Abstract
LMX1A and LMX1B encode two closely related members of the LIM homeobox family of transcription factors (TF). These genes play significant, and frequently overlapping, roles in the development of many structures in the nervous system, including the cerebellum, hindbrain, spinal cord roof plate, sensory systems and dopaminergic midbrain neurons. Little is known about the cis-acting regulatory element (REs) that dictate their temporal and spatial expression or about the regulatory landscape surrounding them. The availability of comparative sequence data and the advent of genomic technologies such as ChIP-Seq have revolutionized our capacity to identify regulatory sequences like enhancers. Despite this wealth of data, the vast majority of loci lack any significant in vivo functional exploration of their non-coding regions. We have completed a significant functional screen of conserved non-coding sequences (putative REs) scattered across these critical human loci, assaying the temporal and spatial control using zebrafish transgenesis. We first identify and describe the LMX1A paralogs lmx1a and lmx1a-like, comparing their expression during embryogenesis with that in mammals, along with lmx1ba and lmx1bb genes. Consistent with their prominent neuronal expression, 47/71 sequences selected within and flanking LMX1A and LMX1B exert spatial control of reporter expression in the central nervous system (CNS) of mosaic zebrafish embryos. Upon germline transmission we identify CNS reporter expression in multiple independent founders for 22 constructs (LMX1A, n=17; LMX1B, n=5). The identified enhancers display significant overlap in their spatial control and represent only a fraction of the conserved non-coding sequences at these critical genes. Our data reveal the abundance of regulatory instruction located near these developmentally important genes.
Introduction
LMX1A and LMX1B encode closely related transcription factors (TFs) that contain LIM-homeodomain (LIM-HD) motifs. They play pivotal roles during nervous system development, specifically in neural tube regionalization, the extension of axonal projections and the acquisition of neurotransmitter phenotypes [1,2,3]. Despite their clinical and developmental importance and the significant inquiry that these LMX1 genes have been subject to, relatively little is known about the sequences that establish the genomic regulatory landscapes required to execute their developmental programs. We set out to provide a significant, though incomplete, characterization of the cis-regulatory landscape of the LMX1A and LMX1B gene intervals by identifying elements that display CNS regulatory control. Such CNS enhancers are considered candidates for LMX1A and LMX1B transcriptional control and thus also may contain variation therein that could compromise their function and underlie disease risk.
Both LMX1A and LMX1B are involved in hindbrain and spinal cord roof plate formation and in directing the development of midbrain dopaminergic (DA) neurons [4,5,6]. LMX1A is an essential regulator of neuronal proliferation and differentiation in the cerebellar rhombic lip and telencephalic cortical hem [4,7]. Additionally, LMX1B plays a role in formation and function of the isthmic organizer (IsO), which directs the establishment of midbrain and hindbrain regional identities [8,9]. It has also been shown to be necessary for serotonergic neuronal specification [10]. Studies in mice first established the impact of Lmx1b deficiency, and lead to the demonstration that LMX1B mutations were responsible for human nail patella syndrome [11,12]. Similarly, Lmx1a mutations were initially described in mouse neurological mutant dreher, which displays defects in cerebellar, hippocampal and cortical development, as well as hindbrain roof plate malformations, short tail and deafness consistent with the patterns of its embryonic expression [13,14].
Instructions encrypted within transcriptional regulatory elements (REs) such as enhancers instruct cell fate determination, and render cells transcriptionally competent to respond to their environment [15]. Although regulatory variation is expected to contribute significantly to disease risk [16,17,18], REs, unlike coding sequences, lack an established vocabulary to facilitate their immediate recognition in primary sequence. The recent emergence of chromatin immunoprecipitation (ChIP)-based strategies coupled to next generation sequencing (ChIP-seq) has facilitated the identification of REs in a sequence agnostic manner. However, these approaches may not be well suited to comprehensive analyses of single genes, particularly those with pleiotropic expression in discrete cell populations that cannot be obtained in sufficient numbers. In situations such as this, evolutionary sequence conservation still provides a powerful tool for the identification of functional sequences, and although conservation alone is unable to discern the biological roles of sequences, one can, through functional analyses, reveal REs with a wide range of regulatory control [15]. When available, sequence intervals may be cross-referenced with pertinent ChIP-Seq / DNase-Seq data to provide additional evidence of regulatory activity and help predict cell-type dependent activity.
We selected 71 human, conserved non-coding DNA regions at LMX1A and LMX1B for preliminary functional evaluation using transgenesis in zebrafish. Of these sequences, 47 (66%) directed reporter expression in the central nervous system (CNS) of mosaic G0 embryos. We identified multiple independent founders for 22/45 enhancers at LMX1A (n=17) and LMX1B (n=5). Each directs expression in aspects of the developing nervous system of zebrafish embryos, consistent with expression of their respective endogenous genes. All 22 enhancers directed reporter expression in the CNS. A subset of these enhancers direct expression in the diencephalon, the cerebellum and at the midbrain-hindbrain (Mb-Hb) boundary, consistent with the critical role of LMX1 factors in the development of hindbrain roof plate and isthmic organizer (IsO) formation. Many also direct expression in the peripheral nervous system (14/22) and non-neuronal tissues such as the otic vesicles, cartilage, pronephros and muscles. This study adds significantly to the number of enhancer elements identified at LMX1A and LMX1B, but perhaps more importantly it reveals the complexity of regulatory control can exist at individual loci.
