, shows the brain plaques peculiar to the pathology of AD ( Herholz et al., 2011). Others, such as cerebrospinal fluid (CSF) or MRI analyses ( Blennow and Zetterberg, 2013; Jack et al., 2012), provide strong indications of AD even when patients do not show any sign of dementia or memory loss.
The dynamic changes in AD biomarkers are known to occur non-linearly. Dynamic models of various neuroimaging biomarkers over time (as the disease progresses) have recently been well characterized ( Frisoni et al., 2010), whereas genetics in combination with imaging biomarkers will soon provide even more diagnostic and prognostic information ( Ramanan et al., 2013). Nevertheless, there is still no multimodal and multiscale approach integrating all the information captured by each single methodology.
The new proposed criteria for AD have already started advocating the multimodal use of brain imaging techniques to examine the inner structure and function of the brain using one biological [e.g., beta-amyloid (Aβ42) or tau protein in CSF] and three imaging markers (e.g., PET amyloid imaging, 18 FDG PET and MRI). A number of validation studies have already been conducted, showing that these new criteria have excellent sensitivity, specificity and accuracy ( Jack, 2012). Although the AD scientific community welcomed the new criteria, they have still not been fully adopted in daily practice ( Frisoni et al., 2011).
In this article, we discuss the strengths and weaknesses of the single-model approach so far used to diagnose (and prognosticate) AD, and we try to define the key issues involved in designing a brain hypermodel, leveraging on the information available at atomic, molecular, cellular and tissue level, with the aim of launching a new multimodal approach to the study of the brain.
This article begins by focusing on the biological marker measurements used to monitor the evolution of AD, before going on to define the problems and limitations of a single-scale, single-modality approach, and finally leading the reader towards a more detailed definition of an initial brain hypermodel based on Bayesian inference and the e-infrastructures needed to implement it.
Biological markers for brain investigation, diagnosis and monitoring
Although it is now clear that AD involves gradual neuron failure, why this happens is still not clear. Experts believe that AD, like other chronic conditions, is the intricate result of multiple factors, rather than of a dominant cause ( Barreto, 2013). Both age and genetics have been identified as the most common risk factors ( Karran et al., 2011). Evidence from cases where the disease runs in families with an autosomal dominant (FAD) mode of transmission indicates that the affected genes – mainly presenilin-1 ( PS1), presenilin-2 ( PS2) , and amyloid precursor protein ( APP) – are involved in the metabolism of beta-amyloid (Aβ) ( Roberson and Mucke, 2006), a small protein of 40 to 42 amino acids ( Fig. 1 ).
Figure 1 .
The amyloid cascade.
Evidence indicates that the amyloid precursor protein (APP) is involved in the metabolism of beta-amyloid (Aβ). The proteolytic processing of APP unfolds through two alternative pathways. In the non-amyloidogenic pathway, APP is processed by an α-secretase. In the amyloidogenic pathway, APP is first cleaved at a β-secretase site and subsequently cleaved by a γ-secretase complex to release the Aβ peptide, which can aggregate into fibrils and cause long-term neuronal injury. The amyloid cascade has led to the identification of potential therapeutic paths: Aβ might be removed by immune-mediated mechanisms induced by vaccination; alternatively the synthesis of Aβ might be blocked by the inhibition of both enzymes involved in the cleavage of Aβ from APP (β-secretase and presenilin-dependent γ-secretase). Finally, the degradation of Aβ might be accelerated by enhancing the activity of Aβ-degrading enzymes. Although the mechanism is still not completely understood, Aβ promotes the deposition of hyperphosphorylated tau, the second pathological marker of the disease.
It is currently believed that Aβ-driven neurotoxicity triggers neurodegeneration, leading to synaptic and neuronal loss. This is indexed by intraneuronal accumulation of abnormally phosphorylated tau, a structural protein allowing the stabilization of microtubules ( George-Hyslop and Rossor, 2001).
Beta-amyloid and tau proteins are the main constituents of senile plaques and neurofibrillary tangles, originally described by Alois Alzheimer in the brains, examined under the microscope, of patients with progressive dementia. Pathology studies then showed that, even though AD symptoms generally develop later in life, Aβ and tau accumulate in the brain decades before the clinical onset of the disease ( Smith, 2002). Pre-existing neural reserve and plastic resources of the brain are thought to compensate, for a long time, for the progressive damage caused by Aβ and tau, until a threshold is passed and symptoms develop ( Fig. 2a ). These acquisitions have allowed the development of new drugs that interfere with Aβ and tau metabolism and accumulation and might halt, or even reverse, the brain damage ( Fig. 2b ). A number of molecules, used in cholinergic, serotonergic, histaminergic, anti-amyloid and tau-related therapies, are currently in phase II and III clinical trials (efficacy studies in patients), and many more are at the preclinical development phase ( Mangialasche et al., 2010; Vellas and Aisen, 2010; Pillai and Cummings 2013).
Figure 2a .
The natural history of clinical and neurobiological changes.
Recent studies have greatly advanced our knowledge of the pathophysiology of AD, as well as the progression of the neurobiological changes over time. The abnormal accumulation of Aβ and tau in the brain leads to neuronal injury, starting years before the first clinical symptoms appear and proceeding with a stereotypical pattern of early medial temporal lobe (entorhinal cortex and hippocampus) involvement, followed by progressive neo-cortical damage. The delay in the development of cognitive symptoms suggests that the toxic effects of tau and/or Aβ progressively erode the ‘brain reserve’ until a clinical threshold is crossed and amnestic symptoms appear. Amnestic mild cognitive impairment (MCI) is the prodromal phase of AD and is characterized by non-disabling memory symptoms. MCI is followed by more widespread cognitive deficits in multiple domains and disability (i.e. lack of self-sufficiency in one or more activities of daily living), when the traditional diagnostic criteria for AD are fulfilled. The case of a person whose brain starts accumulating tau and Aβ at around age 40, experiences MCI at age 74, and is diagnosed with AD at age 78 is depicted. The clinical course of the disease lasts only four years, but the neurobiological course lasts almost 40 years.
Figure 2b .
Tomorrow’s therapeutic strategy.
The natural history of the neurobiological changes in AD suggests that drugs that might potentially delay the progression of cognitive deterioration should be administered as soon as possible in the course of the disease. The earliest time when it is now possible to recognize the disease is at the stage of MCI. An effective disease-modifying drug administered at this stage might keep the patient in the MCI stage, which is associated with a reasonably good quality of life.
The development and potential availability of drugs is generating great hopes and expectations, but at least two major hurdles need to be overcome. First, to be maximally effective anti-amyloid and anti-tau drugs need to be prescribed early in the course of the disease. Indeed, the failure of many anti-amyloid drugs, such as: tramiprosate, tarenflurbil, AN1792, and many others, has been attributed, among other things, to the fact that patients were treated in the overt stages of the disease. Second, researchers working on AD will need to find the right set of meaningful surrogate outcomes, or biological biomarkers, sensitive to disease progression which can be effectively deployed to test drug efficacy in a clinical trial setting. These markers can greatly enhance power, allowing up to 10-fold decreases in sample sizes, and thereby making it possible to test a much larger number of drugs and increasing the chances of finding one that is really effective. This approach has already been proven successful in the case of antiretroviral drugs for acquired immunodeficiency syndrome (AIDS), antihypertensive drugs for stroke, and statins for atherosclerosis, whose success was largely due to the availability of relevant biomarkers (blood CD4+ white cell count, blood pressure values, and serum cholesterol levels respectively).
With the advent of the “omics” technologies (i.e., genomics, transcriptomics, proteomics, metabolomics, and connectomics) we entered a new era of biomedical sciences and biomarker discovery. Two-thirds of the approximately 30,000 genes in the human genome are related to brain function, and up to half of the variance in age-related changes in cognition, brain volume and neuronal function appears to be genetically determined. The Ensembl project [the EBI EMBL genome browser sequencing project – http://www.ensembl.org; ( Flicek et al. 2013)] has produced a genome database for human and other eukaryotic species, freely available online. In addition, our knowledge of and ability to analyze the transcriptome, proteome and metabolome are now advancing at the same rapid pace that characterized the genomic phase. Besides the specific and complex challenge of identifying and characterizing proteins relevant to pathological AD brains, we need to consider that the overall human proteome is currently estimated to be made up of over 1 million proteins, a staggering amount resulting from: i) single-nucleotide polymorphisms, which cause two-thirds of all the human genes to generate alternative isoforms ( Mullikin et al., 2000); and ii) alternative splicing mechanisms of a single gene, which can lead to the codification of different proteins. Since the discovery of these mechanisms, there have been many attempts to catalog the whole human proteome, and in this context, a special mention must be made of HUPO – Human Proteome Organisation – an international institution fostering proteomic initiatives geared at furthering understanding of human disease (http://www.hupo.org). To date, only few studies have correlated human neuroimaging findings with genomic, transcriptomic, proteomic or metabolomic findings, but such correlations are expected soon.
