Abstract
eEF2K is a kinase that controls the rate of peptide chain elongation by phosphorylating eukaryotic Elongation Factor 2 (eEF2), the protein that mediates the movement of the ribosome along the mRNA by promoting translocation from the A to the P site. eEF2K-mediated phosphorylation of eEF2 on Thr56 decreases its affinity for the ribosome, thereby inhibiting elongation. Here we show that in response to genotoxic stress, eEF2K is activated by AMPK-mediated phosphorylation on Ser398. Activated eEF2K phosphorylates eEF2 and induces a temporary ribosomal slowdown at the stage of elongation. Subsequently, during checkpoint silencing, eEF2K is degraded by the ubiquitin-proteasome system via the SCFβTrCP ubiquitin ligase to allow rapid resumption of translation elongation. This event requires eEF2K autophosphorylation on a canonical βTrCP-binding domain. The inability to degrade eEF2K during checkpoint silencing caused sustained phosphorylation of eEF2 on Thr56 and delayed resumption of translation elongation. Our study establishes an important link between DNA damage signaling and translation elongation.
INTRODUCTION
Cells activate surveillance molecular networks known as DNA damage checkpoints to protect their genome from environmental and metabolic genotoxic stress. Depending on the type and extent of DNA lesions and the cellular context, damaged cells with an activated checkpoint can undergo senescence, die by apoptotic cell death, or repair the damaged genome and, following checkpoint termination, resume their physiological functions (1, 2).
Recent findings have shown that, upon genotoxic stress, gene expression is affected much more dramatically at the level of mRNA translation than at the level of transcription (3). This may be due to the fact that protein synthesis requires approximately 40% of the total cellular energy and cells need to couple stress response to their metabolic demands (4). Indeed, it is conceivable that in response to genotoxic stress, cells aim to preserve energy by reducing protein synthesis in order to be able to repair the damage.
Protein synthesis is controlled by the mTOR (mammalian Target of Rapamycin) pathway, a crucial integrator of growth and stress signals. Numerous studies have shown that mTOR regulates several critical components involved in both translation initiation and elongation. The p70S6 kinase (S6K) and eIF4E binding protein 1 (4E-BP1), regulators of translation initiation, are among the best-characterized targets of mTOR (5, 6). In addition, mTOR controls translation elongation by negatively regulating eEF2K, which in turn phosphorylates and inactivates eEF2, a factor that mediates the translocation step of peptide-chain elongation (7-13). eEF2K-mediated phosphorylation of eEF2 on Thr56 reduces the affinity of eEF2 for the ribosome, thereby inhibiting its function (13-23). The activity of eEF2K is controlled under a range of conditions, suggesting that eEF2K is a crucial regulator of translation elongation. Stimuli that induce protein synthesis trigger the inactivation of eEF2K and the subsequent dephosphorylation of eEF2 (24). In contrast, nutrient- and energy-deficiency lead to activation of eEF2K and impairment of translation elongation.
In spite of the major impact of genotoxic stress on mRNA translation, no information is available on how translation elongation is affected by genotoxic stress and, more generally, there have been surprisingly few studies directed to understanding the regulation of protein synthesis by the DNA damage response. It has been shown that DNA damage inhibits the mTOR-S6K axis via p53, a key sensor of genotoxic stress, leading to decreased protein synthesis (25, 26). Moreover, TSC2, a crucial negative regulator of mTOR, has been reported to be induced by p53 (26).
Numerous studies have uncovered fundamental functions of the ubiquitin-proteasome system in the DNA damage response (1, 2, 27). This system involves two discrete and sequential processes: the tagging of substrates by covalent attachment of multiple ubiquitin molecules, and the degradation of poly-ubiquitylated proteins by the 26S proteasome (28). Ubiquitin is transferred and covalently attached to substrates through an enzymatic cascade involving ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3). E3 ubiquitin ligases represent the essential regulators of ubiquitylation because they physically interact with target substrates, linking them to E2 ubiquitin-conjugating enzymes.
SCFβTrCP is a multi-subunit RING-finger type ubiquitin ligase composed of a cullin scaffold, Cul1, which simultaneously interacts with the RING subunit Rbx1 and the adaptor protein Skp1 (29-32). Skp1 in turn binds the F-box protein βTrCP (β-transducin repeat-containing protein), the substrate receptor subunit that recruits specific substrate proteins. Via its WD40 β-propeller structure, βTrCP recognizes a di-phosphorylated motif with the consensus DpSGXX(X)pS in which the serine residues are phosphorylated by specific kinases to allow interaction with βTrCP.
Here we show that upon genotoxic stress, eEF2K is first activated by AMPK-mediated phosphorylation to induce a temporary translational slowdown at the stage of elongation and then, during checkpoint silencing, is targeted for proteasome-dependent degradation by the SCFβTrCP ubiquitin ligase to allow rapid and efficient resumption of translation. These findings establish an important link between DNA damage response and translation of messenger RNAs.
RESULTS
eEF2K specifically interact with βTrCP1 and βTrCP2
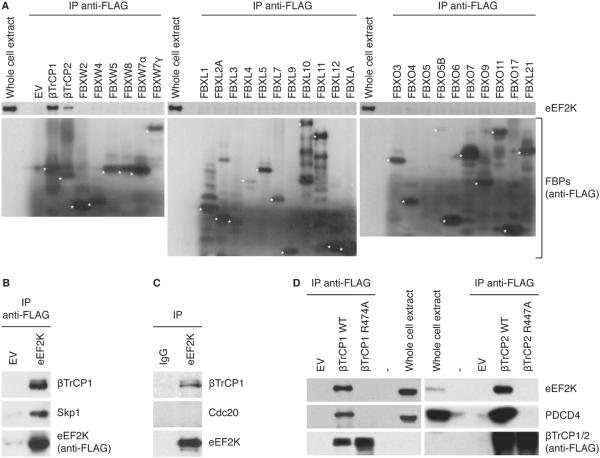
To identify substrates of the SCFβTrCP ubiquitin ligase, we employed immunoaffinity chromatography followed by tandem mass spectrometry. We expressed FLAG-HA epitope-tagged βTrCP2 in HEK293T cells and analyzed by mass spectrometry proteins that co-purify with FLAG-HA-βTrCP2 after sequential FLAG and HA immunoprecipitations. We unambiguously and specifically identified eEF2K, recording 26 spectra corresponding to 9 unique peptides (Figure S1A). To validate the interaction between βTrCP and eEF2K and further test its specificity, we expressed FLAG-tagged versions of 29 F-box proteins in HEK293T cells. After FLAG immunoprecipitation, we examined their binding to endogenous eEF2K. βTrCP1 and βTrCP2 were the only F-box proteins interacting with eEF2K (Figure 1A and S1B). βTrCP1 and βTrCP2 are two F-box proteins sharing identical biochemical properties and substrates; therefore, the term βTrCP will hence refer to both, unless specified otherwise. The βTrCP-eEF2K interaction was not restricted to HEK293T cells, as it was detected also in other cell types (Figures S1C). Moreover, when we followed the reciprocal approach and immunopurified eEF2K expressed in HEK293T cells, we recovered peptides corresponding to βTrCP1 (1 unique peptide), βTrCP2 (4 unique peptides) and the SCF adaptor Skp1 (1 unique peptide; Figure S1D-F). The ability of eEF2K to immunoprecipitate the βTrCP1-Skp1 complex was confirmed by immunoprecipitation, followed by immunoblotting (Figure 1B). The interaction of endogenous βTrCP1 and eEF2K was also observed (Figure 1C).