Results
Evolutionary conservation facilitates identification of zebrafish lmx1a and lmx1b genes
Evaluation of putative LMX1A and LMX1B regulatory sequences in a zebrafish model is aided by an appreciation of the spatial expression of their teleost paralogs. Thus we first set out to identify zebrafish LMX1A and LMX1B paralogs. Approximately 30% of the gene content of Danio rerio remains duplicated subsequent to an ancient genome duplication event in the teleost fish lineage [19]. The zebrafish genome contains two identified LMX1B paralogs (lmx1ba and lmx1bb). However, only one LMX1A paralog (lmx1a) had been identified in the zebrafish genome at the time of these experiments (http://www.ensembl.org/Danio_rerio/Gene/Summary?g=ENSDARG00000020354;r=20:33946947-33964868, Zv9). We performed a BLASTP query of the zebrafish peptide database in GenBank using the human LMX1A RNA sequence (NM_001174069.1) and identified another potential paralog previously annotated with ‘predicted’ status (LIM homeobox transcription factor 1-alpha-like, XP_001922131.3). LMX1A displays 66% identity to LMX1B at the amino acid level, and is 58% and 59% identical to zebrafish Lmx1a and Lmx1a-like, respectively. LMX1B paralogs are even more similar; Lmx1ba is 72% identical and Lmx1bb is 82% identical to the human LMX1B protein sequence (NP_001167617.1). Figure S1 provides a phylogram illustrating the similarity among the amino acid sequences that encode LMX1A, LMX1B and their zebrafish paralogs. The paralogs of LMX1A cluster together, but in a distinct node from their human counterpart. By contrast, LMX1B shares a common node with its zebrafish paralogs.
Zebrafish lmx1 genes are expressed throughout the central nervous system
We performed whole mount in situ hybridizations (ISH) to document the spatial and temporal expression patterns of the endogenous lmx1a, lmx1a-like, lmx1ba and lmx1bb genes, and to determine the level of similarity to the published expression of their mammalian orthologs in mice. Aspects of the early developmental expression of lmx1ba and lmx1bb (formerly called lmx1b.2 and lmxb.1, respectively) have been previously described [20,21,22,23,24]. We present their analysis here to facilitate comparison with expression of lmx1a and lmx1a-like.
lmx1a expression in the central nervous system (CNS) was diffuse, broadly overlapping and extending beyond lmx1ba and lmx1bb expression domains (Figure 1). We detected transcript from lmx1a throughout the brain, including the diencephalon and telencephalon, at both 48 hours post fertilization (hpf) and 72 hpf. Additionally, we detected more localized signal corresponding to lmx1a in the ventral diencephalon, raphe nuclei and otic vesicles at both time points, and at 72 hpf saw specific labeling of the cranial ganglia (Figure 1 A-D). By contrast, lmx1a-like expression was regionally restricted, with distinct labeling of the epiphysis, ventral diencephalon, rhombic lip, and raphe nuclei, closely resembling expression of lmx1ba (Figure 1 E-L).
Figure 1.
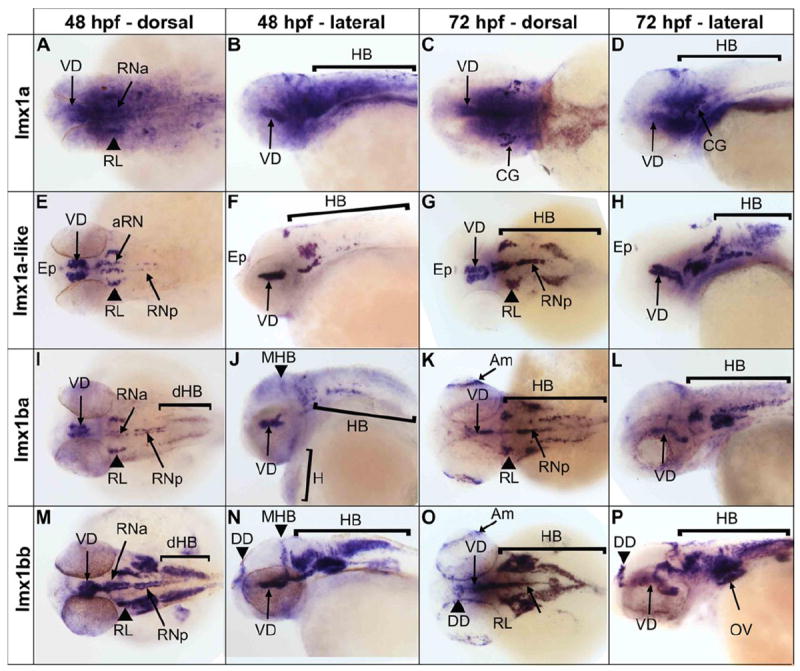
in situ hybridization depicting the expression patterns of endogenous zebrafish LMX1A and LMX1B orthologs. Expression of lmx1a (A-D), lmx1a-like (E-H), lmx1ba (I-L) and lmx1bb (M-P) are shown, assayed at 48 hpf and 72 hpf. Abbreviations: Am – Amacrine neurons, CG – Cranial Ganglia, DD – Dorsal Diencephalon, dHB – Dorsal Hindbrain, Ep – Epiphysis, H – Heart, Hb – Hindbrain, Mb-Hb – midbrain-hindbrain boundary, OV – otic vesicle, RL – rhombic lip, RNa – anterior raphe nuclei, RNp – posterior raphe nuclei, VD – ventral diencephalon. Anterior is shown to the left in each panel.