Proton magnetic resonance spectroscopy (MRS) represents the link with metabolomics, including lipid disorders influencing AD ( Astarita and Piomelli, 2011). In patients at risk of AD, MRS can provide a window onto the biochemical changes associated with the loss of neuronal integrity before cognitive impairment arises ( Graff-Radford and Kantarci, 2013).
“Imaging genetics” is a relatively new branch of neuroimaging which is gaining pace at an unprecedented rate ( Petrella et al., 2008). This methodology exploits an endophenotypes approach in order to identify genes responsible for different cognitive phenotypes. For example, thousands of microarrays containing the genetic markers of people with and without good memory, previously assessed through MRI technology, can be compared simultaneously to identify which genes differ and are linked to poor memory performance. Sequencing an entire genome is currently very expensive, but the National Institutes of Health (NIH) hopes that the total cost can be reduced to $1,000 per genome over the next five years.
Other neuroimaging techniques, including PET, MRI, MRS, and functional MRI (fMRI), allow us to investigate the biological macro effects of genetic alterations. Substantial advances in molecular imaging led to the recent development of PET ligands to track various receptors, neurotransmitters, and proteins, such as Aβ (Pittsburgh compound B, florbetapir, flutemetamol, florbetaben), tau, and acetylcholine ( Bencherif et al., 2002; Klunk and Mathis, 2008; Klunk, 2011; Shin et al., 2011; Clark et al., 2012). Given that cholinergic deficits as well as amyloid and tau deposition are characteristic of AD, these new ligands should be able to refine our understanding of normal and pathological aging.
A final remark should be made about the administration of neuropsychological tests, the most traditional and pervasive approach to describing and characterizing the stages of AD disease. These tests are a useful means of summarizing, in a single final measurement, all the complex interactive processes described above. As we have seen, neuroscientists can now leverage on different instrumental techniques to draw up an overall picture of the disease and we believe that all these instruments must now be played together like the strings of a single guitar. Such a multimodal approach will be instrumental to the success of ambitious scientific initiatives with high societal impact, such as PAD 2020, the Campaign to Prevent Alzheimer’s Disease by 2020 (PAD 2020, http://pad2020.org/).
Modeling dynamic changes of multiple biomarkers over time
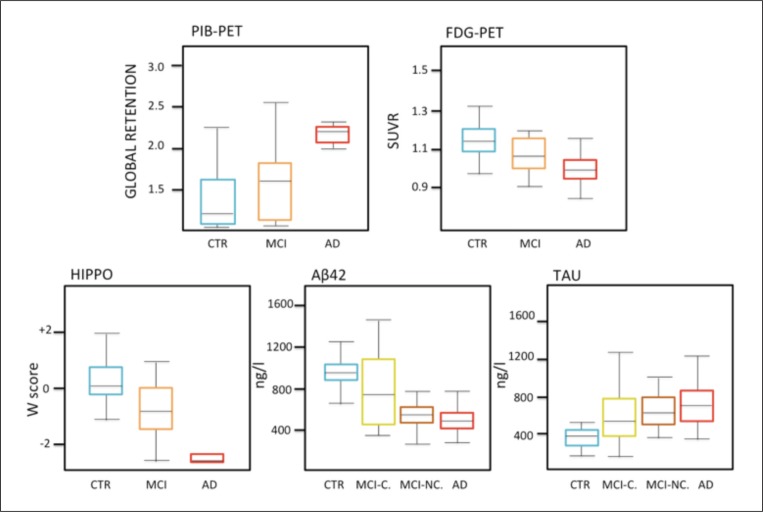
Over the past two decades, the pathological modification pathway of biological markers over the whole course of AD has been defined. Cross-sectional studies ( Hampel et al., 2004; Jack et al., 2008; Furukawa et al., 2010) have demonstrated that by the time a patient shows memory deterioration, markers of Aβ and tau deposition in the brain have changed, as have imaging markers of amyloid deposition detectable through [F]AV45 or [C]PIB PET, metabolic markers on glucose 18 FDG PET, atrophic markers of neurodegeneration on MRI, markers of axonal and myelin integrity on diffusion tensor imaging (DTI), and markers of neuronal and synaptic function on fMRI ( Fig. 3 ). All these biomarkers detect unimodal and single-scale changes, and have poor temporal consistency. Moreover, the percentage difference versus a group of matched cognitively healthy persons could not be taken as a reliable indicator of the earliest phase of change over time due to the different metrics and precision of measurements ( Pepe et al., 2008).
Figure 3 .
Markers of Alzheimer’s disease.
Cross-sectional studies have shown that the pattern of marker abnormality in NC, MCI and AD (see below) is one where controls and AD patients are at the opposite ends (low-high), and MCI patients lie somewhere in the middle. However, the difference between the biomarker level of a patient group and a group of cognitively healthy persons cannot be taken as a point of reference to identify the earliest time of change, since genetic and environmental confounders affect the trajectory of biomarkers. Alzheimer’s pathological modifications occur gradually and the dynamics of biomarker changes over time are complex and often non-linear. Acronyms: PIB=Pittsburgh compound B ( 11 C); PET=positron emission tomography; FDG=fluorodeoxyglucose ( 18 F); HIPPO=hippocampal volume; Aβ42=beta-amyloid protein ending at amino acid 42; CN=Controls, MCI=mild cognitive impairment, MCI-C=subjects that convert from MCI state to probable AD stage; MCI-NC=subjects that do not convert from MCI state to probable AD stage; AD=Alzheimer’s disease; SUVR=standardized uptake value ratio; W score=this is the value from a standard normal distribution corresponding to the observed percentile.
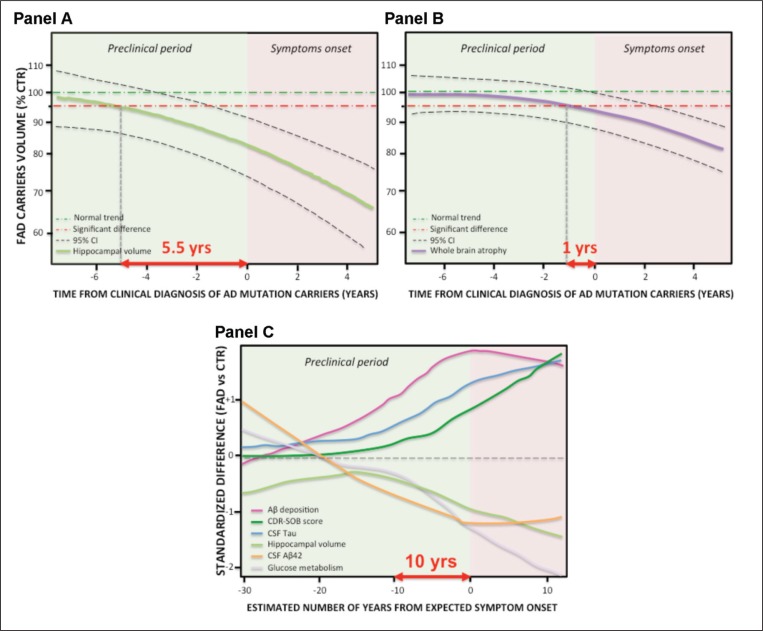
A great incremental advance in the conceptualization of AD has come from serial studies, initially carried out in Europe on mutation carriers coming from families with AD ( Fox et al. 1996) and more recently from large serial studies, in particular the Alzheimer’s Disease Neuroimaging Initiative (i.e.: ADNI-1, -GO and -2) and a related initiative in Australia (Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing – AIBL or “Australian ADNI”). These studies have collected biomarkers serially (generally every 6 to 12 months) in persons with variable degrees of cognitive deterioration, ranging from none, to mild cognitive impairment, through to dementia, allowing neuroscientists to outline the dynamics of the change in biomarkers over time. As an illustrative instance, figure 4 (see over) shows that carriers of fully penetrant mutations (i.e., PSEN1 , PSEN2 and APP ), who will inexorably develop AD, exhibit a deviation from the normal trajectory of hippocampal shrinkage as early as 5.5 years before the diagnosis of dementia (the hippocampus is the brain structure where memories are consolidated and where, in AD, tau pathology and neurodegeneration are particularly severe), while a global indicator of whole-brain shrinkage can be detected no earlier than one year before diagnosis. Moreover, when hippocampal shrinkage emerges, atrophy accelerates at a rate of about 0.3% per year ( Ridha et al., 2006), showing that the evolution of this biomarker is not linear.
Figure 4 .
Dynamics of the change in AD biomarkers over time.
