Abstract
The anaphase promoting complex (APC) is the crucial ubiquitin ligase targeting the regulatory machinery of the cell cycle. Emi1, a major modulator of APC activity, is thought to act competitively as a pseudosubstrate. We show that the modulation of APC activity is more subtle: Emi1 inhibits ubiquitylation at both substrate binding and separately at the step of ubiquitin transfer to APC-bound substrates. The zinc-binding region of Emi1 allows multiple monoubiquitylation of substrates, but preferentially suppresses the ubiquitin chain elongation by UBCH10. Furthermore, the C-terminal tail of Emi1 antagonizes chain elongation by Ube2S, via competitively preventing its binding to APC cullin subunit through electrostatic interaction. Combinatorially, Emi1 effectively stabilizes APC substrates by suppressing ubiquitin chain extension. Deubiquitylating enzymes can then convert inhibited substrates to their basal state. Chain elongation may be a particularly sensitive step for controlling degradation and this study provides the first kinetic evidence for how it is inhibited.
Introduction
The anaphase promoting complex (APC) is a multisubunits ubiquitin ligase that controls cell cycle progression through mitosis to G1 phase1. Various key cell cycle regulators are ubiquitylated by APC, and targeted for destruction by 26S proteasome. The ubiquitylation reaction consists of two steps: multiple monoubiquitylation on lysine residues of substrate, and ubiquitin chain assembly via ubiquitin conjugated on the substrate. Human APC cooperates with two E2 enzymes, UBCH10, which primarily carries out the first step and Ube2S, which exclusively carried out the second step, to catalyze efficient polyubiquitylation preferentially via K11 linkages2, 3.
Various studies have shown that APC activity is tightly regulated at several levels4-7. Given the dynamic features of E2 activity, direct interference with the E2-E3 association, or inhibition of the ubiquitin transfer to substrates should be an effective way to inhibit the ongoing ubiquitylation. Additionally, because degradation of most APC substrates by 26S proteasome requires formation of ubiquitin chains, prevention of the ubiquitin chain elongation alone may suffice to stabilize APC substrates. However, little is known about how natural inhibitors of E3 ligases function by direct modulation of the ubiquitin transfer process.
The primary inhibitor of APC through interphase is early mitotic inhibitor 1 (Emi1), which is required for accumulation of mitotic cyclins during S and G2 phase8. The C-terminal 150 amino acid fragment of Emi1 confers its inhibitory activity toward APC9, 10. This C-terminal fragment of Emi1 contains destruction box (D-box), zinc-binding region (ZBR) 10and its extreme C-terminal tail11, which interact with APC separately. Based on competitive binding assays, it was proposed that Emi1 acts as a pseudosubstrate inhibiting substrates binding to the D-box receptor site on APC10. Considering the total concentration of the more than 100 APC substrates12, it is hard to explain how the Emi1 could possess high enough affinity to occlude the E3-substrate interaction. Moreover, in this model, once a substrate is already bound to APC, Emi1 could do nothing to inhibit its ubiquitylation and degradation. Other mechanisms must explain the efficient inhibition of APC by Emi1. A recent study suggested that Emi2, a meiotic analog of Emi1, may affect the ubiquitin transfer from E2 to substrates13. However, solid kinetic evidence is still needed to further support that argument for either protein.
Using a set of quantitative assays we dissected the inhibitory mechanism of Emi1. Emi1 acts principally to block ubiquitin chain elongation by inhibiting ubiquitin transfer and interrupting E2-E3 association.
Results
Emi1 competes for substrate binding and blocks ubiquitin chain extension
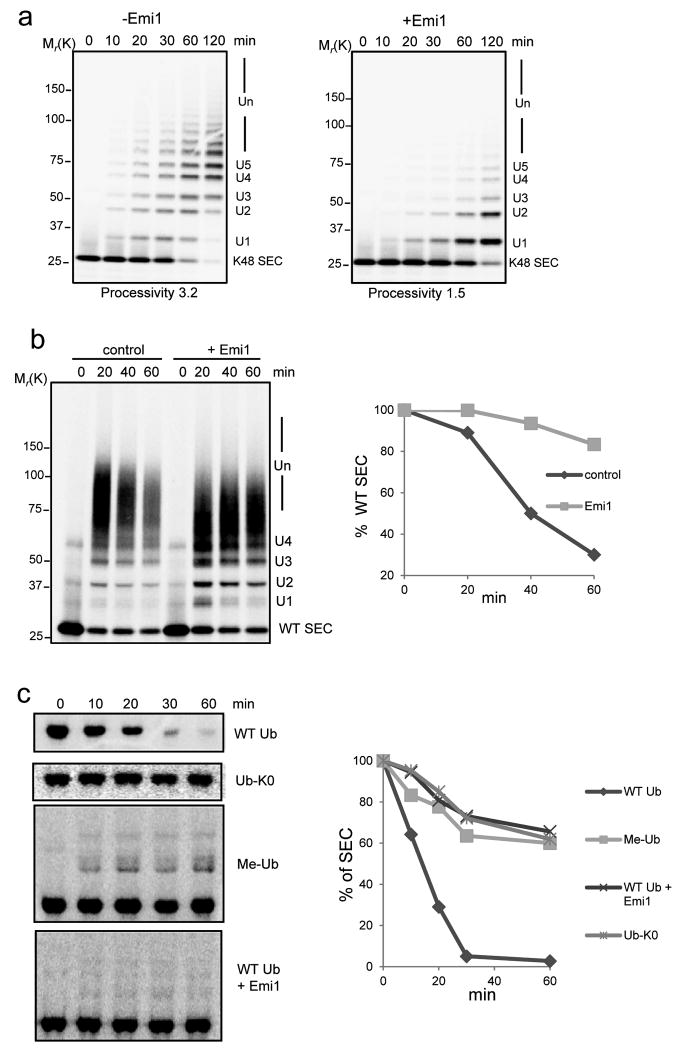
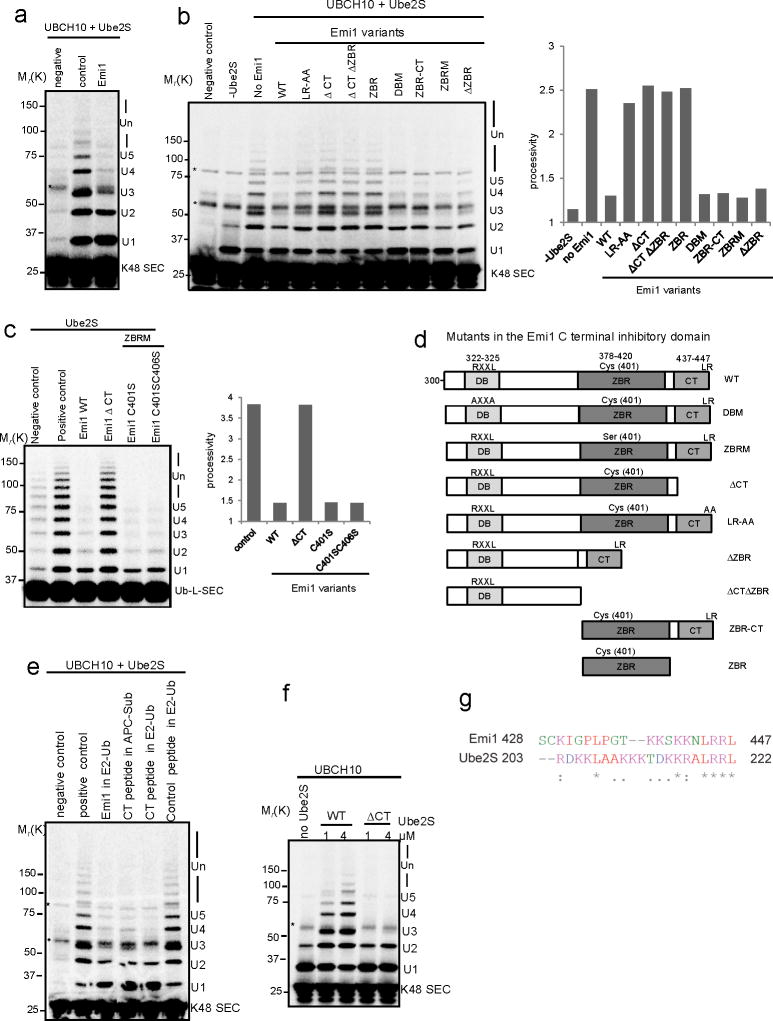