Figure 1. eEF2K specifically associates with βTrCP1 and βTrCP2 in vivo.
(A) HEK293T cells were transfected with the indicated FLAG-tagged F-box proteins (FBPs) or an empty vector (EV). Forty-eight hours after transfection, cells were treated with the proteasome inhibitor MG132, then harvested and lysed. Cell extracts were immunoprecipitated (IP) with anti-FLAG resin, and immunoblotted with the indicated antibodies. (B) HEK293T cells were transfected with FLAG-tagged eEF2K or an empty vector and treated as in (A). (C) Endogenously expressed eEF2K and βTrCP1 interacts in vivo. HEK293T cells were treated with MG132 during the last five hours prior to lysis. Lysates were immunoprecipitated with either an affinity purified polyclonal antibody against eEF2K or affinity purifed IgG and then analyzed by immunoblotting with antibodies to the indicated proteins. (D) The association of βTrCP1 (left panels) and βTrCP2 (right panels) with eEF2K requires the presence of Arg474 in the WD40 repeat of βTrCP1 and Arg447 in the WD40 repeat of βTrCP2. HEK293T cells were transfected as indicated and then treated as in (A). Data are representative of at least three independent experiments.
To obtain insight about the nature of the βTrCP-eEF2K binding, we mutated the arginine residue (Arg474 of human βTrCP1, isoform 2) in the WD40 β-propeller structure of βTrCP1 that interacts with the substrate destruction motif (32) and analyzed its ability to bind eEF2K. Whereas wild type βTrCP1 immunoprecipitated eEF2K and the established βTrCP1 substrate PDCD4, the βTrCP1(R474A) mutant did not (Figure 1D, left panels). Similar results were obtained when we mutated the equivalent arginine residue in βTrCP2 (Arg447 of human βTrCP2, isoform C; Figure 1D right panels).
eEF2K interaction with βTrCP requires a conserved phospho-degron
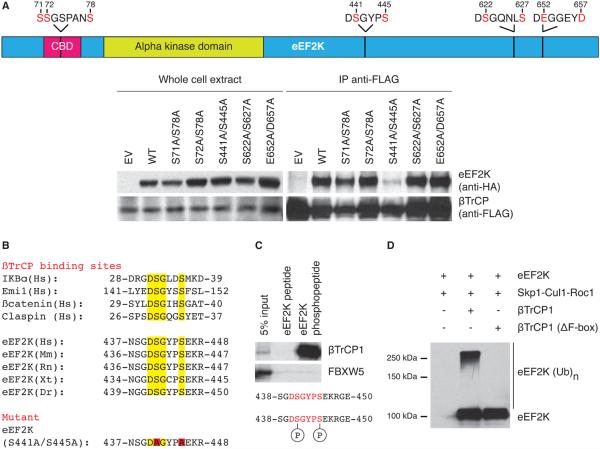
The WD40 β-propeller structure of βTrCP binds its substrates via a di-phosphorylated degradation motif (phospho-degron) with the consensus DpSGXX(X)pS (30-32), but some substrates of βTrCP have one or both serine residues replaced by either aspartic or glutamic acid (33-35). eEF2K has four of these potential βTrCP binding domains (Figure 2A top). To analyze which of these four domains is required for the interaction with βTrCP, we generated a number of serine/aspartic acid/glutamic acid to alanine double mutants, and examined their ability to bind βTrCP. Whereas wild type eEF2K, eEF2K(S71A/S78A), eEF2K(S72A/S78A), eEF2K(S622A/S627A), and eEF2K(E652A/D657A) immunoprecipitated βTrCP1, the eEF2K(S441A/S445A) mutant did not (Figure 2A bottom). The motif surrounding Ser441 and Ser445 is highly conserved in vertebrate orthologs of eEF2K (Figure 2B).
Figure 2. The βTrCP-eEF2K interaction depends on a conserved phospho-degron.
(A) eEF2K Ser441 and Ser445 are required for the association with βTrCP1.
Top: schematic representation of four putative eEF2K phospho-degrons. Bottom: HEK293T cells were transfected with FLAG-tagged βTrCP1 along with an empty vector, HA-tagged eEF2K wild type or mutants (as indicated). Cells were treated with MG132 and lysed. Whole-cell extracts were subjected either to immunoblotting or immunoprecipitation with anti-FLAG resin and subsequent immunoblotting. (B) Alignment of the amino acid regions corresponding to the βTrCP-binding motif (highlighted in yellow) in previously reported βTrCP substrates and eEF2K orthologs (top). Amino acidic sequence of the eEF2K double mutant is shown (bottom). (C) Phosphorylation of Ser441 and Ser445 in eEF2K is required for the interaction with βTrCP1. In vitro translated 35S-labeled βTrCP1 and FBXW5 were incubated with beads coupled to the indicated eEF2K (phospho)peptides. Bound proteins were eluted and subjected to electrophoresis and autoradiography. (D) βTrCP1 ubiquitylates eEF2K in vitro. HEK293T cells were transfected with eEF2K, Skp1, Cul1, Rbx1 together with either FLAG-tagged βTrCP1 or βTrCP1(ΔF-box). After immunopurification with anti-FLAG resin, in vitro ubiquitylation of eEF2K was performed. Samples were analyzed by immunoblotting with an antibody anti-eEF2K. Data represent at least three independent experiments.
Next, we used mass spectrometry to analyze eEF2K phosphorylation in this region (36). FLAG epitope-tagged eEF2K was co-expressed with a Cul1 dominant negative deletion mutant [Cul1-N252 (37)] in HEK293T cells and immunopurified. Analysis of the recovered eEF2K phosphopeptides demonstrated that Ser441 and Ser445 are phosphorylated in vivo (Figure S2, see figure legend for details).
As an additional approach to test whether phosphorylation is required for the interaction of eEF2K with βTrCP, we employed immobilized synthetic peptides comprising the βTrCP binding domain of eEF2K (aa 438-450 in human eEF2K). While a peptide containing phosphorylated Ser441 and Ser445 interacted with βTrCP1 (but not with FBXW5), the corresponding non-phosphorylated peptide was unable to do so (Figure 2C), indicating that phosphorylation of Ser441 and Ser445 directly mediates the association with βTrCP.
To test whether SCFβTrCP was directly responsible for the poly-ubiquitylation of eEF2K, we reconstituted the ubiquitylation of eEF2K in vitro. Immunopurified βTrCP1, but not an inactive βTrCP1(ΔF box) mutant, induced the in vitro ubiquitylation of eEF2K (Figure 2D). Moreover, the eEF2K(S441A/S445A) mutant could not be efficiently ubiquitylated by βTrCP in vitro (Figure S3). Thus, consistently with our findings in cultured cells, the in vitro data show that βTrCP promotes the ubiquitylation of eEF2K in a phosphodegron-dependent manner.
eEF2K is degraded during silencing of the G2 DNA damage checkpoint
We then investigated under which conditions eEF2K is degraded in the cell. Because mRNA translation is regulated by mitogens, we analyzed their effect on the abundance of eEF2K. We serum-starved T98G cells (revertants from T98 glioblastoma cells that acquired the property to accumulate in G0/G1 after serum deprivation) for 72 hours and then reactivated them by addition of serum. We observed a slight decrease in the abundance of eEF2K in response to mitogens (Figure S4A), however this decrease is not mediated by SCFβTrCP, as eEF2K abundance was not affected when we silenced βTrCP by transfecting T98G cells with a double-stranded RNA oligonucleotide that efficiently silence both βTrCP1 and βTrCP2 (Figure S4A) (33, 38-41).