Panel A: Ridha et al. (2006) report that persons carrying mutations for FAD provide evidence of acceleration of brain atrophy with disproportionate hippocampal involvement preceding clinical diagnosis. As patients moved from the pre-symptomatic to MCI and AD stages, the mean of total hippocampal and whole-brain volumes decreased. The estimated difference of hippocampal atrophy between autosomal mutation carriers and control groups becomes significant 5.5 years before clinical diagnosis of AD, while the difference in whole-brain atrophy only around 1 year before diagnosis (Panel B). Moreover, once hippocampal and whole-brain shrinkage has appeared, atrophy rates accelerate, showing a non-linear change over time. Panel C: Jack et al. (2013) report that the Dominantly Inherited Alzheimer’s Network (DIAN) and studies of Colombian kindred carriers of a PS1 mutation support the idea of a protracted preclinical period (10 years or more) during which biomarkers become abnormal sequentially while people remain clinically asymptomatic. Additionally, the DIAN results suggest that CSF Aβ42 might become abnormal before amyloid PET, with CSF Aβ42 initially starting at abnormally high concentrations followed by a progressive decline. DIAN results also suggest that tau becomes abnormal before FDG PET and that FDG PET and MRI become abnormal very close in time. Standardized difference in Panel C is derived from the mutation carrier group and the non-carrier group showing the non-linear changes over time. Abbreviations: Yrs=years; FAD=familial Alzheimer’s disease; CTR=controls; CSF=cerebrospinal fluid; Aβ42=beta-amyloid protein ending at amino acid 42; CDRSOB=Clinical Dementia Rating Scale Sum of Boxes.
Studies with a similar design, investigating the changes in Aβ42 and tau in CSF, [ 11 C]-PIB amyloid tracer uptake, and [ 18 F]-glucose uptake, have outlined a theoretical scenario wherein some markers change earlier and reach a plateau ahead of others, which instead change later and reach a delayed plateau. In both cases, it is clear that biomarkers follow a non-linear, sigmoid curve ( Jack et al., 2010, 2013).
Depending on the degree of abnormality and the slope of change, different biomarkers at different times can be used for diagnosis or monitoring progression over the disease course. The recently conceptualized scenario suggests that the first pathological event consists of the brain changes taking place at the molecular level (toxic amyloid deposition), which lead to the destruction of synaptic functions and axonal integrity; these are followed by neuronal loss, gray and white matter atrophy and, finally, clinical cognitive decline ( Teipel et al., 2013). Modeling the changes in biomarkers over time is useful for many reasons. It allows us to predict which biomarker or combinations of biomarkers are more sensitive to the disease state, with practical implications for the diagnosis, and which to the disease progression, with the possibility of better understanding the efficacy of disease-modifying drugs. Figure 4 shows that hippocampal atrophy might be of poor diagnostic value at the earliest stages of the disease, but might be an accurate and valuable marker of progression later on in its course. On the contrary, amyloid burden might be a good diagnostic marker in the earliest stages of the disease, but might be poorly useful as a marker of disease progression. When modeled biomarkers are used as the basis of a prediction, they allow the formulation of a robust pathogenic hypothesis. For example, according to the modeled rates of atrophy, the medial temporal cortex changes substantially in the early stages of the disease, while rates of atrophy in the frontal cortex are either flat or changes occur at a later stage. What this shows is that neurodegeneration starts earlier in the medial temporal cortex and only later spreads to the frontal cortex.
In the next section, we will conceptualize what we have called the hypermodel of AD pathology. The main point on which the following dissertation hinges is the capability of the brain simulator to estimate the multimodal and multiscale pathological changes over time with the highest possible consistency ( Silvestri et al., 2013). The hypermodel will transform the diagnosis and treatment of brain diseases, providing insights into the organizational complexity of this convoluted organ.
The brain hypermodel: a multiscale and multi-modal dynamic simulator
Definition of the brain hypermodel
Modern neuroscience has afforded deep insights into every level of brain organization – from genes to cognition. Neuroscience today is faced with the compelling need to fit the different levels together, exploiting advanced theoretical models, such as the hypermodel of the brain, in order to capture, through solid mathematical rules, all the deep mechanics of the brain. Thus, our knowledge of AD can be improved ( Ewers et al., 2011). This multiscale challenge is made even more complicated by the temporal and spatial scales at play, which range over nine-ten orders of magnitude ( Table I ). The hyper brain is a mathematical model, based on the Bayesian inference, useful to describe brain activity ranging from the low-level mechanisms up to the large-scale biological processes ( Friston et al., 2002a, b). To understand the Bayesian brain it is necessary to understand the structure and connectivity hierarchically linking the variables considered in the analysis. The general assumption is that genes influence cognition and behavior. Therefore, the first step involves regulation and transcription of genes in many proteins. These proteins eventually influence cell processes and functions through enzymatic reactions. Neurons, forming neural networks, work together in a complex pattern of stimulation and inhibition, along with other interactions, to produce a given cognitive behavior. Taking this into consideration, neuroimaging techniques, including PET, MRI, MRS, and fMRI, allow us to examine the biological effects of genetic alterations. Both at genetic and neuroimaging level, hierarchical Bayesian mixture models have started to be proposed ( Wakefield et al., 2010, Nathoo et al., 2012). The structure and function of the human brain in these models can be studied at multiple temporal and spatial scales.
Table I .
The brain hypermodel relies on multiple biomarkers describing the dynamics of the brain in the different stages of the disease.
| Scale | CTR | MCI | AD | |
|---|---|---|---|---|
| Genomics (SNP analyses) | nm (10 −9 ) | Carriers of protective alleles | Carriers of protective alleles | Carrier of risky alleles |
| Transcriptomics (Alternative splicing & promoter regions analyses) | nm (10 −9 ) | mRNA transcripts from: BACE1, APOE, COMT, GRM3, serotonergic genes and APP protective gene | mRNA transcripts from: BACE1, APOE, APP, PSEN, COMT, BDNF, ACE, NOS, GRM3, KIBRA, IGF, SOD1, PLXNB3, MTHFR genes | mRNA transcripts from: BACE1, APOE, APP, PSEN1-2, COMT, BDNF, ACE, NOS, GRM3, KIBRA, IGF, SOD1, DAPK1, DISCI, MTHFR, BCHE, CADPS2 genes |
| Proteomics (sequence and functions analyses) | 10 nm (10 −8 ) | Systematic characterization of: homocysteine, C-reactive protein, α2-macroglobulin, complement factor H isoforms | Systematic characterization of: homocysteine, C-reactive protein, α2-macroglobulin, fibrinogen, GSK-3, complement factor H isoforms | Systematic characterization of: Homocysteine, C-reactive protein, α2-Macroglobulin, Oxysterols, F2-isoprostanes, DKK-3, complement factor H isoforms |
| CSF/Blood markers | 10 nm (10 −8 ) |
T-TAU ➙ P-TAU ➙ Aβ42 ➙ Inflammation ➙ |
T-TAU ⬆ P-TAU➚ Aβ42 ⬇ Inflammation ➙ |
T-TAU ⬆ P-TAU ⬆ Aβ42 ⬇ Inflammation ⬆ |
| Metabonomics (MRS) | 10 μm (10 −5 ) |
NAA/Cr ratio ⬆ mi/Cr ➘ Cho/Cr ➘ |
NAA/Cr ratio ➘ mi/Cr ➙ Cho/Cr ➙ |
NAA/Cr ratio ⬇ mi/Cr ➚ Cho/Cr ➚ |
| Synaptic functions (FDG PET) | 1/10 mm (10 −4 ) | Glucose consumption unimpaired ➙ | Glucose consumption very mild impaired ➘ | Regional decline of glucose consumption ⬇ |
| Functional (rsfMRI), Structural (T13D) and Connectivity (DTI) | mm (10 −3 ) |
Functional ➙ Structural ➙ Connectivity ➙ |
Functional ➘ Structural ➘ Connectivity ➙ |
Functional ⬇ Structural ⬇ Connectivity ⬇ |
| Cognitive / Behavioral tests | meter (10 1 ) |
|
|
|
Abbreviations: AD=Alzheimer’s disease; CTR=healthy elderly control; MCI=mild cognitive impairment; CSF=cerebrospinal fluid; mRNA=messenger ribonucleic acid; SNP=single nucleotide polymorphism; ACE =gene encoding for angiotensin-converting enzyme; APOE =gene encoding for apolipoprotein E; APP =gene encoding for amyloid precursor protein; BACE1 =gene encoding for beta-secretase1; BCHE =gene involved in butyrylcholinesterase synthesis; BDNF =gene encoding for brain-derived neurotrophic factor; CADPS2 = gene encoding for calcium-binding protein involved in exocytosis of vesicles filled with neurotransmitters and neuropeptides; COMT =gene encoding for catechol-O-methyl transferase; DAPK1 =gene encoding for death-associated protein kinase 1; DISC1 =gene implicated in thought and working memory; GRM3 =gene encoding for metabotropic glutamate receptor; IGF=gene encoding for insulin growth factor; KIBRA =gene involved in hippocampal activation; MTHFR =gene encoding for methyl-tetrahydrofolate reductase; NOS =gene encoding for nitric oxide synthase; PLXNB3 =this gene is a member of the plexin family playing a role in axon guidance; PSEN1-2 = genes encoding for presenilin-1 and 2; SOD1 =gene encoding for superoxide dismutase 1; T-TAU=total tau; P-TAU: phosphorylated tau; AB42=Aβ 1–42 protein in CSF; ↑=increased ↗=slightly increased ↓=decreased ↘=slightly decreased →=stable; MRS=magnetic resonance spectroscopy; Cho=choline; Cr=creatine; mi=myo-inositol; NAA=N-acetyl aspartate; FDG PET= 18 F-fluorodeoxyglucose positron emission tomography; MRI=magnetic resonance imaging; rsfMRI=resting state functional MRI; DTI=diffusion tensor imaging; CDR=Clinical Dementia Rating scale; MMSE=Mini Mental State Examination.
