Abstract
Organelle genomes lose their genes by transfer to host nuclear genomes, but only occasionally are enriched by foreign genes from other sources. In contrast to mitochondria, plastid genomes are especially resistant to such horizontal gene transfer (HGT), and thus every gene acquired in this way is notable. An exceptional case of HGT was recently recognized in the peculiar peridinin plastid genome of dinoflagellates, which is organized in plasmid-like minicircles. Genomic and phylogenetic analyses of Ceratium horridum and Pyrocystis lunula minicircles revealed four genes and one unannotated open reading frame that probably were gained from bacteria belonging to the Bacteroidetes. Such bacteria seem to be a good source of genes because close endosymbiotic associations between them and dinoflagellates have been observed. The HGT-acquired genes are involved in plastid functions characteristic of other photosynthetic eukaryotes, and their arrangement resembles bacterial operons. These studies indicate that the peridinin plastid genome, usually regarded as having resulted from reduction and fragmentation of a typical plastid genome derived from red algae, may have a chimeric origin that includes bacterial contributions. Potential contamination of the Ceratium and Pyrocystis plastid genomes by bacterial sequences and the controversial localization of their minicircles in the nucleus are also discussed.
Keywords: Bacteroidetes, Ceratium, dinoflagellates, endosymbiotic gene transfer, genome, horizontal gene transfer, minicircle, peridinin plastid, Pyrocystis
Rarity of Horizontal Gene Transfers (HGTs) to Plastid Genomes
The process of massive endosymbiotic gene transfer (EGT) from mitochondrial and plastid genomes to host nuclear genomes is responsible for the significant reduction of these genomes in comparison to their α-proteobacterial and cyanobacterial ancestors, respectively (for reviews see refs. 1 and 2). Although EGT is dominant over the uptake of foreign genetic material via HGT by mitochondria and plastids, many examples of HGT to mitochondria have been reported (e.g., refs. 3–5). In contrast, plastid genomes have been transformed extremely rarely by HGT over the course of evolution. The scarceness of such events in plastids may result from: (1) lack of an efficient exogenous DNA-uptake system in plastids, (2) the compact structure of their genomes, which makes them susceptible to disruption upon integration of foreign genetic material, (3) greater restrictions in the process of non-homologous recombination in these organelles, and (4) inability to fuse one plastid with another and thus insufficient propagation of potentially transferred genes.2,6
Extensive inspection of more than 200 plastid-encoded genes resulted in the addition of only one well-documented case of a gene, rpl36, transferred to plastids via HGT;6 the set of plastid-encoded genes derived from HGT includes also genes encoding the large (rbcL) and small subunits (rbcS) of RuBisCO form I that were acquired from a proteobacterium by the primary plastid genome of the common ancestor of red algae.7 The rpl36 gene encodes a ribosomal protein, is derived from a proteobacterium or a planctomycete bacterium and exists in the genomes of four membrane-bound plastids from cryptophytes and haptophytes.6 The other example of HGT to plastids is dnaX, which encodes the τ/γ subunit of DNA polymerase III.8 This gene was transferred from a firmicute bacterium to the complex plastid of the cryptophyte Rhodomonas salina. Two independent HGT transfers also occurred in the genome of a non-photosynthetic plastid, the apicoplast, that is characteristic of apicomplexan parasites. The first of these transfers involved clpC, which encodes a class III ATPase, and occurred from spirochetes.9 In the second case, three ribosomal protein genes (rpl2, rpl14 and rps12) were gained from a mitochondrion.10 In a single HGT event from the mitochondria of an unknown donor, two genes were acquired that cluster in the same inverted repeat in the plastid genome of the green alga Oedogonium cardiacum. One of these genes, int, encodes a site-specific tyrosine recombinase and the second, dpoB, encodes a DNA-directed DNA polymerase of the B family.11
Bacterial Genes Transferred to Dinoflagellate Plastid Minicircles
An interesting new case of HGT was recently identified by us in the genome of the peridinin plastid.12 This plastid, which is surrounded by three membranes, contains chlorophyll c and the carotenoid peridinin.13,14 Its nuclear-encoded proteins carry complex signal peptide-containing presequences, which results in targeting to the plastid via the endoplasmic reticulum and/or Golgi apparatus.15 The peridinin plastid is characteristic of the dinoflagellates, a group of protists that have been classified to Alveolata together with apicomplexans and ciliates.13,14 Dinoflagellates have several unique cellular features including the dinokaryon, a nucleus consisting of permanently condensed chromosomes that lack nucleosomes.16 These protists play important ecological roles as primary producers, parasites and symbionts, e.g., with reef-building corals.13,14 They are also responsible for harmful algal blooms, which exert serious medical and economic consequences.