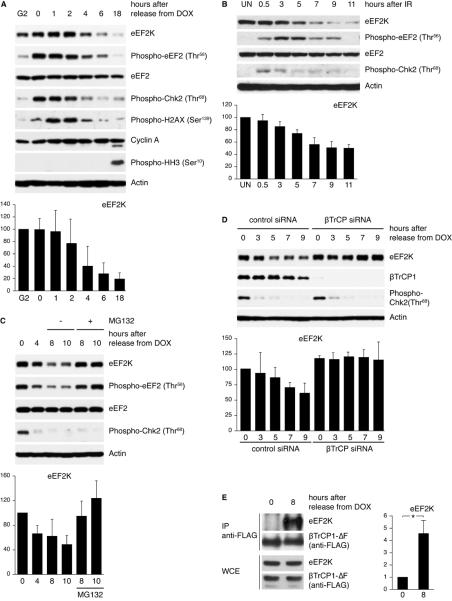
As eEF2K is a crucial inhibitor of translation elongation and since inhibition of protein synthesis is a common response to stress conditions (8, 10, 23, 42, 43), we analyzed the abundance of eEF2K in response to hypoxic and genotoxic stress. Hypoxia (and recovery from it) does not lead to detectable changes in the abundance of eEF2K (Figure S4B). Next, we examined eEF2K abundance in response to genotoxic stress. U2OS cells were synchronized in the G2 phase of the cell cycle and then treated with a low dose of doxorubicin to activate the G2 DNA damage checkpoint. Subsequently, cells were allowed to recover from the DNA damage checkpoint by incubation, after extensive washes, in drug-free medium. The abundance of eEF2K steadily decreased during checkpoint silencing, paralleling the decrease in Chk2 and Histone 2AX phosphorylation on Thr68 and Ser139, respectively (Figure 3A). A similar pattern of eEF2K expression was observed in experiments in which ionizing radiation (Figure 3B) was used. When U2OS cells were released towards mitosis without DNA damage eEF2K abundance did not change (Figure S4C-D). The change in the abundance of eEF2K observed in response to DNA damage was not caused by an indirect cell cycle effect because eEF2K abundance does not oscillate during the cell cycle (Figure S4C-D).
Figure 3. βTrCP-dependent degradation of eEF2K occurs during silencing of the G2 DNA damage checkpoint.
(A) U2OS cells were synchronized in G2 as described in Material and Methods. Cells were then pulsed with doxorubicin (DOX) for 1 hour, washed extensively, and incubated in fresh medium (indicated as time 0). Cells were harvested at the indicated time points and analyzed by immunoblotting. (B) U2OS cells were irradiated (10 Gy), collected at the indicated time points and analyzed by immunoblotting (UN: untreated). (C) U2OS cells were treated as in (A). MG132 was added when indicated. (D) U2OS cells were transfected with control and βTrCP siRNA and then treated as in (A). (E) U2OS cells were transfected with FLAG-tagged βTrCP1(ΔF-box) or an empty vector and then treated as in (A).
Graphs show eEF2K abundance normalized to a loading control and relative to the amount present at G2 (A), in untreated (B) or at time 0 (C, D and E). * P<0.01 compared with time 0 by one-way ANOVA. Data represent at least three independent experiments.
Treatment of U2OS cells with the proteasome inhibitor MG132 blocked the decrease of eEF2K during checkpoint silencing (Figure 3C), indicating that the decrease in eEF2K abundance observed during checkpoint silencing was due to proteasome-dependent degradation.
To test whether βTrCP mediates eEF2K degradation during checkpoint silencing, we knocked down the expression of βTrCP in U2OS cells by RNA interference. We then synchronized U2OS cells in G2 and induced genotoxic stress by doxorubicin treatment as in the experiment shown in Figure 3A. βTrCP knockdown inhibited the degradation of eEF2K during checkpoint silencing (Figure 3D). In agreement with this finding, the eEF2K-βTrCP interaction was strikingly stimulated in U2OS cells during DNA damage checkpoint silencing (Figure 3E).
Finally, to analyze the phosphorylation of the eEF2K degron in response to genotoxic stress, we generated a phospho-specific antibody that recognizes eEF2K only when it is phosphorylated on Ser441 and Ser445 (Figure S5A). Figure S5B shows that phosphorylation of the eEF2K degron is induced by DNA damage in G2 and precedes eEF2K degradation.
Altogether these findings imply that βTrCP targets eEF2K for degradation upon checkpoint silencing.
AMPK-dependent phosphorylation of eEF2K on Ser398 leads to activation of eEF2K in response to genotoxic stress
Numerous studies have demonstrated that eEF2K controls translation elongation by phosphorylating eEF2 on Thr56 (14, 15, 17-19, 21-23, 44-47). These studies have also shown that eEF2K-mediated phosphorylation of eEF2 on Thr56 inhibits the ribosome-eEF2 complex formation by reducing the affinity of eEF2 for the ribosome. Our results indicate that phosphorylation of eEF2 on Thr56 increases upon checkpoint activation and decreases during checkpoint silencing, mirroring the phosphorylation profile of both Chk2 on Thr68 (Figures 3A, 3B, and 6B) and Histone 2AX on Ser139 (Figure 3A). To confirm that the increased phosphorylation of eEF2 on Thr56 in response to DNA damage is mediated by eEF2K, we silenced the expression of eEF2K in U2OS cells by RNA interference. After transfection, cells were synchronized in G2 and then pulsed with doxorubicin for 1 hour to induce DNA damage. Figure S6A shows that eEF2K knockdown blocked the DNA damage-induced phosphorylation of eEF2 on Thr56.
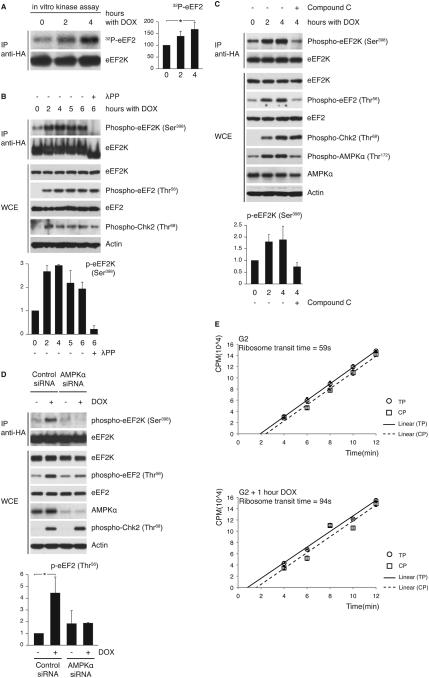
Next, we investigated the mechanism by which eEF2K-dependent phosphorylation of eEF2 is stimulated by genotoxic stress. First, to assess whether genotoxic stress stimulates the activity of eEF2K, we pulled down eEF2K from G2 cells treated with doxorubicin and assayed immunoprecipitated eEF2K activity against in vitro translated eEF2 (Figure 4A). The activity of eEF2K immunoprecipitated from G2 cells treated with doxorubicin was increased when compared to eEF2K activity in G2 cells.