The presence of multiple sites on Emi1 for interacting with APC indicates that APC, Emi1 and substrate could form a ternary complex. Accordingly, we found that Emi1 bound both APC and its substrate simultaneously (Supplementary Fig. S1). To ask how Emi1 regulates APC activity, we simplified the reaction products by assays with a single-lysine securin mutant (K48 SEC), Emi1 decreased the rate of conversion of the substrate to ubiquitin adducts, consistent with its ability to compete with the substrate for APC binding (Fig. 1a). Emi1 also significantly reduced the rate of ubiquitin chain assembly, as reflected in reduced processivity (Fig. 1a). A decrease in processivity cannot be explained by competitive inhibition of substrate binding. This indicates that Emi1 might interfere with the ubiquitylation process also by preventing ubiquitin chain extension.
Figure 1.
Emi1 stabilizes substrates by preventing ubiquitin chain assembly.
(a) 200 nM 33P labeled single-lysine securin (K48 SEC) was ubiquitylated by 4 nM APCCdh1 in the presence (+) or absence (-) of 200 nM Emi1. The reactions were supplemented with UBCH10, Ube2S and WT ubiquitin, and analyzed by autoradiography. (b) Ubiquitylation and degradation of WT securin was examined in the presence or absence of Emi1. 100 nM 33P labeled WT securin, 4 nM purified APCCdh1, 100 nM Emi1 and 8 nM 26S proteasomes were used in the reconstituted reaction, supplemented with UBCH10, Ube2S and WT ubiquitin and analyzed by autoradiography. In the graphs the remaining substrates were quantified and plotted as percentages of input radiolabeled securin at indicated time points. These data are representative of three independent experiments. (c) Degradation of 35S labeled WT securin was examined in cell extracts prepared from HeLa S3 cells synchronized at G1 phase. WT ubiquitin (WT Ub), lysine-free ubiquitin (Ub-K0), methylated ubiquitin (Me-Ub) or Emi1 were added as indicated and analyzed by autoradiography. The amount of remaining securin was quantified and plotted at indicated time points. These data are representative of three independent experiments.
We examined the effects of Emi1 on the degradation of WT securin in a purified system reconstituted with proteasomes (Fig. 1b). Ubiquitylation was still efficient (though somewhat slower) in the presence of Emi1. By contrast, degradation was strongly inhibited (Fig. 1b). When ubiquitylation was restricted to multiple-monoubiquitylations with methylated ubiquitin, the degradation of securin was inhibited (Supplementary Fig. S2). Furthermore, the proteolysis of securin in HeLa cell extracts was also inhibited by Emi1 by either supplementing the extracts with lysine-free ubiquitin or methylated ubiquitin (Fig. 1c). In summary, Emi1 more strongly inhibits degradation than ubiquitylation, by preferentially preventing ubiquitin chain elongation.
The features of UBCH10 and Ube2S, revealed in single encounter assays
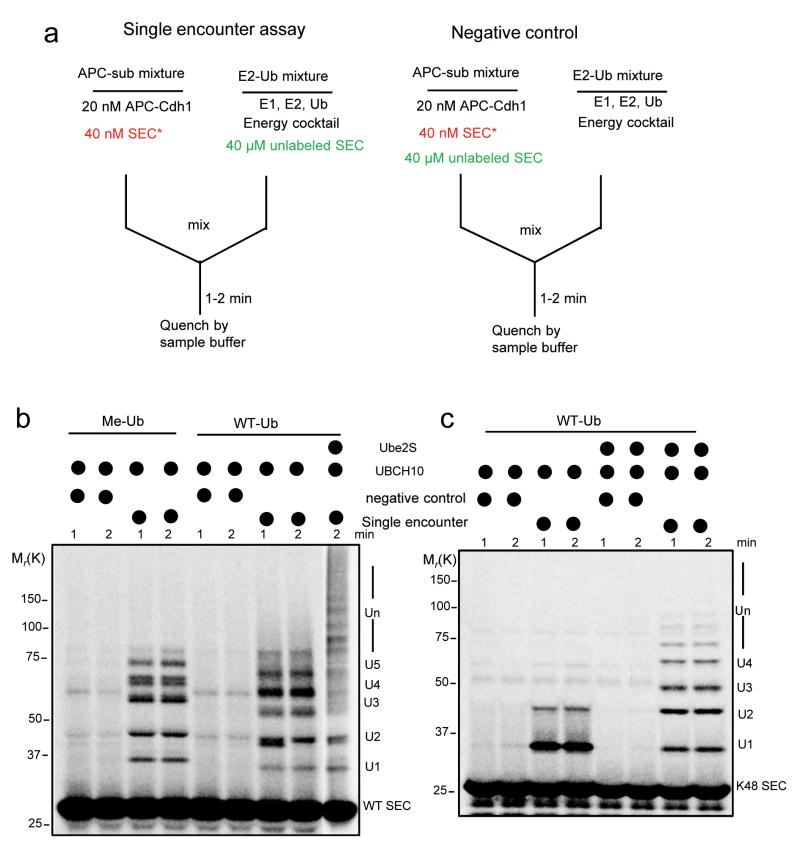
Because Emi1 inhibits APC activity at two levels, it is hard to attribute its effects cleanly to each process by traditional assays. To circumvent this problem, we set up a quantitative single encounter reaction based on a recent SCF study14. As depicted in Fig. 2a, an “APC-Sub” mixture and an “E2-Ub” mixture are pre-incubated separately to allow substrates to pre-bind to APC in one reaction and E2 to be charged with ubiquitin in the other. When the two are mixed, the radiolabeled substrate pre-bound to APC starts to acquire ubiquitins until it dissociates from APC. Rebinding of the radiolabeled substrate to APC is prevented by the excess of cold substrate. Hence the labeled reaction records the ubiquitylation profile from a single APC-substrate binding event. With this assay, we found that with methylated ubiquitin UBCH10-catalyzed multiple monoubiquitylation was very processive, meaning that UBCH10 rapidly transfers ubiquitins to several lysine residues on substrates (Fig. 2b left part). The processivity was only slightly increased with WT ubiquitin (Fig. 2b right part), indicating that UBCH10-catalyzed ubiquitin chain elongation is a slow process. However, processivity increased greatly upon addition of Ube2S, consistent with previous studies suggesting that Ube2S rapidly catalyzes ubiquitin chain assembly3, 15. The properties of these two E2s were confirmed by a single encounter assay with the single-lysine securin (Fig. 2c).
