Summary
The Italian region of Lombardy, with its existing stroke centers and high-technology laboratories, provides a favorable context for studying monogenic diseases associated with stroke. The Lombardia GENS project was set up to create a regional network for the diagnosis of six monogenic diseases associated with stroke: CADASIL, Fabry disease, MELAS, familial and sporadic hemiplegic migraine, hereditary cerebral amyloid angiopathy and Marfan syndrome. The network comprises 36 stroke centers and seven high-technology laboratories, performing molecular analysis. In this context, all stroke/TIA patients fulfilling clinical criteria for monogenic diseases are currently being included in an ongoing study. Demographic, clinical and family data and diagnostic criteria are collected using standardized forms. On the basis of stroke incidence in Lombardy and the reported prevalence of the diseases considered, we expect, during the course of the study, to collect datasets and DNA samples from more than 200 stroke patients suspected of having monogenic diseases. This will allow evaluation of the regional burden and better phenotype characterization of monogenic diseases associated with stroke.
Keywords: cerebrovascular disease, genetics, monogenic disorders, stroke
Introduction
Stroke is a leading cause of mortality and long-term disability. To relieve the heavy burden of stroke, there is a need for better understanding of its pathogenetic mechanisms, in order to improve its prevention and treatment. The genetic contribution to stroke is well established and there is evidence that genetic factors influence stroke occurrence and outcome (1,2). Underlying monogenic diseases account for about 1% to 5% of all strokes, although their incidence is believed to be underestimated. In fact, monogenic disorders can be misdiagnosed simply because physicians fail to include them in the differential diagnosis. Moreover, even when there are elements that may support a genetic cause, such as young age at onset, positive family history, presence of specific associated clinical features, and absence of conventional vascular risk factors, the wide phenotypic spectrum makes it difficult to select patients and decide which disorders to screen for. However, these disorders, although rare, are usually life-threatening or chronically debilitating diseases; because of their low prevalence and high complexity they are also difficult to manage. The diagnosis of these disorders is important both for genetic counseling and therapeutic decision-making (3–5). In this context, the identification of a large number of patients affected by monogenic diseases may help to better clarify stroke pathogenesis and lead to the development of potential new drugs and therapeutic targets. Also, careful selection of phenotypes is considered an essential requirement for all genetic studies (6).
In the Italian region of Lombardy (about 6 million inhabitants) there is a clinical network of neurological centers specialized in stroke diagnosis and care, including 29 stroke units; the region also has seven high-technology laboratories with well-established expertise in stroke genetics. The presence of these centers prompted the creation of a regional stroke genetics network (Lombardia GENS) designed to exploit the databases of the existing clinical stroke network and also benefit from collaboration with laboratories interested in stroke genetics.
The Lombardia GENS project is a prospective multicenter cohort study aimed at organizing a structured service for complete diagnosis of six single-gene disorders associated with stroke [CADASIL, Fabry disease, MELAS, familial and sporadic hemiplegic migraine (FHM/SHM), hereditary cerebral amyloid angiopathy (H-CAA), and Marfan syndrome] through the setting up of a specific diagnostic work-up for patients with clinically suspected genetic diseases. The aim of the project is to create a regional database and DNA biobank of well-phenotyped stroke patients for studies on regional incidence of monogenic diseases in stroke, phenotypic characterization, and therapeutic trials.
Study design and population
The study population consists of a continuous series of patients with stroke or transient ischemic attack (TIA), in whom there is clinical suspicion of one of the following monogenic diseases: CADASIL, Fabry disease, MELAS, FHM/SHM, H-CAA, or Marfan syndrome, referred during the study period to the clinical units participating in the project. Both ischemic and hemorrhagic strokes are included. The coordinating center managing the project is the Neurology Unit at the Ca’ Granda Foundation, Maggiore Policlinico Hospital, IRCCS, University of Milan, Italy, which is supported by a scientific steering committee (SC). The SC comprises members from the Neurology Unit, Ca’ Granda Foundation, Maggiore Policlinico Hospital, IRCCS, University of Milan; the Cerebrovascular Unit, Carlo Besta Institute of Neurology Foundation, IRCCS, Milan; the Emergency Unit, C. Mondino National Institute of Neurology Foundation, IRCCS, Pavia; the Stroke Unit, Niguarda Ca’ Granda Hospital, Milan; the Stroke Unit, Division of Neuroscience & Institute of Experimental Neurology (INSPE), San Raffaele Scientific Foundation, Milan, the CNS Inflammatory Unit, Division of Neuroscience & Institute of Experimental Neurology (INSPE), San Raffaele Scientific Foundation, Milan, the Stroke Unit, San Gerardo Hospital, University of Milan-Bicocca, Monza, and the Department of Clinical and Experimental Sciences, Neurology Clinic, University of Brescia, Italy.
