Abstract
Cardiac myxomas are benign mesenchymal tumors that can present as components of the human autosomal dominant disorder Carney complex. Syndromic cardiac myxomas are associated with spotty pigmentation of the skin and endocrinopathy. Our linkage analysis mapped a Carney complex gene defect to chromosome 17q24. We now demonstrate that the PRKAR1α gene encoding the R1α regulatory subunit of cAMP-dependent protein kinase A (PKA) maps to this chromosome 17q24 locus. Furthermore, we show that PRKAR1α frameshift mutations in three unrelated families result in haploinsufficiency of R1α and cause Carney complex. We did not detect any truncated R1α protein encoded by mutant PRKAR1α. Although cardiac tumorigenesis may require a second somatic mutation, DNA and protein analyses of an atrial myxoma resected from a Carney complex patient with a PRKAR1α deletion revealed that the myxoma cells retain both the wild-type and the mutant PRKAR1α alleles and that wild-type R1α protein is stably expressed. However, in this atrial myxoma, we did observe a reversal of the ratio of R1α to R2β regulatory subunit protein, which may contribute to tumorigenesis. Further investigation will elucidate the cell-specific effects of PRKAR1α haploinsufficiency on PKA activity and the role of PKA in cardiac growth and differentiation.
This article may have been published online in advance of the print edition. The date of publication is available from the JCI website, http://www.jci.org. J. Clin. Invest. 106:R31–R38 (2000).
Introduction
Cardiac myxomas are benign neoplasms that occur in 7 per 10,000 individuals (1). These slowly proliferating lesions arise from subendocardial pluripotent primitive mesenchymal cells, which can differentiate within myxomas along a variety of lineages including epithelial, hematopoietic, and muscular (2, 3). Morbidity and mortality from cardiac myxomas are the result of embolic stroke, heart failure due to intracardiac obstruction, and rheumatologic symptoms attributed (4) to myxoma-mediated production of IL-6. Seven percent of cardiac myxomas (1) are components of a familial autosomal dominant syndrome (Figure 1) that has been variably referred to as LAMB (lentigines, atrial myxoma, mucocutaneous myxoma, blue nevi) and NAME (nevi, atrial myxoma, myxoid neurofibromata, ephelides) and more recently as Carney complex (OMIM #160980, ref. 5; and ref. 6). In Carney complex, autosomal dominant cardiac myxomas are associated with spotty pigmentation of the skin and nonneoplastic hyperfunctioning endocrine states, e.g., primary pigmented nodular adrenocortical hyperplasia. Individuals affected with Carney complex may have extracardiac (most often cutaneous) myxomas, as well as a variety of other benign neoplasms including schwannomas, pituitary adenomas, thyroid adenomas, and breast fibroadenomas. Cardiac myxomas of Carney complex are histologically indistinguishable from more common sporadic cardiac myxomas and, like the latter, most often arise in the left atrium at the fossa ovalis (7). However, unlike sporadic cardiac myxomas, which most often occur as isolated single lesions in middle-aged women and which are usually amenable to surgical resection, syndromic cardiac myxomas exhibit no age or sex preference and may present as multiple concurrent lesions in any cardiac chamber. Affected individuals may have multiple recurrences at any cardiac location despite adequate surgical margins (7).
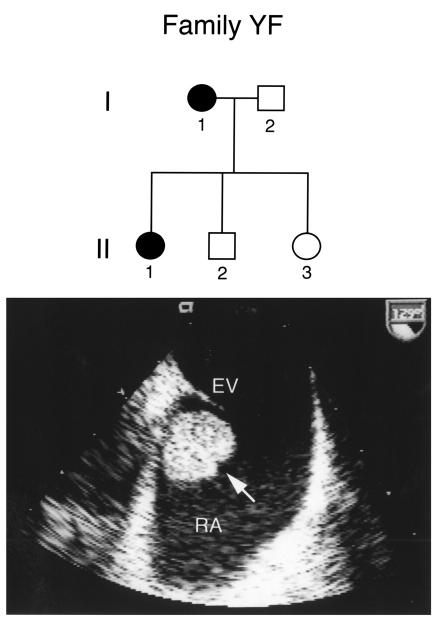
Figure 1.