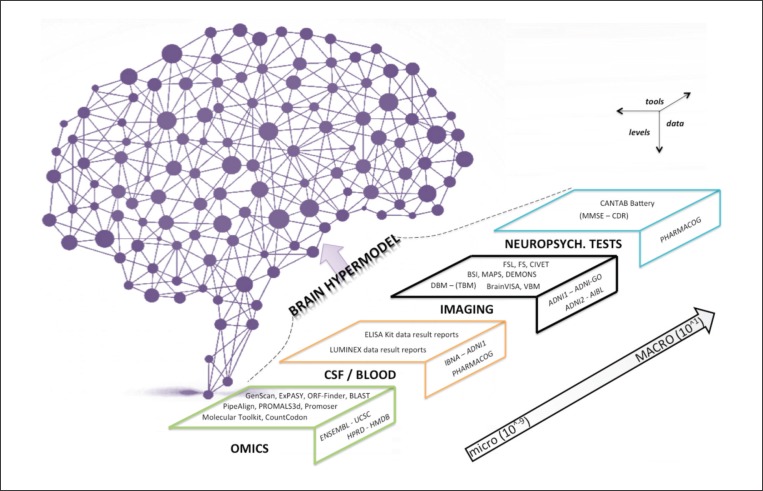
BOX 1 should help to familiarize the reader with the basic Bayesian components adopted by the brain hypermodel. To develop the brain hypermodel of AD based on genetic, clinical, imaging and behavioral data, a large number of postprocessing tools are required ( Fig. 6 , over) in order to generate inputs that feed the Bayesian network. BOX 2 will help the reader to understand the programs and information and communication technology (ITC) tools used by the brain hypermodel.
BOX 1 . The brain hypermodel: basic concepts .
Hierarchical model : The model is described by several parameters that vary at more than one level. The hierarchical model is suitable in cases of nested data (e.g., omics, imaging, clinical, patient’s neuropsychologial data, etc.). In this hierarchical analysis, the estimated elements come from subjects randomly selected from a larger population.
Bayesian model : A typical Bayesian model concerns the probabilistic relationships between diseases and symptoms. Given a set of symptoms, a Bayesian network can be used to compute the probabilities of the presence of a certain disease. Brain hypermodels typically rely on a Bayesian network, a probabilistic model expressed via a graph ( Fig. 5 ): here, every node of the graph represents random variables (e.g., observable quantities, unknown parameters or hypotheses), edges represent dependencies and those nodes which are not connected represent independent variables. Each node is associated with a probability function that takes a particular set of values from the node’s parent as input and gives the probability of the variable represented by the node as output. The Bayesian model relies on additional concepts, i.e.: (A) Prior probability : this is the probability distribution that confers the uncertainty on an uncertain quantity defined as “p” (e.g.: suppose “p” is the number of voxels that will be activated for a specific task in an fMRI experiment) before the data are taken into account (in this case, the results of the experiment obtained via independent component analysis); (B) Likelihood : this is synonymous with probability, albeit with some differences. Probability is used when describing a function of the outcome given a fixed parameter value, and it can be described as follows: “if a transcript of a messenger ribonucleic acid (mRNA) is expressed 100 times and this mRNA is not affected by errors from the RNA polymerase, what is its probability of expressing a fully functional protein?”. The term likelihood, instead, is used when describing a function of a parameter given an outcome. For example: “if an mRNA is translated 100 times and it encoded for an active protein 100 times, what is the likelihood of the mRNA being unaffected by errors?”; (C) Posterior probability : this measures the likelihood that an event will occur given that a related event has already occurred. An example can be given by calculating the probability of a case of MCI converting to AD, given that the level of Aβ42 in the CSF has risen. Let A be the event that MCI converts to AD, and the probability that MCI will convert is 75% (P(A) = 0.75). Let B be the event that the level of Aβ42 rises, with a probability of 80% (P(B) = 0.80). Finally, let the likelihood that Aβ42 will rise, given that MCI converts to AD, be 99% (P(BIA) = 0.99). The probability that MCI will convert to AD given that Aβ42 rises can be determined by plugging these values into the Bayes’ Theorem, giving P(AIB) = = 0.92. This means that in this hypothetical situation if the CSF Aβ42 level is rising, MCI has a 92% chance of converting.
Multivariate analysis : This statistical technique is based on observation and analysis of more than one outcome variable at a time. The technique is used to perform studies across multiple dimensions while taking into account the effects of all variables on the responses of interest.
Figure 6 .
Tools and data exploited by the brain hypermodel.
The scheme represents algorithms and datasets structured by different levels of depth and scale.
Explanations: Genscan=program to identify the gene structure from the DNA strand (http://genes.mit.edu/GENSCANinfo.html); ExPASy=Web-portal to obtain user access to proteomics, genomics, phylogeny, systems biology, population genetics and transcriptomics (http://www.expasy.org/); ORF-Finder=Open Reading Frame finder in RNA coding strand (http://www.ncbi.nlm.nih.gov/projects/gorf/); BLAST=algorithm able to find regions of similarity between biological sequences (http://blast.ncbi.nlm.nih.gov/); PipeAlign=toolkit for protein family analysis (http://bips.u-strasbg.fr/PipeAlign/); PROMALS3d=multiple protein sequence and structure alignment tool (http://pro-data.swmed.edu/promals3d/promals3d.php); Promoser=tool for transcription regulation analysis (http://cagt.bu.edu/page/Promoser_about); Molecular Toolkit=tool for manipulation of nucleic acids and protein (http://www.vivo.colostate.edu/molkit/); CountCodon=on-line tool to count codons in mRNA (http://www.kazusa.or.jp/codon/countcodon.html); FSL=complete library for the analysis of fMRI, MRI, DTI data (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/); FS=FreeSurfer, a set of automated tools for reconstruction of the brain’s cortical surface from structural MRI data (https://surfer.nmr.mgh.harvard.edu/); CIVET=tool for the segmentation of the cerebral cortex (http://cbrain.mcgill.ca/); BSI= Boundary Shift Integral (http://idealab.ucdavis.edu/software/bbsi.php); MAPS=multiple-atlas propagation and segmentation tool; DEMONS=diffeomorphic registration algorithm (http://www.insight-journal.org/browse/publication/154); DBM – (TBM)=deformation based morphometry; BrainVISA=complete set of tools and libraries to process brain image data (http://brainvisa.info/); VBM=voxel-based morphometry (http://www.fil.ion.ucl.ac.uk/spm/); CANTAB=Cambridge Neuropsychological Test Automated Battery; ENSEMBL=EBI EMBL genome browser sequencing project (www.ensembl.org9; UCSC=University of California Santa Cruz genome and transcriptome browser (http://genome.ucsc.edu/); HPRD=Human Protein Reference Database (http://www.hprd.org/); HMDB=Human metabolome database (http://www.hmdb.ca/); IBNA=Italian Brain Normative Archive; ADNI (1-GO-2)=Alzheimer’s Disease Neuroimaging Initiative (http://adni.loni.usc.edu/); AIBL=Australian ADNI (http://www.aibl.csiro.au/); PHARMACOG=European ADNI dataset (http://www.imi.europa.eu/content/pharma-cog). OMICS=of or pertaining to related measurements or data from fields such as genomics, proteomics, transcriptomics, metabolomics; CSF=cerebrospinal fluid; NEUROPSYCH. TESTS=neuropsychological tests.
BOX 2 . ITC tools needed for the brain hypermodel .
“OMICS” tools : Being concerned with strands of nucleotides [i.e., deoxyribonucleic acid (DNA) or ribonucleic acid (RNA)] and chains of amino acids (i.e., oligomers or proteins), the hypermodel should handle inputs coming from sequence analysis tools (e.g. Genscan, ExPASY, ORF-finder), sequence alignment programs (e.g., BLAST, PipeAlign, PROMALS3d), and monitoring protein expression algorithms (e.g., CountCodon, Molecular Toolkit, Promoser).