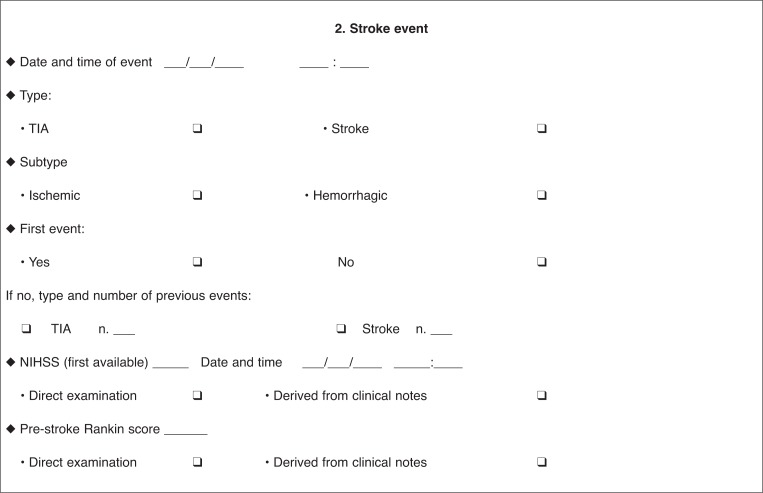
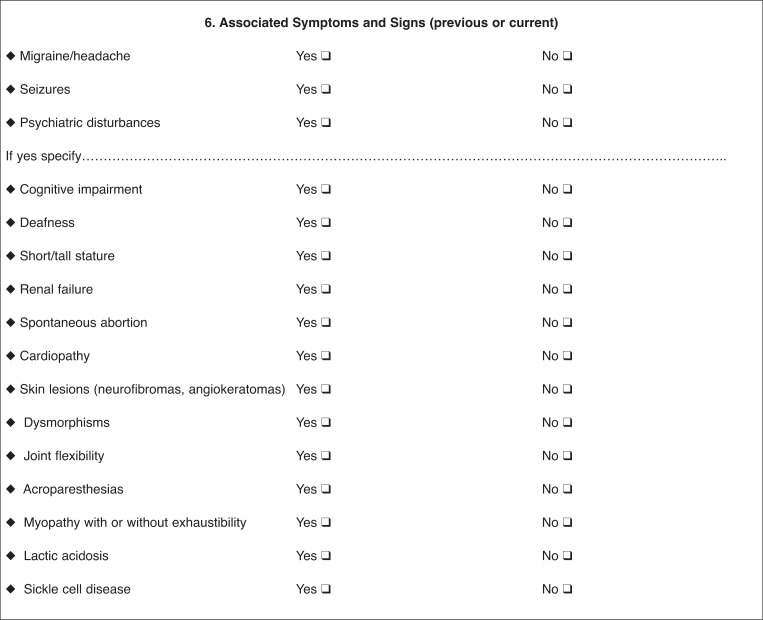
Clinical centers that hospitalize more than 100 patients for stroke annually are included. The project envisages a two-step diagnostic pathway. Through a standardized form, stroke physicians in the participating units collect the demographic and clinical data of all stroke patients with a suspected genetic cause (‘probable cases’). The form includes demographic data, family antecedents, racial descent, current pathologies, and stroke risk factors. Strokes are classified according to established standardized criteria (hemorrhagic, ischemic, TOAST, and Oxfordshire classification). Findings from neuroimaging and instrumental examinations are also recorded, as are other associated neurological or systemic clinical features. It is also envisaged that physicians fill in clinical and radiological diagnostic algorithms, developed from literature data and specific for each monogenic disease in order to address the disease suspicion (‘suspected case’).