Pedigree of family YF and cardiac myxoma in a proband. The subject number and disease status of each family member analyzed are indicated. Squares denote male family members, and circles females. Affected and unaffected individuals are represented by closed and open symbols, respectively. Carney complex is transmitted in an autosomal dominant fashion. Transesophageal echocardiography of individual I-1 revealed an unusual posterior mass (arrow) in the right atrium (RA) below the eustachian valve (EV). Pathologic analysis at surgery revealed that the mass was a cardiac myxoma.
Clinical evaluation and genetic linkage analysis of families affected by Carney complex (8, 9) suggested two human chromosomal loci for disease genes: chromosome 2p16 and chromosome 17q24. Linkage to the chromosome 17q24 locus (CAR) was observed in five families affected by Carney complex (8). We have now refined the CAR interval and used a positional cloning strategy to demonstrate that familial cardiac myxomas and Carney complex are caused by mutations in the gene (PRKAR1α) encoding the R1α regulatory subunit of cAMP-dependent protein kinase A (PKA) on chromosome 17q24. We further propose that PRKAR1α acts as a tumor suppressor gene to regulate cell proliferation within the human heart.
Methods
Clinical evaluation.
Informed consent was obtained from all participants in accordance with the Weill Medical College of Cornell University Committee on Human Rights in Research. All family members were evaluated by a thorough history and physical exam without knowledge of genotype status. If there was any evidence of dermatologic, cardiac, or endocrine disease suggestive of Carney complex, patients were further evaluated by electrocardiography, transthoracic echocardiography, and serum chemistry.
Genetic analyses.
Peripheral blood was obtained from each family member, and lymphoblastoid lines were established by transformation with the Epstein-Barr virus (8). Genomic DNA and RNA were isolated from peripheral lymphocytes, lymphoblasts, or tumor cells with QIAamp columns (QIAGEN Inc., Valencia, California, USA). Polymorphic short tandem repeats (STRs) were amplified by PCR with published nucleotide primer sequences (10), analyzed on denaturing polyacrylamide gels (8), and visualized by autoradiography or on an ABI 377 sequencer (Perkin-Elmer Inc., Norwalk, Connecticut, USA). Cytogenetic analysis of tumor and lymphocyte samples was performed as previously described (11).
Cloning and sequence analysis of GNAS13.
Oligonucleotides (5′-CTATTCTGCATGACAACCTGAAGC-3′ / 5′-TCTAATTCTGGTTGTAAACTGCTA-3′) corresponding to the 3′ portion of the GNAS13 gene (12), including untranslated sequence, were used to screen, by PCR, arrayed pools of the Roswell Park Cancer Institute (Buffalo, New York, USA) human genomic PAC library. These oligonucleotides were also used to amplify a GNAS13 gene segment from human genomic DNA. This product was random hexamer–radiolabeled and used as a probe to screen membrane arrays of the Roswell Park Cancer Institute human genomic BAC library. BAC and PAC DNA was prepared with the QIAGEN Large Construct kit (QIAGEN Inc.) and was sequenced directly with oligonucleotides randomly designed from GNAS13 coding sequence on an ABI 377 sequencer (Perkin-Elmer Inc.). Primers were designed as follows from intronic sequence flanking exons 2, 3, and 4: Exon 2: 5′-GAGTCAGTTCGCTGGTTCC-3′ / 5′-TGCCCTTAACCCCGGCCCCATTC-3′ Exon 3: 5′-AAGGTTTGTCTAGGTTAATTC-3′ / 5′-TAGCCTTTCAGTCTGTTCCTCCA-3′ Exon 4: 5′-GAGTCAGTTCGCTGGTTCC-3′ / 5′-TGCCCTTAACCCCGGCCCCATTC-3′ Exon 5 was amplified in two overlapping segments using primer pairs: 5′-AGGCATGACCACCATGTCTGACCA-3′ / 5′-GATTGGTCAGTCGATCTTCCA-3′ 5′-GTTTCGACAGTGTGACATCAATAC-3′ / 5′-TCTAATTCTGGTTGTAAACTGCTA-3′
Each exon was PCR-amplified with the appropriate primer pair from human genomic DNA samples with reaction conditions as follows: [94°C × 20 s, 57°C × 30 s, 72°C × 45 s] × 35 cycles. In all cases except exon 2 amplification, standard PCR reagents were used with AmpliTaq Gold (Perkin-Elmer Inc.). Exon 2 was amplified with the QIAGEN Taq DNA Polymerase Kit (QIAGEN Inc.). Amplified products were sequenced in both directions on an ABI 377 automated sequencer using BigDye Terminator Cycle Sequencing reagents (Perkin-Elmer Inc.).