Imaging tools : The model needs specific algorithms to properly register different brain image modalities in different subjects at different time points into the same space. Modeling the course of brain changes in neurodegenerative disorders requires spatial consistency at multiple spatial scales. The Boundary Shift Integral (BSI) is one of the tools available to segment and register brain scans at multiple time points ( Leung et al., 2012). Other single image analysis tools for AD are: the Multi-Atlas Propagation Segmentation (MAPS) for the hippocampus, which combines atlas-based segmentation and multi-feature pattern recognition ( Leung et al., 2011); the PIB uptake model ( Scheinin et al., 2009); the multi-atlas based anatomical segmentation tool ( Wolz et al., 2010; Lötjönen et al., 2011); DEMONS, a deformation-based patient normalization and follow-up method ( Vercauteren et al., 2009); and the 4D longitudinal brain atrophy simulation tools to predict brain atrophy. New insights into microstructural changes of the white matter should be assessed through deformation-based morphometry ( Zhang et al., 2010). All these algorithms can play an important role in the definition of the hypermodel. However, the aforementioned tools are needed to pre-process multimodal data that must then be analyzed through advanced imaging libraries and tools to assess the final biomarkers. Therefore, additional algorithms to be adapted and plugged in the model are: FSL ( Woolrich et al., 2009), FreeSurfer ( Fischl, 2012; Bernal-Rusiel et al., 2012), Civet ( Kim et al., 2005), BrainCSI for MRS, Voxel/Bayesian based morphometry and BrainVISA ( Chaari et al., 2010).
Neuropsychological tools : the integration of the neuropsychological data can be done by interfacing computerized assessment tools such as CANTAB ( Egerházi et al., 2007).
The level of complexity implicit in connecting and integrating all these single-modality tools into a consistent multimodal framework (the hypermodel) is such as to drive the evolution of the research framework from the traditional models, such as a generalized linear model, to hierarchical (mixed effects) models.
Brain hypermodel axioms
The brain hypermodel needs to be based on specific fundamental principles:
Current observation depends on past observation.
The distribution (i.e.: prior, likelihood and posterior) of every biomarker has to be derived according to Jack (2013) .
Multi-level descriptions of the brain space, ranging from genes to proteins, from microcircuits to voxels, from small tissues to global regions of interest, must come from large serial datasets, such as: ENSEMBL – (www.ensembl.org); UCSC – University of California Santa Cruz genome and transcriptome browser (http://genome.ucsc.edu/); HPRD – Human Protein Reference Database (http://www.hprd.org/); HMDB – Human metaboloma database (http://www.hmdb.ca/); and ADNI – Alzheimer’s Disease Neuroimaging Initiative (which comprises a huge number of image modalities e.g.: [ 11 C]PIB PET, [ 18 F]-AV45 PET, [ 18 F]FDG PET, resting fMRI, DTI and structural MRI) (http://adni.loni.usc.edu/). Additional datasets might need to be added for further refinement of the brain hypermodel.
Brain hypermodel statistical pillars
The most obvious approach to the modeling of dynamic, multimodal and longitudinal measurements is through hierarchical (or random effect) models, as used in many recent publications ( Ridha et al., 2006). A hierarchical model offers the advantage of modeling the spatial dependence of variables at neighboring locations using multilevel descriptions of the space at scales ranging from local (nucleotide) to global (voxel regions of interest or even lobes).
The use of hierarchical models, with empirical Bayes estimation, in the field of neuroimaging was initially proposed by Friston et al. in the context of fMRI data analysis, as a way of overcoming some constraints and limitations of the classical statistical parametric mapping approach. In the analysis conducted by Friston et al., this technique made it possible, in contrast with the classical statistical approach, to move from a local (e.g. at voxel level) to a global (at whole-brain level) estimation, with the tangible benefit of increasing the power in the detection of statistically significant results.
At all spatial scales, however, a statistical issue may arise that needs to be taken into account, namely, the false positive detection rate due to multiple comparisons. However, techniques for a posteriori correction of results are available, based on both parametric (i.e., Bonferroni, false discovery rate, family-wise error) ( Hochberg and Benjamini, 1990; Friston, 1995; Genovese et al., 2002) and non-parametric assumptions (i.e., bootstrap, permutation tests) ( Nichols and Holmes, 2002).
The hierarchical formulation might benefit from the so-called multivariate exchangeability assumption. This approach allows missing data to be substituted in order to promptly estimate the subject’s parameters. What this means is that, if necessary, the individual estimated subject parameter can gain in consistency thanks to an increased weight, assumed from the estimation gathered from the entire population, and thus move from a poor subject estimate to a wider and well-defined population perspective. This is one of the key advantages offered by hierarchical Bayesian modeling as opposed to the classical regression approach.
Flexibility and added value of the brain hypermodel
The hypermodel might be considered a high-order marker of disease progression that could be highly representative of all the data. While many parameters will provide direct information about the progression of the disease, others might give “clues” as to the right direction to explore, and provide new insights for a better explanation of data. A statistical analysis of the hypermodel, considering the correlation and redundancy between the variables, could identify significant spatial patterns and time trends.
Unfortunately, the brain hypermodel can still be hampered by a very large number of variables. In the practical clinical setting (e.g. in clinical trials and for early/differential diagnosis), a reduced or simplified model might be used for more predictive and individualized healthcare. Examples of this model have been proposed showing incredibly high diagnostic and prognostic power ( Soininen et al., 2012). Predict-AD (http://www.predictad.eu/), a recently EU-funded research project, has developed and adopted objective and efficient methods to enable earlier diagnosis of AD through a holistic view of patients which combines information from several sources, such as blood samples, imaging and clinical tests ( Antila et al., 2013).
Additionally, the hypermodel could estimate the deviation from the “experienced curve” of neurodegeneration during a clinical trial with a disease-modifying agent, a deviation translatable into a measure of treatment efficacy. Modeling the adverse effects of a treatment will allow researchers to assess the safety of new drugs, which is a critical step on their route to market and an aspect that in the past has proved to be a common cause of expensive failures ( López-Arrieta and Schneider, 2006; Qizilbash et al., 2007). There is evidence that side effects of new AD drugs might include micro-bleeds and inflammation ( Cordonnier and van der Flier, 2011). Even if subjective assessment of radiological images can be used to detect these kinds of side effects, these measures are relatively crude and lack quantification. In this regard, the hypermodel might make a significant contribution to imaging safety in the context of biomarker quantifications.
Finally, the brain hypermodel might help to overcome current limitations in early detection and clinical management of dementia due to lack of sensitive and specific biomarkers for classification and prediction. Specifically, the hypermodel could locate a given patient, studied at one point in time, on the appropriate trajectory (e.g. healthy or AD), and from there predict past and future points (e.g. five years before symptoms, one year after symptoms, etc.) according to the specific pattern of his/her disease marker evolutions.
The computational engine
To overcome the high computational needs required by a multimodal and multiscale brain model, we describe here the most well-equipped e-infrastructures available worldwide that can host the Bayesian model and its processing tools, to perform ad hoc brain hyper simulations on real data. First, what is an e-infrastructure? An e-infrastructure offers neuroscientists advanced image analysis algorithms, powerful resources, 3D visualization tools, quality control services as well as statistical tools, a fertile ground for brain hypermodels. An e-infrastructure allows neuroimaging experiments to be conducted using dedicated computational resources such as: grids, high-performance computing (HPC) systems, and clouds. The remarkable growth, accessibility and availability of imaging and non-imaging data from people affected by neurodegenerative conditions have recently fostered the development of many of these computational e-infrastructures.
Table II (over) summarizes core features, datasets and tools of the three leading e-infrastructures available in the field of neuroimaging. Amongst these, neuGRID (www.neugrid4you.eu) is the leading European e-infrastructure, developed with the aim of overcoming those hurdles that each neuroscientist has to face daily when trying to set up an advanced experiment on computational neuroimaging. Here, neuroscientists can find core resources for their analyses. The neuGRID platform offers access to 500 processing cores, 25 terabytes of effective storage and it has established a connection with external computing resources to double its capacity on demand. From a bandwidth point of view, neuGRID leverages on the pan-European research and education network GEANT (www.geant.net). Although originally designed for neuroscientists working on AD, neuGRID has, in a second phase, been expanded to deal with a wider range of brain diseases, such as white matter disease and psychiatric diseases. NeuGRID also includes tools useful for clinical use, sensitive to the departure of single cases from a normative reference image database ( Fig. 7 , over).
Table II .