We detected expression of lmx1a-like in the anterio-dorso-lateral hindbrain and in the ventro-midline, corresponding with the cerebellar rhombic lip and serotonergic raphe nuclei, respectively (Figure 1 E-H). Both lmx1a and lmx1a-like appear to be more highly expressed in the anterior raphe nuclei at 48 hpf (Figure 1 A, B, E, and F), while both lmx1b transcripts are detected approximately equally in both raphe nuclei populations (Figure 1 I, J, M, and L). These data are consistent with both Lmx1a and Lmx1b mammalian counterparts, which are also expressed in the developing cerebellum and serotonergic neurons. lmx1a-like has very little dorsal hindbrain expression at 48 hpf but by 72 hpf transcripts are detected strongly in the posterior dorsal hindbrain (Figure 1 G and H). This pattern is unique to lmx1a-like while some expression domains overlap expression of lmx1ba and lmx1bb in the ventral diencephalon, rhombic lip, serotonergic raphe nuclei and faintly in the otic vesicles (Figure 1 I, J, M and N) [21,24]. Notably, strong expression is seen for all transcripts in the ventral diencephalon through 72 hpf, the area where main clusters of dopaminergic (DA) neurons are formed, consistent with their role in the induction of midbrain DA neurons [4,6].
The patterns of expression are similar between lmx1ba and lmx1bb with common domains in the ventral diencephalon, raphe nuclei, rhombic lip, and dorsal hindbrain, as well as the amacrine neurons of the retina at 72 hpf (Figure 1 I-P). lmx1bb, and to a lesser extent lmx1ba, show additional expression in the dorsal diencephalon that is not seen for the lmx1a transcripts (Figure 1 I-P). Overall lmx1bb shows broader domains of expression than lmx1ba throughout the CNS, however lmx1ba transcript is also unexpectedly detected in the heart (Figure 1 I-L). All probes were designed to exclude the potential for cross-hybridization with other lmx1 family members, and with other unrelated transcripts (Methods).
Selection of non-coding sequences at human LMX1A and LMX1B genomic loci
The human LMX1A gene comprises 7 exons, encompassing 154 kb on chromosome 1q24 and is flanked by PBX1 and RXRG (Figure 2A). LMX1B includes 8 exons that encompass 112 kb of chromosome 9q33.3 and is flanked by FAM125B and ZBTB43 (Figure 2B). Candidate intervals for functional analysis were selected from the sequence contained between their respective flanking genes, therefore providing LMX1A and LMX1B regions of 554 kb and 298 kb, respectively. Although this search space was not exhaustively explored, we prioritized conserved noncoding sequences for assay using Genomic Evolutionary Rate Profiling (GERP) [25], successfully PCR amplifying 71 non-coding DNA sequence intervals (see methods; 43 sequences at LMX1A and 28 at LMX1B). The putative REs were cloned into pGWcfos: EGFP [37,38] and injected into fertilized zebrafish embryos. All zebrafish embryos showing mosaic EGFP reporter expression (33 LMX1A (77%) and 14 LMX1B (50%) elements) were separated to be raised for germline transmission analysis.
Figure 2.
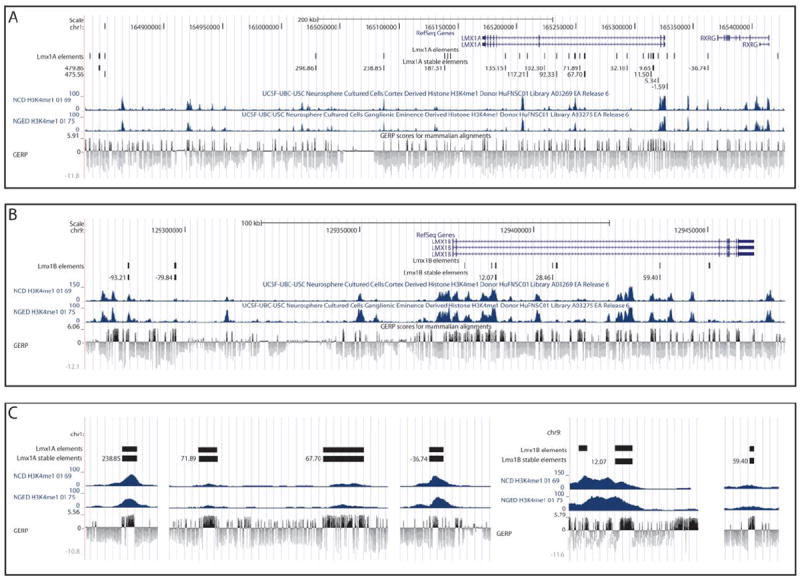
LMX1A (A) and LMX1B (B) genomic loci displaying the selected sequences and their corresponding GERP sequence conservation tracks. H3K4me1 ChIP-seq signal is included from two types of cultured neurospheres, cortex derived and ganglionic eminence derived, showing substantial overlap between conservation and high H3K4me1 signal intensity. Panel C provides enlarged example intervals to indicate local sequence conservation within amplicons. The names of REs indicate approximate distance in kb from the start codon of the gene.
Assayed sequences display LMX1A and LMX1B-appropriate neuronal enhancer activities
Of the assayed sequences, 37 displayed reporter expression in the CNS upon passage through the germline. We identified two or more founders with concordant expression in 22/37 (59%) of these sequences (LMX1A, n=17 and LMX1B n=5). Those lines, for which we were unable to identify multiple founders, in general suffered from poor survival and fecundity. Those elements were therefore most often excluded, not because of divergent expression patterns but due to the inability to obtain a sufficient number of fertilized embryos. All 22 sequences displayed spatial control in the CNS in a manner consistent with aspects of LMX1A [14] and LMX1B [11] and with endogenous zebrafish patterns of expression described above. This includes directing reporter expression within discrete regions of the diencephalon, telencephalon and hindbrain. Additionally, we identify enhancers at LMX1A that display regulatory control of reporter expression resembling the more diffuse expression of lmx1a in the CNS (LMX1A_-1.59). Consistent with their neuronal activity in our synthetic assay, the majority of endogenous sequence intervals corresponding to our assayed sequences display enrichment for histone 3 Lysine 4 monomethylation (H3K4me1), a histone mark enriched at enhancers, in cultured neurospheres derived from human neuronal cells: NGED (Neurosphere Cultured Cells, Ganglionic Eminence Derived) and NCD (Neurosphere Cultured Cells, Cortex Derived) (Figure 2A-C) [26]. Indeed, despite lacking a positive GERP alignment score, sequence LMX1A_36.74 displayed strong H3K4me1 binding in both NGED and NCD cells and was also validated in our zebrafish assay.
