Polycyclic aromatic hydrocarbons (PAH) are ubiquitous environmental carcinogens (Connell et al. 1997; Conney, 1982; Dipple, 1985; Lesko, 1984). They are produced during forest fire, volcanic eruption, incomplete burning of fuel and other materials, tobacco smoke, and food processing (Baum, 1978; Connell et al. 1997). Exposure to PAHs has been linked to the development of skin and lung cancers as it is summarized in the 8th Report on Carcinogens (National Toxicology Program, 1998). PAHs are considered relatively nontoxic themselves, but they can be activated after entering the cell. The first activation pathway is metabolism. PAH metabolic products, diol-epoxides or diones, are known to be carcinogenic through DNA covalent product formation (Connell et al. 1997; Conney, 1982; Devanesan et al. 1996; Dipple, 1985; Lesko, 1984). The diol-epoxides can alkylate DNA, usually form a bond to the exocyclic amino group of the guanine residue in duplex DNA (Geacintov et al. 1997). The diones are able to oxidatively damage DNA by generation of free radicals, which either cause DNA damages or form DNA covalent products (Chen et al. 1996; Devanesan et al. 1996).
Another pathway that enhances PAH toxicity is light activation. There have been studies on the photo-induced toxicity of PAH mixtures or individuals in the marine sediment toward micro-organisms and plants in the aquatic systems (Pelletier et al. 1997; Swartz et al. 1997). It is found that PAHs are generally more toxic when the system is exposed to the simulated solar radiation (ssr) than if it is kept in the dark. The increase in toxicity due to ssr can exceed 100 times (Swartz et al., 1997). It is suggested that PAHs act as photosensitizers (Pelletier et al. 1997). After absorbing UV light energy, PAHs in the excited-state may transfer its energy to molecular oxygen to produce reactive oxygen species that can cause a variety of damages to the cell. The phototoxicity can also be due to DNA covalent product formation from earlier studies. These works showed that, under light irradiation, benzo[a]pyrene can form DNA covalent adducts or cause DNA strand breakage (Blackburn et al. 1977; Brooks and Lawley, 1964; Hoard et al. 1981; Santamaria et al. 1966; Striste et al. 1980). The presence of benzo[a]pyrene can also increase the formation of 8-hydroxy-2′-deoxyguanine (Liu et al., 1998), a compound generated by oxidative damage of DNA. These DNA damage mechanisms have been suggested to relate to tumor induction and other adverse effects (Brooks and Lawley, 1964; Camalier et al. 1981; Santamaria et al. 1966). However, the studies so far have mostly focused on benzo[a]pyrene alone. In this research, we will examine light-induced DNA cleavage by the environmental contaminants: a series of 3, 4, 5-ring PAHs and their derivatives.
METERIAL AND METHODS
Polycyclic Aromatic Hydrocarbons (PAHs, structures are listed in Scheme 1) were purchased from Aldrich Chemical Co. (Milwaukee, WI). All of these compounds were in their highest purity grade and were used without further purification. UV-Vis spectra of these compounds were recorded on a Varian CARY 300E UV-Vis Spectrophotometer to calculate the extinction coefficient for each individual molecule. The extinction coefficients were used to determine the concentration of the PAHs. PAH stock solutions, generally 1 mM, were prepared in DMSO and stored with brown container in the refrigerator to exclude light. ΦX-174 Phage DNA (80–90% supercoiled RF-1 DNA) with a molecular weight of 3.6 × 106 Da and approximately 5386 base pairs (bp) was purchased from Sigma Chemical Co. (St. Louis, MO). Agarose, sodium dihydrogen-phosphate, TRIS base, boric acid, and EDTA were from Fisher Scientific. All solvents used were spectroscopic grade. The water used (18Ω) for buffer preparation was deionized by a Barnstead Nanopure Infinity water deionization system.
SCHEME 1.