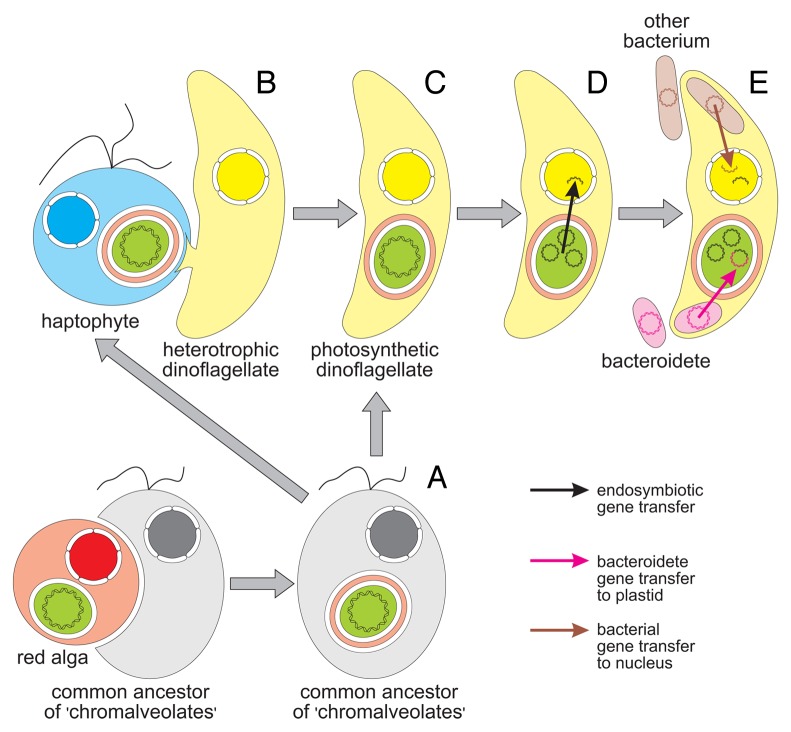
The evolutionary origin of the peridinin plastid is still controversial. According to the traditional view, known as the chromalveolate hypothesis, the plastid was acquired from a red alga via secondary endosymbiosis by a common ancestor of all alveolates and other protists classified formerly in the kingdom Chromista, such as cryptophytes, haptophytes and stramenopiles (Fig. 1).17 However, several lines of evidence indicate that the peridinin plastid evolved from a haptophyte via tertiary endosymbiosis (e.g., refs. 18 and 19).
Figure 1. Evolution of the peridinin plastid in dinoflagellates and bacterial gene transfers to the dinoflagellate plastid and nuclear genomes. (A) The origin of the peridinin plastid is usually explained in the framework of the chromalveolate hypothesis, which assumes that all chromalveolates (including haptophytes and dinoflagellates) inherited plastids from their common ancestor, which contained a red alga-derived secondary plastid. (B) However, it seems more probable that this plastid was acquired by a heterotrophic dinoflagellate from a haptophyte via tertiary endosymbiosis. (C) During the initial stages of tertiary endosymbiosis, the plastid lost redundant membranes and was finally transformed into the three-membrane state; the ancestral genome of the peridinin plastid initially consisted of typical plastid chromosomes. (D) These chromosomes disintegrated and were reduced to small plasmid-like minicircles. The plastid genome was also subjected to massive escape of its genes to the nuclear genome via endosymbiotic gene transfer. (E) Recent analysis of minicircles in Ceratium and Pyrocystis uncovered a process opposite to EGT in which the plastid genome gained the ycf16, ycf24, rpl28 and rpl33 genes and an open reading frame homologous to ftsY from bacteria related to the Bacteroidetes. The nuclear genome was also enriched with foreign genes from bacteria belonging to distinct groups; these bacteria may have been retained as endosymbionts, acquired as prey, or invaded the host cell as parasites.
In contrast to typical plastids with 100–200-kb circular genomes, the dinoflagellate plastid genome is extremely reduced and is organized into numerous 0.4- to 10-kb (usually 2- to 3-kb) plasmid-like chromosomes called minicircles (Fig. 1) (for a review see ref. 20). Every minicircle can encode 0–5 (usually 1 or 2) genes. This peculiar genome has been subjected to recombination and fragmentation of a typical plastid chromosome with multiple transposed replication initiation sites, or to independent deletions in many typical plastid chromosomes driven by tandem repeats.21 It seems that dinoflagellate hosts are able to evolve unusual fragmented organelle genomes because their mitochondrial genome is also organized into many small chromosomes encoding only three proteins (Cob, Cox1 and Cox3) and two fragmented rRNAs.22 Additionally, an independent fragmentation process has occurred in the plastid genome of Karlodinium veneficum, which harbors a fucoxanthin plastid derived from a haptophyte alga.23
Our previous study of C. horridum AF490364 and P. lunula AF490367 minicircles12 indicated that their genes (or even whole minichromosomes) may be of bacterial origin, thus adding an extra level of complexity to our view of the evolution of the peridinin plastid genome. Of 103 studied sequences, these minicircles were the only ones that lacked similarity to regions of other dinoflagellate minicircles. The Ceratium minicircle contains ycf16 and ycf24, which correspond to bacterial sufC and sufB, respectively.12 These bacterial genes belong to the sufABCDSE operon, which encodes proteins that contribute to assembly and repair of iron-sulfur clusters under oxidative stress. The Pyrocystis minicircle includes rpl28 and rpl33, whose bacterial counterparts, rpmB and rpmG, respectively, encode proteins related to the ribosomal large subunit.12 In contrast to the genes of red algae and their secondary descendants, these genes are adjacent on the Pyrocystis minicircle and resemble the bacterial rpmBG operon. Additionally, the Pyrocystis minicircle includes an unannotated open reading frame (ORF) that encodes a SRP54 N domain with α-helices typical of FtsY proteins; the ORF is located downstream of and close to rpl33, as in many bacterial genomes.12 FtsY plays a role in co-translational transport of proteins as a receptor for the signal recognition particle associated with synthesized polypeptides and ribosomes.