Figure 4. AMPK-mediated phosphorylation of eEF2K on Ser398 causes eEF2K activation in response to genotoxic stress.
(A) eEF2K was immunoprecipitated from doxorubicin-treated G2 U2OS cells and assayed for activity against in vitro translated eEF2. The graph shows the amount of phosphorylated eEF2 relative to time 0. *P<0.05 compared to time 0 by one-way ANOVA and Dunnett’s post-hoc test. (B) G2 U2OS cells were treated with doxorubicin for indicated times. One IP sample (λPP) was treated with λ-phosphatase prior to immunoblotting. (C and D) U2OS cells were treated with compound-C (C) or siRNA oligonucleotides targeting AMPKα (D). Graphs for (B) and (C) illustrate the amount of phospho-eEF2K(Ser398) normalized to eEF2K and relative to time 0. The graph in (D) shows abundance of phospho-eEF2(Thr56) normalized to eEF2 and relative to untreated control siRNA sample. *P<0.01 compared to all other samples by one-way ANOVA and Bonferroni’s post-hoc test. (E) Ribosome transit time for G2 cells and doxorubicin-treated G2 cells. [35S]methionine incorporation into total proteins (TP; circles) or into completed peptides released from ribosomes (CP; squares) is shown (CPM: counts per minute). Comparison of ribosome transit time for G2 and doxorubicin-treated cells by Student’s t-test gives P<0.005. Data represent at least three independent experiments.
Proud and co-workers have shown that eEF2K is phosphorylated on Ser398 by the AMP-activated protein kinase (AMPK) and that this phosphorylation leads to eEF2K activation (48). Intriguingly, it has recently been reported that AMPK is activated by the p53 target genes Sestrin1 and Sestrin2 in response to genotoxic stress (49). To test whether genotoxic stress leads to phosphorylation of eEF2K on Ser398, we used a previously characterized phosphospecific antibody that can recognize eEF2K phosphorylated on Ser398 (48). U2OS cells were synchronized in G2 as described above and then treated with doxorubicin. We found that phosphorylation of eEF2K on Ser398 increased in response to doxorubicin treatment and is associated with increased phosphorylation of eEF2 on Thr56 (Figure 4B). To examine the possibility that AMPK phosphorylates eEF2K on Ser398 in response to genotoxic stress, we used the AMPK inhibitor compound C (49, 50). U2OS cells in G2 were treated with doxorubicin with or without compound C. As shown in Figure 4C, treatment of U2OS cells with compound C blocked the increase in eEF2K phosphorylation on Ser398 and eEF2 phosphorylation on Thr56 in response to genotoxic stress.
To rule out non-specific effects of compound C, we silenced the expression of AMPKα by RNA interference and observed that AMPKα knockdown inhibited the increase in eEF2 phosphorylation on Thr56 in response to genotoxic stress (Figure 4D).
Taken together, these data indicate that genotoxic stress in G2 cells stimulates AMPK, which phosphorylates eEF2K on Ser398 causing its activation. In turn, activated eEF2K phosphoryates eEF2 on Thr56 blocking its activity.
Moreover, the above results suggest that translation elongation slows down upon activation of the DNA damage checkpoint. Indeed, G2 cells pulsed with doxorubicin exhibited increased ribosome transit time (94 seconds), i.e., slower rates of elongation, when compared with untreated G2 cells (59 seconds; Figure 4E).
The observed slowdown in translation elongation upon activation of the DNA damage checkpoint is associated with an eEF2K-dependent decrease in global protein synthesis as indicated by metabolic labeling of G2 U2OS cells following genotoxic stress (Figure S6B).
Autophosphorylation of the eEF2K degron is required for its interaction with βTrCP
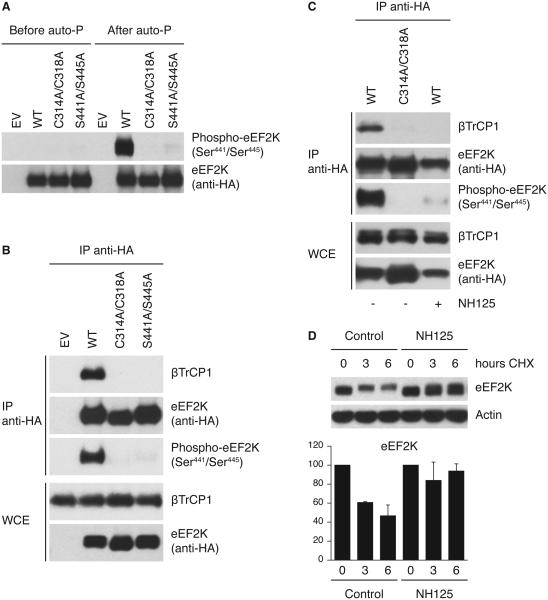
The above results indicate that in response to genotoxic stress, eEF2K is first activated by AMPK-mediated phosphorylation and, subsequently, during checkpoint silencing, is targeted for degradation by the SCFβTrCP ubiquitin ligase. This sequence of events suggests a coupled activation-destruction mechanism by which eEF2K is targeted for destruction when its activity is sustained. As it has been shown that eEF2K has autophosphorylation activity (20, 51), we hypothesized that eEF2K, once activated, may autophosphorylate on its degron triggering eEF2K binding to βTrCP. First, to assess whether eEF2K can phosphorylate itself on Ser441 and Ser445, eEF2K was immunopurified from U2OS cells, dephosphorylated, and then subjected to autophosphorylation in vitro. As a control, we used eEF2K(C314A/C318A), a kinase-dead eEF2K mutant that can not phosphorylate either eEF2 or itself (51). eEF2K phosphorylation on Ser441 and Ser445 was then analyzed by immunoblotting using the phosphospecific antibody that recognizes eEF2K only when it is phosphorylated on Ser441 and Ser445. Figure 5A shows that the eEF2K degron is autophosphorylated in wild type eEF2K but not in the kinase-dead eEF2K mutant. Similar results were obtained when we used eEF2K purified from E. coli (Figure S7).
Figure 5. eEF2K autophosphorylation on the degron triggers its binding to βTrCP.
(A) In vitro autophosphorylation of the eEF2K degron. U2OS cells were transfected with either HA-tagged wild type eEF2K, eEF2K(C314A/C318A), or eEF2K(S441A/S445A). eEF2K was purified, dephosphorylated by λ-phosphatase treatment, and then allowed to autophosphorylate in vitro. Samples were then analyzed by immunoblotting. Auto-P: autophosphorylation reaction. (B) Kinase-dead eEF2K mutant does not bind βTrCP1. U2OS cells were transfected with an empty vector (EV), HA-tagged wild type eEF2K, eEF2K(C314A/C318A), or eEF2K(S441A/S445A). After transfection, cells were synchronized in G2 as described in Figure 3A, pulse-treated with doxorubicin, and collected after 8 hours. Three hours before harvesting, cells were treated with MG132. (C) The βTrCP1-eEF2K interaction is blocked by an eEF2K inhibitor. U2OS cells were transfected with an empty vector (EV) or HA-tagged eEF2K. Cells were treated as in (B) and, when indicated, treated with the eEF2K inhibitor NH125. (D) NH125 induces the stabilization of eEF2K. Cells were pulse-treated with doxorubicin and then treated with NH125. eEF2K turnover was analyzed by cycloheximide chase (100 μg/ml for indicated times). The graph illustrates the quantification of eEF2K abundance relative to the amount at time 0. Data are representative of at least three independent experiments.