Figure 2.
A single encounter assay for APC catalyzed ubiquitylation by UBCH10 and Ube2S.
(a) As shown in this design diagram, In the “APC-Sub” mixture, 20 nM APCCdh1 and 40 nM 33P radiolabeled securin were pre-incubated; in the “E2-Ub” mixture, E1, E2, ubiquitin, energy regenerating cocktail and 40 μM unlabeled substrate were pre-incubated. The reaction was started by combining the two mixtures, quenched with SDS sample buffer, and analyzed by autoradiography. As a negative control, 40 μM unlabeled substrate was added to “APC-Sub” complex to measure its efficacy for competing for APC binding. (b) Single encounter assay was performed to test the ubiquitylation of WT securin. Methylated ubiquitin (Me-Ub) or WT ubiquitin (WT-Ub), Ube2S and UBCH10 were supplemented as indicated. (c) Single encounter assay was performed to test the ubiquitin chain assembly on the single-lysine securin (K48 SEC). WT ubiquitin was used in this assay. UBCH10 and Ube2S were added as indicated. These reactions were analyzed by autoradiography.
The domains of Emi1 responsible for inhibiting association of substrates with APC
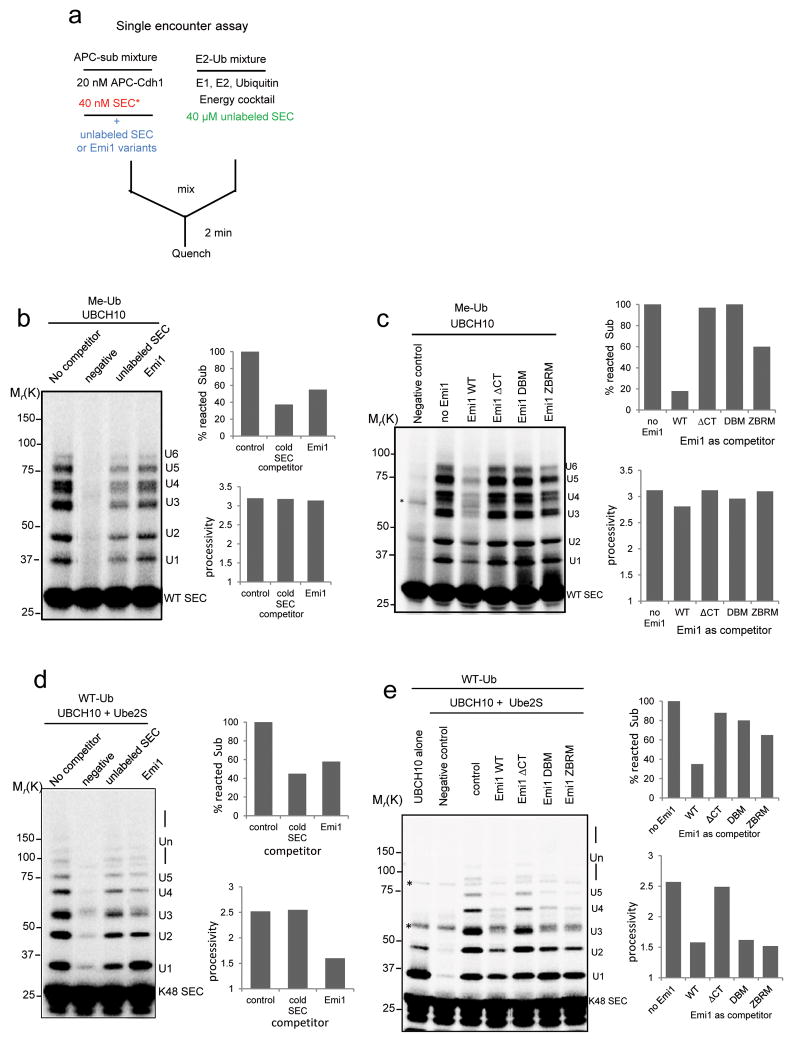
To assess the effectiveness of Emi1 as a pseudosubstrate, we measured its ability to inhibit the association of securin to APC (Fig. 3a). Both cold securin and Emi1 competitively reduced the binding of labeled substrate to APC, as reflected in the decreased substrate conversion rates, with cold securin slightly more potent (Fig. 3b). This suggests that though Emi1 competes with substrates for APC binding, the effect is weak; its activity is no better than that of a good substrate. To identify the domains of Emi1 responsible for this weak competitive binding, we mutated known APC interaction domains9-11. Mutation of D-box (DBM), zinc binding region (ZBRM), or deletion of the C-terminal tail (ΔCT), all reduced the competitive ability of Emi1 (Supplementary Fig. S3a). When a 25 fold excess (1 μM) of Emi1 mutants were pre-incubated with “APC-Sub” mixture (Fig. 3c), The DBM and ΔCT could not compete for substrate binding even at this saturating concentration. In contrast, ZBRM still retained some potency for substrate competition. These data imply that the C-terminal tail is indispensable for recruitment and position of Emi1 to the catalytic site of APC, whereas the ZBR may help orient Emi1 for optimal interaction with the D-box receptor.
