Abstract
Natural genetic transformation and restriction-modification (R–M) systems play potentially antagonistic roles in bacteria. R–M systems, degrading foreign DNA to protect the cell from bacteriophage, can interfere with transformation, which relies on foreign DNA to promote genetic diversity. Here we describe how the human pathogen Streptococcus pneumoniae, which is naturally transformable, yet possesses either of two R–M systems, DpnI or DpnII, accommodates these conflicting processes. In addition to the classic restrictase and double-stranded DNA methylase, the DpnII system possesses an unusual single-stranded (ss) DNA methylase, DpnA, which is specifically induced during competence for genetic transformation. We provide further insight into our recent discovery that DpnA, which protects transforming foreign ssDNA from restriction, is crucial for acquisition of pathogenicity islands.
Keywords: Streptococcus pneumoniae, DpnA ssDNA methylase, capsule switch, competence, genetic transformation, heterologous DNA transfer, pathogenicity island transfer, restriction–modification
Natural genetic transformation has been proposed as the bacterial equivalent of eukaryotic sexual reproduction, promoting genetic diversity.1 Transformation involves internalization of foreign DNA in the form of single strands (ss), generated from a double-stranded (ds) substrate, which are recombined into the host genome by homology. Transformation is a widespread process2 which contributes to genetic diversity in the human pathogen Streptococcus pneumoniae (the pneumococcus).3 Transformation is thus crucial for pneumococcal vaccine escape, promoting switching of capsule loci between isolates,4,5 where over 90 capsular types exist6 but only ~10–15% are targeted by current conjugate vaccines. On the other hand, many bacteria possess restriction-modification (R–M) systems, which are suggested to act as analogs of the vertebrate immune system,7 protecting the cell from attack by foreign DNA. R–M systems, which classically encode a restrictase to degrade foreign DNA and a dsDNA methylase to protect the host genome, are seen as limiting genetic diversity.8,9 S. pneumoniae cells possess one of two R–M systems, DpnI or DpnII, which respectively restrict dsDNA which is me+ is methylated (me+) on the adenine base of the GATC sequence and unmethylated (me0).10 The DpnII system, encoded by the dpnMAB operon, besides a restrictase (DpnII encoded by dpnB) and dsDNA methylase (DpnM), possesses an unusual methylase of ssDNA, DpnA. Re-evaluating the role of this ssDNA methylase in genetic transformation was the focus of our recent study, which led to the discovery that DpnA is crucial for acquisition of me0 pathogenicity islands by transformation.11 In this article, we first provide a concise review and discussion of our previous findings. We then genetically investigate the potential effect of the transformation-dedicated ssDNA-binding protein SsbB12 on DpnA methylation of ssDNA during chromosomal or plasmid transformation. Finally, we evaluate and discuss the activity of both DpnI and DpnII restriction enzymes against transforming plasmid DNA.
The DpnI/DpnII Complementary System Defends Pneumococci Against Phage Attack
The DpnI and DpnII systems protect the cells from me+ or me0 dsDNA bacteriophage, respectively.13 This complementary system is believed to be of protective value in the event of phage attack on mixed pneumococcal populations. Thus, DpnII isolates will survive attack by me0 bacteriophage produced through infection and lysis of DpnI cells (Fig. 1). Conversely DpnI isolates would survive an attack by me+ bacteriophage progeny from DpnII cells. As a result, a part of the mixed population will survive either of these bacteriophage attacks. The existence of this complementary R–M system is thus regarded as increasing the likelihood for species survival in phage-containing environment.
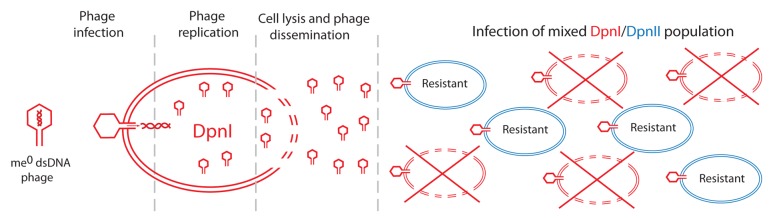
Figure 1. Advantage of the DpnI/DpnII system for survival of mixed pneumococcal population facing phage attack. Infection of a mixed DpnI/DpnII population by me0 dsDNA bacteriophage. While a DpnII isolate infected by a me0 phage (red hexagonal shape) survives because it is protected by restriction, infection of a DpnI isolate results in phage replication and cell lysis, releasing me0 progeny phage (left part of figure). These progeny can infect DpnI (red) or DpnII (blue) cells in the population (right part of figure). DpnI cells will lyse (represented by large red cross) as DpnI cannot restrict me0 DNA, while DpnII cells will survive due to restriction of me0 phage dsDNA by DpnII upon infection. Conversely, infection of mixed DpnI/DpnII population by me+ dsDNA bacteriophage leads to killing of DpnII cells and release of me+ progeny phage, while DpnI cells survive (not shown).