Main features of the three e-infrastructures in terms of (i) Image data sets available; (ii) Image-processing algorithms, suites and tools available; (iii) resources and connectivity.
| neuGRID | LONI | CBRAIN | |
|---|---|---|---|
| HARDWARE AND CONNECTIVITY FEATURES | |||
| Infra topology | Distributed using the GRID and CLOUD paradigm | Centralized using HPC | Distributed using the GRID and HPC paradigm |
| Storage capacity | 25 TB + distributed storage | 12 PB (at USC/LONI) | 140 TB + distributed storage |
| Core resources | 500 CPU cores (plus EGEE external 10,000 cores) | 10,000 CPU cores | Over 50,000 CPU cores (plus JUROPA external 50,000 HPC cores) |
| Network provider | GEANT | Internet2 | CANet |
| Bandwidth | 1 GB/s | 2*10 Gb/s (load balanced) | 10 GB/s |
| Platform link | www.neugrid4vou.eu | http://pipeline.loni.usc.edu/ | https://portal.cbrain.mcgill.ca/login |
| ALGORITHMS & PIPELINES | |||
| Target community | AD, WMD, PSY | AD, PSY | AD |
| Processing algorithms | Imaging tools for structural (T13D, T2), functional (PET, rsfMRI) and diffusion (DTI) imaging analysis. | Imaging tools for structural (T13D, PD, T2), functional (PET, rsfMRI, ASL) and diffusion (DTI) imaging analysis. Bioinformatic tools for conducting analysis on GWAS, BLAST, sequence alignment tools. | Imaging tools for structural (T13D) and functional (PET) imaging analysis |
| Statistical tools | R, Octave | Many different tools covering data classification, linear and non-linear regression, feature selection, and multivariate analysis | R and the Rminc package. Integrated voxel-based statistics and voxel wise-linear models. |
| DBMS | Cristal | SQL | MySQL |
| IMAGE DATASETS | |||
| AD | OASIS; ADNI (through LONI); 1000 Functional Connectomes - INDI; ADHD-2DD; PharmaCOG | ADNI1; ADNI-GO; ADNI2; AIBL; ABIDE; BRIN; PAD/CRYO | ADDNEUROMED |
| WMD | LADIS; EDSD | ||
| PSY | FBIRN; ELUDE | ABIDE | NIHPD |
Abbreviations: TB=terabytes; PB=petabytes; EGEE=Enabling Grids for E-Science in Europe. This is a public resource expanding the computational power of the neuGRID platform; LONI=Laboratory of Neuro Imaging; USC=University of Southern California, Los Angeles; CPU=central processing unit; GB/s=gigabytes per second; AD=Alzheimer’s disease; WMD=white matter disease; PSY=psychiatric disease; T13D=volumetric sequence weighted in T1; T2=MRI sequence weighted in T2; PD=proton density-weighted image; PET= positron emission tomography; rsfMRI: resting-state functional MRI; DTI=diffusion tensor imaging; GWAS=genome wide association study; BLAST=basic local alignment search tool; ADNI= Alzheimer’s Disease Neuroimaging Initiative; AIBL=The Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing; ABIDE=Autism Brain Imaging Data Exchange; BRIN=Brain Info; PAD/CRYO=Public Anonymized Dataset/Cryosection; ADDNEUROMED=the AddNeuroMed study; NIHPD: National Institutes of Health Pediatric Database; OASIS=Open Access Series of Imaging Studies; 1000 Functional Connectomes – INDI=1000 functional connectomes International Neuroimaging Data Sharing Initiative project; LADIS=Leukoaraiosis And DISability; EDSD=European diffusion tensor imaging study in dementia; FBIRN=The Functional Bioinformatics Research Network; ELUDE=Efficient Longitudinal Upload of Depression in the Elderly.
Figure 7 .
The N4U infrastructure.
- An on line web-portal (https://neugrid4you.eu) to provide facilities for users to interact with the neuGRID services. Users can leverage on the Online Help Desk, a one-stop assistance facility with which every neuroscientist can interact in order to learn about neuGRID and how to use it.
- A set of data resources. Data can be integrated in N4U upon users’ request. In neuGRID, all data are indexed and registered creating a user-friendly atlas. All data, pipelines and experimental results can be browsed and queried.
- An analysis work area. Here neuroscientists can define new pipelines or configure existing algorithms to be run against selected datasets. At the end, results can be visualized.
- Access to the quality control and statistical tool environment providing neuroscientists with informative reports on the execution of their pipelines.
- Tier 1: this is the real core of the infrastructure. It is composed of a number of sites providing computing resources and integrated services. The sites are located in Italy, France, Sweden, Switzerland and The Netherlands.
- Tier 2: this attaches additional public facilities augmenting N4U’s capacity (e.g. LONI, CBRAIN, ESFRI).
- Tier 3: this adds private cloud computing resources from external providers.
LONI (Laboratory Of Neuro Imaging – http://www.loni.usc.edu/) focuses on the development of image analysis methods and their application to health issues. The LONI e-infrastructure is the longest-established platform among those available in the field of neuroimaging ( Dinov et al., 2009). It responds to the needs of a wide range of users, offering specific services (data and algorithms) to both neuroscientists and neurobiologists. LONI hosts the ADNI databases, which comprise clinical data and information from genetic scans from older people with AD (400 mild AD), people with mild cognitive impairment (350 early MCI, 400 MCI and 150 late MCI), and healthy elders (350 CTR). The LONI imaging portfolio comprises high-resolution structural MRI (T13D MPRAGE, T2, PD), 18 F-FDG PET, amyloid PET (AV45 PET), fMRI and DTI. Algorithms for data analysis are available via the LONI Pipeline graphical interface, a user-friendly workflow management system that makes it possible to automatize the measurement of functional, tractographic and morphometric analyses, to dynamically assess volume and shape features, and to extract and associate cognitive, genetic, clinical, behavioral and imaging biomarkers. LONI provides access to a large, centralized HPC infrastructure – located at the University of Southern California, Los Angeles (USC) – for computationally intensive analyses. External researchers are granted access to the LONI HPC resources on the basis of ad hoc scientific agreements. CBRAIN (http://cbrain.mcgill.ca/) is a network of five Canadian brain imaging research centers, connected to HPC centers in Canada and Europe. The CBRAIN e-infrastructure offers advanced networking, transparent access to computing resources, a wide range of tools as well as web-based results visualization, all thanks to a comprehensive and well organized web portal. CBRAIN is a distributed environment connected through a high-speed wide area network bandwidth.
NeuGRID is expanding its platform internationally, bridging with the other e-infrastructures, with the ultimate aim of delivering an authentic virtual laboratory, integrating the widest range of available analysis services with a specific support center for end users. This will create a virtual space accessible to the user via web no matter where he/she is physically located.
The above facts and figures support the notion that einfrastructures are today the most advanced and the best equipped platforms to support the deployment and distribution of the hypermodel of the brain. In this way, a neuroscientist would be just a click of his/her fingertips away from all he/she needs to start a simulation. Along the same lines, the recent EU FET Flagship initiative called The Human Brain Project (HBP: http://www.humanbrainproject.eu/) as well as the American Brain Activity Map project (BAM: http://www.nih.gov/science/brain/) will widely exploit the larger scale of data and the huge power of the resources available through NeuGRID, LONI and CBRAIN to characterize, build and test the respective in silico brain models.
Concluding remarks
According to the latest EU estimates, the global prevalence of AD is predicted to quadruple to reach 105 million by 2050. To tackle this social emergency and improve AD diagnosis over the next 15 years, clinical practice will need to rely more and more on multimodal methodologies, using an integrated approach based on genetic, biological, imaging methods, as well as neuropsychological and cognitive tests. Indeed, modern neurobiology and neuroscience have gained deep insights into every level of brain organization, and this will help us to move closer to the real cause of the disease rather than just looking at its symptoms: however, to date, there is no clear consensus on how to fit the different levels together. In this review, we have described a possible method, based on a theoretical approach in which use is made of virtual laboratories concretely capable of implementing these notions: it is our belief that this approach could succeed in making sense of the deep mechanics that govern the underlying processes of the brain, thus helping neuroscientists in their daily work.
With such a complete model of the human brain, four main objectives could be addressed: i) earlier and more accurate detection of AD; ii) new surrogate outcomes for clinical trials; iii) faster development of drugs aimed at delaying or halting the neurodegeneration; iv) the development of a reference model that could also be used in other multimodal neurodegenerative brain diseases and research communities.
Finally, the idea of defining a multiscale and multi-modal approach to further understanding of the complex pathophysiology of AD has recently been turned into major projects. In Europe, a billion-euro initiative, the HBP (mentioned earlier) is currently under way ( D’Angelo et al., 2013; Markram, 2013; Calimera et al., 2013), wherein all existing knowledge about the human brain is to be pulled together in order to build up a model of the brain, piece by piece, with advanced ITC simulations. This idea has also prompted a similar initiative, the BAM, this time in the US. The ultimate aim of this project is to map the activity of every single neuron in the human brain. This historical moment will certainly be destined to leave a large footprint in our community and in the way we conduct (neuro)science. As Henry Makram, HBP principal investigator, has claimed: “It is not impossible to build a human brain and we can do it in 10 years” reconstructing deeply the brain’s every circuit and process. Therefore, there is nothing left to do but work!
Figure 5 .
Definition of the brain hypermodel.
This is represented as a set of mathematical relations expressed in terms of random variables and associated probability distributions with the aim of describing the observations of brain atrophy merging with different levels of information. These levels are dynamically linked through a directed acyclic graph. The relations among the nodes of the graph describe the mutual biomarker variations as well as their temporal and spatial interactions from the lowest to the highest scale.