Structure and abreviated names of PAHs used in this study
UVA-light induced plasmid DNA cleavage by PAHs were done as following: Solutions (a total of 60 μL for each sample) containing ΦX174 Phage DNA and various amounts of PAHs were filled into the wells of a 3×8 well flat-bottomed Titertek™ plate (ICN Biochemicals). The DNA concentration was about 27 μM in base pairs in all experiments. The solution was buffered with 10 mM sodium phosphate at pH 7.1 and contained 4% dimetheylformamide (DMF) for better PAH solubility. Other solvent systems such as 10% methanol or 20% DMF, 5% DMSO were also tried. We found the 4% DMF delivered the best results. The Titertek plate was tightly covered with a piece of glass and placed onto a Pyrex glass support, which is placed on an O-ring secured on a ring stand. The Pyrex glass served as a light filter to efficiently cut off any light below 300 nm that can potentially cleave DNA (when the Pyrex glass was checked on the Varian CARY 300 UV-Vis spectrometer, over 99% of light below 300 nm are filtered off). A 100 W UV lamp (type B, UVP Inc., Upland, CA) was placed beneath the Pyrex glass and the light was applied through the bottom of the Titertek plate from a fixed distance of 6.5 cm. The intensity of the light output was 170 J/cm2 measured with a UVA detector (Model PMA 2100, Solar Light Co., Inc., Philadelphia, PA). A stream of cold air blowing through the bottom of the Pyrex glass was used to eliminate any heat generated by the light source.
After irradiation for one hour, each well was added with 10 μL of a gel-loading dye solution (bromophenol blue and xylencyanol in 50% glycerol). Then 10 μL of the sample was loaded into the wells of a 1% agarose gel. The gel was run in 1× TBE buffer (pH=8.27) at 105 volts for 70–90 min at room temperature. Following electrophoresis, the gel was stained with ethidium bromide (2 mg/L) for 30 min and analyzed by a NucleoVision Gel-Documentation System (NucleoTech Inc., CA). The amount of the supercoiled Form I DNA (sc-DNA) and the relaxed open circular Form II DNA (oc-DNA) were quantified by the total fluorescence intensity of the bands after subtracting a common background. A coefficient of 1.66 was used to correct the lower efficiency of ethidium bromide binding to the sc-DNA than to the oc-DNA (Ciulla et al. 1989). The fraction of sc-DNA, Xsc, was calculated by equation 1.
| (1) |
where Asc and Aoc are the total fluorescence intensities for the sc-DNA and oc-DNA bands, respectively. Then the percent of sc-DNA cleavage or percent of sc-DNA converted into oc-DNA is calculated by equation 2.
| (2) |
where Xsco is the initial percentage of the sc-DNA before cleavage. Both the Xsc and Xsco are calculated from equation 1. A plot of the percent of sc-DNA cleavage versus PAH concentration was used to figure out C25, the PAH concentration that causes 25% sc-DNA cleavage. The C25 values are used for comparing the potency of each PAH in terms of causing light-induced DNA cleavage. The concentration range was carefully chosen for each individual PAH based on preliminary experiments done in a much larger concentration range. These experiments served as the first step to figure out an approximate C25. Then an experiment with the concentration of a PAH around the approximate C25 was be done to determine a more accurate C25 value. All experiments are a triplicate. The reason we chose C25 instead of the more traditional C50 is: sc-DNA needs only a single cut to be converted into oc-DNA. As the concentration of the oc-DNA increases due to this conversion, oc-DNA will also be cut by PAH, this will definitely leads to errors in determining the effective concentration of the PAH that only cuts the sc-DNA.
Time course of light-induced ΦX-174 phage DNA cleavage was done in a quartz cuvette containing the DNA and a PAH sitting on the Pyrex glass. The light source was set the same way as described above. Samples were taken after a period of irradiation and kept immediately in the dark. After all samples were taken, they were treated and subjected to electrophoresis and quantified as described above.
RESULTS AND DISCUSSION
UVA-light induced DNA cleavage by PAHs is both PAH concentration and irradiation time dependent. Figure 1 shows the UVA dose dependence of the DNA cleavage induced by 6 μM 1-AP. The bottom band is the sc-DNA and the top band is the oc-DNA. Below the picture of the gel is the quantified band intensity for the oc-DNA. Lane 1 is the sample taken just before irradiation and it represents the original constitution of the DNA with the sc-DNA versus oc-DNA ratio of 80% to 20%. As irradiation time increases, the amount of sc-DNA decreases as the amount of the oc-DNA increases, signifying that the sc-DNA was converted to the oc-DNA. This conversion must be due to DNA single stranded cleavage caused by 1-AP and light because it has been known that a knick or a single strand cut of the sc-DNA converts the sc-DNA to oc-DNA The absence of a band for the linear form-III DNA suggests that the DNA cleavage is a single, not a double stranded cut. The linear DNA band would have appeared between the sc-DNA and the oc-DNA. Other PAHs tested also had the similar irradiation time dependence.
Figure 1.