The use of these standardized forms and a centralized training procedure for all clinicians participating in the patient data collection ensures standardized and homogeneous data collection across the participating centers. Furthermore, the validity of the screening procedure is periodically validated at meetings of representatives from the centers and laboratories and the group of experts belonging to the scientific steering committee. All data are stored in a computer database (Access format). On receipt of forms from the centers and laboratories, data are entered centrally by the staff at the coordinating center, who also apply quality evaluation processes. Two advisors (Prof. Hugh Markus, Centre for Clinical Neuroscience, St George’s University of London, and Dr Caspar Grond Ginsbach, Neurologische Klinik Universität, Heidelberg) collaborated on the project design and thus provided a further guarantee of the quality of the project. Nine trained clinical monitors, who visit all the participating clinical centers every two weeks, are working to guarantee the recruitment of a continuous series of suspected cases, as well as the accuracy and completeness of the data. Periodic monitoring of the participating centers reduces the risk of missed cases.
Genetic analysis
Blood samples for DNA analysis are collected during each patient’s hospital stay as well as during outpatient activity, after obtaining patient informed consent according to the local ethics regulations. All centers guarantee correct acquisition of informed consent; this includes providing patients with a full description of the project’s goals as well as informing them of the possibility that biological material collected may be used for future analyses still to be defined at the time of the acquisition of consent. The use and sharing of biospecimens and associated clinical data complies with all applicable privacy and human subjects’ protection regulations. All samples are processed by automatic extraction to obtain high purity DNA and stored locally at −80°C. Blood samples are sent together with the clinical and criteria forms to the specific laboratories for the genetic diagnosis. The genetic diagnosis is obtained by performing the following evaluations:
for CADASIL, screening of exons 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, and 23 of the NOTCH3 gene on chromosome 19;
for Fabry disease, α-galactosidase activity initially in all suspected men; in suspected women and in men with reduced α-galactosidase activity, a genetic screening of the seven most frequently mutated exons of the α-GAL A (Xq 22.1) gene;
for MELAS, screening for point mutations in the tRNALeu gene on mtDNA; in highly suspected cases or in cases with a muscular biopsy consistent with mitochondrial myopathy, screening for other mtDNA mutations, e.g. in the MTTL1 gene and other tRNA genes (MTTF, MTTv, MTTQ) as well as in other subunits of complex 1 such as MTND1, MTND5, and MTND6;
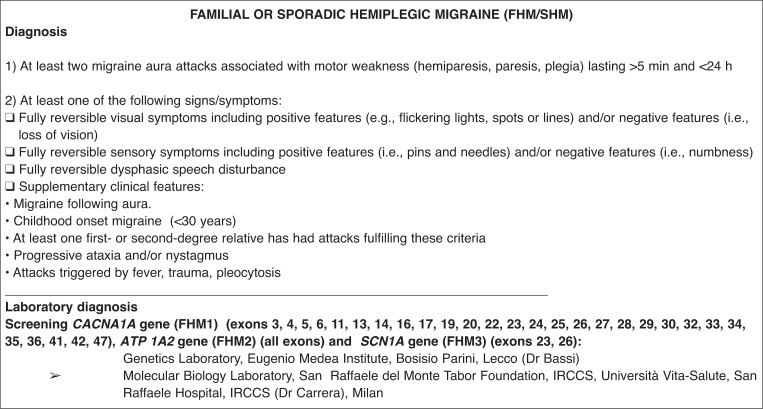
for FHM/SHM, screening of all exons of the ATP1A2 gene; exons 3, 4, 5, 6, 11, 13, 14, 16, 17, 19, 20, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33, 34, 35, 36, 41, 42, and 47 of the CACNA1A gene; and exons 23 and 26 of the SCN1A gene;