Identification of LMX1A enhancers with telencephalic and diencephalic regulatory control
Consistent with the endogenous expression of the mouse Lmx1a mammalian ortholog [14], we identified many LMX1A enhancers displaying overlapping control in the telencephalon (Figure 3 A-C, D, E; Figure S2 and Table 1). Telencephalic expression displayed by these sequences is consistent with the function of LMX1 genes in cortical hem development (Figure 3 A-E) [7,8,9]. Telencephalic expression is also evident for the zebrafish lmx1a, although diffuse and not at significant levels (Figure 1A-D). This observation may thus reflect mammalian (LMX1A/Lmx1a) control alone in this structure. Additionally, we identify multiple sequences at LMX1A that direct expression in the diencephalon (Figure 3A, D-F; Figure S2 and Table 1 e.g. LMX1A_238.85, LMX1A_-36.74 and LMX1A_9.65). These populations may include portions of the catecholaminergic diencephalic cluster, consistent with the established role of Lmx1a in mouse catecholaminergic neurogenesis [6].
Figure 3.
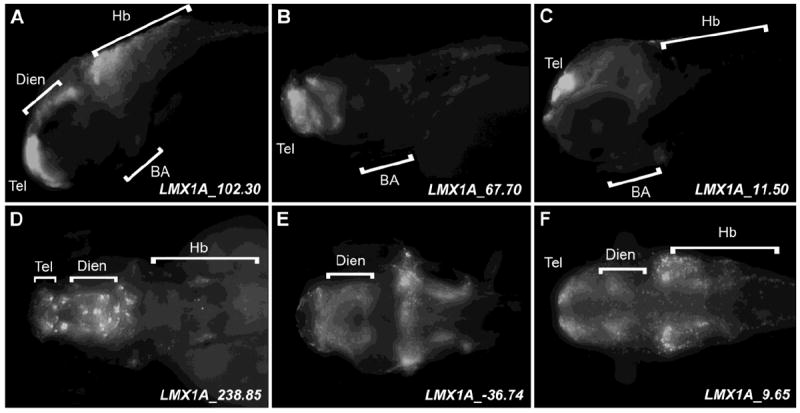
Expression of EGFP reporter in the diencephalon and telencephalon of 6 representative transgenic zebrafish lines. Zebrafish embryos were fixed at 72 hpf and stained with anti-GFP antibody. Anterior is shown to the left in each panel. A-C, lateral images. D-F, dorsal images. Abbreviations: BA – branchial arches, Hb – hindbrain, Dien – diencephalon, Hb – hindbrain, Tel – telencephalon.
Table 1.
Systematic annotation of LMX1A and LMX1B enhancers activity in zebrafish body structures
Annotation of observed enhancer activity in zebrafish embryos.
| Enhancer | Telen | Dien | Mesen | Rhomben | Sp Co | Mb/Hb | PNS | Other |
|---|---|---|---|---|---|---|---|---|
| LMX1A_-36.74 | + | + | + | + | ++ | + | ||
| LMX1A_-1.59 | ++ | ++ | ++ | ++ | ++ | + | + | |
| LMX1A_5.34 | + | ++ | + | NTC | ||||
| LMX1A_9.65 | + | + | + | + | OV | |||
| LMX1A_11.50 | + | + | + | + | + | + | ||
| LMX1A_32.09 | + | + | + | + | Pn, L | |||
| LMX1A_67.70 | ++ | + | + | + | NTC | |||
| LMX1A_71.89 | + | + | + | + | + | C, F, H, R, OV | ||
| LMX1A_92. | + | + | OV, C, F | |||||
| LMX1A_102.30 | ++ | + | + | ++ | + | + | ||
| LMX1A_117.21 | + | + | + | + | + | + | + | H, C |
| LMX1A_135.15 | + | + | + | + | + | ++ | + | |
| LMX1A_187.31 | + | + | + | + | + | ++ | ||
| LMX1A_238.85 | + | + | ++ | ++ | ++ | |||
| LMX1A_296.85 | ++ | + | ++ | + | ||||
| LMX1A_475.56 | + | + | + | + | + | + | OV | |
| LMX1A_479.56 | + | + | + | ++ | C | |||
| LMX1B_-93.21 | + | + | H, C | |||||
| LMX1B_-79.84 | ++ | + | + | ++ | + | |||
| LMX1B_12.07 | + | + | + | + | + | + | + | U |
| LMX1B_28.46 | + | + | + | + | + | + | L | |
| LMX1B_59.40 | + | + | + | + | + | + |
Telen, Telecephalon; Dien, Diencephalon; Mesen, Mesencephalon; Rhomben, Rhombencephalon; SpCo, Spinal Cord; Mb/Hb, Midbrain/Hindbrain; PNS, Peripheral Nervous System; H, Heart; C, Cartilage; OV, Otic Vesicle; L, Lens; Ub, Ubiquitous; NTC, Notochord, F, Fins, Pn, Pronephros; R, Retina, B, Blood.