Next, we tested the ability of eEF2K(C314A/C318A) to bind βTrCP. Figure 5B shows that the kinase-dead eEF2K mutant is unable to interact with endogenous βTrCP, analogously to the eEF2K(S441A/S445A) phosphodegron mutant. Accordingly, the eEF2K degron is found phosphorylated in vivo in wild type eEF2K but not in kinase-dead eEF2K (Figure 5B).
To exclude the possibility that the inability of eEF2K(C314A/C318A) to interact with βTrCP is indirectly caused by structural changes in eEF2K, we examined the binding of wild type eEF2K in the presence of NH125, a specific inhibitor of the eEF2K kinase activity (52). We found that treatment of U2OS with NH125 abolished both the phosphorylation of the eEF2K degron and βTrCP binding to eEF2K (Figure 5C). Accordingly, NH125 treatment resulted in eEF2K stabilization (Figure 5D).
Altogether, these data strongly suggest that AMPK-mediated activation of eEF2K in response to genotoxic stress is required for eEF2K degradation during checkpoint silencing. Indeed, inhibition of AMPK by treatment with compound C prevented eEF2K degradation during checkpoint silencing (Figure S8). Although necessary, AMPK-mediated activation of eEF2K is not sufficient for eEF2K destruction, as serum starvation, a well-characterized stimulus that activates AMPK, did not trigger the degradation of eEF2K (Figure S9 and Discussion).
Defective degradation of eEF2K delays the resumption of translation elongation during checkpoint silencing
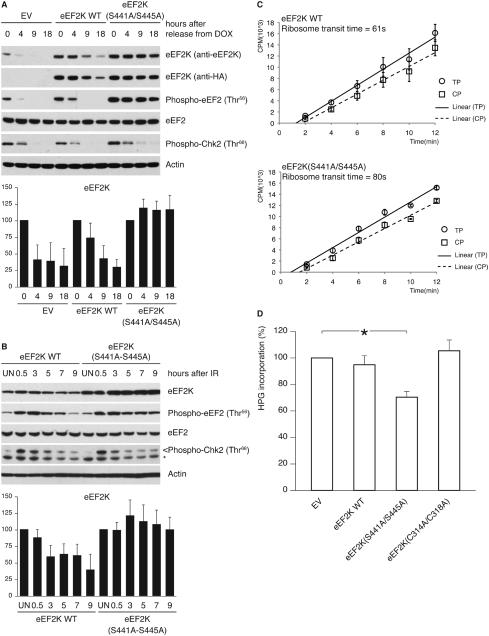
We then asked whether βTrCP-mediated degradation of eEF2K is required to resume protein synthesis upon checkpoint silencing. U2OS cells were retrovirally transduced with HA-tagged wild type eEF2K or HA-tagged eEF2K(S441A/S445A), synchronized in G2 as described above, and subsequently pulsed with doxorubicin for 1 hour, released and then collected at the indicated times. Compared to cells expressing wild type eEF2K and untransduced cells, cells expressing the stable eEF2K mutant unable to bind βTrCP displayed a much slower decrease of eEF2 phosphorylation on Thr56 upon silencing of the DNA damage checkpoint (Figure 6A). Similar results were obtained in cells recovering from genotoxic stress induced by ionizing irradiation (Figure 6B). Moreover, the kinase-dead eEF2K mutant was not downregulated during checkpoint silencing (Figure S10).
Figure 6. Inability to degrade eEF2K delays the resumption of translation elongation during silencing of the DNA damage checkpoint.
(A) U2OS cells, transduced with retroviruses expressing HA-tagged wild type eEF2K or eEF2K(S441A/S445A), were synchronized in G2, doxorubicin-pulsed and collected at the indicated times. Cell extracts were analyzed by immunoblotting. (B) U2OS cells, transduced as in (A), were exposed to ionizing radiation. Cell extracts were analyzed by immunoblotting. The graphs illustrate the quantification by densitometry of eEF2K abundance relative to the amount at time 0 (A) or in untreated sample (B). (C) U2OS cells, transduced as in (A), were collected 12 hours after ionizing radiation. Incorporation of [35S]methionine into total proteins (TP; circles) or into completed peptides released from ribosomes (CP; squares) is shown. Comparison of ribosome transit time for eEF2K WT and eEF2K(S441A/S445A) expressing cells by Student’s t-test gives P<0.005.
(D) Transduced U2OS cells were treated as in (A). Sixteen hours after the doxorubicin pulse, cells were labeled with L-homopropargylglycine. Incorporation of L-homopropargylglycine was detected using Click-iT technology. Results are the mean ± SD (n=3; *P<0.01 comparing to EV by one-way ANOVA and Dunnett’s post-hoc test). Data represent at least three independent experiments.
Notably, during silencing of checkpoint signaling, cells expressing eEF2K(S441A/S445A) displayed a longer ribosomal transit time when compared with cells expressing wild type eEF2K, indicating a defective resumption of the normal translation elongation rate when βTrCP-mediated degradation of eEF2K is impaired (Figure 6C).
Next, we tested whether defective degradation of eEF2K affects global protein synthesis during checkpoint silencing. Cells expressing eEF2K(S441A/S445A) displayed a decreased global protein synthesis during checkpoint silencing, when compared to control cells, as measured by metabolic labeling (Figure 6D). This difference was not observed in untreated cells (Figure S11), indicating that eEF2K degradation is required for efficient resumption of global protein synthesis upon silencing of the DNA damage checkpoint.
DISCUSSION
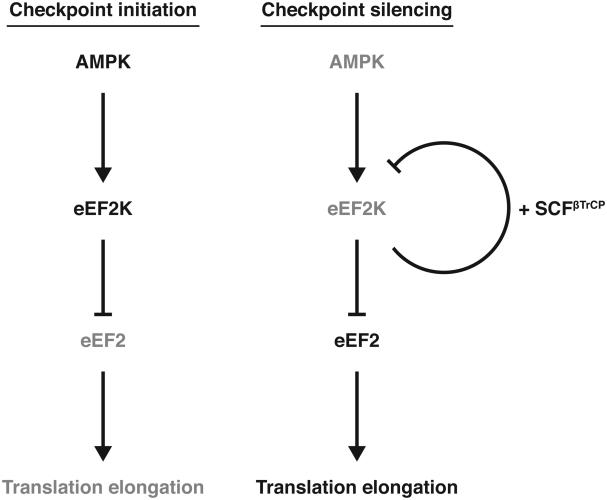
eEF2K is a ubiquitous protein kinase which plays a crucial role in the regulation of translational elongation by phosphorylating and inhibiting eEF2, a factor that promotes ribosomal translocation during the elongation phase of protein synthesis. In agreement with its key function, eEF2K activity is tightly controlled by different upstream protein kinases, which either block or stimulate its function in response to different stimuli (24). In addition to this level of regulation, eEF2K abundance is controlled by the ubiquitin-proteasome system (53), however the biological significance of the ubiquitylation and degradation of eEF2K and the identity of the E3 ubiquitin ligase involved are unknown. In the present report we show that following the activation of the DNA damage checkpoint, AMPK mediates the activation of eEF2K, which in turn phosphorylates eEF2 leading to a decrease in translation elongation rates. Subsequently, eEF2K autophosphorylation generates a phospho-degron for the recruitment of the SCFβTrCP ubiquitin ligase. This event triggers the ubiquitylation of eEF2K and its proteasome-mediated degradation, which releases the inhibitory effect on eEF2 and translation elongation (Figure 7). The coupled activation-destruction of eEF2K represents a mechanism that generates a pulse of kinase activity to dynamically tune the rate of elongation in response to genotoxic stress. This mechanism, by which eEF2K is targeted for degradation when its activity is sustained, would imply that any condition that induces the activity of eEF2K would inevitably trigger eEF2K destruction. However, βTrCP-mediated degradation of eEF2K, which occurs specifically in response to DNA damage, suggests that additional regulatory mechanisms must exist in order to protect eEF2K from being degraded following other activating conditions. Possible protective roles of protein phosphatases (counteracting eEF2K autophosphorylation), deubiquitinating enzymes (cleaving ubiquitin chains conjugated on eEF2K via SCFβTrCP), and chaperons (shielding eEF2K from ubiquitin conjugation) in response to stress conditions different from DNA damage need to be investigated. Interestingly, it has been observed that eEF2K is chaperoned by Hsp90 and that cellular abundance of eEF2K is controlled by a balance between association with Hsp90 and degradation by the ubiquitin-proteasome system (53).