PRKAR1α sequence analysis.
Oligonucleotides (as below) were designed based on intronic sequence flanking each PRKAR1α exon (3–11) and were used to PCR-amplify them from human genomic DNA samples. PCR conditions were [94°C × 20 s, 57°C × 30 s, 72°C × 45 s] × 35 cycles with standard PCR reagents and AmpliTaq Gold (Perkin-Elmer Inc.). Exon 3: 5′-GAATTGGTGTTTTCCTCTTAACTT-3′ / 5′-TATGATTCATTCATCAAAGGAGAC-3′ Exon 4: 5′-AATGTTTTTGGTTTATGGAATTGT-3′ / 5′-CACACCCTTACTTGAAAAATAGTG-3′
Exon 5: 5′-GACAGTCTGGGGTCTTTAATTCTA-3′ / 5′-TCAAAGAGGAAAACAAACTTCAAT-3′ Exon 6: 5′-TTTCTTTAATTTGGAATATGCTTC-3′ / 5′-ATCTGACATACAAGGGATGTAATG-3′ Exon 7: 5′-TTTTTAAAACAAAGTTCAGGATTG-3′ / 5′-CTAAATCACACTCTCAAACACCAT-3′ Exon 8: 5′-ATTATTCCATAGCATTATGTGGTG-3′ / 5′-AGTCACAGAGGAAATAACTGTGAA-3′ Exon 9: 5′-GGCTATTTGTTGAATCTCTTTAT-3′ / 5′-TGAGTTCTTTACCTCTAAAATTCAA-3′ Exon 10: 5′-TTGTTTAGCTTTTTGG GATTTTA-3′ / 5’GGAGAAGACAAAA TATGGAAGAC-3′ Exon 11: 5′-TATTGTCTTCTTTCTCAGAAGTGC-3′ / 5′-GTGCAATAAAAGCAACTTTCAATA-3′
Exons 1 and 2 were amplified by RT-PCR from patient lymphocyte or lymphoblast RNA samples with the Access RT-PCR kit (Promega Corp., Madison, Wisconsin, USA) and primers:
5′-GCTGGGAGCAAAGCGCTGAGGGA-3′ / 5′-TGTAGACCTCAGCGCTGATAGC-3′
Amplified products were sequenced in both directions on an ABI 377 automated sequencer using BigDye Terminator Cycle Sequencing reagents (Perkin-Elmer Inc.). Two novel STRs, an AG28 dinucleotide repeat and an ATT11 trinucleotide repeat, were noted in the PRKAR1α 5′ untranslated region and 21 kb 3′ to the gene, respectively. PCR primers used to amplify using these repeats for genotyping studies were: AG: 5′-CTGCAGGAAGAGGGAGAAATGGGAA-3′ / 5′-AGAAGTTAAGTGACTTGCCCATGGC-3′ ATT: 5′-TTTCAGCCTCAGTATCTTTATTTG-3′ / 5′-ACCTAAAGTCTTTCTGCCAGTTAT-3′
Western blot analysis.