Weak;
Moderate;
Strong expression (Relative determination)
Many identified LMX1A and LMX1B enhancers display regulatory control at the midbrain/hindbrain boundary and in the hindbrain
Multiple REs from both loci are able to drive expression in the midbrain-hindbrain boundary region that includes the IsO and anterior cerebellum (Figure 4, S2 and Table 1; e.g. LMX1A_-36.74, LMX1A_41.10, LMX1A_479.86, LMX1A_135.15 and LMX1B_-79.84). These data corroborate with the role of mammalian LMX1 genes in IsO and cerebellum development and function [8,9]. Additionally, many assayed sequences directed expression in the hindbrain (Figure 4, S2 and Table 1; e.g. LMX1A_475.57, LMX1A_238.85, LMX1A_5.34, LMX1A_-1.59 and LMX1B_28.46), including the roof plate (LMX1A_102.3) and the spinal cord (Figure S2 and Table 1; e.g. LMX1A_-1.59, LMX1A_238.85 and LMX1B_28.46) consistent with the endogenous expression of their corresponding zebrafish paralogous transcripts.
Figure 4.
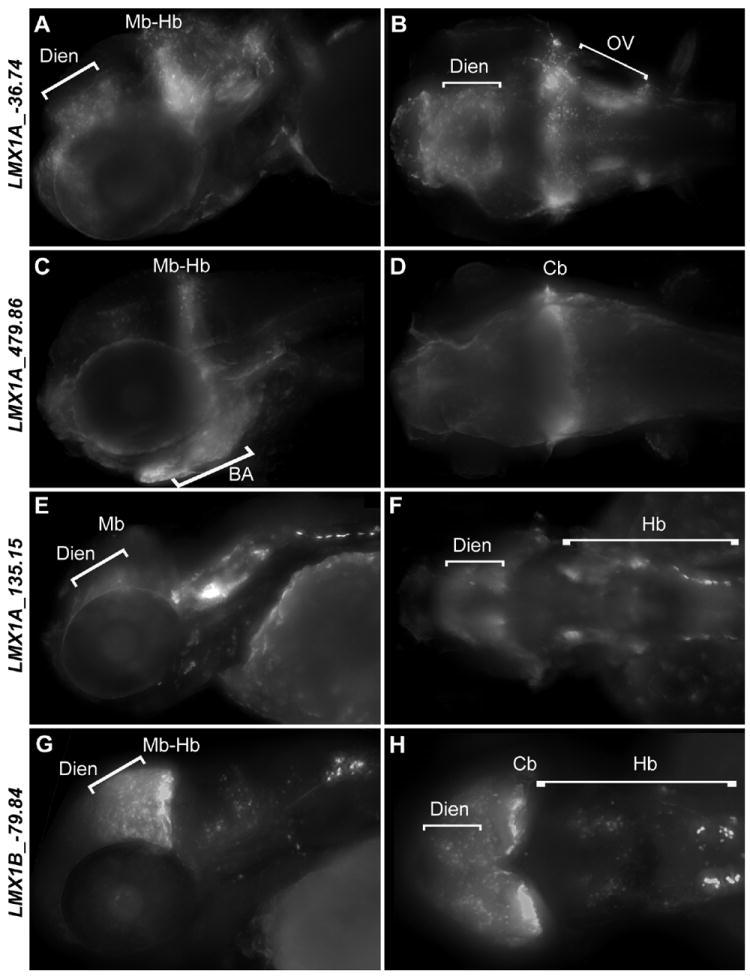
Expression of EGFP reporter in the hindbrain and midbrain-hindbrain boundary of 4 representative transgenic zebrafish lines. Zebrafish embryos were fixed at 72 hpf and stained with anti-GFP antibody. Anterior is shown to the left in each panel. A,C,E,G are lateral images. B,D,F,H are dorsal images. Abbreviations Cb – cerebellum, Dien – diencephalon, Hb – hindbrain, Mb – midbrain, Mb-Hb – midbrain-hindbrain boundary, OV – otic vesicle.
LMX1 enhancers display regulatory control in peripheral neuronal as well as non-neuronal cell populations
We identified several LMX1A and LMX1B sequences that direct expression in the otic vesicle (Figure 3 B, S2 and Table 1, e.g. LMX1A_-36.74, LMX1A_92.33 and LMX1A_117.21, LMX1B_-93.21), consistent with the expression of Lmx1a [14]. Furthermore, mice deficient in Lmx1a display abnormal ear development and deafness [13,28]. Multiple lines display reporter expression in PNS structures, like motor neurons (Figure S2 and Table 1; LMX1A_238.85) or sympathetic chain (Figure S2 and Table 1; e.g. LMX1A_475.57). In contrast to the largely neuronal roles of LMX1A and LMX1B, many identified enhancers also drive reporter expression in non-neuronal tissues such as the branchial arches, (Table 1, Figure 3 A-C, Figure S2) which corresponds to documented mouse expression [11,14]. One LMX1B enhancer also displays expression in the heart (LMX1B_-93.21) consistent with the endogenous expression of lmx1ba. The biological significance of this expression has not yet been determined but may, in part, correspond to the aortic arch neurons where expression of the catecholaminergic marker tyrosine hydroxylase has been previously reported [27].