Figure 7. Model of eEF2K regulation in response to genotoxic stress.
Black lines represent activated proteins, gray indicates inactive or degraded proteins. See text for details.
We cannot rule out that only one of the two serine residues within the eEF2K degron (Ser441 and Ser445) is a site of autophosphorylation. Indeed, liquid-chromatography tandem mass spectrometry analysis of the eEF2K degron after in vitro autophosphorylation revealed that Ser445 is the prevalent phosphorylated site, implying the role of an additional kinase targeting Ser441. According to this model the specific degradation of eEF2K in response to DNA damage would be triggered by the cooperative actions of two kinases. The combination of eEF2K autophosphorylation (targeting Ser445) and a yet to be identified kinase (targeting Ser441) would be needed to generate the eEF2K phosphodegron specifically in response to DNA damage.
Regulation of translation elongation in response to DNA damage might have several advantages over controlling translation initiation. Indeed, elongation inhibition during checkpoint activation avoids the disassembly of polysomes, which, by stalling ribosomes on the mRNAs, might allow for mRNA protection from degradation or sequestration into, e.g., stress granules. This mechanism will also ensure that translation can rapidly resume when the checkpoint is turned off.
General inhibition of translation coupled with stimulation of translation of specific mRNA pools has been demonstrated both at the level of initiation and elongation. For instance, DNA-PKcs-mediated translation reprogramming in response to UV radiation allows selective synthesis of DNA damage response proteins and is based on upstream open reading frames in the 5′ untranslated region of these mRNAs (54). Other studies have shown that eEF2K-dependent phosphorylation of eEF2 acts to slow down the elongation step of translation and inhibits general protein synthesis but simultaneously increases the translation of Arc/Arg3.1, a neuronal immediate early gene involved in both synapse plasticity and long-term potentiation (55). Microarray analysis of polysomes- and monosome-associated mRNA pools will be required to identify mRNAs specifically translated during the DNA damage checkpoint.
Recent findings have implicated SCFβTrCP in the degradation of several substrates during the recovery from DNA damage and replication stress checkpoints. Indeed, βTrCP is required to reactivate Cdk1 by targeting Claspin and Wee1 for proteasome-dependent degradation (35, 41, 56-60) and turn off the DNA repair machinery by causing the destruction of the Fanconi anemia protein FANCM (61). Our findings show that βTrCP is also needed during checkpoint silencing to resume protein synthesis by triggering eEFK proteolysis, suggesting an orchestrating role for the SCFβTrCP ubiquitin ligase in coordinating different processes (i.e. cell cycle progression, DNA repair and protein synthesis) that are critical for checkpoint termination.
MATERIALS AND METHODS
Cell culture, synchronization and drug treatment
HEK293T, 293GP2 and U2OS cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% fetal calf serum. HCT116, SW480 and MCF7 cells were maintained in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) containing 10% fetal calf serum. Synchronizations and flow analysis were performed as described (38). MG132 (10 μM) was added when indicated.
Biochemical methods
Extract preparation, immunoprecipitation, and immunoblotting were previously described (33, 40).
Purification of βTrCP2 interactors
HEK293T cells were transfected with pcDNA3-FLAG-HA-βTrCP2 and treated with 10 μM MG132 for 5 hours. Cells were harvested and subsequently lysed in lysis buffer (LB: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP40, plus protease and phosphatase inhibitors). βTrCP2 was immunopurified with anti-FLAG agarose resin (Sigma). After washing, proteins were eluted by competition with FLAG peptide (Sigma). The eluate was then subject to a second immunopurification with anti-HA resin (12CA5 monoclonal antibody crosslinked to protein G Sepharose; Invitrogen) prior to elution in Laemmli sample buffer. The final eluate was separated by SDS-PAGE, and proteins were visualized by Coomassie colloidal blue. Bands were sliced out from the gels and subjected to in-gel digestion. Gel pieces were then reduced, alkylated and digested according to a published protocol (62). For mass spectrometric analysis, peptides recovered from in-gel digestion were separated with a C18 column and introduced by nano-electrospray into the LTQ Orbitrap XL (Thermo Fisher, Bremen). Peak lists were generated from the MS/MS spectra using MaxQuant (Cox and Mann, 2008), and then searched against the IPI Human database using Mascot search engine (Matrix Science). Carbaminomethylation (+57 Da) was set as fixed modification and protein N-terminal acetylation and methionine oxidation as variable modifications. Peptide tolerance was set to 7 ppm and fragment ion tolerance was set to 0.5 Da, allowing 2 missed cleavages with trypsin enzyme.
Antibodies
Mouse monoclonal antibodies were from Invitrogen (Cul1), Sigma (FLAG, eEF2K), Santa Cruz Biotechnology (actin), BD (β-catenin, p27) and Covance (HA). Rabbit polyclonal antibodies were from Bethyl (PDCD4), Cell Signaling [βTrCP1, eEF2K, eEF2, phosho-eEF2(Thr56), phospho-Chk2(Thr68)], Millipore [(phospho-Histone H3(Ser10), phospho-Histone H2AX(Ser139)], Sigma (FLAG, USP8), Novus Biologicals (HIF1α) and Santa Cruz (Cyclin A, Cdc20, Skp1 and HA).
Plasmids
eEF2K mutants were generated using the QuickChange Site-directed Mutagenesis kit (Stratagene). For retrovirus production, both wild type eEF2K and eEF2K mutants were subcloned into the retroviral vector pBABEpuro. All cDNAs were sequenced.
Transient transfections and retrovirus-mediated gene transfer
HEK293T cells were transfected using the calcium phosphate method as described (33, 40). Retrovirus-mediated gene transfer was previously described (33, 40).
Gene silencing by small interfering RNA
The sequence and validation of the oligonucleotides corresponding to βTrCP1 and βTrCP2 were previously published (33, 40, 41, 63). Both eEF2K and AMPK□ SMARTpool siRNA oligonucleotides were from Dharmacon. Cells were transfected with the oligos twice (24 and 48 hours after plating) using Oligofectamine (Invitrogen) according to manufacture’s recommendations. Forty-eight hours after the last transfection lysates were prepared and analyzed by SDS-PAGE and immunoblotting.