Figure 3.
Single encounter assays to study the competition of Emi1 for substrate binding to APC.
(a) Experimental design to test the relative competitiveness of securin and Emi1 variants on ubiquitylation. (b) 100 nM cold securin or Emi1 was pre-incubated with APCCdh1 and 33P radiolabeled WT securin in the single encounter reaction. (c) 1μM Emi1 and its variants were pre-incubated with “APC-WT SEC” In the single encounter reaction and ubiquitylation was assayed. In both (b) and (c), methylated ubiquitin (Me-Ub) was used to restrict the reactions to multiple monoubiquitylation. (d) 50 nM cold securin or WT Emi1 was pre-incubated with APCCdh1 and radiolabeled single-lysine securin (K48 SEC) in the single encounter assay supplemented with Ube2S. (e) Emi1 variants were pre-incubated with “APC-K48 SEC” mixture in the single encounter assay involving Ube2S. Effectiveness of Emi1 variants as inhibitors was measured and compared. In (d) and (e), WT ubiquitin (WT-Ub) was used to support ubiquitin chain assembly. All the reactions above were analyzed by autoradiography, and substrate conversion rate and processivity was calculated. * indicates nonspecific band.
Despite weak competitive binding as a pseudosubstrate, Emi1 efficiently inhibits ubiquitin chain elongation
Since competitive inhibition at the substrate binding site cannot explain the strong inhibition exerted by Emi1 on substrate degradation, we tested if it has its major effect on ubiquitin chain assembly. We used single-lysine securin in single encounter reactions with a full complement of UBCH10, Ube2S and WT ubiquitin. As noted above, both Emi1 and securin competed with substrates for APC binding, reducing the substrate conversion rate (Fig. 3d). However, Emi1, but not securin, significantly reduced the processivity of ubiquitin chain elongation, confirming that Emi1 is not simply a competitive inhibitor, but has an effect on the process of ubiquitylation.
Ubiquitin chain elongation, but not multiple monoubiquitylation by UBCH10, is subject to the effective inhibition by Emi1 via the ZBR domain
Among the three mutants, only deletion of the C-terminal tail had an obvious effect in abrogating the inhibition of chain assembly primarily catalyzed by Ube2S (Fig. 3e). This result alone implies that suppression of chain elongation by Emi1 involves a mechanism different from the pseudosubstrate model.
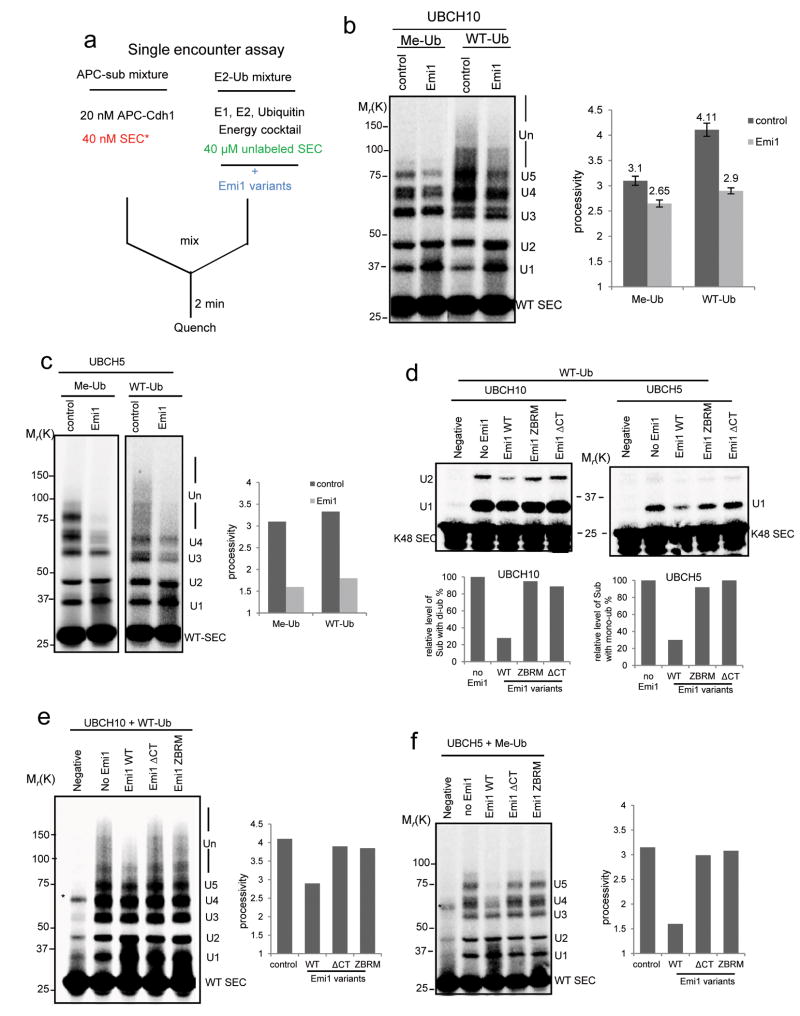
To separate the role of Emi1 as an inhibitor of ubiquitin elongation from its role as pseudosubstrate, we pre-incubated Emi1 variants with “E2-Ub” mixture before starting the reaction (Fig. 4a). In this setting, Emi1 does not compete with radiolabeled substrate for binding to APC. Thus, if Emi1 confers any inhibitory effect on ubiquitylation, it should be entirely due to its effect on the process of ubiquitin transfer. Using this approach, we first addressed how Emi1 inhibits ubiquitylation of WT securin by UBCH10 (without Ube2S). Whereas the processivity contributed by multiple monoubiquitylation was only slightly reduced (Fig. 4b left), the processivity contributed by chain extension was greatly reduced upon addition of Emi1 (Fig. 4b right), indicating that the chain extension by UBCH10 is preferentially inhibited. This specific effect of Emi1 on UBCH10 activity was also confirmed when single-lysine securin was used (Fig. 4d left panel). Moreover, titration assays with Emi1 at various concentrations (Supplementary Fig. S3d and Fig. S3e) further reinforced the conclusion Emi1 more effectively inhibits UBCH10 catalyzed chain formation.
Figure 4.
Emi1 inhibits the slow process of ubiquitin transfer from charged UBCH10 or UBCH5.