Lymphoblast and tumor samples were solubilized in 2% SDS. Equivalent protein concentrations of lysate were electrophoresed on 10% acrylamide denaturing gels and transferred to PVDF membrane. Western blotting was performed with anti-actin (Sigma Chemical Co., St. Louis, Missouri, USA) and with primary mAb’s to the R1α and R2β PKA subunits (Becton Dickinson Transduction Laboratories, Lexington, Kentucky, USA). Antigenic epitopes in the PKA regulatory subunits are in the NH2-terminal hinge region (13), and positive controls supplied by Becton Dickinson Transduction Laboratories were rat cerebrum lysate (R1α) and human aortic endothelial lysate (R2β). Bound primary antibody was detected by ECL (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA). Densitometry was performed with a Fluor-S imaging system and MultiAnalyst software (Bio-Rad Laboratories Inc., Hercules, California, USA). An equivalent amount of total cellular protein for lymphoblast samples was loaded on each gel, based on actin expression. As estimated relative to actin expression, the amount of myxoma lysate cellular protein loaded was 10% of lymphoblast lysate cellular protein.
Results
Genetic and physical mapping of the CAR locus.
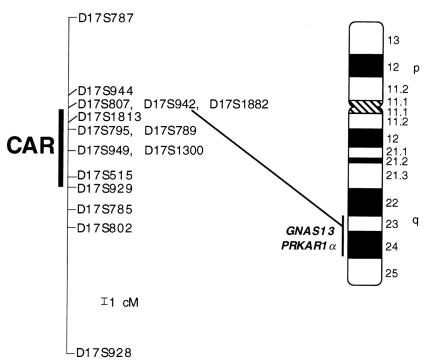
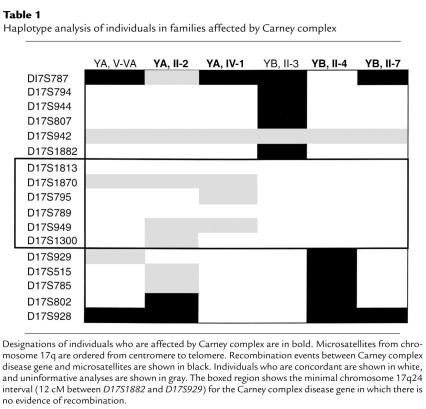
Our previous analyses demonstrated that the Carney complex disease gene is located in the 17-cM interval between D17S807 and D17S785. Analysis of high-density genetic and physical maps (10, 12) of this locus (Figure 2) permitted the fine ordering of ten additional microsatellite polymorphisms (Figure 2, Table 1) within this interval, which were used to genotype families YA and YB. Haplotype analysis revealed recombination events between the Carney complex disease gene and D17S1882 and D17S929 at the centromeric and telomeric ends, respectively, of the CAR locus. Thus, these genetic analyses refined the CAR locus and established a minimal genetic interval of about 12 cM between D17S1882 and D17S929 for the CAR locus.
Figure 2.
Ideogram of chromosome 17 with Giemsa banding pattern showing localization of the Carney complex locus and the PRKAR1α gene. Positive bands are black and the pericentromeric region is gray. Numbers indicate cytogenetic designations for bands. The genetic map locations of polymorphic loci analyzed from 17q are given. One centimorgan (cM) is indicated. Linkage data suggested that the gene responsible for Carney complex is located in the 12-cM interval between D17S1882 and D17S929 (CAR). Both GNAS13 and PRKAR1α map cytogenetically to chromosome 17q23-q24.
Table 1.
Haplotype analysis of individuals in families affected by Carney complex
Exclusion of GNAS13.
Computerized analysis of the Human Genome Project/GenBank database (12) revealed that the D17S2016 sequence-tagged site (STS) was included within the centromeric portion of the CAR locus. BLAST analysis of this STS demonstrated that the sequence corresponded to the coding sequence of the GNAS13 gene (GenBank accession no. L22075) encoding the GTP-binding protein, Gα13. Mutations in another GTP-binding protein involved in intracellular signal transduction, Gsα, cause McCune-Albright syndrome, which shares several phenotypic features with Carney complex, including abnormal skin pigmentation and endocrinopathy (5), but Gsα mutations are not found in Carney complex patients (14). Given the homologies between Gα13 and Gsα and the chromosomal location of the GNAS13 gene, we searched for mutations in the GNAS13 gene in five unrelated Carney complex patients.