The Atypical DpnI Restrictase Intrinsically Provides Defense without Compromising Chromosomal Transformation
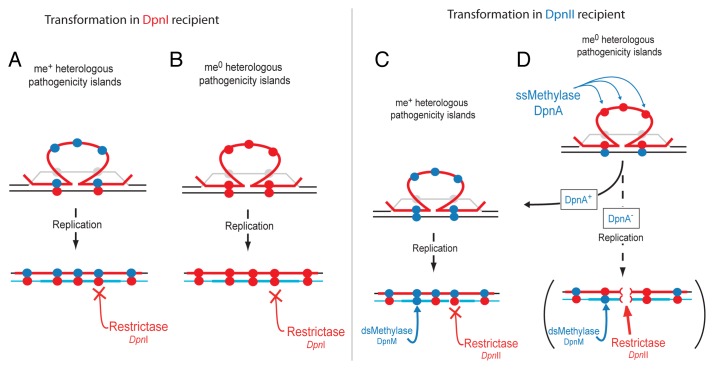
The DpnI restrictase is atypical in specifically targeting methylated dsDNA, which is normally the form of DNA offering protection against restriction. As a result, the DpnI system protects the cell from me+ bacteriophage attack. However, the DpnI system does not limit chromosomal transformation, as fully me+ dsDNA is never created in the chromosome. Integration of homologous me+ ssDNA (from DpnII donor) into a DpnI chromosome produces intrinsically resistant me+/0 dsDNA. When a heterologous pathogenicity island, which has no homology in the host chromosome, is transferred, its integration occurs via flanking homology. The pathogenicity island DNA thus remains in ss form in the transformation heteroduplex until passage of the chromosomal replication fork, which then produces DpnI-resistant me+/0 dsDNA (Fig. 2A). On the other hand, integration of me0 ssDNA (from DpnI donor) produces me0 dsDNA which is resistant to DpnI (Fig. 2B). Thus, DpnI pneumococcal isolates can readily interchange pathogenicity islands by transformation irrespective of the Dpn status of the donor cells.
Figure 2. Differential impact of DpnI and DpnII R–M systems on transformation. (A) DpnI does not interfere with transformation of a pathogenicity island on me+ (closed blue circles) DNA. Transforming me+ ssDNA (red line) pairing with homologous DNA on host chromosome (black line) displaces the complementary strand (gray line). The pathogenicity island sequence remains in the form of ssDNA due to lack of homology. After replication, me+/0 sites are produced by synthesis of complementary neosynthesized DNA (light blue), which are not sensitive to restriction by the DpnI restrictase. (B) DpnI does not interfere with transformation of a pathogenicity island on me0 (closed red circles) DNA. DNA identity as in (A). (C) DpnII does not interfere with transformation of a pathogenicity island on me+ DNA. DNA identity as in (A). After replication, dsDNA is either fully me+ or me+/0 depending on the presence and action of methylases, and therefore resistant to DpnII. (D) Transformation of a pathogenicity island on me0 (closed red circles) transforming DNA into a DpnII strain in the presence or absence of ssDNA methylase DpnA. DNA identity as in (A). In the presence of DpnA, the ssDNA of the pathogenicity island in the transformation heteroduplex is methylated, and thus protected from DpnII restriction after replication, as in (C) (DpnA+ arrow). In the unnatural absence of DpnA, the ssDNA of the pathogenicity island remains me0 in the transformation heteroduplex. After replication (DpnA− arrow and brackets), me0 sites in the heterologous transforming DNA are paired with me0 sites in the neosynthesized DNA. Resulting dsDNA me0 sites in the chromosome are sensitive to DpnII, which can restrict the chromosome (red brackets in DNA) and kill the cell.
The DpnII System is Tuned to Avoid Compromising Genetic Exchange
The DpnII restrictase is orthodox in that it restricts me0 dsDNA. R–M systems, which classically contain a restrictase of me0 dsDNA and a dsDNA methylase, have been suggested to antagonize genetic exchange.8,9 During the transformation of a pathogenicity island from me0 donor DNA, the integrated pathogenicity island sequence is rendered fully me0 after replication, with neosynthesized me0 DNA paired with me0 donor DNA. Once this DNA is produced in the chromosome, the restrictase and methylase compete for access to me0 sites, with restriction degrading the chromosome and resulting in loss of the transformant cell. However, the DpnII system is unorthodox in that a third enzyme, the unusual ssDNA methylase DpnA, is also expressed.