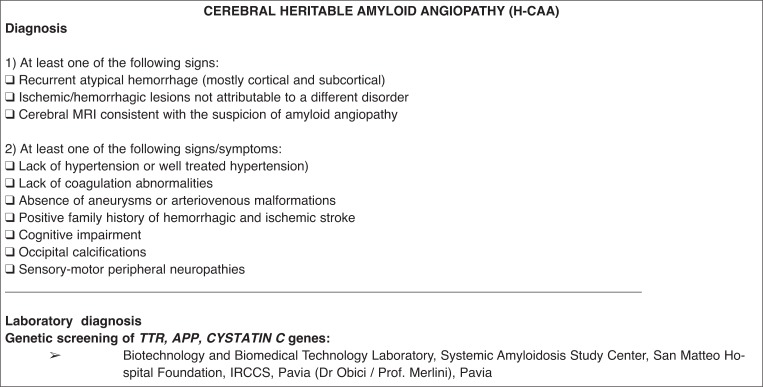
for H-CAA, sequencing in suspected cases of all exons of TRANSTHYRETIN, CYSTATIN C, and AMYLOID PRECURSOR PROTEIN gene; and
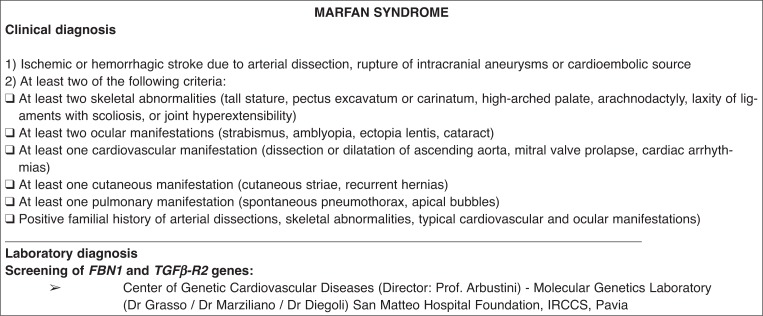
for Marfan syndrome, because the diagnosis is established on clinical grounds (5), molecular analysis consisting of whole-exon screening of the FB1 and TGFβ-R2 genes will be performed only in highly suspected cases. The project involves the use of clinical and personal data and of blood samples for DNA extraction. The source of genetic material is venous blood collected from the patients by standard blood-drawing methods. No personal risks are envisaged for the patients involved since they are not exposed to any treatment or challenge. Italian national rules on data protection are complied with, and patient anonymity is guaranteed. The results of genetic tests are communicated to physicians in charge of the cases, who are responsible for informing the patient, or next of kin, of diagnostic test results and of the resulting diagnosis, according to local ethics regulations.
For data analysis, a database encryption strategy has been developed. Since results from single individuals would not be informative, information is extracted from group data. Thus, all published data will be at group, not individual, level.
Statistical analysis
Before the planned results analysis, a quality evaluation will be performed and the validity of the screening procedures assessed. The analysis will be carried out centrally, by the coordinating center, using the full dataset. Correlation analysis of phenotype and genotype characteristics will be performed on the total population as well as on different subgroups. A subgroup analysis of stroke subtypes is also planned. An analysis of monogenic disease incidence in the regional stroke population, stratified by sex and age, will also be performed.
Expected results
Through the present study, which involves the creation of an integrated regional network of clinical centers and laboratories and the coordination of their activity, and its results, we expect to provide a good practical example for the implementation of similar projects and for the diagnosis of other monogenic disorders. In addition, our network is expected to be a useful tool for supporting research excellence in stroke genetics. A cohort of well-phenotyped and well-characterized stroke patients, homogeneously collected, is expected to provide good-quality results in terms of: i) better knowledge of the natural history of monogenic disorders; ii) more accurate definitions of phenotypes associated with monogenic disorders; iii) identification of possible genotype-phenotype correlations; iv) identification of additional environmental and risk factors modulating the disease process or variably contributing to the full disease phenotype; and v) the creation of a proven network for future collaboration in therapeutic clinical trials targeting prevention of disease progression or symptom relief.