Discussion
In order to better understand the regulatory landscape of LMX1A and LMX1B we undertook a functional study of conserved, noncoding sequences (putative REs) at these loci, using zebrafish transgenesis. We first established the identity of two zebrafish paralogs for each human LMX1 gene. We then demonstrate that their endogenous expression closely resembles the previously characterized expression of their mammalian counterparts, including expression in the areas of presumptive catecholaminergic neurons, cerebellum, raphe nuclei and otic vesicles. Next we used comparative sequence analyses to identify conserved, non-coding sequences at the human LMX1A and LMX1B loci successfully amplifying 71 putative REs for functional evaluation. Of these 45 directed CNS reporter expression in mosaic zebrafish embryos. We further described the reporter expression of 22 sequences in stable transgenic lines (LMX1A, n=17; LMX1B, n=5). All 22 display consistent CNS enhancer function (n≥2 independent founders) that overlaps, at least in part, with the endogenous transcripts. The majority of these sequences display enrichment for H3K4me1, a modification known to be enriched at enhancers [29], in cultured neurospheres [26], consistent with their neuronal activity in our synthetic assay, and providing evidence supporting their likely cis-regulatory role in their endogenous context.
The diencephalon, telencephalon and midbrain-hindbrain boundary were among the most common structures marked by reporter expression for REs identified at both loci (Figure 3, Table 1, Figure S2). Many enhancers similarly directed broad expression in the midbrain (Figure 3, Table 1, Figure S2) and more discrete expression in the hindbrain e.g. in single rhombomeres, area postrema (Figure 4, Table 1, Figure S2, LMX1B_-79.84) or hindbrain roof plate (Figure 3, Table 1, Figure S2, LMX1A_102.3). These sites of expression overlap known domains of Lmx1a and Lmx1b expression in mammals and teleosts.
The expression directed in the midbrain-hindbrain boundary, cerebellum and posterior rhombomeres is consistent with the important function of both LMX1A and LMX1B in the development of the cerebellum rhombic lip and hindbrain roof plate [4,7]. Furthermore, LMX1B is known to be instrumental for proper functioning of the IsO [8,9]. We speculate that the forebrain expression of LMX1A enhancers might reflect its role in early development of cortical hem in mammals.
Many sequences also direct expression in the PNS and some non-neuronal tissues, consistent with the endogenous expression of these TFs and their critical role in the differentiation and maintenance of a range of populations. The most common non-neuronal sites of reporter expression is in the otic vesicles, which is consistent with LMX1A/B biology [13,14,28]. We also see some enhancers driving expression in the heart that may correspond to peripheral neuronal populations [27].
Importantly, transgenic assays provide an approximation of how a regulatory sequence can behave in a model system and may not capture every nuance that the corresponding sequence may display in context. Furthermore their correspondence to spatial expression of lmx1 genes does not definitively demonstrate their control (exclusive or shared with neighbors) of these genes. In particular, we recognize that aspects of CNS regulatory control displayed by enhancers isolated at LMX1A and LMX1B may equally also be considered consistent with flanking gene expression. In particular expression of the zebrafish pbx1a paralog includes many domains that also show expression of lmx1a/lmx1a-like [39], including discrete expression in the telencephalon. Thus firm conclusions regarding enhancer driven reporter expression and their direct relation to LMX1-expressing neuronal populations or those of their flanking genes will require additional experimental determination of possible physical interaction between enhancer and one or more cognate promoter. Despite these caveats, these assays can and do provide significant insight into the biological relevance of assayed sequences.
When it comes to genome annotation, there is no satisfactory “one size fits all” approach. We demonstrate how a range of available data types may be integrated in the exploration of the genomic information content of sequence encompassing two critical human genes. This work describes the endogenous expression patterns of zebrafish LMX1A paralogs, identifies 22 previously unknown enhancers and sheds light on previously unknown transcriptional regulatory landscape at the LMX1A and LMX1B loci. If one accounts for the presence of additional conserved and/or histone-marked sequences in the genomic intervals under consideration, these enhancers may represent only a fraction of the conserved non-coding elements at these loci. We hypothesize that many enhancers may be required in combination to orchestrate regulatory control of these genes. The pleiotropy of neuronal subsets marked by these identified enhancers may highlight additional complexity in this regulatory control or reflect position effects. These data reinforce the value of targeted screens in the analysis of human disease loci integrating comparative sequence analysis, chromatin modifications and functional validation using zebrafish transgenesis in the identification of transcriptional regulatory sequences.
Methods
Fish maintenance
Zebrafish were kept and bred under standard conditions at 28.5°C [32]. Embryos were staged and fixed at 48 and 72 hpf using 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; pH 7.2) as described elsewhere [33]. To better visualize in situ hybridization and EGFP reporter results, embryos were grown in 0.2 mM 1-phenyl-2-thiourea (Sigma) to inhibit pigment formation [32].
Whole mount in situ hybridization
Digoxigenin labeled riboprobes complementary to lmx1a, lmx1a-like, lmx1ba or lmx1bb mRNAs were generated by linearization of pCR II TOPO TA vectors containing partial ORFs of the genes (for probe sequences see Figure S3). Plasmids were linearized with EcoRV (New England Biolabs) and subsequently labeled riboprobes were transcribed using SP6 polymerase and the DIG RNA Labeling Kit (T7/SP6) (Roche). Probes were synthesized for 2 hours at 37°C, followed by the addition of 1 μl of RNAse free DNAse I for DNA template digestion. Subsequently, probes were purified using SigmaSpin columns (Sigma-Aldrich). Whole mount in situ hybridization reactions were performed using 1:4000 dilutions of rioboprobes at 70°C as previously described [34, 35] – see http://zfin.org/zf_info/zfbook/chapt9/9.82.html for detailed protocol. Probe sequences were selected to avoid cross-hybridization with lmx1 family members and unrelated transcripts by using pairwise alignment of lmx1 transcripts to find unique stretches of mRNA. Sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). All probe sequences and corresponding oligonucleotides for their amplification are provided in supplemental data (Figure S4 and S5).