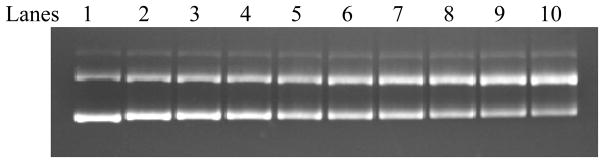
Irradiation time dependent DNA cleavage by 1-AP induced by UVA light. The ΦX 174 DNA and 1-AP concentrations are 27 μM and 6 μM in all samples, respectively. Lane 1 is the control sample before irradiation. Lanes 2–10 are after 2, 4, 6, 10, 15, 20, 30, 40, 60 min of irradiation by the 100 W UVA lamp.
The PAH concentration-dependence of DNA cleavage was tested by mixing the phage DNA with various amounts of PAHs in the buffer with 4% DMF. Figure 2 shows the gel for the ΦX-174 Phage DNA treated with various concentrations of 1-AP after 1 hr of irradiation. The bands representing the oc-DNA were quantified and shown as a bar graph below the gel. Control experiments show that the percent of sc-DNA remains about the same as the original DNA’s 80% whether the DNA is irradiated by light in the absence of 1-AP (lane 1) or kept in the dark in the presence of 8 μM 1-AP (lane 2). As the concentration of added 1-AP increases from 2–8 μM in lanes 3–8, the amount of sc-DNA decreases and the amount of oc-DNA increases due to single stranded cleavages caused by 1-AP and light. All other PAHs tested here also have the same concentration dependence.
Figure 2.
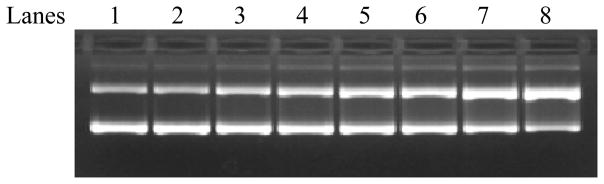
1-AP concentration DNA cleavage after 1 hr irradiation using a 100 W UVA lamp. The ΦX 174 DNA concentration is 27 μM in all samples. Controls: lane 1: no 1-AP but under light; lane 2: 8 μM 1-AP + DNA, no light; lanes 3–8 are with 2, 3, 4, 5, 6, 8 μM and light.
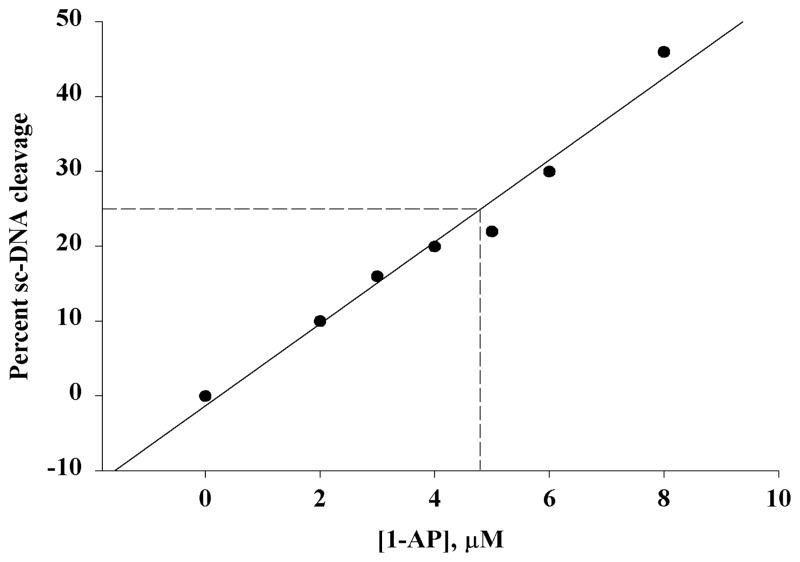
The relative DNA photocleavage efficiency expressed as C25, the PAH concentration at which 25% of the original sc-DNA was converted into oc-DNA, was determined by plotting the percent of sc-DNA cleavage versus the PAH concentration as shown in Figure 3 for 1-AP. All PAHs examined were able to cause light-induced DNA cleavage and generate a linear graph as shown for 1-AP. The C25 was obtained directly from the graph by determining the concentration of the PAH that corresponds to a 25% sc-DNA cleavage. Table 1 lists the C25 data for the PAHs examined in three solvent systems: 10 mM phosphate buffers with 4% DMF, 10% MeOH and 20% DMF.
Figure 3.
Determination of C25. The concentration at 25% cleavage (C25) is determined on the graph as shown.
Table 1.