Preliminary results
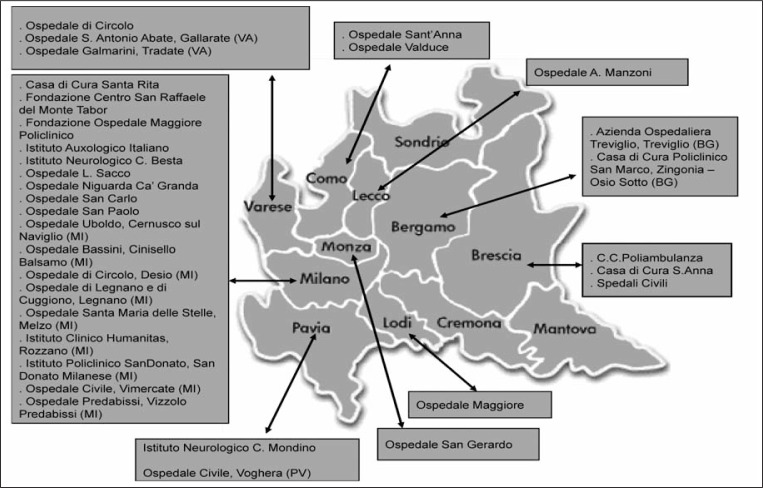
Thirty-three clinical units among the region’s centers hospitalizing more than 100 stroke patients each year are participating in the project (Fig. 1). These centers cumulatively hospitalize about 10,000 stroke patients annually, representing about 90% of all stroke cases in Lombardy. Although the recruiting period was intended to start in January 2008, patient data collection began later than the anticipated date because most centers were waiting for local ethics committee approval. Moreover, clinicians required a start-up period, which meant that the initial recruitment phase moved slowly. After April, 2008 recruitment became steady and improved further following the introduction of clinical monitors in December 2008.
Figure 1.
Lombardia GENS recruiting centers
In about three years of the project, data for about 200 patients with a suspected monogenic disease have been collected. Two diagnostic steps provide first for the selection of suspected phenotypes and then for the identification of patients with a positive genetic test for each specific disease. Of the patients included so far, 39% were screened for CADASIL, 13% for Fabry disease, 27% for H-CAA, 6% for MELAS, 11% for FHM/SHM and 3% for Marfan syndrome. The final expectation is that physicians will be able to apply, in their everyday clinical practice, the diagnostic tools for monogenic disease acquired through their participation in this project.
Discussion
The Lombardia GENS project will provide a database of well-phenotyped stroke patients with diagnoses of monogenic disorders. Despite the fact that stroke-associated monogenic diseases are rare and most strokes are believed to be polygenic, diagnosis of single-gene disorders is needed because some have proven amenable to disease-specific treatments. Moreover, a correct diagnosis may provide a concrete benefit to affected individuals in terms of prognostic evaluation, genetic counseling, and specific management measures (3,4).
Most of the available studies on single-gene disorders are underpowered because they included only a small number of cases and were conducted at individual centers. Moreover, given ongoing results in terms of the genetic factors underlying polygenic stroke pathogenesis, careful phenotype characterization and accurate genotype-phenotype correlations, in addition to further insight into the pathogenesis of monogenic disorders, may help to clarify some pathogenic bases of heritable stroke (3,7–9).
Unlike previous studies in this field, which focused on only one monogenic disease (10–18), Lombardia GENS is, to our knowledge, the largest genetic network, allowing the recruitment of a high number of patients suspected of having monogenic disorders, in which the cerebrovascular event is the mandatory inclusion criterion. It has a high probability of success as it favors the collaboration of people (physicians, geneticists, biologists) involved in stroke genetics research. The participation of clinicians in the project facilitates collection of well-characterized populations and the translation of research outcomes into clinical practice, with the aim of identifying possible etiological factors involved in stroke pathogenesis and new treatment modalities for all stroke subtypes. Through Lombardia GENS, stroke genetics laboratories are offered an opportunity to standardize procedures and to increase their research potential, avoiding the risk of diluting effort and delaying outcome. Moreover, the project will help to promote adequate education and training across the health professions, raising awareness of the existence of these diseases and of the resources available for their diagnosis and treatment.