In vitro ubiquitylation assay
eEF2K ubiquitylation was performed in a volume of 10 μl containing SCFβTrCP-eEF2K immunocomplexes, 50 mM Tris pH 7.6, 5 mM MgCl2, 0.6 mM DTT, 2 mM ATP, 2 μl in vitro translated unlabeled βTrCP1, 1.5 ng/μl E1 (Boston Biochem), 10 ng/μl Ubc3, 2.5 μg/μl ubiquitin (Sigma), 1 μM ubiquitin aldehyde. The reactions were incubated at 30°C for the indicated times and analyzed by immunoblotting. In the assay shown in Figure S3 35S-labeled in vitro translated eEF2K [wild type or eEF2K(S441A/S445A)] was used instead of immunoprecipitated eEF2K. The reactions were analyzed by SDS-PAGE and autoradiography.
Phosphorylation analysis of the eEF2K degron in vivo
FLAG-eEF2K was co-expressed with the Cul1 dominant negative deletion mutant CUL1-N252, and immunopurified. FLAG immunocomplexes were eluted with 8 M urea. Samples were then reduced with 10 mM DTT for 30 minutes at 60°C, followed by addition of iodoacetamide to 20 mM followed by 30-minute incubation in the dark at room temperature. The first digestion was performed using Lys-C for 4 hours at 37°C. Subsequently, the digest was diluted 5-fold using 50 mM ammonium bicarbonate to a final urea concentration of less than 2 M, and a second digestion with trypsin was performed overnight at 37°C. Finally, the digestion was quenched by addition of formic acid to a final concentration of 0.1% (vol/vol). The resulting solution was desalted using 200 mg Sep-Pak C18 cartridges (Waters Corporation, Milford, MA), lyophilized and stored at −20°C.
Ti-IMAC microcolumns were prepared as previously described (64). Trypsinized peptides were loaded onto pre-conditioned microcolumns, which were later sequentially washed with 60 μl of loading buffer, followed by 60 μl of 50% ACN/0.5% TFA containing 200 mM NaCl and 60 μl of 50% ACN/0.1% TFA. The bound peptides were eluted by 20 μl of 5% ammonia into 20 μl of 10% formic acid and then stored at −20°C prior to LC-MS/MS analysis. MS spectra assignment was performed with MaxQuant Version 1.1.1.25.
In vitro kinase assay
eEF2K kinase activity was assayed as described (48).
Ribosome transit time measurements
The method for ribosome transit time measurements is adapted from (45, 65). In brief, cells were incubated in methionine-free medium for 30 minutes, prior to adding labeling medium containing 10 μCi/ml L-[35S]Met (NEG709A, Perkin-Elmer). Samples were harvested in PBS containing 100 μg/ml cycloheximide at 2, 4, 6, 8, 10, 12 minutes after labeling and pelleted. Cell lysis and pelleting of polysomes were executed as described (65). Polysomes were pelleted at 55,000 μ g for 20 min at 4°C in a Beckman Sw55Ti rotor.
Fifty microliters of the total protein fraction (post-mitochondrial supernatant) and completed protein fraction (post-ribosomal supernatant), were loaded on 3MM paper and precipitated by consecutive incubations in cold 10% TCA and boiling 10% TCA (2,5 minutes per step) and briefly washing in acetone and ethanol. Filters were air-dried and incorporation of [35S]-methionine was measured by liquid scintillation counting (three measurements per sample).
To calculate ribosome transit times obtained values were analyzed as described (66). In this analysis, we measure the lag between the linear incorporation of labeled methionine in total protein (TP) and the linear incorporation in complete protein (CP), which represents the time it takes for all nascent peptides to get labeled. This lag is seen in the graph as the difference in x-intercepts of the two extrapolated lines and represents the half transit time (half because the average length of polypeptides on a polyribosome is always half of the full length of completed peptides). To obtain the ribosome transit time, the difference in intercepts is doubled.
Metabolic labeling
U2OS cells were culture in methionine-free medium for 60 minutes and then incubated in 50 μM Click-iT HPG (L-homopropargylglycine) for 90 minutes. Cells were fixed in 70% ethanol, stained with Alexa Fluor 488 azide and analyzed by FACS.
Statistical analysis
All data shown are from one representative experiment of at least three performed. Statistical tests used are specified in the legends of relevant figures.
Supplementary Material
Acknowledgements
We thank W.N. Hait, M. Innocenti, N.V. Dorrello, R.H. Giles and H. Zhou for their contributions, R.H. Medema and D. Frescas for critically reading the manuscript. Funding: Work in D.G.’s laboratory is supported by the Royal Dutch Academy of Arts and Sciences (KNAW), the Dutch Cancer Society (KWF), the Cancer Genomics Centre and the European Union under Marie Curie Actions (FP7). Work in M.P.’s laboratory is supported by grants from the National Institutes of Health. M.P. is an investigator of the Howard Hughes Medical Institute. T.Y.L., S.M., and A.J.R.H. are supported by the Netherlands Proteomics Center (NPC). C.G.P. is funded by the Wellcome Trust.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and material availability: Scaffold files containing geLC-MSMS data can be downloaded from ProteomeCommons.org Tranche (https://proteomecommons.org/tranche) using the hash key and passphrase provided below.