We screened the Roswell Park Cancer Institute BAC and PAC human genomic DNA libraries and identified six BAC (472J20, 484A6, 583F2, 489G5, 657B12, 752A23) and four PAC (704J11556, 704K19308, 704A09613, 704A10613) clones for GNAS13. Comparison of BAC/PAC sequences with GNAS13 cDNA sequence established the genomic structure of the human GNAS13 gene: 5 exons with an open reading frame beginning in exon 2. Exons 2–5 (GNAS13 coding sequence) were amplified from patient DNA samples and subjected to automated sequencing. Although a GGC7 trinucleotide STR within the 5′ untranslated sequence in exon 2 of GNAS13 (3 bp 5′ to the translation start site) was found to be polymorphic, no mutations were founding within the GNAS13 coding sequence.
Identification of PRKAR1α mutations.
Interrogation of the Human Genome Project databases further revealed that the PRKAR1α gene (accession no. NM002374) encoding the R1α regulatory subunit of cAMP-dependent PKA was also included within the CAR locus minimal interval (Figure 2). PRKAR1α had previously been mapped (as the Tissue-Specific Extinguisher 1 [TSE1] locus) to chromosome 17q23-q24 by somatic cell hybrid analysis (15, 16). Additionally, we noted that the 155-kb human genomic BAC clone RP11-62F10 (accession no. AC005799.1) contained sequences corresponding to both PRKAR1α and the anonymous STR D17S789, which exhibited no recombination with the Carney complex disease gene in the families that we had studied. Therefore, the PRKAR1α chromosomal location, the known role of PKA in signal transduction and cell growth, and the ubiquitous expression pattern of the R1α subunit (16–18) all suggested that PRKAR1α was a candidate disease gene for Carney complex.
The coding sequence of PRKAR1α is 1.146 kb in length. Comparison of the genomic sequence derived from BAC RP1162F10 with PRKAR1α cDNA sequence elucidated the intron-exon boundaries and showed that the gene was comprised of 11 exons (Figure 3), not 10 as previously reported (19). However, no novel genomic sequence was available in the Human Genome Project database corresponding to exons 1 and 2 of the PRKAR1α gene to compare with the previously described genomic structure (accession no. Y07641). Analysis of the genomic sequence of a PRKAR1α pseudogene contained within BAC clone RP4-621B10 (ref. 19; accession no. AL13883), which maps to chromosome 1 and begins with sequence 5′ to the open reading frame, raised the possibility that the proposed boundary (19) between exons 1 and 2 might be imprecise.
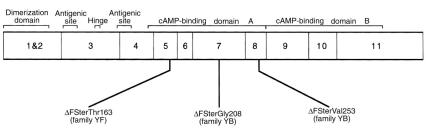
Figure 3.
Three frameshift mutations in PRKAR1α that cause Carney complex. A schematic representation of PRKAR1α cDNA shows exons 1–11. Regions that encode functional domains (dimerization domain, antigenic sites, hinge/pseudophosphorylation site, cAMP-binding domains A and B) of R1α are denoted (13). Location of the PRKAR1α deletion mutations ΔFSterGly208 in family YA, ΔFSterVal253 in family YB, and ΔFSterThr163 in family YF are shown. Mutations are denoted by ΔFSter to indicate a deletion with resultant frameshift and premature stop codon, and by the first amino acid residue affected by the deletion.