Selection and amplification of human non-coding sequences
To select regions to test for potential enhancer activity, genomic intervals encompassing LMX1A and LMX1B loci were considered up to the neighboring genes, set as boundaries, (LMX1A, chr1: 163,082,934 - 163,636,974 bp; LMX1B, chr9: 128,309,140 – 128,607,182 bp). This study is not intended to be exhaustive. The genomic intervals encompassing these genes are very large. Thus sequences were prioritized for selection based upon proximity to the LMX loci and conservation but no ranking of conservation was applied. Consequently this approach is likely to have identified only a subset of enhancers regulating LMX1A and LMX1B. Using Galaxy computational interface [36] and UCSC genome browser, we chose conserved non-coding vertebrate elements with positive GERP alignment scores [25]. The GERP algorithm identifies constrained sequences in genomic alignments by determining whether a paucity of substitutions exists at each point in an alignment compared to what one expects of the neutral rate of evolution. Selected intervals positioned less than 500 bp apart were merged into single amplicons. DNA region coordinates and primer sequences used for amplification are listed in table S2. Sequences were also selected to avoid clustering and are distributed across these loci (Figure 1A). Amplicons were PCR amplified from human genomic DNA, TA cloned into pCR8 (Invitrogen) and then cloned using the Gateway system (Invitrogen) into pGW_cfosEGFP as previously described [37,38].
Embryo injection and analysis
EGFP reporter constructs were injected into AB background G0 embryos (n≥200) at the one to two cell stage with tol2 transposase as previously described [37,38]. Injected embryos were evaluated for EGFP expression between 24 and 72 hpf. As negative controls EGFP reporter constructs containing only the cFos promoter were injected. Nonspecific expression from the cfos minimal promoter is occasionally observed in the myotome and no other nonspecific expression was detected (data not shown). Embryos showing consistent EGFP expression were selected and raised for further analysis when signal was observed in ≥10% of injected embryos. Mosaic fish were subsequently crossed to identify those constructs that passed through the germline transmission, better facilitating spatial evaluation of corresponding EGFP expression. Instances where we do not report expression reflect failure to identify more than one founder transgenic line and not inconsistencies among multiple lines for a single construct. Re-injection and additional screening may help resolve the neuronal regulatory control of additional constructs we have generated at these loci but whose activities we do not report upon here. Embryos were imaged using a Carl Zeiss Lumar V12 Stereo microscope with AxioVision software (version 4.5).
Immunocytochemistry
Embryos were anesthetized with tricaine (10 μg/ml) in embryo medium [32] and fixed in 4% PFA in phosphate-buffered saline (PBS; pH 7.2) for 2 hours. They were then rinsed four times in PBST (PBS/0.1% Triton X-100), incubated in Proteinase K for 1h at room temperature, washed 5 × 5 minutes in PBST, and incubated for 2 hours in blocking solution (10% goat serum, 1% bovine serum albumin (BSA), in PBST). Embryos were then incubated overnight at room temperature in primary antibody (anti-GFP, Invitrogen 1:2000), rinsed 6 × 45 minutes in PBST 1% goat serum, and incubated overnight at room temperature in secondary antibody (Alexa-Fluor, 488, Invitrogen 1:1000). They were then rinsed 5 × 10 minutes in PBST and transferred to 50% glycerol in PBS for imaging.
Supplementary Material
Phylogram showing relatedness between LMX1A, LMX1B and their zebrafish orthologous proteins.
EGFP expression patterns driven by all identified regulatory elements from LMX1A and LMX1B loci in zebrafish at 72 hpf. Each of the line is represented by 3 images (from the left): lateral, low magnification view; lateral high magnification of the head; high magnification dorsal view of the head. Anterior is shown to the left in each panel.
Alignment of sequences of in situ probes used in the study; Multiple Sequence Alignment by Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The sequence of each probe is highlighted.
Primer sequences used in the generation of RNA in situ probe fragments
lmx1 RNA in situ probe sequences in FASTA format
Genomic coordinates of tested elements and primer sequences used for amplification of all assayed sequences
Acknowledgments
The authors gratefully acknowledge the support of the McKusick Nathans Institute of Genetic Medicine Center for Functional Investigation in Zebrafish (FINZ). This research was supported in part by the National Institute of Neurological Disease and Stroke (R01 NS062972; NINDS, NIH) to ASM. XR was also supported by NIH pre-doctoral training grant 5T32GM07814. SM was supported by funds from the National Institute of Standards and Technology.
Certain commercial equipment, instruments, materials, or companies are identified in this paper to specify adequately the experimental procedure. Such identification does not imply recommendation nor endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are the best available for the purpose. *Official contribution of NIST; not subject to copyright.
Footnotes
Ethics
All zebrafish work was performed under an approved protocol (FI10M369), reviewed by the Johns Hopkins Institutional Animal Care and Use Committee.