Hash:
7vUEg7l5ShPgDh6HCaZBbNznpCi63nXCv/SCS5tWq1hc6pW8vNjDigrJ4pvDdzoUcbTBAFR+PXw5M49Ih2CUElxXnD0AAAAAAAAChg==
Passphrase: NyCbEVxg1q3Mahvlh0JM
REFERENCES AND NOTES
- 1.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Current opinion in cell biology. 2007;19:238. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW, Elledge SJ. The DNA damage response: ten years after. Molecular cell. 2007;28:739. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Lu X, de la Pena L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052. doi: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- 4.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312(Pt 1):163. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22:169. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 8.Browne GJ, Proud CG. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol. 2004;24:2986. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly E, Braunstein S, Formenti S, Schneider RJ. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol Cell Biol. 2006;26:3955. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hait WN, Wu H, Jin S, Yang JM. Elongation factor-2 kinase: its role in protein synthesis and autophagy. Autophagy. 2006;2:294. doi: 10.4161/auto.2857. [DOI] [PubMed] [Google Scholar]
- 11.Proud CG. Regulation of mammalian translation factors by nutrients. Eur J Biochem. 2002;269:5338. doi: 10.1046/j.1432-1033.2002.03292.x. [DOI] [PubMed] [Google Scholar]
- 12.Proud CG. Control of the translational machinery in mammalian cells. Eur J Biochem. 2002;269:5337. doi: 10.1046/j.1432-1033.2002.03289.x. [DOI] [PubMed] [Google Scholar]
- 13.Redpath NT, Price NT, Proud CG. Cloning and expression of cDNA encoding protein synthesis elongation factor-2 kinase. J Biol Chem. 1996;271:17547. [PubMed] [Google Scholar]
- 14.Carlberg U, Nilsson A, Nygard O. Functional properties of phosphorylated elongation factor 2. Eur J Biochem. 1990;191:639. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- 15.Mitsui K, Brady M, Palfrey HC, Nairn AC. Purification and characterization of calmodulin-dependent protein kinase III from rabbit reticulocytes and rat pancreas. J Biol Chem. 1993;268:13422. [PubMed] [Google Scholar]
- 16.Nairn AC, Bhagat B, Palfrey HC. Identification of calmodulin-dependent protein kinase III and its major Mr 100,000 substrate in mammalian tissues. Proc Natl Acad Sci U S A. 1985;82:7939. doi: 10.1073/pnas.82.23.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nairn AC, Matsushita M, Nastiuk K, Horiuchi A, Mitsui K, Shimizu Y, Palfrey HC. Elongation factor-2 phosphorylation and the regulation of protein synthesis by calcium. Prog Mol Subcell Biol. 2001;27:91. doi: 10.1007/978-3-662-09889-9_4. [DOI] [PubMed] [Google Scholar]
- 18.Ovchinnikov LP, Motuz LP, Natapov PG, Averbuch LJ, Wettenhall RE, Szyszka R, Kramer G, Hardesty B. Three phosphorylation sites in elongation factor 2. FEBS Lett. 1990;275:209. doi: 10.1016/0014-5793(90)81473-2. [DOI] [PubMed] [Google Scholar]
- 19.Price NT, Redpath NT, Severinov KV, Campbell DG, Russell JM, Proud CG. Identification of the phosphorylation sites in elongation factor-2 from rabbit reticulocytes. FEBS Lett. 1991;282:253. doi: 10.1016/0014-5793(91)80489-p. [DOI] [PubMed] [Google Scholar]
- 20.Redpath NT, Proud CG. Purification and phosphorylation of elongation factor-2 kinase from rabbit reticulocytes. Eur J Biochem. 1993;212:511. doi: 10.1111/j.1432-1033.1993.tb17688.x. [DOI] [PubMed] [Google Scholar]
- 21.Ryazanov AG, Natapov PG, Shestakova EA, Severin FF, Spirin AS. Phosphorylation of the elongation factor 2: the fifth Ca2+/calmodulin-dependent system of protein phosphorylation. Biochimie. 1988;70:619. doi: 10.1016/0300-9084(88)90245-3. [DOI] [PubMed] [Google Scholar]
- 22.Ryazanov AG, Shestakova EA, Natapov PG. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- 23.Ryazanov AG, Ward MD, Mendola CE, Pavur KS, Dorovkov MV, Wiedmann M, Erdjument-Bromage H, Tempst P, Parmer TG, Prostko CR, Germino FJ, Hait WN. Identification of a new class of protein kinases represented by eukaryotic elongation factor-2 kinase. Proc Natl Acad Sci U S A. 1997;94:4884. doi: 10.1073/pnas.94.10.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006;21:362. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 25.Horton LE, Bushell M, Barth-Baus D, Tilleray VJ, Clemens MJ, Hensold JO. p53 activation results in rapid dephosphorylation of the eIF4E-binding protein 4E-BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation. Oncogene. 2002;21:5325. doi: 10.1038/sj.onc.1205662. [DOI] [PubMed] [Google Scholar]
- 26.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8204. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 28.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 29.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 30.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 33.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Chang S, Hernando E, Pagano M. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanemori Y, Uto K, Sagata N. Beta-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc Natl Acad Sci U S A. 2005;102:6279. [Google Scholar]
- 35.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boersema PJ, Mohammed S, Heck AJ. Phosphopeptide fragmentation and analysis by mass spectrometry. J Mass Spectrom. 2009;44:861. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 37.Piva R, Liu J, Chiarle R, Podda A, Pagano M, Inghirami G. In vivo interference with skp1 function leads to genetic instability and neoplastic transformation. Mol Cell Biol. 2002;22:8375. doi: 10.1128/MCB.22.23.8375-8387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, Margottin-Goguet F, Jackson PK, Yamasaki L, Pagano M. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev Cell. 2003;4:799. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 39.Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 40.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 41.Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27:6639. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagaglio DM, Cheng EH, Gorelick FS, Mitsui K, Nairn AC, Hait WN. Phosphorylation of elongation factor 2 in normal and malignant rat glial cells. Cancer Res. 1993;53:2260. [PubMed] [Google Scholar]
- 45.Redpath NT, Foulstone EJ, Proud CG. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. EMBO J. 1996;15:2291. [PMC free article] [PubMed] [Google Scholar]
- 46.Redpath NT, Price NT, Severinov KV, Proud CG. Regulation of elongation factor-2 by multisite phosphorylation. Eur J Biochem. 1993;213:689. doi: 10.1111/j.1432-1033.1993.tb17809.x. [DOI] [PubMed] [Google Scholar]
- 47.Ryazanov AG, Davydova EK. Mechanism of elongation factor 2 (EF-2) inactivation upon phosphorylation. Phosphorylated EF-2 is unable to catalyze translocation. FEBS Lett. 1989;251:187. doi: 10.1016/0014-5793(89)81452-8. [DOI] [PubMed] [Google Scholar]
- 48.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 49.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, Musi N, Hirshman MF, Goodyear LJ, Moller DE. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diggle TA, Seehra CK, Hase S, Redpath NT. Analysis of the domain structure of elongation factor-2 kinase by mutagenesis. FEBS letters. 1999;457:189. doi: 10.1016/s0014-5793(99)01034-0. [DOI] [PubMed] [Google Scholar]
- 52.Arora S, Yang JM, Kinzy TG, Utsumi R, Okamoto T, Kitayama T, Ortiz PA, Hait WN. Identification and characterization of an inhibitor of eukaryotic elongation factor 2 kinase against human cancer cell lines. Cancer Res. 2003;63:6894. [PubMed] [Google Scholar]
- 53.Arora S, Yang JM, Hait WN. Identification of the ubiquitin-proteasome pathway in the regulation of the stability of eukaryotic elongation factor-2 kinase. Cancer Res. 2005;65:3806. doi: 10.1158/0008-5472.CAN-04-4036. [DOI] [PubMed] [Google Scholar]
- 54.Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC, Wek R, Hayes C, Gant TW, Spriggs KA, Bushell M, Willis AE. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freire R, van Vugt MA, Mamely I, Medema RH. Claspin: timing the cell cycle arrest when the genome is damaged. Cell Cycle. 2006;5:2831. doi: 10.4161/cc.5.24.3559. [DOI] [PubMed] [Google Scholar]
- 57.Mailand N, Bekker-Jensen S, Bartek J, Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell. 2006;23:307. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 58.Mamely I, van Vugt MA, Smits VA, Semple JI, Lemmens B, Perrakis A, Medema RH, Freire R. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr Biol. 2006;16:1950. doi: 10.1016/j.cub.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 59.Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe N, Arai H, Iwasaki J, Shiina M, Ogata K, Hunter T, Osada H. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc Natl Acad Sci U S A. 2005;102:11663. doi: 10.1073/pnas.0500410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kee Y, Kim JM, D’Andrea AD. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev. 2009;23:555. doi: 10.1101/gad.1761309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 63.Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou H, Ye M, Dong J, Han G, Jiang X, Wu R, Zou H. Specific phosphopeptide enrichment with immobilized titanium ion affinity chromatography adsorbent for phosphoproteome analysis. J Proteome Res. 2008;7:3957. doi: 10.1021/pr800223m. [DOI] [PubMed] [Google Scholar]
- 65.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes & development. 2005;19:2199. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen PJ, McConkey EH. Evidence for control of protein synthesis in HeLa cells via the elongation rate. J Cell Physiol. 1980;104:269. doi: 10.1002/jcp.1041040302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.