Acknowledgments
The authors thank all the partners in the FP7 neuGRID project (N4U: www.neugrid4you.eu) and in the Human Brain Project (HBP: http://www.humanbrainproject.eu/). Special thanks to Cristina Bagnoli and Chiara Barattieri di San Pietro for their efforts in editing the manuscript. G.B. Frisoni and D. Manset are supported by FP7 neuGRID4You funded by the European Commission (FP7/2007–2013) under grant agreement no. 283562.
References
- Alzheimer A , Stelzmann RA , Schnitzlein HN , et al. An English translation of Alzheimer’s 1907 paper, “Uber eine eigenartige Erkankung der Hirnrinde” . Clin Anat . 1995 ; 8 : 429 – 431 . doi: 10.1002/ca.980080612. [DOI] [PubMed] [Google Scholar]
- Antila K , Lötjönen J , Thurfjell L , et al. The PredictAD project: development of novel biomarkers and analysis software for early diagnosis of the Alzheimer’s disease . Interface Focus . 2013 ; 3 : 20120072 . doi: 10.1098/rsfs.2012.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarita G , Piomelli D . Towards a whole-body systems [multi-organ] lipidomics in Alzheimer’s disease . Prostaglandins Leukot Essent Fatty Acids . 2011 ; 85 : 197 – 203 . doi: 10.1016/j.plefa.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto P de S . Alzheimer’s disease: learning from the past, looking to the future . Am J Alzheimers Dis Other Demen . 2013 ; 28 : 304 – 305 . doi: 10.1177/1533317513488926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencherif B , Endres CJ , Musachio JL , et al. PET imaging of brain acetylcholinesterase using [11C]CP-126,998, a brain selective enzyme inhibitor . Synapse . 2002 ; 45 : 1 – 9 . doi: 10.1002/syn.10072. [DOI] [PubMed] [Google Scholar]
- Bernal-Rusiel JL , Greve DN , Reuter M , et al. Statistical analysis of longitudinal neuroimage data with Linear Mixed Effects models . Neuroimage. 2012 ;66C: 249 – 260 . doi: 10.1016/j.neuroimage.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K , Zetterberg H . The application of cerebrospinal fluid biomarkers in early diagnosis of Alzheimer disease . Med Clin North Am . 2013 ; 97 : 369 – 376 . doi: 10.1016/j.mcna.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Calimera A , Macii E , Poncino M . The Human Brain Project and neuromorphic computing . Funct Neurol . 2013 ; 28 : 191 – 196 . [PMC free article] [PubMed] [Google Scholar]
- Chaari L , Pesquet J-C , Tourneret J-Y, et al. A hierarchical Bayesian model for frame representation. IEEE Transactions on Signal Processing. 2010 ; 58 : 5560 – 5571 . [Google Scholar]
- Clark CM , Pontecorvo MJ , Beach TG , et al. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-beta plaques: a prospective cohort study . Lancet Neurol . 2012 ; 11 : 669 – 678 . doi: 10.1016/S1474-4422(12)70142-4. [DOI] [PubMed] [Google Scholar]
- Cordonnier C , van der Flier WM . Brain microbleeds and Alzheimer’s disease: innocent observation or key player? . Brain . 2011 ; 134 : 335 – 344 . doi: 10.1093/brain/awq321. [DOI] [PubMed] [Google Scholar]
- D’Angelo E , Solinas S , Garrido J , et al. Realistic modeling of neurons and networks: towards brain simulation . Funct Neurol . 2013 ; 28 : 153 – 166 . [PMC free article] [PubMed] [Google Scholar]
- Dinov ID , Van Horn JD , Lozev KM , et al. Efficient, distributed and interactive neuroimaging data analysis using the LONI pipeline . Front Neuroinform . 2009 ; 3 : 22 . doi: 10.3389/neuro.11.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerházi A , Berecz R , Bartók E , et al. Automated Neuropsychological Test Battery (CANTAB) in mild cognitive impairment and in Alzheimer’s disease . Prog Neuropsychopharmacol Biol Psychiatry . 2007 ; 31 : 746 – 751 . doi: 10.1016/j.pnpbp.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Ewers M , Frisoni GB , Teipel SJ , et al. Staging Alzheimer’s disease progression with multimodality neuroimaging . Prog Neurobiol . 2011 ; 95 : 535 – 546 . doi: 10.1016/j.pneurobio.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B . FreeSurfer . NeuroImage . 2012 ; 62 : 774 – 781 . doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P , Ahmed I , Amode MR , et al. Ensembl 2013. Nucleic Acids Res. 2013 ;41(Database issue): D48 – 55 . doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NC , Warrington EK , Freeborough PA , et al. Presymptomatic hippocampal atrophy in Alzheimer’s disease. A longitudinal MRI study . Brain . 1996 ; 119 : 2001 – 2007 . doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- Frisoni GB , Fox NC , Jack CR , Jr , et al. The clinical use of structural MRI in Alzheimer disease . Nat Rev Neurol . 2010 ; 6 : 67 – 77 . doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB , Redolfi A , Manset D , et al. Virtual imaging laboratories for marker discovery in neurodegenerative diseases . Nat Rev Neurol . 2011 ; 7 : 429 – 438 . doi: 10.1038/nrneurol.2011.99. [DOI] [PubMed] [Google Scholar]
- Frisoni GB , Winblad B , O’Brien JT . Revised NIA-AA criteria for the diagnosis of Alzheimer’s disease: a step forward but not yet ready for widespread clinical use . Int Psychogeriatr . 2011 ; 23 : 1191 – 1196 . doi: 10.1017/S1041610211001220. [DOI] [PubMed] [Google Scholar]
- Friston KJ . Commentary and opinion: II. Statistical parametric mapping: ontology and current issues . J Cereb Blood Flow Metab . 1995 ; 15 : 361 – 370 . doi: 10.1038/jcbfm.1995.45. [DOI] [PubMed] [Google Scholar]
- Friston KJ , Glaser DE , Henson RN , et al. Classical and Bayesian inference in neuroimaging: applications . Neuroimage . 2002a ; 16 : 484 – 512 . doi: 10.1006/nimg.2002.1091. [DOI] [PubMed] [Google Scholar]
- Friston KJ , Penny W , Phillips C , et al. Classical and Bayesian inference in neuroimaging: theory . Neuroimage . 2002b ; 16 : 465 – 483 . doi: 10.1006/nimg.2002.1090. [DOI] [PubMed] [Google Scholar]
- Furukawa K , Okamura N , Tashiro M , et al. Amyloid PET in mild cognitive impairment and Alzheimer’s disease with BF-227: comparison to FDG-PET . J Neurol . 2010 ; 257 : 721 – 727 . doi: 10.1007/s00415-009-5396-8. [DOI] [PubMed] [Google Scholar]
- Genovese CR , Lazar NA , Nichols T . Thresholding of statistical maps in functional neuroimaging using the false discovery rate . Neuroimage . 2002 ; 15 : 870 – 878 . doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- George-Hyslop PS , Rossor M . Alzheimer’s disease. Unravelling the disease process. Lancet. 2001 ;358(Suppl):S1. doi: 10.1016/s0140-6736(01)07014-3. [DOI] [PubMed] [Google Scholar]
- Graff-Radford J , Kantarci K . Magnetic resonance spectroscopy in Alzheimer’s disease . Neuropsychiatr Dis Treat . 2013 ; 9 : 687 – 696 . doi: 10.2147/NDT.S35440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H , Teipel SJ , Fuchsberger T , et al. Value of CSF beta-amyloid1-42 and tau as predictors of Alzheimer’s disease in patients with mild cognitive impairment . Mol Psychiatry . 2004 ; 9 : 705 – 710 . doi: 10.1038/sj.mp.4001473. [DOI] [PubMed] [Google Scholar]
- Herholz K , Westwood S , Haense C , et al. Evaluation of a calibrated (18)F-FDG PET score as a biomarker for progression in Alzheimer disease and mild cognitive impairment . J Nucl Med . 2011 ; 52 : 1218 – 1226 . doi: 10.2967/jnumed.111.090902. [DOI] [PubMed] [Google Scholar]
- Herskovits EH , Peng H , Davatzikos C . A Bayesian morphometry algorithm . IEEE Trans Med Imaging . 2004 ; 23 : 723 – 737 . doi: 10.1109/tmi.2004.826949. [DOI] [PubMed] [Google Scholar]