The main role of DpnA, which is specifically induced during pneumococcal competence for genetic transformation,14 was suggested to be promotion of plasmid transfer by transformation, as a dpnA− mutant showed a deficit in transformation of me0 plasmids, but not of an me0 chromosomal point mutation.14,15 However, plasmids are rare in S. pneumoniae16,17 and their transfer poorly efficient.18 Furthermore, recent results clearly indicated that the competence-induced SSB paralogue SsbB, which protects transforming ssDNA and promotes chromosomal transformation, antagonizes plasmid transformation, suggesting that pneumococcal transformation has not been tuned to favor plasmid exchange.12 These considerations made it unlikely that DpnA was recruited to the competence regulon simply to promote plasmid transfer. We suggested that DpnA should be crucial for acquisition of me0 pathogenicity islands by DpnII isolates.3 Logically, DpnA should not be required for transfer of me+ pathogenicity islands, as the transforming DNA is already methylated, protecting the chromosomes of resulting transformants from restriction (Fig. 2C). For the transfer of a me0 pathogenicity island, when DpnA is present, it methylates the transforming ssDNA, so that resulting chromosomal dsDNA is hemimethylated (me+/0) post-replication, and thus protected from DpnII (Fig. 2C and D). In the absence of DpnA, me0 pathogenicity islands, which remain in ss form in the heteroduplex, cannot be methylated prior to replication (Fig. 2D). After replication, complementary neosynthesized DNA is produced forming fully me0 dsDNA in the chromosome. This DNA can be degraded by DpnII, destroying the potential transformant (Fig. 2D). We used transformation studies to validate this hypothesis, showing that acquisition of foreign me0 plasticity islands (e.g., switch of capsule locus from DpnI isolates) was severely depleted in a dpnA− mutant, while no effect was observed for acquisition of isogenic me+ islands.11 By transforming these strains with heterologous cassettes containing varying numbers of GATC sites, we showed that increasing the number of GATC sites in the heterologous region increased the dependency on DpnA for protection. As well as being induced during competence, DpnA is also expressed constitutively from a promoter upstream of dpnM. By mutating the competence-induced promoter in front of dpnA (Pcin), showed that the majority of DpnA is produced during competence, and this specific induction is crucial for full protection of transforming me0 pathogenicity islands. We concluded that the main role of DpnA and of its induction during competence is to promote acquisition of foreign me0 pathogenicity islands by transformation. Extrapolating from this, DpnA should be critical to vaccine escape of DpnII isolates, which most likely occurs via exchange of capsule loci by transformation.
Our results suggest two important roles for the DpnII R–M system. First, the DpnII restrictase plays an important role in protecting the cell from me0 bacteriophage attack, a role which is mirrored by the protection against me+ bacteriophage by the complementary DpnI restrictase.13 Second, the presence of the unusual ssDNA methylase DpnA maintains the plasticity potential of the bacterium by methylating foreign DNA, promoting acquisition of me0 pathogenicity islands by protecting transformant chromosomes from DpnII. In the absence of this protection, the DpnII R–M system would limit genetic diversity, as is the case for other R–M systems,8,9 by degrading the chromosomes of transformant clones. The genetic organization of the dpnMAB operon allowing co-expression of the three genes and the co-induction of only dpnA and dpnB at competence constitutes a remarkably economical and elegant set-up ensuring simultaneously increased protection against bacteriophage throughout the competence window (i.e., during a period when cells are physiologically at risk)19 and negation of any antagonizing effect on genetic transformation. DpnA-like ssDNA methylases appear rare, although the DpnII locus is also present in the closely-related Streptococcus mitis species. Similar methylases are also present in other Streptococci such as Streptococcus suis20 and Streptococcus mutans. These enzymes remain uncharacterized, although it is tempting to speculate that they may play a similar role of maintenance of genetic plasticity in these members of the diverse Streptococcal genus.21-23
Interplay between DpnA and SsbB in the Processing of Internalized ssDNA
As described, transformation proceeds through internalization and integration of ssDNA into the host chromosome via flanking homology. Directly after uptake, exogenous ssDNA is presumably coated by tetramers of the ssDNA-binding protein SsbB.24 This paralogue of the essential house-keeping SSB, SsbA, is specifically induced during competence,25 and creates a reservoir of transforming ssDNA, protected from degradation by endogenous nucleases.12 SsbB-coated ssDNA was thus shown to be resistant to nuclease digestion in vitro.26,27 SsbB could therefore compete with DpnA for access to transforming ssDNA. Alternatively, SsbB may actively recruit DpnA to transforming DNA, promoting methylation. This hypothesis is based on the observation that SsbB possesses a carboxy terminus enriched in acidic amino acids, reminiscent of the acidic tail of Escherichia coli and Bacillus subtilis SSB proteins, which is the site for specific protein–protein interactions with various partners involved in DNA metabolism and enables their recruitment to the replication fork.28 To investigate the interplay between SsbB and DpnA, we explored the effect of the inactivation of the ssbB gene as well as of the deletion of the 7 acidic carboxy terminal amino acids of SsbB (ssbB∆7 mutant).12 Neither the lack of SsbB nor the absence of its acidic tail altered chromosomal transformation of plasticity islands (Fig. 3A), suggesting that the ssDNA-binding protein neither competes with or recruits DpnA to internalized ssDNA in the reservoir.