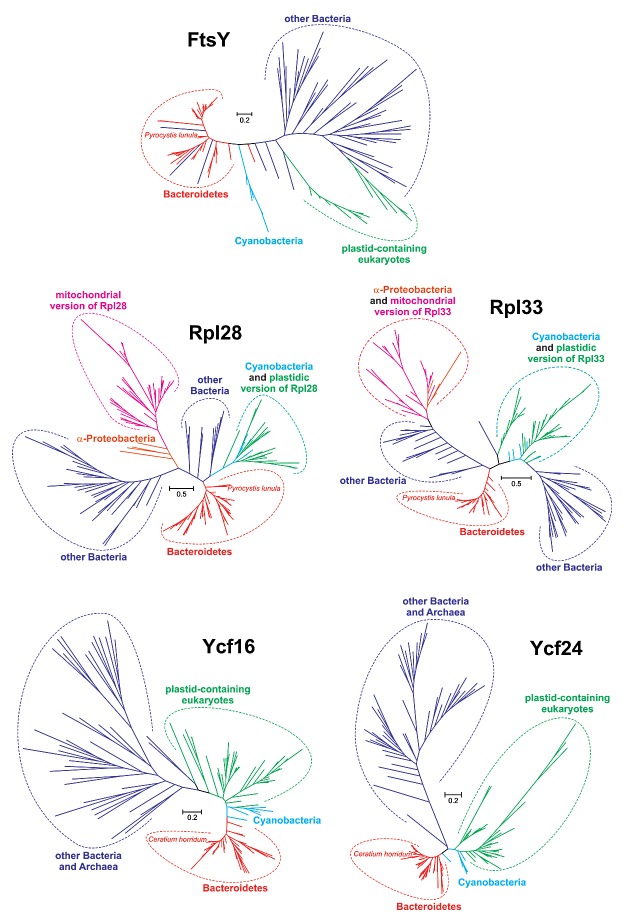
Although eukaryotic homologs to these five bacterial genes are dispersed among the plastid, mitochondrial and/or nuclear genomes of various algae and plants, our previous phylogenetic analyses showed that the sequences of genes on the Ceratium and Pyrocystis minicircles are most closely related to genes from the Bacteroidete genera Algoriphagus and/or Cytophaga than to eukaryotic homologs.12 Since these studies were based on quite small sequence samples and did not include FtsY phylogeny, we performed more extensive and varied analyses involving, in each phylogeny, more than 100 sequences selected from several thousand homologs to represent all prokaryotic and eukaryotic groups, including dinoflagellate and alveolate sequences not analyzed previously (see Supplementary File for details). The new data sets were also significantly enriched with novel Bacteroidete sequences. All applied methods undoubtedly placed the minicircle gene sequences within the Bacteroidete clade (Fig. 2). Pyrocystis Rpl28 and Rpl33 grouped with bacteria from class Sphingobacteria, whereas Ceratium Ycf16 and Ycf24 grouped with the Cytophagia (Fig. S1). Even the very short sequence of the Pyrocystis ORF was located with members from one of these two Bacteroidete groups (depending on the method used). In addition, tree topology tests significantly rejected alternative hypotheses in which the four minicircle gene sequences clustered with other sequences from dinoflagellates or their relatives, such as the apicomplexans, chromerids, or perkinsids (see Supplementary File). The placement of the ORF with its homologs from plastid-containing eukaryotes (green plants, cryptophytes, red algae and stramenopiles) was also significantly worse than its location with Bacteroidetes. All of these findings clearly indicate that the genes and the ORF located on the Ceratium and Pyrocystis minicircles resulted from HGT from a Bacteroidete donor (Fig. 1).
Figure 2. Phylogenetic trees for FtsY, Rpl28, Rpl33, Ycf16 and Ycf24 sequences inferred in PhyloBayes under the LG+Γ(5) model. The sequences localized to Pyrocystis lunula AF490367 and Ceratium horridum AF490364 minicircles branch clearly with sequences from Bacteroidetes rather than eukaryotes, indicating horizontal transfer of these genes from the bacteria to the peridinin plastid of dinoflagellates.