- Hochberg Y , Benjamini Y . More powerful procedures for multiple significance testing . Stat Med . 1990 ; 9 : 811 – 818 . doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Jack CR ., Jr Alzheimer disease: new concepts on its neurobiology and the clinical role imaging will play . Radiology . 2012 ; 263 : 344 – 361 . doi: 10.1148/radiol.12110433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR , Jr , Knopman DS , Jagust WJ , et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers . Lancet Neurol . 2013 ; 12 : 207 – 216 . doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR , Jr , Knopman DS , Jagust WJ , et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade . Lancet Neurol . 2010 ; 9 : 119 – 128 . doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR , Jr , Knopman DS , Weigand SD , et al. An operational approach to National Institute on Aging-Alzheimer’s Association criteria for preclinical Alzheimer disease . Ann Neurol . 2012 ; 71 : 765 – 775 . doi: 10.1002/ana.22628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR , Jr , Lowe VJ , Senjem ML , et al. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer’s disease and amnestic mild cognitive impairment . Brain . 2008 ; 131 : 665 – 680 . doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P , Mattsson N , Hansson O , et al. Cerebrospinal fluid biomarkers for Alzheimer’s disease: diagnostic performance in a homogeneous mono-center population . J Alzheimers Dis . 2011 ; 24 : 537 – 546 . doi: 10.3233/JAD-2011-101878. [DOI] [PubMed] [Google Scholar]
- Karran E , Mercken M , De Strooper B . The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics . Nat Rev Drug Discov . 2011 ; 10 : 698 – 712 . doi: 10.1038/nrd3505. [DOI] [PubMed] [Google Scholar]
- Kim JS , Singh V , Lee JK , et al. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification . Neuroimage . 2005 ; 27 : 210 – 221 . doi: 10.1016/j.neuroimage.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Klunk WE . Amyloid imaging as a biomarker for cerebral beta-amyloidosis and risk prediction for Alzheimer dementia . Neurobiol Aging. 2011 ;32(Suppl 1): S20 – 36 . doi: 10.1016/j.neurobiolaging.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE , Mathis CA . The future of amyloid-beta imaging: a tale of radionuclides and tracer proliferation . Curr Opin Neurol . 2008 ; 21 : 683 – 687 . doi: 10.1097/WCO.0b013e3283168e1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK , Barnes J , Modat M , et al. Brain MAPS: an automated, accurate and robust brain extraction technique using a template library . Neuroimage . 2011 ; 55 : 1091 – 1108 . doi: 10.1016/j.neuroimage.2010.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK , Ridgway GR , Ourselin S , et al. Consistent multi-time-point brain atrophy estimation from the boundary shift integral . Neuroimage . 2012 ; 59 : 3995 – 4005 . doi: 10.1016/j.neuroimage.2011.10.068. [DOI] [PubMed] [Google Scholar]
- López-Arrieta JM , Schneider L . Metrifonate for Alzheimer’s disease . Cochrane Database Syst Rev. 2006 ;2:CD003155. doi: 10.1002/14651858.CD003155.pub3. [DOI] [PubMed] [Google Scholar]
- Lötjönen J , Wolz R , Koikkalainen J , et al. Fast and robust extraction of hippocampus from MR images for diagnostics of Alzheimer’s disease . Neuroimage . 2011 ; 56 : 185 – 196 . doi: 10.1016/j.neuroimage.2011.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangialasche F , Solomon A , Winblad B , et al. Alzheimer’s disease: clinical trials and drug development . Lancet Neurol . 2010 ; 9 : 702 – 716 . doi: 10.1016/S1474-4422(10)70119-8. [DOI] [PubMed] [Google Scholar]
- Markram H . Seven challenges for neuroscience . Funct Neurol . 2013 ; 28 : 145 – 151 . [PMC free article] [PubMed] [Google Scholar]
- McKhann GM , Knopman DS , Chertkow H , et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease . Alzheimers Dement . 2011 ; 7 : 263 – 269 . doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullikin JC , Hunt SE , Cole CG , et al. An SNP map of human chromosome 22 . Nature . 2000 ; 407 : 516 – 520 . doi: 10.1038/35035089. [DOI] [PubMed] [Google Scholar]
- Nathoo FS , Lesperance M , Lawson A , et al. Comparing variational Bayes with Markov chain Monte Carlo for Bayesian computation in neuroimaging . Stat Methods Med Res . 2012 ; 22 : 398 – 423 . doi: 10.1177/0962280212448973. [DOI] [PubMed] [Google Scholar]
- Nichols TE , Holmes AP . Nonparametric permutation tests for functional neuroimaging: a primer with examples . Human Brain Mapp . 2002 ; 15 : 1 – 25 . doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS , Feng Z , Janes H , et al. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design . J Natl Cancer Inst . 2008 ; 100 : 1432 – 1438 . doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC . New clinical criteria for the Alzheimer’s disease spectrum . Minn Med . 2012 ; 95 : 42 – 45 . [PubMed] [Google Scholar]
- Petrella JR , Mattay VS , Doraiswamy PM . Imaging genetics of brain longevity and mental wellness: the next frontier? . Radiology . 2008 ; 246 : 20 – 32 . doi: 10.1148/radiol.2461061994. [DOI] [PubMed] [Google Scholar]
- Pillai JA , Cummings JL . Clinical trials in predementia stages of Alzheimer disease . Med Clin North Am . 2013 ; 97 : 439 – 457 . doi: 10.1016/j.mcna.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Qizilbash N , Birks J , Lopez Arrieta J , et al. WITHDRAWN: Tacrine for Alzheimer’s disease. Cochrane Database Syst Rev. 2007 ;(3): CD000202 . doi: 10.1002/14651858.CD000202. [DOI] [PubMed] [Google Scholar]
- Ramanan VK , Risacher SL , Nho K , et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study . Mol Psychiatry . 2013 doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridha BH , Barnes J , Bartlett JW , et al. Tracking atrophy progression in familial Alzheimer’s disease: a serial MRI study . Lancet Neurol . 2006 ; 5 : 828 – 834 . doi: 10.1016/S1474-4422(06)70550-6. [DOI] [PubMed] [Google Scholar]
- Roberson ED , Mucke L . 100 years and counting: prospects for defeating Alzheimer’s disease . Science . 2006 ; 314 : 781 – 784 . doi: 10.1126/science.1132813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinin NM , Aalto S , Koikkalainen J , et al. Follow-up of [11C]PIB uptake and brain volume in patients with Alzheimer disease and controls . Neurology . 2009 ; 73 : 1186 – 1192 . doi: 10.1212/WNL.0b013e3181bacf1b. [DOI] [PubMed] [Google Scholar]
- Shin J , Kepe V , Barrio JR , et al. The merits of FDDNPPET imaging in Alzheimer’s disease . J Alzheimers Dis. 2011 ;26(Suppl 3): 135 – 145 . doi: 10.3233/JAD-2011-0008. [DOI] [PubMed] [Google Scholar]
- Silvestri L , Sacconi L , Pavone FS . The connectomics challenge . Funct Neurol . 2013 ; 28 : 167 – 173 . [PMC free article] [PubMed] [Google Scholar]
- Smith AD . Imaging the progression of Alzheimer pathology through the brain . Proc Natl Acad Sci U S A . 2002 ; 99 : 4135 – 4137 . doi: 10.1073/pnas.082107399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soininen H , Mattila J , Koikkalainen J , et al. Software tool for improved prediction of Alzheimer’s disease . Neurodegener Dis . 2012 ; 10 : 149 – 152 . doi: 10.1159/000332600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R , Johnson K . Biomarkers of Alzheimer disease: current and future applications to diagnostic criteria . Continuum (Minneap Minn) 2013 ; 19 : 325 – 338 . doi: 10.1212/01.CON.0000429181.60095.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel SJ , Sabri O , Grothe M , et al. Perspectives for multimodal neurochemical and imaging biomarkers in Alzheimer’s disease . J Alzheimers Dis. 2013 ;33(Suppl 1): S329 – 347 . doi: 10.3233/JAD-2012-129030. [DOI] [PubMed] [Google Scholar]
- Vellas B , Aisen PS . Early Alzheimer’s trials: new developments . J Nutr Health Aging . 2010 ; 14 : 293 . doi: 10.1007/s12603-010-0064-3. [DOI] [PubMed] [Google Scholar]
- Vercauteren T , Pennec X , Perchant A , et al. Diffeomorphic demons: efficient non-parametric image registration . Neuroimage. 2009 ;45(1 Suppl): S61 – 72 . doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Wakefield J , De Vocht F , Hung RJ . Bayesian mixture modeling of gene-environment and gene-gene interactions . Genetic Epidemiol . 2010 ; 34 : 16 – 25 . doi: 10.1002/gepi.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R , Aljabar P , Hajnal JV , et al. LEAP: learning embeddings for atlas propagation . Neuroimage . 2010 ; 49 : 1316 – 1325 . doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolrich MW , Jbabdi S , Patenaude B , et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 2009 ;45(1Suppl): S173 – 186 . doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- Zhang H , Awate SP , Das SR , et al. A tract-specific framework for white matter morphometry combining macroscopic and microscopic tract features . Med Image Anal . 2010 ; 14 : 666 – 673 . doi: 10.1016/j.media.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]