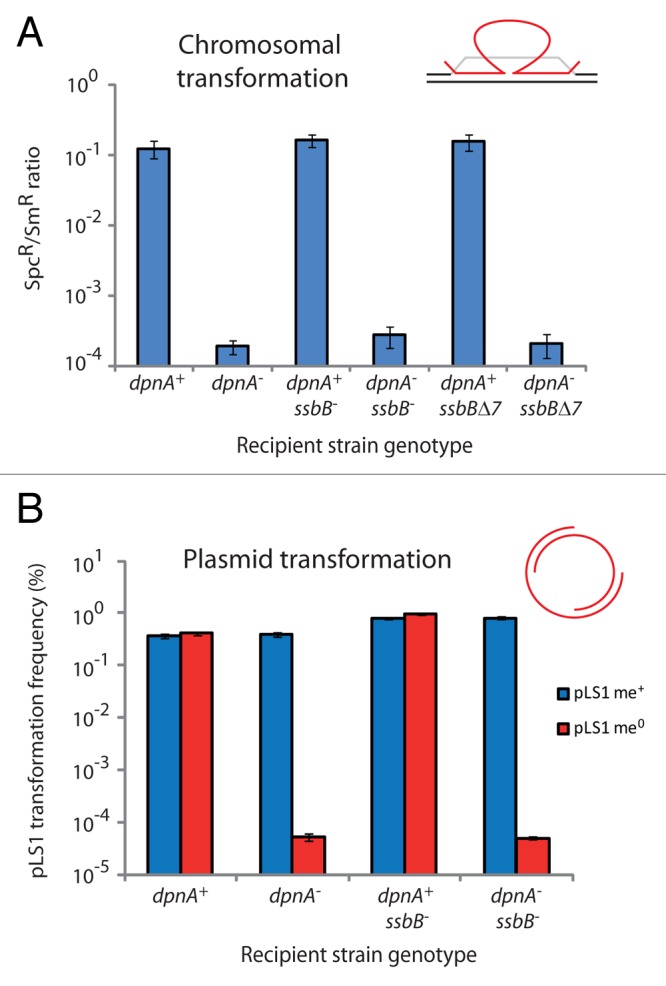
Figure 3. Alteration of SsbB does not affect methylation of chromosomal or plasmid ssDNA by DpnA. (A) Transformation efficiency of ciaR::spc22C cassette into DpnII strains lacking dpnA or ssbB, or possessing the ssbB∆7 mutation. Efficiency represented as a ratio of SpcR transformants to SmR transformants, selecting the rpsL41 point mutation present on the same donor DNA (R1173), as described in reference 11. Error bars calculated from triplicate repeats. (B) Transformation efficiency of pLS1 plasmids (me+ and me0) into same strains as in (A), represented as a percentage of TetR transformants. Red bars, pLS1 me0; blue bars, pLS1 me+. Error bars calculated from triplicate repeats.
This conclusion was further confirmed through testing of the effect of ssbB inactivation on replicative plasmid transformation in the presence or absence of DpnA. Essentially no difference was observed between transformation efficiency of me+ or me0 plasmids into ssbB+ or ssbB− strains, whether they be dpnA+ or dpnA− (Fig. 3B). An increase in transformation efficiency was observed in ssbB− strains, as previously observed for high plasmid concentrations.12 However, there was no other difference in transformation efficiencies between the tested strains, suggesting that although SsbB antagonizes plasmid transformation,12 it does not alter the ability of DpnA to methylate transforming plasmid DNA.
Where Does Methylation of Foreign DNA by DpnA Occur?
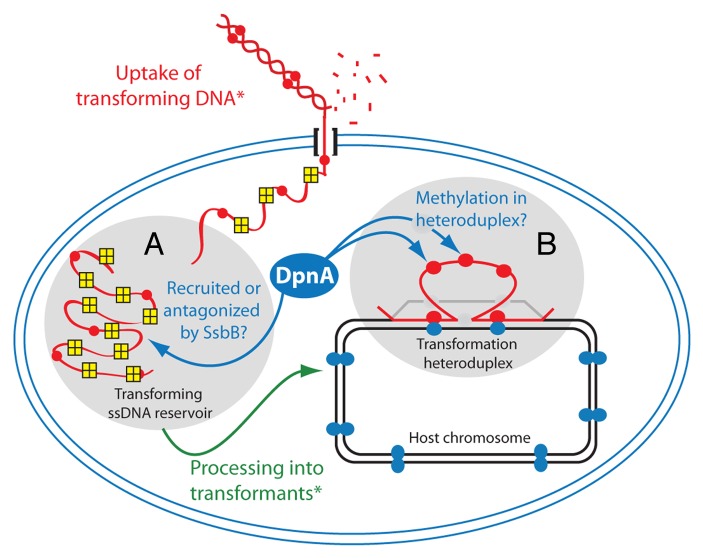
One aspect not discussed in our previous study was the location of ssDNA methylation by DpnA. This may occur at two distinct stages during genetic transformation. First, DpnA may be able to methylate me0 ssDNA within the SsbB-protected reservoir (Fig. 4A). Second, after formation of the transformation heteroduplex in the host chromosome, heterologous DNA such as a pathogenicity island remains in ss form due to lack of homology (Fig. 2). DpnA may access the ssDNA present in the heteroduplex and methylate it here, prior to replication (Fig. 4B), thus maintaining protection of resulting transformants from DpnII restriction (Fig. 2D).
Figure 4. Potential subcellular sites of methylation of me0 heterologous transforming ssDNA by DpnA. Transforming me0 dsDNA (red line with red circles) is internalized in ss form through the DNA entry pore*, with one strand of the dsDNA substrate degraded outside of the cell (short red lines). SsbB tetramers (yellow squares) coat internalized ssDNA and protect it from degradation, producing a reservoir for transformation (gray oval A). Processing of ssDNA into transformants* (green arrow) involves formation of a transformation heteroduplex (as in Fig. 2), with the ss loop maintained in the heteroduplex until replication (see Fig. 2). During this process, DpnA (blue oval) could methylate transforming ssDNA in the reservoir (gray oval A) or in the heteroduplex of transformation (gray oval B). *For a review of transformation processes and proteins involved in S. pneumoniae, see reference 28.