Bacteroidetes as Good Donors of Genetic Material to Dinoflagellates
Bacteroidetes seem to be an especially good source of genes due to their close ecological relationships with toxic and non-toxic dinoflagellates. These bacteria, which are associated with the surface of algal cells, play an important role in: (1) supplying iron and vitamins, (2) cycling excreted organic matter, (3) inducing sexual stages, (4) exhibiting algicidal activity or protection from other algicidal bacteria and (5) metabolizing algal toxins.24 Associations between Bacteroidetes and at least eight dinoflagellate genera have been reported (for a review see ref. 25). Interestingly, three of these dinoflagellates belong to the same order (Gonyaulacales) as Ceratium and Pyrocystis. Some of the observed associations may consist of long-term ecological relationships, since Bacteroidetes were identified as endosymbionts in Heterocapsa circularisquama26 and in Alexandrium minutum.27 Although Gyrodinium instriatum probably preys on cytoplasmic Bacteroidetes, the possibility of an intimate association, such as endosymbiosis or parasitism, cannot be excluded.28 Such intracellular bacteria are excellent candidates for donors of genetic material into plastids, and may have donated their genes to C. horridum and P. lunula. Since the minicircle genes from Ceratium and Pyrocystis clustered with different Bacteroidete classes (Cytophagia and Sphingobacteria respectively), it seems probable that the postulated HGT occurred independently in these dinoflagellates, which belong to different families.29
Evolution of the Investigated Minicircle Genes in Dinoflagellates: A Case of Endosymbiotic Gene Replacement?
Homologs of Ycf16, Ycf24, Rpl28 and Rpl33 are present in expressed sequence tag databases of other peridinin dinoflagellates (e.g., Alexandrium and Amphidinium), but were found in none of 101 dinoflagellate minicircles, indicating that these genes are located on dinoflagellate nuclear genomes; these genes group also with other nuclear-encoded sequences from various eukaryotic lineages (ref. 12 and Fig. S1). In eukaryotes these genes were originally located in the plastid genome, as they persist in red algae, which have served as direct or indirect donors of plastids to numerous protist lineages. This observation indicates that these genes must have been transferred via EGT to the nuclear genome in many eukaryotic groups during plastid endosymbioses (e.g., refs. 30 and 31). Unfortunately, Ceratium and Pyrocystis expressed sequence tag databases are sparse, and so it is not yet possible to distinguish between two alternatives: are the additional copies of these genes still present in the nuclear genomes, or were they lost from these genomes?
Both scenarios have interesting implications. In the first case, the Bacteroidete-derived and plastid-located genes should have adapted to cooperate with the nuclear copies already present, or were subjected to functional differentiation (neo- or subfunctionalization). However, our analysis (using all available methods in MEGA32) of the four genes and the ORF minicircle sequences with their closest bacterial homologs did not reveal any positive selection. If the nuclear copies were lost, HGT from bacteria to the peridinin plastid would be associated with a peculiar example of endosymbiotic gene replacement of nuclear copies by organelle genes. Full expressed sequence tag libraries for these dinoflagellates are necessary to answer these questions.
Potential Contamination of Ceratium and Pyrocystis Plastid Genomes by Bacterial Sequences
Although the described data are most consistent with gene transfer from Bacteroidetes to the two dinoflagellate minicircles, potential contamination of dinoflagellate DNA by bacterial sequences of external origin should be taken into account. In the case of contamination we would expect different types of contaminating sequences, but all analyzed dinoflagellate minicircle genes have clear relationships only to plastid functions and possess homologs in the plastids of other eukaryotes.12 If bacterial contamination were a major issue, more genes unconnected with plastid function should be reported because Bacteroidete sequences related to those found in dinoflagellates such as Flexibacter and Flavobacterium26 are gene-rich and encode as many as 2,500–5,000 proteins. Moreover, sequences from Ceratium AF490364 and Pyrocystis AF490367 minicircles were isolated with other molecules encoding typical minicircle- or plastid-specific genes.33 These minicircles are also similar in length (1,954 bp and 1,149 bp, respectively) to typical dinoflagellate minicircles, and, in the case of AF490364, we identified palindromes and inverted repeats characteristic of dinoflagellate minicircles.20,21
Public sequence databases such as NCBI contain 2,504,077 protein sequences and 392 genome sequences (including 94 completely sequenced) from the Bacteroidetes. These data sets also include Bacteroidete species or close relatives that have been reported in dinoflagellate cultures. Therefore, if contamination had occurred, we expect that the minicircle genes analyzed by us would be more similar to their homologs in these databases. The maximal identity that we observed between the studied gene and ORF sequences and their homologs was only 72.5–78% in local alignments. There were 0.17–0.27 amino acid substitutions per site in the analyzed protein sequences between their minicircle versions and their closest homologs from Bacteroidetes. This number of substitutions corresponds to the value between their homologs in the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana (0.14–0.42; 0.14–0.22 when excluding the very short FtsY and mitochondrial Rpl proteins) or between the dicots Arabidopsis thaliana and Vitis vinifera (0.05–0.26; 0.11–0.23 when excluding FtsY and the mitochondrial Rpl proteins). The ratio of the number of substitutions between minicircle proteins and their closest Bacteroidete homologs to the number of substitutions between P. tricornutum and T. pseudonana for corresponding proteins is on average 0.8 (1.0 when excluding FtsY and the mitochondrial Rpl proteins). The ratio involving A. thaliana and V. vinifera is even higher at 1.7 (1.4 when excluding FtsY and the mitochondrial Rpl proteins). The two diatoms diverged approximately 90 million years ago,30 and the dicots diverged 115 million years ago.34 Even assuming a higher substitution rate in bacteria and dinoflagellate sequences, a long evolutionary history would be necessary since their split, while we would expect much lower genetic distances if contamination were present.