(a) Assay was designed for testing how Emi1 affects ubiquitin transfer from charged E2. Emi1 variants were pre-incubated with “E2-Ub” mixture in the single encounter assay to study the possible function of Emi1 against E2 activity. (b) 2 μM Emi1 was pre-incubated with “E2-Ub” to test how Emi1 affects the ubiquitylation of 33P radiolabeled WT securin catalyzed by 1μM UBCH10. Methylated ubiquitin (Me-Ub) was used to measure the processivity of multiple-monoubiquitylation; Wild type ubiquitin (WT-Ub) was used to measure the processivity from both monoubiquitylation and chain extension. The processivity of reactions was calculated, and the mean value ± s.d. of five independent experiments is shown. Note that the processivity contributed by chain extension in control and Emi1 (+) group is 1.01 (4.11-3.1) and 0.25 (2.9-2.65) respectively. (c) 2 μM Emi1 was pre-incubated with “E2-methylated Ub” to test how Emi1 affects the ubiquitylation of 33P radiolabeled WT securin catalyzed by 1 μM UBCH5. (d) Emi1 variants were pre-incubated with “E2-WT Ub” to evaluate their effects on ubiquitylation of the single-lysine securin (K48 SEC) by either UBCH10 or UBCH5. The reaction products by UBCH10 with di-ubiquitin and products by UBCH5 with mono-ubiquitin were quantified respectively. (e) Emi1 variants were pre-incubated with “UBCH10-WT Ub” to examine their inhibitory activities on the polyubiquitylation of WT securin. (f) Emi1 variants were pre-incubated with “UBCH5-methylated Ub” to study their inhibitory activities on multiple monoubiquitylation of WT securin. All the reactions above were analyzed by autoradiography. * indicates nonspecific band.
How does Emi1 distinguish between these two steps of ubiquitylation catalyzed by the same E2? Based on the observations above that UBCH10 catalyzes rapid monoubiquitylation but is relatively slow to elongate the chains, we hypothesized that the ubiquitylation with a low reaction rate is more sensitive to Emi1 inhibition. If so, slowing down the rate of monoubiquitylation should render it susceptible to Emi1 activity. UBCH5, an E2 previously reported to support APC activity in vitro2, catalyzes multiple monoubiquitylation at a slower rate than UbcH10 (Supplementary Fig. S3b). It turned out that both the processivity of multiple monoubiquitylation of WT securin (Fig. 4c) and the rate of monoubiquitylation of K48 securin (Fig. 4d right panel) by UBCH5 were significantly reduced by Emi1. This supports the conclusion that Emi1 specifically targets the slow process of ubiquitylations catalyzed by UBCH10 or UBCH5.
To ask which domain inhibits the slow process of ubiquitin transfer, we examined several mutants. Previous study indicated that the zinc binding region (ZBR) antagonizes APC E3 ligase activity10. Using both WT securin (Fig. 4e and 4f) and single-lysine securin (Fig. 4d), we found that ZBR mutant (ZBRM) failed to inhibit these ubiquitylations: ubiquitin chain assembly by UBCH10 or monoubiquitylation by UBCH5. This indicates that the primary role of ZBR is to inhibit ubiquitylation of APC substrates. Interestingly, C-terminal tail deficient mutant also failed to inhibit the ubiquitylation by either UBCH10 or UBCH5 (Fig. 4d, 4e and 4f). This may suggest that the C-terminal tail of Emi1 positions the ZBR, allowing it to interfere with ubiquitin transfer to substrates.
Rapid chain elongation by Ube2S is competitively inhibited by the C-terminal tail of Emi1, which blocks its association with APC
To study the regulation of Ube2S by Emi1 on assembling ubiquitin chains, we again used the single-lysine securin (K48 SEC) to simplify analysis of the products. Pre-incubation of Emi1 in “E2-Ub” containing UbcH10 and Ube2S in the single encounter assay greatly reduced this processivity (Fig. 5a). Since Ube2S mediates chain assembly at very high rate, and we showed above that ZBR only efficiently inhibits the slow process of ubiquitylation, we posited that this inhibition of Ube2S activity is not attributed to ZBR function. To prove this, we employed a chimeric protein (Ub-L-securin) with ubiquitin fused to the N-terminus of securin. Ube2S catalyzed very processive chain formation on Ub-L-securin without initiation by UBCH10 (Fig. 5c). As shown in Fig. 5c, WT Emi1 profoundly reduced the processivity of the reaction, and the two ZBR mutants (C401S and C401SC406S) retained the full inhibitory activity against chain assembly, indicating that ZBR has very little effect on Ube2S activity. To address the additional mechanism accounting for inhibiting Ube2S activity, single encounter assays with K48 SEC were performed at various levels of Ube2S, and the processivity contributed by Ube2S was calculated (Supplementary Fig. S4). Emi1 did not affect the Vmax, but significantly increased the Km of the reaction, indicating that Emi1 acts as competitive inhibitor for antagonizing Ube2S activity. To define exact domains responsible for this inhibitory activity, various Emi1 mutants were tested by the single encounter assay with K48 SEC (Fig. 5b and 5d). Interestingly, all the variants with mutations of the C-terminal tail (ΔCT, ΔCTΔZBR, ZBR and LR-AA) lost inhibitory activity, whereas those with intact C-terminal tail (ZBRM, DBM and ZBR-CT) efficiently inhibited Ube2S-catalyzed chain elongation, suggesting that the C-terminal tail of Emi1 alone may be sufficient to suppress Ube2S activity. Consistently, the C-terminal tail deficient mutant (ΔCT) totally lost the inhibitory activity against chain formation upon Ub-L-securin by Ube2S (Fig. 5c). Furthermore, the C-terminal tail peptide (residues 428-447) effectively prevented ubiquitin chain elongation (Fig. 5e). The C-terminal tails of Ube2S and Emi1 are very similar, especially in the last 4 amino acid (Fig. 5g). Ubiquitylation by Ube2S without its C-terminal tail is not processive in the single encounter assay (Fig. 5f), suggesting the C-terminal tail is required for Ube2S activity, consistent with a previous report3. Thus, we concluded that Emi1 antagonizes the activity of Ube2S by competitively preventing the association of this E2 to APC via the C-terminal tail.
Figure 5.