The simplest interpretation of the observation that DpnA is equally efficient in protecting plasmid and chromosomal DNA (Fig. 3) is that DpnA methylates internalized ssDNA in the reservoir. This would imply that DpnA has the ability to displace SsbB to methylate GATC sites, despite the previously documented effect of SsbB binding resulting in protection of ssDNA from DNase I, Neurospora endonuclease, nuclease P1, and pneumococcal EndA nuclease.26 If this is the case, the displacement of SsbB by DpnA does not depend on the acidic tail of SsbB, which is thought to recruit functional partners of SSB proteins (Fig. 3A).12 Could there be a utility for homologous transformation in methylation of ssDNA by DpnA in the reservoir, since such DNA could be methylated by DpnM in the form of dsDNA once integrated into the heteroduplex? This could be useful during transformation of homologous DNA with high numbers of methylation sites. In this situation, were the replication fork to pass over the me+/0 heteroduplex before DpnM had rendered every site me+, me0 dsDNA could be produced prompting restriction of the newly replicated transformant chromosome. Thus, any prior methylation of transforming ssDNA by DpnA would lighten the workload of DpnM after heteroduplex formation, and could thus also favor acquisition of point mutations in this situation.
However, the alternative possibility that DpnA cannot displace SsbB and therefore does not access ssDNA in the reservoir can by no means be excluded. As concerns chromosomal transformation, DpnA should then methylate ssDNA in the transformation heteroduplex, i.e., in the heterologous ssDNA loop (Fig. 2D). This hypothesis necessarily implies that SsbB is not covering the ssDNA loop, possibly because it is displaced during heteroduplex formation. It is of note that such a mechanism would reduce the load on DpnA activity as after heteroduplex formation, only heterologous me0 sites remain in the form of ssDNA, whereas if acting on ssDNA in the reservoir, DpnA should methylate me0 sites on all internalized ssDNA molecules. For plasmid transformation, which does not involve heteroduplex formation, DpnA should act at some stage during reconstitution of the plasmid replicon, which presumably occurs through annealing of partially overlapping complementary ssDNA strands (Fig. 3B).18 This hypothesis would also imply that SsbB is displaced during the annealing reaction to provide access to DpnA for methylation of me0 ssDNA regions before repair/replication restores a fully ds plasmid molecule that would be sensitive to DpnII.
Re-Evaluating Efficiency of Restriction by DpnI and DpnII During Plasmid Transfer
While methylation status of transforming ssDNA integrated into the chromosome is irrelevant in DpnI isolates (see above), the situation differs for transformation of me+ plasmids as strands of ssDNA will be internalized, with dsDNA formed by annealing of complementary molecules18 to form a molecule composed of both ssDNA and dsDNA components, with ds me+ sites in the overlapping dsDNA regions (Fig. 3B, diagram). In an attempt to directly compare the restrictive activity of DpnI and DpnII on plasmids, we compared efficiency of transformation of isogenic me+ or me0 replicative plasmids into wild-type DpnI or DpnII dpnA-recipient cells. Since DpnI strains do not have an equivalent to DpnA to protect internalized ssDNA, comparison of transformation efficiency between DpnI and DpnII dpnA− strains gives a truer comparison of the activity of the restriction enzymes on plasmid transfer. As expected, a large loss of efficiency was observed for me0 plasmids in a DpnII dpnA− strain (Fig. 5A), mirroring results observed previously15 and confirming that methylation of internalized ssDNA by DpnA is important for plasmid transfer. In comparison, me+ plasmids transformed into DpnI strains with only 10-fold less efficiency than isogenic me0 plasmids (Fig. 5A). No difference in transformation efficiency was observed in control DpnII or Dpn0 strains (results not shown). Plasmid transfer in Dpn strains was previously explored in two studies.15,29 We observe very similar results to those published for the DpnII dpnA− strain. However, we first show a greater loss of me+ plasmid transfer in DpnI strains (~10-fold compared with ~3-fold). This could be due to the number and position of GATC sites within the plasmids used. Second, while we observed no difference in efficiency of plasmid transfer in DpnII strains, authors observed losses of me+ plasmid efficiency ranging from ~3-fold ~25-fold in different experiments. Oddly, such differences were also observed for transfer into a Dpn0 strain containing neither DpnI nor DpnII system, which should readily accept both plasmids. These differences may result from the non-isogenic nature of the donor strains used. Our study employs more precise transformation conditions, and isogenic donor and recipient strains giving more precise and reproducible ratios of transformation efficiency.