Controversy About the Nuclear Localization of Ceratium and Pyrocystis Minicircles
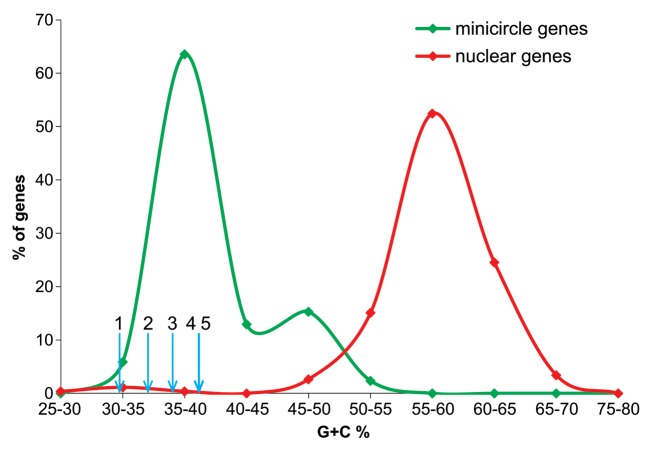
The sequence of the C. horridum AF490364 minicircle was published by Laatsch et al.,33 who assumed the presence of the minicircle and possibly other minicircles only in the nuclear compartment. Annotation of the Pyrocystis AF490367 minicircle suggests such localization as well, although this hypothesis has not been confirmed by any published investigation. However, several lines of evidence, as discussed by us12 put in doubt the nuclear location of minicircles. For example, the nuclear location of the minicircles was deduced by Laatsch et al.33 based on one gene (psbB) from the Ceratium minicircle other than two minicircles considered here, whereas their Southern blot data and in situ hybridization experiments revealed a weak psbB signal inside the plastid. Moreover, none of the genes located on the Ceratium and Pyrocystis minicircles encodes the complex presequence necessary for targeting of their products to the peridinin plastid. Therefore, the genes likely are carried by the plastid to ensure activity of their protein products in the proper compartment. In further support of this conclusion, the G+C% of the four Ceratium and Pyrocystis minicircle genes and the new ORF are very close to those of other minicircle genes and significantly lower than genes from dinoflagellate nuclear genomes.12 The nuclear genes are much richer in G+C% than known minicircle genes, to which the four minicircle genes and the ORF undoubtedly belong (Fig. 3).
Figure 3. Comparison of G+C% calculated for ycf16, ycf24, rpl28, rpl33 and the ORF homologous to ftsY from the dinoflagellate minicircles with the G+C% of other minicircle genes and genes located in nuclear dinoflagellate genomes. Blue short arrows indicate G+C% content of (1) the unannotated ORF homologous to ftsY, (2) rpl33, (3) rpl28, (4) ycf24 and (5) ycf16. Genes 1–3 are located on the P. lunula AF490367 minicircle, whereas genes 4 and 5 are on the C. horridum AF490364 minicircle. The distribution for minicircle genes was calculated based on 85 sequences downloaded from GenBank, and the nuclear gene distribution was obtained from 265 sequences published by Kii et al.35 and McEwan et al.36
Nevertheless, the possibility of dual plastid-nuclear localization of the minicircle-specific psbB probe, as deduced from the studies of Laatsch et al.,33 remains intriguing. It seems probable that the minicircle DNA in the nuclear fraction represents an ongoing process of EGT because the psbB probe occurs between chromosomes in the nuclear matrix, as discussed by us.12 It is well documented that EGT from the peridinin plastid genome to the nucleus in dinoflagellates has proceeded on a massive scale (Fig. 1), and has been more extensive than in any other group of organisms.37,38 As a result of this process, the total coding capacity of all minicircles was reduced to only 16 protein- and 5 RNA-coding genes.20 However, it is still unknown whether this gene escape resulted from the selective pressure to avoid the mutational effects of free radicals of oxygen or to avoid Muller’s ratchet of nonrecombining genomes.38,39 We cannot exclude the possibility that selective pressure for minimization of replication time and metabolic costs led to extreme reduction of the peridinin plastid genome.38,39
Other Bacterial Gene Transfers to Dinoflagellates
The HGT from bacteria to the dinoflagellate plastid genome observed by us is probably only a byproduct of much larger gene transfers directed mainly to the dinoflagellate nuclear genome (Fig. 1). An interesting example is the histone-like proteins, which play a role in the organization of the unique chromosome structure in dinoflagellates.40 After their acquisition from proteobacteria, these genes were subjected to several rounds of duplication. In one HGT event, two shikimate biosynthetic genes encoding the enzymes 3-dehydroquinate synthase and O-methyltransferase were transferred from a cyanobacterial source into the dinoflagellate lineage.41 These two genes fused and now encode a novel plastid-targeted protein, probably introducing new biochemical functionalities to the plastid. Interestingly, the fused gene reverted to two separate genes in the dinoflagellate Karlodinium, which contains the fucoxanthin plastid. Both of these genes maintained their plastid localization, but the gene encoding O-methyltransferase must have evolved its own plastid-targeting presequence. In another case, the gene encoding acetolactate synthase was transferred relatively recently from an α-proteobacterium to Heterocapsa triquetra.42