References
- 1.Dai JX, Johnson RL, Ding YQ. Manifold functions of the Nail-Patella Syndrome gene Lmx1b in vertebrate development. Dev Growth Differ. 2009;51:241–250. doi: 10.1111/j.1440-169X.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- 2.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 3.Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 4.Mishima Y, Lindgren AG, Chizhikov VV, Johnson RL, Millen KJ. Overlapping function of Lmx1a and Lmx1b in anterior hindbrain roof plate formation and cerebellar growth. J Neurosci. 2009;29:11377–11384. doi: 10.1523/JNEUROSCI.0969-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatani T, Kumai M, Mizuhara E, Minaki Y, Ono Y. Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol. 2010;339:101–113. doi: 10.1016/j.ydbio.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Yan CH, Levesque M, Claxton S, Johnson RL, Ang SL. Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci. 2011;31:12413–12425. doi: 10.1523/JNEUROSCI.1077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chizhikov VV, Lindgren AG, Mishima Y, Roberts RW, Aldinger KA, et al. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc Natl Acad Sci U S A. 2010;107:10725–10730. doi: 10.1073/pnas.0910786107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams KA, Maida JM, Golden JA, Riddle RD. The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development. 2000;127:1857–1867. doi: 10.1242/dev.127.9.1857. [DOI] [PubMed] [Google Scholar]
- 9.Guo C, Qiu HY, Huang Y, Chen H, Yang RQ, et al. Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development. 2007;134:317–325. doi: 10.1242/dev.02745. [DOI] [PubMed] [Google Scholar]
- 10.Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, et al. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Mao T, Dai Y. Experimental study of artificial bone composite of bicoral, rhBMP-2 and PLA in repairing calvarial defects. Hua Xi Kou Qiang Yi Xue Za Zhi. 2003;21:474–476. [PubMed] [Google Scholar]
- 12.Vollrath D, Jaramillo-Babb VL, Clough MV, McIntosh I, Scott KM, et al. Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail-patella syndrome. Hum Mol Genet. 1998;7:1091–1098. doi: 10.1093/hmg/7.7.1091. [DOI] [PubMed] [Google Scholar]
- 13.Millonig JH, Millen KJ, Hatten ME. The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature. 2000;403:764–769. doi: 10.1038/35001573. [DOI] [PubMed] [Google Scholar]
- 14.Failli V, Bachy I, Retaux S. Expression of the LIM-homeodomain gene Lmx1a (dreher) during development of the mouse nervous system. Mech Dev. 2002;118:225–228. doi: 10.1016/s0925-4773(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 15.Noonan JP, McCallion AS. Genomics of Long-Range Regulatory Elements. In: Chakravarti A, Green ED, editors. Annual Review of Genomics and Human Genetics: Annual Reviews. 2011. [DOI] [PubMed] [Google Scholar]
- 16.Kleinjan DJ, Coutinho P. Cis-ruption mechanisms: disruption of cis-regulatory control as a cause of human genetic disease. Brief Funct Genomic Proteomic. 2009;8:317–332. doi: 10.1093/bfgp/elp022. [DOI] [PubMed] [Google Scholar]
- 17.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amores A, Force A, Yan YL, Joly L, Amemiya C, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 20.O’Hara FP, Beck E, Barr LK, Wong LL, Kessler DS, Riddle RD. Zebrafish Lmx1b.1 and Lmx1b.2 are required for maintenance of the isthmic organizer. Development. 2005;132(14):3163–3173. doi: 10.1242/dev.01898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng CW, Yan CH, Choy SW, Hui MN, Hui CC, Cheng SH. Zebrafish homologue irx1a is required for the differentiation of serotonergic neurons. Dev Dyn. 2007;236(9):2661–2667. doi: 10.1002/dvdy.21272. [DOI] [PubMed] [Google Scholar]
- 22.Elsen GE, Choi LY, Millen KJ, Grinblat Y, Prince VE. Zic1 and Zic4 regulate zebrafish roof plate specification and hindbrain ventricle morphogenesis. Dev Biol. 2008;314(2):376–392. doi: 10.1016/j.ydbio.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMahon C, Gestri G, Wilson SW, Link BA. Lmx1b is essential for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish. Dev Biol. 2009;332:287–298. doi: 10.1016/j.ydbio.2009.05.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filippi A, Mahler J, Schweitzer J, Driever W. Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. J Comp Neurol. 2010;518:423–438. doi: 10.1002/cne.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen L, Wei W, Gu W, Huang P, Ren X, Zhang Z, Zhu Z, Lin S, Zhang B. Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev Biol. 2008;314(1):84–92. doi: 10.1016/j.ydbio.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Huang M, Sage C, Li H, Xiang M, Heller S, et al. Diverse expression patterns of LIM-homeodomain transcription factors (LIM-HDs) in mammalian inner ear development. Dev Dyn. 2008;237:3305–3312. doi: 10.1002/dvdy.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 30.Koo SK, Hill JK, Hwang CH, Lin ZS, Millen KJ, et al. Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev Biol. 2009;333:14–25. doi: 10.1016/j.ydbio.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichols DH, Pauley S, Jahan I, Beisel KW, Millen KJ, et al. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res. 2008;334:339–358. doi: 10.1007/s00441-008-0709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westerfield M. The Zebrafish Book: A Guide for the Laboratory USe of Zebrafish (Danio rerio) Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- 33.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 34.Thisse C, Degrave A, Kryukov GV, Gladyshev VN, Obrecht-Pflumio S, et al. Spatial and temporal expression patterns of selenoprotein genes during embryogenesis in zebrafish. Gene Expr Patterns. 2003;3:525–532. doi: 10.1016/s1567-133x(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 35.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 36.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 38.Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 39.Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 ( http://zfin.org)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogram showing relatedness between LMX1A, LMX1B and their zebrafish orthologous proteins.
EGFP expression patterns driven by all identified regulatory elements from LMX1A and LMX1B loci in zebrafish at 72 hpf. Each of the line is represented by 3 images (from the left): lateral, low magnification view; lateral high magnification of the head; high magnification dorsal view of the head. Anterior is shown to the left in each panel.
Alignment of sequences of in situ probes used in the study; Multiple Sequence Alignment by Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The sequence of each probe is highlighted.
Primer sequences used in the generation of RNA in situ probe fragments
lmx1 RNA in situ probe sequences in FASTA format
Genomic coordinates of tested elements and primer sequences used for amplification of all assayed sequences