The project will also give stroke patients with rare diseases increased access to care, resources, and expertise, reducing misdiagnosis and non-diagnosis of monogenic diseases and improving their quality of life. In addition, it will lead to the identification of centers of expertise throughout the country and organized healthcare pathways for patients suffering from monogenic diseases. The large patient dataset planned for this project may help to make the diagnosis of monogenic diseases more straightforward, increasing the accuracy of epidemiologic data on the incidence of rare diseases in stroke and knowledge of disease phenotypes, and harnessing the power needed to make significant genotype-phenotype correlations.
The project does, however, present some methodological limits. First, although the participating clinical centers hospitalize most stroke patients in Lombardy, the cases are recruited through selected hospitals which do not provide full coverage of the geographical area. Moreover, hospital-based sampling frames do not allow a true measure of population incidence. Second, the cases are recruited through stroke centers and the presence of stroke or TIA is an inclusion criterion; the data are biased by these criteria which allow the inclusion only of symptomatic patients. The objective of recruiting about 300 cases overall across all subgroups will also limit multiple risk factor analysis studies.
Nevertheless, the project could continue in future years and yield a network model that could be extended to other regions and countries. Because rare diseases continue to be a priority of the European Community, mostly with regard to developing new diagnostics and treatments, as well as performing epidemiological research into these disorders, multicountry or multicenter comprehensive approaches to increase the number of patients included for study are considered to be highly valuable.
Acknowledgments
The Lombardia GENS project has received funding from the Regione Lombardia government as an Independent Research Project (DGR n°VIII/006128-12/12/2007). Lombardia GENS is an investigator-driven, academic, non-profit consortium and is publicly funded.
Appendix 1. Characteristics of patients with suspected monogenic disease
LOMBARDIA GENS
Appendix 2. Characteristics of patients with suspected monogenic disease
LOMBARDIA GENS
† Lombardia GENS investigators: Ospedale S. Antonio Abate, Gallarate (VA) - Zarcone Davide, Mazzucchelli Francesca; Ospedale di Circolo, Saronno (VA) - Grampa Giampiero, Fusi Laura; Ospedale Galmarini, Tradate (VA) - Ghiringhelli Paolo, Giarracca Valentina; Ospedale Valduce, Como - Guidotti Mario, Checcarelli Nicoletta, Muscia Francesco; Ospedale Bassini, Cinisello Balsamo (MI) - Albizzati Maria Grazia, Proserpio Paola; Ospedale Maggiore, Lodi - Riva Maurizio, Iurlaro Simona; Ospedale di Circolo, Desio (MI) - Colombo Antonio, Bordo Bianca Maria; Ospedale Santa Maria delle Stelle, Melzo (MI), Mascherpa Mauro, Molini Graziella Emanuela, Marsile Claudia; Ospedale Predabissi, Vizzolo Predabissi (MI) - Sasanelli Francesco, Molini Graziella Emanuela, Marsile Claudia; Ospedale Civile, Vimercate (MI) - Crespi Vittorio, Braga Massimiliano; Ospedale Uboldo, Cernusco sul Naviglio (MI) - Tel Aldo, Molini Graziella Emanuela, Marsile Claudia; Casa di Cura Santa Rita, Milano - Tadeo Carlo