To analyze the coding sequence of the PRKAR1α gene for mutations that might cause Carney complex, we designed PCR primers from the intron sequence flanking exons 3–11 and amplified these exons from human genomic DNA samples. Because of ambiguity about the genomic organization of PRKAR1α exons 1 and 2, we designed oligonucleotides based on coding sequence from exon 3 and on 5′ untranslated sequence within exon 1 to amplify, via RT-PCR, exons 1 and 2 from lymphocyte/lymphoblast RNA samples. PRKAR1α exons 1–11 amplified from five Carney complex unrelated probands were then subjected to sequence analysis. Two probands were from families that had been linked to chromosome 17q24 (families YA and YB), and the others (YE, YF, and SY2) were from families that were too small to definitively establish linkage to any chromosomal locus. Clinical features of families YA and YB have been previously described (8). Probands from the other three families all exhibit spotty pigmentation of the skin and have had cardiac myxomas.
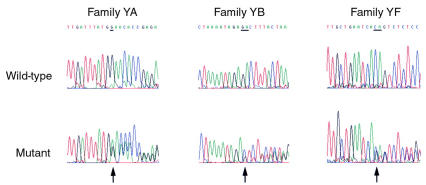
We observed heterozygous PRKAR1α gene mutations (Figures 3 and 4) in three Carney complex probands, including the two probands from chromosome 17q24–linked families (YA and YB). Bidirectional sequence analysis demonstrated the presence of a 1-bp deletion (G) of nucleotide 710 at Gly208 in affected individuals but not unaffected individuals in family YA with a consequent frameshift and premature stop 13 codons later. A 2-bp deletion (TC) of nucleotides 845–846 at Val253 was observed in affected individuals but not unaffected individuals from family YB with a consequent frameshift and premature stop 15 codons later. A 2-bp deletion (TG) of nucleotides 576–577 at Thr163 was present in all affected individuals but not in unaffected individuals in family YF with a resultant frameshift leading to a premature stop 6 codons later. Mutations (ΔFSterGly208 in family YA, ΔFSterVal253 in family YB, ΔFSterThr163 in family YF) were confirmed by sense and antisense sequence analysis and by gel electrophoresis of PCR products to demonstrate the different lengths of the alleles. None of these sequence variants was observed in 100 normal unrelated chromosomes. We also observed a single base pair thymine insertion in a poly-T tract in intron 3 that is 9 nucleotides 5′ to the exon 4 splice acceptor site. This insertion was a common polymorphism that was present in 50% of individuals genotyped.
Figure 4.
Mutational analysis of PRKAR1α in Carney complex families YA, YB, and YF. Automated sequence analyses of wild-type and mutant exons 7, 8, and 5 amplified from normal and affected individuals in families YA, YB, and YF, respectively, are shown. Sequence of the sense strand is shown for family YA, and the antisense strand for families YB and YF. A heterozygous 1-bp deletion is noted in family YA, and 2-bp deletions in families YB and YF (arrows; deleted bases are underlined). These mutations all result in a frameshift with consequent premature stop codons.
PRKAR1α analysis in a cardiac myxoma.