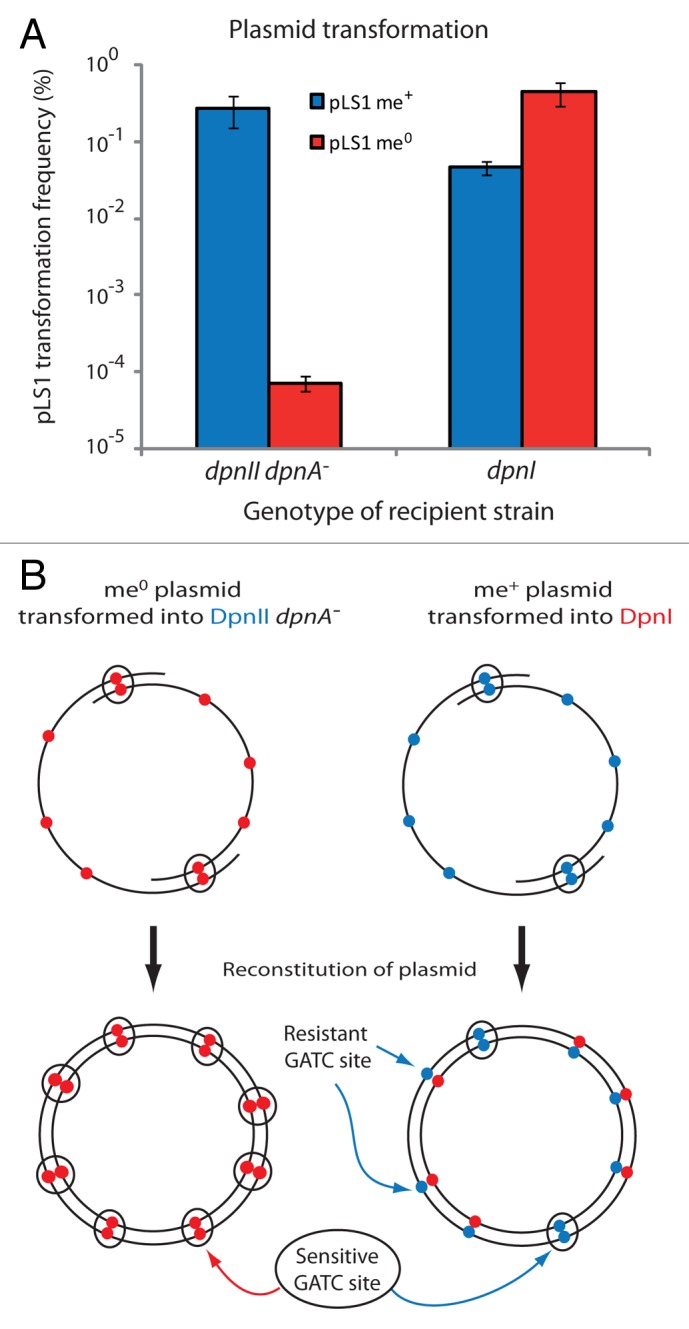
Figure 5. Comparing restrictive activity of DpnI and DpnII on pLS1 plasmid transformation. (A) Transformation of pLS1 plasmid (me+ and me0) into DpnII dpnA− and DpnI strains, represented as a percentage of TetR transformants. Red bars, pLS1 me0; blue bars, pLS1 me+. Error bars calculated from triplicate repeats. (B) Schematic representation of reconstitution of me0 and me+ plasmid replicons during transformation, showing regions of plasmid susceptible to Dpn restriction enzyme during and after reconstitution. Closed red circles, me0 GATC sites; closed blue circles, me+ GATC sites. Single circle, ssDNA; circle pairs, dsDNA. GATC sites sensitive to restriction are enclosed in black circles in each case.
Unprotected plasmids were transformed with over 100-fold more efficiency in DpnI than DpnII dpnA− strains (Fig. 5A), suggesting that restriction of reconstituted plasmids by DpnII is more efficient than by DpnI. We suggest that the explanation for this difference lies in the nature of the reconstitution process of plasmid replicons. Upon annealing between two linear plasmid molecules to reconstitute the plasmid, only regions of annealing between the two ssDNA plasmid molecules will initially be dsDNA and thus sensitive to restriction (Fig. 5B). However, neosynthesis of complement to the ssDNA to create a fully ds plasmid molecule has differing outcomes for sensitivity to DpnI or DpnII (Fig. 5B). In the case of DpnII, neosynthesis creates fully me0 GATC sites in the plasmid and DpnII can restrict any GATC site on the plasmid, in competition with the dsDNA methylase DpnM. In the case of DpnI, the donor plasmid is me+ while the neosynthesized complement is me0, creating resistant me+/0 dsDNA. As a result, me+ DNA is only found at the regions of initial annealing between the two ssDNA molecules, limiting the number of target GATC sites for DpnI, and likely producing a fraction of fully resistant plasmids where no GATC sites are present in this region. This could explain why DpnI appears less able to restrict plasmids than DpnII. On the other hand, it is possible that the DpnII restrictase may simply be more efficient than DpnI at restricting GATC sites in vivo. This could be due to the specific co-induction of dpnB (encoding DpnII) with dpnA during competence, which may lead to DpnII concentration increase in competent cells similar to that observed for DpnA.11 Alternatively, access of restriction enzymes to reconstituted plasmid replicons may differ, with DpnII readily able to access the me0 GATC sites, and DpnI less so.
DpnA and the Raison d’être of DNA Uptake in S. pneumoniae
There has long been debate as to the reason that bacteria actively take up exogenous DNA.19,30-32 Two credible suggestions have emerged, suggesting that internalized ssDNA is used for genome maintenance via template-directed repair, and for genetic diversity via chromosomal integration of exogenous DNA. Template-directed repair involves the repair of dsDNA breaks in the chromosome by recombination, thus requiring specifically homologous transforming DNA as a template. Conversely, promotion of genetic diversity involves acquisition of heterologous, foreign DNA sequences, although these must be flanked by homologous sequence to allow classic recombination to occur. By uncovering the role of DpnA in me0 pathogenicity island transfer, we show that protection of foreign, heterologous DNA is a mechanism programmed by the host cell. The only reason we can see to have such a programmed mechanism of protection of foreign DNA is to promote genetic diversity, as foreign, heterologous DNA should be of no use for genome maintenance. Our results thus provide the first concrete evidence that S. pneumoniae takes up DNA with the specific goal of promoting genetic diversity.