The C-terminal tail of Emi1 suppresses Ube2S activity by competitively blocking the interaction between Ube2S and APC
a) 2 μM Emi1 was pre-incubated with “E2-WT Ub” mixture in the single encounter assay. 1μM UBCH10 and 1μM Ube2S were supplemented to fully support ubiquitin chain assembly on the single-lysine securin (K48 SEC). (b) Emi1 variants were pre-incubated with “E2-WT Ub” mixture in the single encounter assay to evaluate their effects on ubiquitin chain assembly on the single-lysine securin (K48 SEC). UBCH10 and Ube2S were added in this assay. The processivity of reactions was quantified. (c) Inhibitory activity of Emi1 variants on ubiquitylation of Ub-L-securin was examined. Ub-L-securin is a chimeric protein with ubiquitin fused to the N-terminus of WT securin. Emi1 variants were pre-incubated with “E2-WT Ub” mixture in the single encounter reaction supplemented only with Ube2S. The processivity of reactions was quantified. (d) Schematic representation of Emi1 mutants in the C terminal APC inhibitory domain (residues 300-447). The amino acid positions of destruction box (DB), zinc binding region (ZBR) and C-terminal tail (CT) were shown. (e) 10 μM Emi1 C-terminal tail peptide (CT peptide) (residues 428-447) was pre-incubated either with “APC-K48 SEC” or with “E2-WT Ub” mixture in the single encounter assay. The effects of the peptide and the Emi1 protein on Ube2S activity were compared. 10 μM 3X FLAG peptide (23 aa) was used as control peptide. (f) WT Ube2S or Ube2S deficient in the C-terminal tail (ΔCT) was applied in the single encounter assay with single-lysine securin (K48 SEC). (g) The C-terminal tails of human Emi1 and Ube2S were aligned with ClustalX. Basic residues are marked with pink color. These reactions above were analyzed by autoradiography. * indicates nonspecific band.
The C-terminal tails of both Emi1 and Ube2S associate with the cullin subunit of APC by electrostatic interactions
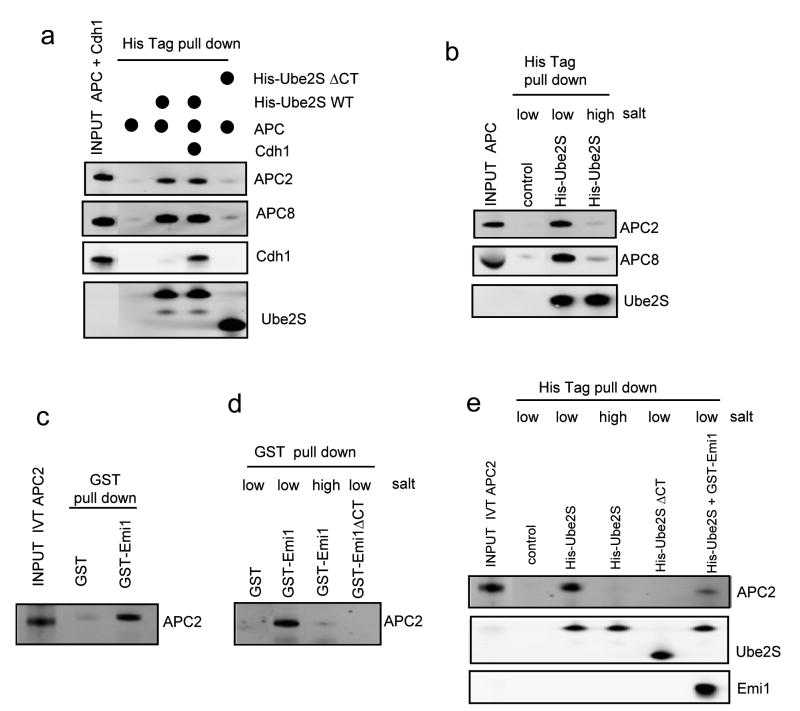
Consistent with kinetic assays, Ube2S associated with the APC complex and deletion of the C-terminal tail abolished this interaction (Fig. 6a and Supplementary Fig. S5a). These results suggest that the C-terminal tail may be the only domain on Ube2S responsible for APC binding with high affinity. By comparison, the Emi1 mutant without the C-terminal tail still associated with APC complex (Supplementary Fig. S5b). Moreover, the binding of Ube2S to APC was not dependent on Cdh1 (Fig. 6a), suggesting that some APC core subunit directly associates with this E2. The C-terminal tail is very rich in basic residues (Fig. 5g), implying this E2-E3 association may be driven by electrostatic force. Protein-protein association through electrostatic interactions is sensitive to salt, which blocks the attraction between opposite charges. High salt washing disrupted the APC-Ube2S binding (Fig. 6b), consistent with a previous study3.
Figure 6.
The C-terminal tail of Emi1 and Ube2S associate with APC2 through electrostatic interaction.
(a) His tagged WT Ube2S or Ube2S without C-terminal tail (ΔCT) was incubated with purified APC complex and recombinant Cdh1 as indicated. Mixtures were subjected to a His-tag pull-down assay, followed by western blot for respective proteins. (b) His tagged Ube2S was incubated with purified APC complex, followed by His-tag pull-down with Ni-NTA beads. The beads were then washed with high salt buffer (with 300 mM NaCl) or low salt buffer (with 5 mM NaCl) before they were subjected to Western blot for APC2, APC8 and Ube2S. (c) GST tagged Emi1 or GST was incubated with APC2 expressed in rabbit reticulocyte lysates and subjected to a GST pull-down, followed by a Western blot with anti APC2 antibody. (d) GST, GST tagged Emi1 or GST tagged Emi1 without C-terminal tail (ΔCT) was incubated with APC2 expressed in rabbit reticulocytes respectively, subjected to GST pull-down, washed with high salt buffer (with 300 mM NaCl) or low salt buffer (with 5 mM NaCl) as indicated, and analyzed by Western blot for APC2. (e) His tagged Ube2S or His tagged Ube2S without C-terminal tail (ΔCT) was incubated with APC2, subjected to His-tag pull-down, washed with low salt (5 mM NaCl) or high salt (300 mM NaCl) buffer, and analyzed by western blot for APC2 and Ube2S. The competition of Emi1 with Ube2S for binding to APC2 was also tested. Uncropped images are presented in Supplementary Fig. S7.
It has been noted that the processive ubiquitylation catalyzed by SCF necessitates the electrostatic interactions between the cullin subunit of SCF and the C-terminal tail of its cognate E2, cdc3416. We guessed that Ube2S may act in a similar fashion. Both Emi1 (Fig. 6c and Supplementary Fig. S5c) and Ube2S (Fig. 6e) directly associate with APC2. Moreover, the interaction between APC2 and Ube2S or Emi1 is abolished either by washing with high salt buffer, or by deletion of their C-terminal tails (Fig. 6d and 6e). These results suggest that Emi1 interferes with the activity of Ube2S mainly through disrupting the dynamic electrostatic association between the E2's C-terminal tail and APC2.