Many examples of HGT from bacteria to dinoflagellates were identified by Nosenko and Bhattacharya.43 Genes encoding an α-tubulin suppressor, a metal-dependent hydrolase of the TIM-barrel fold (COG3618), and a hypothetical protein (COG3022) were acquired from proteobacteria. NAD-dependent sugar nucleotide epimerase/dehydratase, iron-containing alcohol dehydrogenase, NAD-dependent aldehyde dehydrogenase, and pyridoxal phosphate biosynthetic protein are also of bacterial origin, but the precise donor is not known. Interestingly, members of the Bacteroidetes may have donated arylsulfatase A and SAM-dependent methyltransferase (COG3129), which supports the hypothesis of HGT from these bacteria to dinoflagellate minicircles.
Recent wide phylogenomic studies of expressed sequence tags from the dinoflagellate Alexandrium tamarense suggest the scale of HGT from bacterial donors.44 Of 9,765 predicted proteins, 111 shared exclusive similarity and phylogenetic relationships with sequences from distinct lineages of bacteria, including Bacteroidetes, indicating HGT to dinoflagellates from many bacterial sources. The actual scale of transfer is most likely higher than that indicated by the analyses, which assumed very conservative thresholds for sequence similarity and phylogenetic relationships.
It should be noted that HGT of the gene encoding form II RuBisCO from proteobacteria to dinoflagellates was long regarded to be a unique feature of dinoflagellates.45 However, the presence of this gene in Chromera velia and strain CCMP3155, photosynthetic relatives of parasitic apicomplexans, indicates that this transfer took place in the common ancestor of dinoflagellates and apicomplexans.46
The described cases of HGT are in agreement with the previously discussed associations between various dinoflagellates and bacteria (e.g., refs. 27, and 47–49). In addition to the Bacteroidetes, cyanobacteria, α-, β- and γ-proteobacteria have also been reported to form these relationships. Interestingly, some γ-proteobacteria localize to the nuclear surface,26 and some β-proteobacteria have been observed within the nucleus.28 Such localization undoubtedly increases the probability of bacterial DNA uptake and integration into the host nuclear genome (Fig. 1).
Finally, it is worth mentioning the case of the bacteria-like endosymbionts found in the plastid stroma of the dinoflagellate Woloszynskia pascheri.50 These structures are surrounded by a two-membrane envelope and contain a chromosome as well as prokaryotic-sized ribosomes. More than 100 such endosymbionts were observed in some of these plastids. They are most probably transferred to the dinoflagellate’s offspring, as they were also identified in planozygotes. Although the advantage provided to the host by these endosymbionts is unknown, they are probably involved in a stable relationship with this dinoflagellate. This example nicely supports the results presented here about the transfer of genetic material to the dinoflagellate plastid. It is possible that some Bacteroidetes also resided in this organelle in Ceratium and Pyrocystis ancestors as endosymbionts and became donors of the minicircle genes considered here.
Conclusion
Our previous analysis12 and the analyses presented here of the Ceratium and Pyrocystis minicircles have revealed for the first time: (1) HGT to the dinoflagellate plastid genome, (2) Bacteroidetes as donors of genetic material to eukaryotic plastids, and (3) HGT of genes associated with non-photosynthetic metabolic functions of plastids (such as sulfur metabolism) to eukaryotic alga-derived plastids. These data also imply that the peridinin plastid genome may have derived from distinct sources, and that the influence of foreign DNA molecules on the evolution of the plastid genome may be greater than commonly assumed. Given that dinoflagellates have developed close associations with many bacteria, prokaryote-to-eukaryote gene transfers may occur on a much larger scale. Additional genomic and experimental studies are necessary to fully assess the influence of foreign genes on the evolution and function of dinoflagellates.