How do R–M Systems Antagonize Transformation in Other Species?
Although a number of studies had shown previously that R–M systems were capable of antagonizing genetic transformation,8,9 the mechanisms involved had remained unclear. The enigma was that transforming DNA is in the form of ss, while restriction enzymes tend to act exclusively on dsDNA. As a result, authors were unable to provide a concrete hypothesis as to the mechanisms involved, suggesting that restriction may occur after integration into me+/0 DNA transiently produced immediately downstream of the replication fork8 or even extracellularly prior to conversion of dsDNA template to transforming ssDNA.33 The first of these hypotheses appears far-fetched, as to prevent the acquisition of heterologous cassettes in this manner, a transformation heteroduplex would need to form, and since the heterologous region lacks complement in the genome and thus remains ss, the homologous flanking regions would need to be restricted. As admitted by the authors, the large deficit in transformation observed would necessitate that almost every heteroduplex formed downstream of the replication fork and involved only the transiently me0 newly replicated strand, which seems highly unlikely.8 The second hypothesis is also unlikely, since nothing exists to suggest that restriction enzymes are present extracellularly. Recently, the situation has been further complicated by model figures in large audience reviews showing restriction enzymes attacking transforming DNA immediately after internalization, when it is in ss form and thus resistant to most restriction enzymes.33,34 The models elaborated in our study provide a simple explanation for the observed antagonization, suggesting that the restrictases do not act on transforming ssDNA per se, but rather on the post-replicative transformant chromosomes themselves, where fully me0 methylation sites will be present (in the absence of a DpnA analog, Fig. 2B). Our results are fully consistent with these models, and thus solve a long-standing conundrum in the field.
Materials and Methods
Bacterial strains, plasmids, growth and transformation conditions
S. pneumoniae strain growth and transformation were performed as described.35 Strain and plasmid information can be found in Table 1. Recipient strains were rendered hex− by insertion of the hexA::ermAM cassette as described,36 negating any effect of the mismatch repair system on transformation efficiencies.38 Antibiotics were used at the following concentrations; Spectinomycin (Spc) 200 µg ml−1, Streptomycin (Sm) 200 µg ml−1, Tetracycline (Tc) 1.5 µg ml−1.
Table 1. Strains and plasmids used in this study.
|
S. pneumoniae strain |
Genotype/description | Ref. |
|---|---|---|
| R246 |
R800 hexA::ermAM, dpnI; EryR |
36 |
| R1173 |
R800 ΔcomC, rpsL1, ciaR::spc119A, endA::kan6C, dpnI; SmR, SpcR, KanR |
11 |
| R2888 |
R800 dpnC::Janus (dpn0), rpsL1; KanR, SmS |
11 |
| R3087 |
R800 hexA::ermAM, dpnII; EryR |
11 |
| R3088 |
R800 hexA::ermAM, dpnII, dpnA−; EryR |
11 |
| R3089 |
R800 hexA::ermAM, ssbB::kan, dpnII; EryR, KanR |
This study |
| R3090 |
R800 hexA::ermAM, ssbB::kan, dpnII, dpnA−; EryR, KanR |
This study |
| R3091 |
R800 hexA::ermAM, ssbB∆7, dpnII; EryR, CmR |
This study |
| R3092 |
R800 hexA::ermAM, ssbB∆7, dpnII, dpnA−; EryR, CmR |
This study |
|
Plasmid |
Identity; isolation |
|
| pLS1 me0 |
Multicopy pneumococcal plasmid; unmethylated plasmid purified from R246 DpnI strain; TcR |
37 |
| pLS1 me+ | As above; methylated plasmid purified from R3087 DpnII strain; TcR | This study |
S/RSensitivity/Resistance; Cm, chloramphenicol; Ery, erythromycin; Kan, kanamycin
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/25582
References
- 1.Smith JM, Dowson CG, Spratt BG. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 2.Johnsborg O, Eldholm V, Håvarstein LS. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol. 2007;158:767–78. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Claverys JP, Prudhomme M, Mortier-Barrière I, Martin B. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol Microbiol. 2000;35:251–9. doi: 10.1046/j.1365-2958.2000.01718.x. [DOI] [PubMed] [Google Scholar]
- 4.Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007;3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickle TA. Restricting restriction. Mol Microbiol. 2004;51:3–5. doi: 10.1046/j.1365-2958.2003.03846.x. [DOI] [PubMed] [Google Scholar]
- 8.Berndt C, Meier P, Wackernagel W. DNA restriction is a barrier to natural transformation in Pseudomonas stutzeri JM300. Microbiology. 2003;149:895–901. doi: 10.1099/mic.0.26033-0. [DOI] [PubMed] [Google Scholar]