Sebastiano; Istituto Policlinico San Donato, San Donato Milanese (MI) - Meola Giovanni, Bet Lucia-no; Azienda Ospedaliera Treviglio, Treviglio (BG) - Lanza Ezio, Carpo Marinella; Casa di Cura Policlinico San Marco, Zingonia - Osio Sotto (BG) - Camerlingo Massimo; Ospedale Civile, Voghera (PV) - Magrotti Emilio, Valenti Graziana; Ospedale di Legnano e di Cuggiono, Legnano (MI) - Perrone Patrizia, Calloni Maria Vittoria; C.C. Poliambulanza, Brescia - Donati Edoardo; Ospedale di Circolo, Varese - Bono Giorgio, Delodovoci Marialuisa; Ospedale Sant’Anna, Como - Arnaboldi Marco, Tancredi Lucia, Vidale Simone; Ospedale A. Manzoni, Lecco - Agostoni Elio, Longoni Marco; Spedali Civili, Brescia - Magoni Mauro, Costa Angelo, Padovani Alessandro, Pezzini Alessandro; Ospedale Carlo Poma, Mantova - Previdi Paolo; Ospedale San Gerardo, Monza (MI) - Ferrarese Carlo, Santoro Patrizia, Beretta Simone; Ospedale Niguarda Ca’ Granda, Milano - Sterzi Roberto, Motto Cristina; Ospedale San Paolo, Milano - Capitani Erminio, Belvedere Daniela; Ospedale San Carlo, Milano - Bassi Pietro, Lattuada Patrizia; Ospedale L. Sacco, Milano - Mariani Claudio, Gambaro Paola; Istituto Neurologico C. Besta, Milano - Parati Eugenio, Boncoraglio Giorgio, Ballabio Elena; Fondazione Ospedale Maggiore Policlinico, Milano - Bresolin Nereo, Baron Pierluigi, Bersano Anna; Fondazione Centro San Raffaele del Monte Tabor, Milano - Comi Giancarlo, Martinelli Boneschi Filippo, Sessa Maria; Istituto Auxologico Italiano, Milano - Silani Vincenzo, Addobbati Laura, Stramba-Badiale Marco, Michailidis Georgios; Fondazione Istituto Neurologico Nazionale C. Mondino, IRCCS, Pavia - Micieli Giuseppe, Cavallini Anna, Persico Alessandra; Istituto Clinico Humanitas, Rozzano (MI) - Marcheselli Simona, Corato Manue, Incorvaia Barbara. Lombardia GENS monitors: Borellini Linda, Carozzo Mattia, del Zotto Elisabetta, Di Cristofori Andrea, Di Pietro Davide, Gambini Chiara, Grasso Alessandra, Lanzani Francesca, Spinelli Maria Carmela, Susani Emanuela, Valcarenghi Caterina. Lombardia GENS laboratories: Laboratorio di Biochimica e Genetica, Dipartimento di Scienze Neurologiche, IRCCS Fondazione Ospedale Maggiore Policlinico, Mangiagalli e Regina Elena, Milano - Comi Giacomo Pietro; Laboratorio di Genetica, Azienda Ospedaliera Niguarda Ca’ Granda, Milano - Penco Silvana; Laboratorio di Biologia Molecolare, IRCCS Fondazione San Raffaele del Monte Tabor, Università Vita e Salute, Ospedale San Raffaele, Milano - Carrera Paola, Calzavara Silvia, Montrasio Cristina; Laboratorio di Biochimica e Genetica, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano - Taroni Franco, Gellera Cinzia, Rimordi Marco; Istituto Eugenio Medea, Bosisio Parini (LC) - Bassi Maria Teresa; Laboratorio di Biotecnologie e Tecnologie Biomediche, Centro per lo Studio delle Amiloidosi Sistemiche, Fondazione IRCCS Policlinico San Matteo, Pavia - Merlini Giampaolo, Obici Laura; Centro Malattie Genetiche Cardiovascolari, Laboratorio di Genetica Molecolare, Fondazione IRCCS Policlinico San Matteo, Pavia - Arbustini Eloisa, Grasso Maurizia, Diegoli Marta; U.O. di Neuropatologia, Fondazione IRCCS Istituto Nazionale Neurologico C. Besta - Tagliavini Fabrizio, Morbin Michela; Laboratorio di Dermatologia Pediatrica, Fondazione Ospedale Maggiore Policlinico, Mangiagalli e Regina Elena, Milano - Tadini Gianluca.