Supplementary Material
Glossary
Abbreviations:
- EGT
endosymbiotic gene transfer
- HGT
horizontal gene transfer
- ORF
open reading frame
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/nucleus/article/25845.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/25845
References
- 1.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–35. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 2.Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–38. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- 3.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 4.Wołoszyńska M, Bocer T, Mackiewicz P, Jańska H. A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots, to mitochondrial genome of Phaseolus. Plant Mol Biol. 2004;56:811–20. doi: 10.1007/s11103-004-5183-y. [DOI] [PubMed] [Google Scholar]
- 5.Davis CC, Anderson WR, Wurdack KJ. Gene transfer from a parasitic flowering plant to a fern. Proc Biol Sci. 2005;272:2237–42. doi: 10.1098/rspb.2005.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delwiche CF, Palmer JD. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol Evol. 1996;13:873–82. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- 8.Khan H, Parks N, Kozera C, Curtis BA, Parsons BJ, Bowman S, et al. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 2007;24:1832–42. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- 9.Zhu XY. Phylogenetic analysis indicates bacteria-to-apicoplast lateral gene transfer. Yi Chuan Xue Bao. 2004;31:1316–20. [PubMed] [Google Scholar]
- 10.Oborník M, Van de Peer Y, Hypsa V, Frickey T, Slapeta JR, Meyer A, et al. Phylogenetic analyses suggest lateral gene transfer from the mitochondrion to the apicoplast. Gene. 2002;285:109–18. doi: 10.1016/S0378-1119(02)00427-4. [DOI] [PubMed] [Google Scholar]
- 11.Brouard JS, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9:290. doi: 10.1186/1471-2164-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moszczyński K, Mackiewicz P, Bodył A. Evidence for horizontal gene transfer from bacteroidetes bacteria to dinoflagellate minicircles. Mol Biol Evol. 2012;29:887–92. doi: 10.1093/molbev/msr276. [DOI] [PubMed] [Google Scholar]
- 13.Hackett JD, Anderson DM, Erdner DL, Bhattacharya D. Dinoflagellates: a remarkable evolutionary experiment. Am J Bot. 2004;91:1523–34. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- 14.Delwiche CF. The origin and evolution of dinoflagellates. In: Falkowski PG, Knoll AH, eds. Evolution of primary producers in the sea. Elsevier Academic Press, 2007:191-205 [Google Scholar]
- 15.Patron NJ, Waller RF, Archibald JM, Keeling PJ. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 2005;348:1015–24. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Moreno Díaz de la Espina S, Alverca E, Cuadrado A, Franca S. Organization of the genome and gene expression in a nuclear environment lacking histones and nucleosomes: the amazing dinoflagellates. Eur J Cell Biol. 2005;84:137–49. doi: 10.1016/j.ejcb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Keeling PJ. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 18.Shalchian-Tabrizi K, Skånseng M, Ronquist F, Klaveness D, Bachvaroff TR, Delwiche CF, et al. Heterotachy processes in rhodophyte-derived secondhand plastid genes: Implications for addressing the origin and evolution of dinoflagellate plastids. Mol Biol Evol. 2006;23:1504–15. doi: 10.1093/molbev/msl011. [DOI] [PubMed] [Google Scholar]
- 19.Bodył A, Moszczyński K. Did the peridinin plastid evolve through tertiary endosymbiosis? A hypothesis. Eur J Phycol. 2006;41:435–48. doi: 10.1080/09670260600961080. [DOI] [Google Scholar]
- 20.Howe CJ, Nisbet RE, Barbrook AC. The remarkable chloroplast genome of dinoflagellates. J Exp Bot. 2008;59:1035–45. doi: 10.1093/jxb/erm292. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Cavalier-Smith T, Green BR. Evolution of dinoflagellate unigenic minicircles and the partially concerted divergence of their putative replicon origins. Mol Biol Evol. 2002;19:489–500. doi: 10.1093/oxfordjournals.molbev.a004104. [DOI] [PubMed] [Google Scholar]
- 22.Waller RF, Jackson CJ. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. Bioessays. 2009;31:237–45. doi: 10.1002/bies.200800164. [DOI] [PubMed] [Google Scholar]
- 23.Espelund M, Minge MA, Gabrielsen TM, Nederbragt AJ, Shalchian-Tabrizi K, Otis C, et al. Genome fragmentation is not confined to the peridinin plastid in dinoflagellates. PLoS One. 2012;7:e38809. doi: 10.1371/journal.pone.0038809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodama M, Doucette GK, Green DH. Relationships between bacteria and harmful algae. Springer-Verlag Berlin Heidelberg, 2006:243-55 [Google Scholar]
- 25.Garcés E, Vila M, Reñé A, Alonso-Sáez L, Anglès S, Lugliè A, et al. Natural bacterioplankton assemblage composition during blooms of Alexandrium spp. (Dinophyceae) in NW Mediterranean coastal waters. Aquat Microb Ecol. 2007;46:55–70. doi: 10.3354/ame046055. [DOI] [Google Scholar]