- 9.Humbert O, Dorer MS, Salama NR. Characterization of Helicobacter pylori factors that control transformation frequency and integration length during inter-strain DNA recombination. Mol Microbiol. 2011;79:387–401. doi: 10.1111/j.1365-2958.2010.07456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacks SA, Mannarelli BM, Springhorn SS, Greenberg B. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell. 1986;46:993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- 11.Johnston C, Martin B, Granadel C, Polard P, Claverys JP. Programmed protection of foreign DNA from restriction allows pathogenicity island exchange during pneumococcal transformation. PLoS Pathog. 2013;9:e1003178. doi: 10.1371/journal.ppat.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attaiech L, Olivier A, Mortier-Barrière I, Soulet AL, Granadel C, Martin B, et al. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity. PLoS Genet. 2011;7:e1002156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muckerman CC, Springhorn SS, Greenberg B, Lacks SA. Transformation of restriction endonuclease phenotype in Streptococcus pneumoniae. J Bacteriol. 1982;152:183–90. doi: 10.1128/jb.152.1.183-190.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacks SA, Ayalew S, de la Campa AG, Greenberg B. Regulation of competence for genetic transformation in Streptococcus pneumoniae: expression of dpnA, a late competence gene encoding a DNA methyltransferase of the DpnII restriction system. Mol Microbiol. 2000;35:1089–98. doi: 10.1046/j.1365-2958.2000.01777.x. [DOI] [PubMed] [Google Scholar]
- 15.Cerritelli S, Springhorn SS, Lacks SA. DpnA, a methylase for single-strand DNA in the Dpn II restriction system, and its biological function. Proc Natl Acad Sci U S A. 1989;86:9223–7. doi: 10.1073/pnas.86.23.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry AM, Glare EM, Hansman D, Paton JC. Presence of a small plasmid in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol Lett. 1989;53:275–8. doi: 10.1111/j.1574-6968.1989.tb03673.x. [DOI] [PubMed] [Google Scholar]
- 17.Sibold C, Markiewicz Z, Latorre C, Hakenbeck R. Novel plasmids in clinical strains of Streptococcus pneumoniae. FEMS Microbiol Lett. 1991;61:91–5. doi: 10.1111/j.1574-6968.1991.tb04327.x. [DOI] [PubMed] [Google Scholar]
- 18.Saunders CW, Guild WR. Pathway of plasmid transformation in Pneumococcus: open circular and linear molecules are active. J Bacteriol. 1981;146:517–26. doi: 10.1128/jb.146.2.517-526.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–75. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 20.Sekizaki T, Otani Y, Osaki M, Takamatsu D, Shimoji Y. Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J Bacteriol. 2001;183:500–11. doi: 10.1128/JB.183.2.500-511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilian M, Poulsen K, Blomqvist T, Håvarstein LS, Bek-Thomsen M, Tettelin H, et al. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One. 2008;3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefébure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornejo OE, Lefébure T, Bitar PD, Lang P, Richards VP, Eilertson K, et al. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol. 2013;30:881–93. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grove DE, Willcox S, Griffith JD, Bryant FR. Differential single-stranded DNA binding properties of the paralogous SsbA and SsbB proteins from Streptococcus pneumoniae. J Biol Chem. 2005;280:11067–73. doi: 10.1074/jbc.M414057200. [DOI] [PubMed] [Google Scholar]
- 25.Campbell EA, Choi SY, Masure HR. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–39. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 26.Morrison DA, Mannarelli B. Transformation in pneumococcus: nuclease resistance of deoxyribonucleic acid in the eclipse complex. J Bacteriol. 1979;140:655–65. doi: 10.1128/jb.140.2.655-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison DA, Mortier-Barrière I, Attaiech L, Claverys JP. Identification of the major protein component of the pneumococcal eclipse complex. J Bacteriol. 2007;189:6497–500. doi: 10.1128/JB.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claverys JP, Martin B, Polard P. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev. 2009;33:643–56. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 29.Lacks SA, Springhorn SS. Transfer of recombinant plasmids containing the gene for DpnII DNA methylase into strains of Streptococcus pneumoniae that produce DpnI or DpnII restriction endonucleases. J Bacteriol. 1984;158:905–9. doi: 10.1128/jb.158.3.905-909.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–44. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 31.Redfield RJ. Do bacteria have sex? Nat Rev Genet. 2001;2:634–9. doi: 10.1038/35084593. [DOI] [PubMed] [Google Scholar]
- 32.Charpentier X, Polard P, Claverys JP. Induction of competence for genetic transformation by antibiotics: convergent evolution of stress responses in distant bacterial species lacking SOS? Curr Opin Microbiol. 2012;15:570–6. doi: 10.1016/j.mib.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Dorer MS, Sessler TH, Salama NR. Recombination and DNA repair in Helicobacter pylori. Annu Rev Microbiol. 2011;65:329–48. doi: 10.1146/annurev-micro-090110-102931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev. 2013;37:336–63. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 35.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol. 2000;38:867–78. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 36.Mortier-Barrière I, de Saizieu A, Claverys JP, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol. 1998;27:159–70. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 37.Stassi DL, López P, Espinosa M, Lacks SA. Cloning of chromosomal genes in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1981;78:7028–32. doi: 10.1073/pnas.78.11.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claverys JP, Lacks SA. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986;50:133–65. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]