References
- 1.Hassan A, Markus HS. Genetics and ischaemic stroke. Brain. 2000;123:1784–1812. doi: 10.1093/brain/123.9.1784. [DOI] [PubMed] [Google Scholar]
- 2.Rubattu S, Giliberti R, Volpe M. Etiology and pathophysiology of stroke as a complex trait. Am J Hypertens. 2000;13:1139–1148. doi: 10.1016/s0895-7061(00)01249-8. [DOI] [PubMed] [Google Scholar]
- 3.Meschia JF, Worrall BB, Rich SS. Genetic susceptibility to ischemic stroke. Nat Rev Neurol. 2011;7:369–78. doi: 10.1038/nrneurol.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballabio E, Bersano A, Bresolin N, Candelise L. Monogenic vessel diseases related to ischemic stroke: a clinical approach. J Cereb Blood Flow Metab. 2007;27:1649–1662. doi: 10.1038/sj.jcbfm.9600520. [DOI] [PubMed] [Google Scholar]
- 5.Razvi SS, Bone I. Single gene disorders causing ischaemic stroke. J Neurol. 2006;253:685–700. doi: 10.1007/s00415-006-0048-8. [DOI] [PubMed] [Google Scholar]
- 6.Flossmann E, Schulz UG, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35:212–227. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- 7.Markus HS. Stroke genetics. Hum Mol Genet. 2011;20(R2):R124–31. doi: 10.1093/hmg/ddr345. [DOI] [PubMed] [Google Scholar]
- 8.Meschia JF, Singleton A, Nalls MA, Rich SS, Sharma P, Ferrucci L, Matarin M, Hernandez DG, Pearce K, Brott TG, Brown RD, Jr, Hardy J, Worrall BB. Genomic risk profiling of ischemic stroke: results of an international genome-wide association meta-analysis. PLoS One. 2011;6:e23161. doi: 10.1371/journal.pone.0023161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bevan S, Markus HS. Genetics of common polygenic ischaemic stroke: current understanding and future challenges. Stroke Res Treat. 2011;2011:179061. doi: 10.4061/2011/179061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalimo H, Ruchoux MM, Viitanen M, Kalaria RN. CADASIL: a common form of hereditary arteriopathy causing brain infarcts and dementia. Brain Pathol. 2002;12:371–84. doi: 10.1111/j.1750-3639.2002.tb00451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Razvi SS, Davidson R, Bone I, Muir KW. The prevalence of cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL) in the west of Scotland. J Neurol Neurosurg Psychiatry. 2005;76:739–741. doi: 10.1136/jnnp.2004.051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narayan SK, Gorman G, Kalaria RN, Ford GA, Chinnery PF. The minimum prevalence of CADASIL in northeast England. Neurology. 2012;78:1025–7. doi: 10.1212/WNL.0b013e31824d586c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolfs A, Böttcher T, Zschiesche M, et al. Prevalence of Fabry disease in patients with cryptogenic stroke: a prospective study. Lancet. 2005;366:1794–1796. doi: 10.1016/S0140-6736(05)67635-0. [DOI] [PubMed] [Google Scholar]
- 14.Baptista MV, Ferreira S, Pinho-E-Melo T, et al. PORTuguese Young STROKE Investigators Mutations of the GLA gene in young patients with stroke: the PORTYSTROKE study – screening genetic conditions in Portuguese young stroke patients. Stroke. 2010;41:431–436. doi: 10.1161/STROKEAHA.109.570499. [DOI] [PubMed] [Google Scholar]
- 15.Rolfs A, Martus P, Heuschmann PU, et al. sifap1 Investigators Protocol and methodology of the Stroke in Young Fabry Patients (sifap1) study: a prospective multicenter European study of 5,024 young stroke patients aged 18–55 years. Cerebrovasc Dis. 2011;31:253–262. doi: 10.1159/000322153. [DOI] [PubMed] [Google Scholar]
- 16.Wozniak MA, Kittner SJ, Tuhrim S, et al. Frequency of unrecognized Fabry disease among young European-American and African-American men with first ischemic stroke. Stroke. 2010;41:78–81. doi: 10.1161/STROKEAHA.109.558320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manwaring N, Jones MM, Wang J, et al. Population prevalence of the MELAS A3243G mutation. Mitochondrion. 2007;7:230–233. doi: 10.1016/j.mito.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Wityk R, Zanferrari C, Oppenheimer S. Neurovascular complications of Marfan syndrome: a retrospective, hospital-based study. Stroke. 2002;33:680–684. doi: 10.1161/hs0302.103816. [DOI] [PubMed] [Google Scholar]