Relative DNA photocleavage efficiency of PAH expressed by C25 (μM)
| PAHs | 4% DMF | 10% MeOH | 20% DMF |
|---|---|---|---|
| Benzo[a]pyrene | 6.0 | 4.9 | |
| Pyrene | 51 | ||
| 1-Aminopyrene | 4.2 | 1.9 | 4.4 |
| 1-Hydroxypyrene | 0.36 | 0.38 | 4.8 |
| 1-Nitropyrene | 9.9 | na | |
| 1-Bromopyrene | 4.0 | 11 | |
| DHBP | 15 | 19 | |
| Chrysene | 5.2 | 4.1 | |
| 6-Aminochrysene | 60 | 4.2 | |
| 1,4-Chrysenequinone | 5.8 | 10 | |
| 3-Aminofluoranthene | 32 | 4.8 | |
| Anthracene | 8.3 | ||
| 9-Nitro-anthracene | 0.83 | ||
| 2-Hydroxycarbazole | na | 57 |
Comparing the C25 values for the PAHs in different solvents, it is obvious that solvents could have a profound effect on the light-induced DNA cleavage by PAHs. For example, the C25 values for the amino substituted PAHs, 1-AP, 3-AFA, and 6-AC are lowered by 2–14 times going from 4% DMF to 10% methanol as solvent, while it increases 13 fold for 1-HOP going from 4% DMF or 10% MeOH to 20% DMF. The effect by solvent on the C25 is different for different PAHs. While the effect on the C25 is large for the amino substituted PAHs going from 4% DMF to 10% methanol as solvent, the C25 values for 1-HOP, chrysene and 1,4-CQ are only slightly affected. While the C25 value is increaed 13 folds for 1-HOP going from 4% DMF or 10% MeOH to 20% DMF, it only increases slightly for other PAHs. This difference in solvent effects on the light-induced DNA cleavage efficiency might be due to different mechanisms that cause the DNA cleavage by individual PAHs. Mechanistic study on the light-induced DNA cleavage will be published elsewhere (Dong, et. el. 1999).
If all compounds tested are compared within a solvent system (4% DMF), 1-HOP and 9-NA are the two most potent compounds toward light-induced DNA cleavage with C25 values of 0.36 and 0.83 μM, respectively. Eight of the 13 tested compounds belong to the next group that have C25 values of 4.0–15 μM, one order of magnitude higher than 1-HOP and 9-NA. The C25 values for pyrene, 6-AC, and 3-AFA are another order of magnitude higher, while the C25 for HOCZ is too high to be determined.
It is suggested that the phototoxicity of PAHs is due to light-induced generation of reactive oxygen species that further damage biomolecules (Newsted and Giesy, 1987; Pelletier et al., 1997; Swartz et al., 1997). The light-induced DNA cleavage results for the 14 tested PAHs here suggest that one possible phototoxicity source for these environmental contaminants is to cause DNA cleavage. However, acute and genotoxicity data for these PAHs are needed to understand whether the phototoxicity of these compounds relates to their light-induced DNA cleavage. Since the damage is to DNA, it is possible that the light-induced effect of PAHs is genotoxic, in addition to being acute toxic. While there are some phototoxicity data for the general PAHs (Pelletier et al., 1997; Swartz et al., 1997; Takeda et al., 1984), there are not much data available for the phototoxicity of the substituted PAHs.
Acknowledgments
The authors wish to thank the National Institutes of Health for the generous financial supports through grants: NIH-RCMI 1G12RR12459-01 and NIH-MBRS SO6GM08047.