- 26.Maki T, Yoshinaga I, Katanozaka N, Imai I. Phylogenetic analysis of intracellular bacteria of a harmful marine microalga, Heterocapsa circularisquama (Dinophyceae) Aquat Microb Ecol. 2004;36:123–35. doi: 10.3354/ame036123. [DOI] [Google Scholar]
- 27.Palacios L, Marín I. Enzymatic permeabilization of the thecate dinoflagellate Alexandrium minutum (Dinophyceae) yields detection of intracellularly associated bacteria via catalyzed reporter deposition-fluorescence in situ hybridization. Appl Environ Microbiol. 2008;74:2244–7. doi: 10.1128/AEM.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alverca E, Biegala IC, Kennaway GM, Lewis J, Franca S. In situ identification and localization of bacteria associated with Gyrodinium instriatum (Gymnodiniales, Dinophyceae) by electron and confocal microscopy. Eur J Phycol. 2002;37:523–30. doi: 10.1017/S0967026202003955. [DOI] [Google Scholar]
- 29.Gómez F. A checklist and classification of living Dinoflagellates (Dinoflagellata, Alveolata) CICIMAR Oceánides. 2012;27:65–140. [Google Scholar]
- 30.Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–44. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 31.Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laatsch T, Zauner S, Stoebe-Maier B, Kowallik KV, Maier UG. Plastid-derived single gene minicircles of the dinoflagellate Ceratium horridum are localized in the nucleus. Mol Biol Evol. 2004;21:1318–22. doi: 10.1093/molbev/msh127. [DOI] [PubMed] [Google Scholar]
- 34.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci U S A. 2009;106:5737–42. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kii S-I, Tanaka J, Watanabe T. Guanine-cytosine contents of the host and symbiont cDNA in a symbiotic coral. Fish Sci. 2007;73:1362–72. [Google Scholar]
- 36.McEwan M, Humayun R, Slamovits CH, Keeling PJ. Nuclear genome sequence survey of the dinoflagellate Heterocapsa triquetra. J Eukaryot Microbiol. 2008;55:530–5. doi: 10.1111/j.1550-7408.2008.00357.x. [DOI] [PubMed] [Google Scholar]
- 37.Bachvaroff TR, Concepcion GT, Rogers CR, Herman EM, Delwiche CF. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist. 2004;155:65–78. doi: 10.1078/1434461000165. [DOI] [PubMed] [Google Scholar]
- 38.Hackett JD, Yoon HS, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, et al. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr Biol. 2004;14:213–8. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 39.Selosse M, Albert B, Godelle B. Reducing the genome size of organelles favours gene transfer to the nucleus. Trends Ecol Evol. 2001;16:135–41. doi: 10.1016/S0169-5347(00)02084-X. [DOI] [PubMed] [Google Scholar]
- 40.Hackett JD, Scheetz TE, Yoon HS, Soares MB, Bonaldo MF, Casavant TL, et al. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genomics. 2005;6:80. doi: 10.1186/1471-2164-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waller RF, Slamovits CH, Keeling PJ. Lateral gene transfer of a multigene region from cyanobacteria to dinoflagellates resulting in a novel plastid-targeted fusion protein. Mol Biol Evol. 2006;23:1437–43. doi: 10.1093/molbev/msl008. [DOI] [PubMed] [Google Scholar]
- 42.Waller RF, Patron NJ, Keeling PJ. Phylogenetic history of plastid-targeted proteins in the peridinin-containing dinoflagellate Heterocapsa triquetra. Int J Syst Evol Microbiol. 2006;56:1439–47. doi: 10.1099/ijs.0.64061-0. [DOI] [PubMed] [Google Scholar]
- 43.Nosenko T, Bhattacharya D. Horizontal gene transfer in chromalveolates. BMC Evol Biol. 2007;7:173. doi: 10.1186/1471-2148-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan CX, Soares MB, Bonaldo MF, Wisecaver JH, Hackett JD, Anderson DM, et al. Analysis of Alexandrium tamarense (Dinophyceae) genes reveals the complex evolutionary history of a microbial eukaryote. J Phycol. 2012;48:1130–42. doi: 10.1111/j.1529-8817.2012.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morse D, Salois P, Markovic P, Hastings JW. A nuclear-encoded form II RuBisCO in dinoflagellates. Science. 1995;268:1622–4. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 46.Janouskovec J, Horák A, Oborník M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010;107:10949–54. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seibold A, Wichels A, Schutt C. Diversity of endocytic bacteria in the dinoflagellate Noctiluca scintillans. Aquat Microb Ecol. 2001;25:229–35. doi: 10.3354/ame025229. [DOI] [Google Scholar]
- 48.Schweikert M, Meyer B. Characterization of intracellular bacteria in the freshwater dinoflagellate Peridinium cinctum. Protoplasma. 2001;217:177–84. doi: 10.1007/BF01283398. [DOI] [PubMed] [Google Scholar]
- 49.Escalera L, Reguera B, Takishita K, Yoshimatsu S, Koike K, Koike K. Cyanobacterial endosymbionts in the benthic dinoflagellate Sinophysis canaliculata (Dinophysiales, Dinophyceae) Protist. 2011;162:304–14. doi: 10.1016/j.protis.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Wilcox LW. Prokaryotic endosymbionts in the chloroplast stroma of the dinoflagellate Woloszynskia pascheiri. Protoplasma. 1986;135:71–9. doi: 10.1007/BF01277000. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.