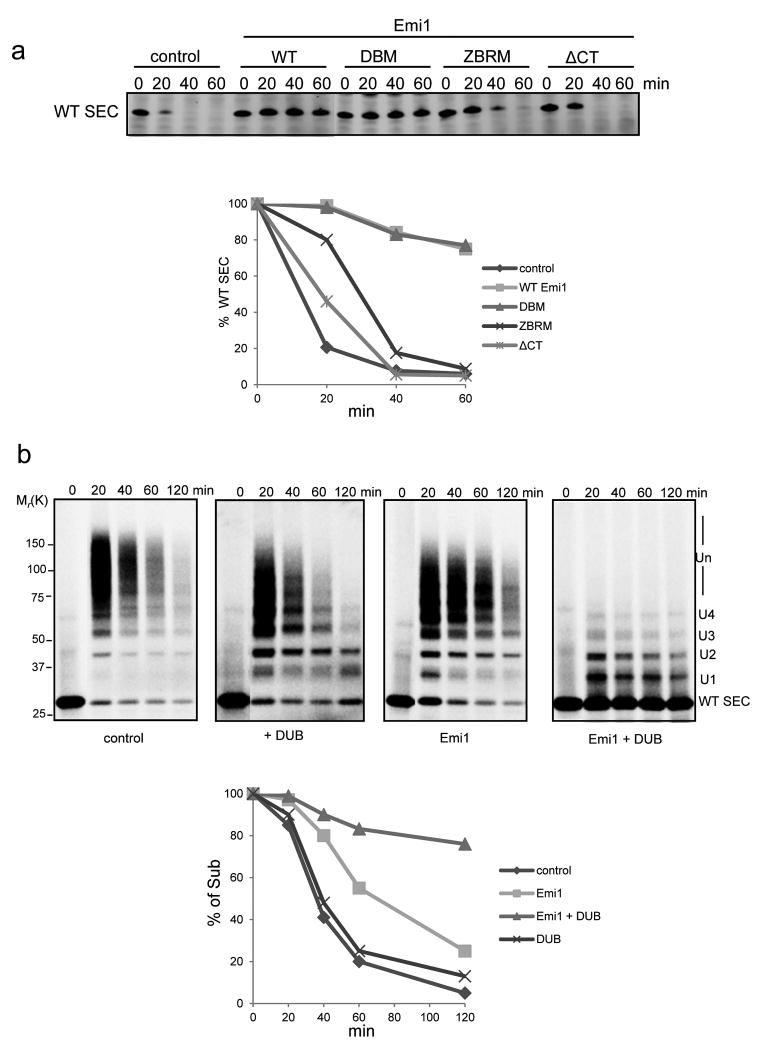
To evaluate contributions of these domains to Emi1 function in a more complex system approximating the cell and in particular containing the complete degradation machinery, we compared the inhibitory activity of Emi1 mutants on securin degradation in highly concentrated HeLa cell extracts (Fig. 7a). In extracts the D-box mutant (DBM) of Emi1 at relatively high concentration (2 μM) efficiently inhibited the degradation of securin, suggesting that Emi1 must have a role, other than as pseudosubstrate. In contrast, mutation of either ZBR or C-terminal tail, the two domains responsible for suppressing chain elongation, largely abolished the inhibitory activity against protein degradation. Thus, we concluded that Emi1 inhibits the degradation of APC substrate primarily through suppression of ubiquitin chain elongation.
Figure 7.
The inhibitory activity of Emi1 against securin degradation in the presence of deubiquitylating enzymes.
(a) Degradation of 400 nM WT securin was performed in cell extracts prepared from HeLa S3 cells synchronized at G1 phase. 2 μM Emi1 variants were supplemented as indicated. Samples were analyzed by Western Blot for securin. In the graphs the remaining substrates were quantified and plotted as percentages of input securin at indicated time points. These data are representative of three independent experiments. Uncropped images are presented in Supplementary Fig. S7. (b) Ubiquitylation and degradation of 100 nM 33P labeled WT securin was examined in a reconstituted assay in the presence of 4 nM APCCdh1 and 8 nM 26S proteasome. WT ubiquitin, UBCH10 and Ube2S were added in the reactions. The deubiquitylating enzyme (DUB), USP2 catalytic domain, and Emi1 were supplemented as indicated. Emi1 at relatively low concentration was added to the reaction, ensuring that the degradation of securin was only partially inhibited, in order to evaluate the impact of DUB on Emi1 inhibitory activity. These reactions were analyzed by autoradiography. The amount of remaining substrates at indicated time points was calculated, and plotted as percentages of input radiolabeled securin. These data are representative of three independent experiments.
Deubiquitylation activity enhances the inhibitory effect of Emi1
Studies in cell extracts were qualitatively similar to those in the purified system that we presented here. However, there was a notable difference. Multiple-monoubiquitylated substrates were easily detected in the reconstituted but purified degradation reactions (Fig. 1b and Supplementary Fig. S2), but ubiquitylated substrates were barely detectable in cell extracts that were inhibited by Emi1 (Fig. 1c). This difference suggests that the non-degradable ubiquitylated proteins were converted back to their basal states by deubiquitylating enzymes (DUBs) in cell extracts. In a complete kinetic scheme, DUBs could play a kinetic proofreading role in distinguishing substrates and determining a precise order of degradation5. Therefore, we tested in a reconstituted system whether DUBs synergized with Emi1 in blocking degradation. USP2 catalytic domain can efficiently remove monoubiquitin from APC substrate (Supplementary Fig. S6). As shown in Fig. 7b, addition of USP2 alone had little effect on substrate degradation. Under these conditions Emi1 at relatively low concentration only partially stabilized the substrate. However, addition of both the DUB and Emi1 showed much greater inhibition, strongly indicating that DUBs can enhance the effect of Emi1 on stabilizing APC substrates.
Discussion
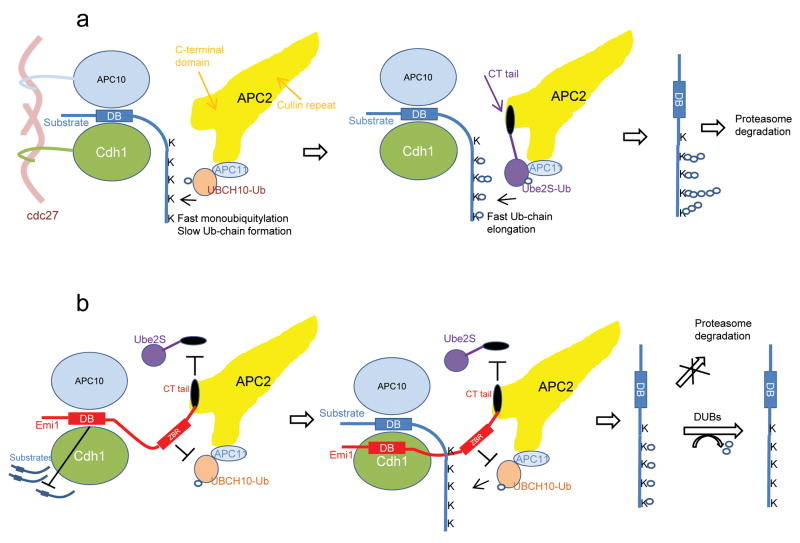
These studies support a model in which Emi1 is a multipurpose inhibitor of APC (Fig. 8). In the absence of Emi1, a substrate is recruited to APC by binding to the D-box co-receptor formed by Cdh1 and APC1017, the concerted activities of UBCH10 and Ube2S enable APC to rapidly polyubiquitylate the substrate (Fig. 8a). When Emi1 is expressed, the D-box of Emi1 can compete with substrates for binding to the D-box receptor (Fig. 8b). When the D-box receptor is occupied by a substrate, the ZBR domain of Emi1 allows the initial monoubiquitylation of the substrate, but strongly inhibits chain formation by UBCH10; meanwhile, the C-terminal tail of Emi1 also antagonizes Ube2S activity by competitively preventing its binding to APC2. By its combinatorial action Emi1 effectively inhibits ubiquitin chain extension. Products without ubiquitin chains cannot easily be degraded by the proteasome, and are reverted by DUBs to their original state.