References
- Baum E. Gelboin & Ts’O, editor. Polycyclic aromatic hydrocarbons and cancer. Vol. 1. Academic Press; New York: 1978. pp. 45–70. [Google Scholar]
- Blackburn GM, Fenwick RG, Lockwood G, Williams GM. Photoproducts from DNA pyrimidine bases and polycyclic aromatic hydrocarbons. Nucleic Acids Res. 1977;4:2487–2494. doi: 10.1093/nar/4.7.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P, Lawley PD. Evidence for the binding of polynuclear aromatic hydrocarbons to the nucleic acids of mouse skin: Relation between carcinogenic power of hydrocarbons and their binding to deoxyribonucleic acid. Nature. 1964;202:781–784. doi: 10.1038/202781a0. [DOI] [PubMed] [Google Scholar]
- Camalier R, Gantt R, Price F, Stephens E, Baeck A, Taylor W, Sanford K. Effect of visible light on benzo(a)pyrene binding to DNA of cultured human skin epithelial cells. Cancer Res. 1981;41:1789–1793. [PubMed] [Google Scholar]
- Chen L, Devanesan PD, Higginbotham S, Ariese F, Jankowiak R, Small GJ, Rogan EG, Cavalieri EL. Expanded analysis of benzo[a]pyrene-DNA adducts formed in vitro and in mouse skin: Their significance in tumor initiation. Chem Res Toxicol. 1996;9:897–903. doi: 10.1021/tx960004a. [DOI] [PubMed] [Google Scholar]
- Ciulla T, Van Camp J, Rosenfeld E, Kochevar I. Photosensitization of single strand breaks in pBR322 DNA by rose bengal. Photochem Photobiol. 1989;49:293–298. doi: 10.1111/j.1751-1097.1989.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Connell DW, Hawker DW, Warne MJ, Vowles PP. Polycyclic aromatic hydrocarbons (PAHs) In: McCombs Ken, Starkweather Albert W., editors. Introduction into Environmental Chemistry. CRC Press LLC; Boca Raton, FL: 1997. pp. 205–217. [Google Scholar]
- Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- Devanesan PD, Higginbotham S, Ariese F, Jankoviak R, Suh M, Small GJ, Cavalieri EL, Rogan EG. Depurinating and stable benzo[a]pyrene-DNA adducts formed in isolated rat liver nuclei. Chem Res Toxicol. 1996;9:1113–1116. doi: 10.1021/tx9600513. [DOI] [PubMed] [Google Scholar]
- Dipple A. In: Polycyclic aromatic hydrocarbons and carcinogenesis. Harvey RG, editor. Ameraican Chemical Society; Washington, DC: 1985. pp. 1–17. [Google Scholar]
- Dong S, Hwang HM, Shi X, Holloway L, Yu H. UVA Light-induced DNA single stranded cleavage and DNA covalent adduct formation by 1-hydroxypyrene. 1999 doi: 10.1021/tx990199x. to be submitted to Photochem Photobiol. [DOI] [PubMed] [Google Scholar]
- Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structure of stereoisomeric covalent polycyclic aromatic carcinogen-DNA adducts: Principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- Hoard DE, Ratliff RL, Bingham JM, Strniste GF. Reaction induced in vitro between model DNA and benzo[a]pyrene by ultraviolet radiation. Chem-Biol Interact. 1981;33:179–194. doi: 10.1016/0009-2797(81)90039-9. [DOI] [PubMed] [Google Scholar]
- Lesko SA. Chemical Carcinogenesis: Benzopyrene System. Methods in Enzymology. 1984;105:539–550. doi: 10.1016/s0076-6879(84)05074-6. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lu Y, Rosenstein B, Lebwohl M, Wei H. Benzo[a]pyrene enhances the formation of 8-hydroxy-2′-deoxyguanosine by ultraviolet A radiation in calf thymus DNA and human epidermoid carcinoma. Biochemistry. 1998;37:10307–10312. doi: 10.1021/bi980606o. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program, Public Health Service, US Department of Health and Human Services. 8th Report on Carcinogens. Integrated Laboratory Systems, Inc; Research Triangle Park, NC: 1998. pp. 178–181. [Google Scholar]
- Newsted JL, Giesy JP. Predictive models for photoinduced acute toxicity of polycyclic aromatic hydrocarbons to Daphnia magna, Strauss (Cladocera, Crustacea) Environ Toxicol Chem. 1987;6:445–461. [Google Scholar]
- Pelletier MC, Burgess RM, Ho KT, Kuhn A, McKinney RA, Ryba SA. Photoxicity of individual polycyclic aromatic hydrocarbons and petroleum to marine invertebrae lavae and juveniles. Envin Toxicol Chem. 1997;16:2190–2199. [Google Scholar]
- Santamaria L, Giordano GG, Alfisi M, Cascione F. Effects of light on 3,4-benzpyrene carcinogenesis. Nature. 1966;210:824–825. doi: 10.1038/210824a0. [DOI] [PubMed] [Google Scholar]
- Striste GF, Martinez E, Martinez AM, Brake RJ. Photo-induced reactions of benzo[a]pyrene with DNA in vitro. Cancer Res. 1980;40:245–252. [PubMed] [Google Scholar]
- Swartz RC, Ferraro SP, Lamberson JO, Cole FA, Ozretich RJ, Boese BL, Schults DW, Behrenfeld M, Ankley GT. Photoactivation and toxicity of mixtures of polycyclic aromatic hydrocarbon compounds in marine sediment. Environ Toxicol Chem. 1997;16:2151–2157. [Google Scholar]
- Takeda N, Teranishi K, Hamada K. Mutagenicity of the sunlight-exposed sample of pyrene in Salmonella typhimurium TA98. Bull Envirn Contam Toxicol. 1984;33:410–417. doi: 10.1007/BF01625563. [DOI] [PubMed] [Google Scholar]