Figure 8. Model for inhibition of APC by Emi1.
(a) In the absence of Emi1, the processive multiple monoubiquitylation by UBCH10 and processive ubiquitin chain elongation by Ube2S enable the substrate to be rapidly polyubiquitylated by APC and targeted for degradation by 26S proteasome.
(b) Emi1 inhibits the degradation of APC substrates at multiple levels. The C-terminal tail enables Emi1 to closely approach APC catalytic site through electrostatic interaction. The D-box of Emi1 competitively blocks access of substrates to the D-box co-receptor formed by APC10 and Cdh1. When a substrate is bound to APC, the activity of ZBR of Emi1 permits the rapid multiple monoubiquitylation of the substrate, but suppresses the slow ubiquitin chain elongation catalyzed by UBCH10. Meanwhile, the C-terminal tail of Emi1 inhibits the processive ubiquitin chain assembly by Ube2S through competitively occluding its binding to APC2. As a result, APC substrates without sufficient ubiquitin chains cannot be efficiently degraded by proteasome, and are converted to their original status by deubiquitylating enzymes (DUBs).
Reconsideration of the standard model of Emi1 activity as a pseudosubstrate
Currently, it is thought that inhibitors of APC, such as Emi1, Acm1 and BubR1 act as pseudosubstrates10, 18, 19. Effective blockage of substrate binding to APC requires a very high affinity interaction between Emi1 and the substrate binding site. Our competition assays show that Emi1 is not a particularly good competitor. To support the pseudosubstrate model, it was shown previously that cyclin B poorly competes for Emi1 binding to the APC10. Indeed, based on our finding, addition of saturating amount of cyclin B would lead to the formation of cyclin B-APC-Emi1 complex, rather than competitive dissociation. The function of Emi1 as a pseudosubstrate only partially explains its inhibitory effect.
The key function of the C-terminal tail of Emi1 in its inhibitory activity
Our data strongly suggest that the C-terminal tail of Emi1 plays a pivotal role in various aspects of Emi1 activity: First, The tail facilitates recruiting Emi1 to the catalytic site of APC, for optimal binding to the D-box receptor; second, it positions ZBR to inhibit ubiquitin transfer; third, it competitively blocks the binding of Ube2S to APC. Moreover, the electrostatic interaction with APC2 could provide very high kon, enabling Emi1 to interfere with APC activity at any point of the ubiquitylation process.
The effect of DUBs on Emi1 inhibitory activity
DUBs have been proposed to confer greater selectivity on substrate selection. Distributive substrates, which cannot obtain sufficient ubiquitins during a single E3-substrate encounter are converted by DUBs to basal status5. Thus, they cannot be degraded until processive substrates are first degraded, freeing APC. By this means, Emi1 theoretically transforms all APC substrates into distributive substrates. Furthermore, it was recently shown that many DUBs have preference for monoubiquitylated substrates but are inefficient in removing ubiquitin chains20. Therefore, DUBs are more effective in acting on ubiquitylated substrates, when chain elongation is prevented by Emi1, explaining how DUBs can magnify the inhibitory effect of Emi1.
In summary, this study provides the first kinetic evidence showing that natural inhibitors of E3 ligase exert their effects by direct interference with E2 activity. We also show that, paired with the activities of DUBs, suppression of the 2nd step of ubiquitylation (chain elongation) alone is sufficient to stabilize E3 substrates. These mechanistic features may provide new ways to think about therapeutic targets affecting critical E3 ligases in various diseases.
Supplementary Material
Acknowledgments
We wish to thank the members of the Kirschner Laboratory for advice, especially Tao Wu and Ying Lu for discussions and insightful suggestions. The work was supported by a grant from the National Institute of General Medical Sciences, R01 GM039023.
Footnotes
Author Contributions: W.W. and M.W.K planned the project. W.W. designed, carried out and analyzed all experiments. W.W. and M.W.K. interpreted the data. The manuscript was written by W.W. and M.W.K.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 2.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu T, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song L, Rape M. Substrate-specific regulation of ubiquitination by the anaphase-promoting complex. Cell Cycle. 2011;10:52–56. doi: 10.4161/cc.10.1.14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 6.Lai LA, Morabito L, Holloway SL. A novel yeast mutant that is defective in regulation of the Anaphase-Promoting Complex by the spindle damage checkpoint. Mol Genet Genomics. 2003;270:156–164. doi: 10.1007/s00438-003-0912-5. [DOI] [PubMed] [Google Scholar]
- 7.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- 9.Reimann JD, et al. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 10.Miller JJ, et al. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohe M, et al. Emi2 inhibition of the anaphase-promoting complex/cyclosome absolutely requires Emi2 binding via the C-terminal RL tail. Mol Biol Cell. 2010;21:905–913. doi: 10.1091/mbc.E09-11-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer HJ, Rape M. Processive ubiquitin chain formation by the anaphase-promoting complex. Semin Cell Dev Biol. 2011;22:544–550. doi: 10.1016/j.semcdb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang W, et al. Emi2-mediated inhibition of E2-substrate ubiquitin transfer by the anaphase-promoting complex/cyclosome through a D-box-independent mechanism. Mol Biol Cell. 2010;21:2589–2597. doi: 10.1091/mbc.E09-08-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi E, Dial JM, Jeong DE, Hall MC. Unique D box and KEN box sequences limit ubiquitination of Acm1 and promote pseudosubstrate inhibition of the anaphase-promoting complex. J Biol Chem. 2008;283:23701–23710. doi: 10.1074/jbc.M803695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer JB, Morgan DO. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem. 2011;286:45186–45196. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.