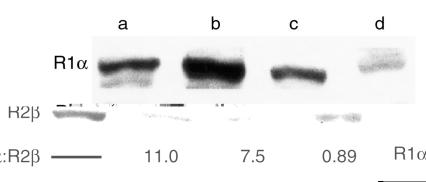
The frameshift PRKAR1α mutations observed in families YA, YB, and YF would all yield truncated protein products lacking the cAMP-binding domains (Figure 3) if the mutant alleles were transcribed and translated. Alternatively, these mutant alleles might be null alleles due to degradation of the mutant mRNA via nonsense-mediated decay (20). To distinguish between these possibilities, expression of the R1α protein was determined with specific antibody to the NH2-terminus of R1α by Western blot (Figure 5) of lymphoblasts derived from a normal individual, lymphoblasts derived from an affected individual (family YA, IV-1 with the Gly208 deletion/frameshift mutation described above), and a left atrial myxoma from this affected individual. Only full-length R1α protein was present in all three samples without any evidence of truncated product. Densitometry showed that the amount of R1α protein in Carney complex lymphocytes was 60% less than that in normal lymphocytes, consistent with haploinsufficiency of PRKAR1α in Carney complex. Full-length R1α was also present in the atrial myxoma sample, suggesting that the wild-type allele is preserved in the tumor. Cytogenetic analysis of this atrial myxoma revealed a normal 46,XX karyotype and failed to demonstrate any abnormalities at chromosome 17q23-24. Genotyping with STR D17S789, which is closely genetically and physically linked to the PRKAR1α gene as described above, as well as with an ATT trinucleotide repeat 21 kb 3′ to the PRKAR1α coding sequence demonstrated no loss of heterozygosity at this locus. (Genotyping with an AG dinucleotide repeat in the 5′ untranslated PRKAR1α sequence was uninformative.) Sequence analysis of PRKAR1α exon 7 shows that both the wild-type and deleted mutant alleles are present in this tumor. Finally, to date, we have not detected any additional sequence variants in PRKAR1α amplified from this atrial myxoma. Densitometry of Western blot analysis (Figure 5) of the R2β regulatory subunit in the same samples (normal lymphoblasts, Carney complex lymphoblasts, and atrial myxoma) analyzed for R1α protein expression demonstrated that R2β expression was similar in normal and affected lymphocytes, with a decrease in the R1α:R2β ratio secondary to decreased R1α expression consistent with PRKAR1α haploinsufficiency. However, in the tumor sample there was a reversal in the R1α:R2β ratio that may relate to further decreased R1α and/or increased R2β (Figure 5).
Figure 5.
Western blot analysis of R1α and R2β in normal lymphoblasts, Carney complex lymphoblasts, and Carney complex cardiac myxoma. (a) Positive controls for antibodies to R1α and R2β were electrophoresed and Western blotted as described in Methods. (b, c) Protein lysates of lymphoblasts from (b) a normal individual and (c) an individual affected by Carney complex (family YA, IV-1) were electrophoresed and analyzed. Densitometry revealed similar levels of R2β protein in the two samples but a 60% decrease in R1α levels. (d) R1α and R2β were analyzed in lysate from a left atrial myxoma resected from individual IV-1 in family YA. Densitometry was used to determine R1α and R2β expression in all samples, and the ratio of R1α:R2β was calculated. A reversal of the R1α:R2β ratio is observed in the tumor sample compared with affected and unaffected lymphocytes.
Discussion
We have demonstrated that mutations in the PRKAR1α gene encoding the R1α regulatory subunit cause familial cardiac myxomas in autosomal dominant Carney complex. Individuals at risk for cardiac myxomas who are affected by Carney complex develop disease based on PRKAR1α haploinsufficiency secondary to heterozygous constitutional frameshift mutation of PRKAR1α that results in a nonexpressed allele. Moreover, PRKAR1α acts as a tumor suppressor gene in the heart and other tissues, presumably by regulation of PKA activity. PRKAR1α function as a tumor suppressor gene is consistent with findings (21) demonstrating that PRKAR1α accounts for tissue-specific inhibition of gene transcription at the TSE1 locus on chromosome 17q24. Haploinsufficiency of another kinase, STK11, followed by somatic mutation, causes the phenotypically related Peutz-Jeghers syndrome, characterized by benign neoplasms and spotty pigmentation of the skin (22). Similarly, constitutional mutation of PRKAR1α followed by somatic mutation of PRKAR1α or other melanocyte genes likely accounts for the spotty skin pigmentation in Carney complex. We have not observed a second acquired somatic mutation of the PRKAR1α wild-type allele in the tumor analyzed here, but a “two-hit” mechanism of tumorigenesis (23) remains possible for other syndromic and nonsyndromic cardiac myxomas. Although our Western blot analyses detected no mutant, truncated R1α protein in a cardiac myxoma, failure to recognize truncated protein could be due to low levels of protein or to conformational abnormalities with consequent loss of antigenicity. Thus, a dominant-negative contribution of a mutant protein remains a formal possibility. Our findings do show that if a second “hit” is necessary for cardiac tumorigenesis, the acquired mutation is not required to occur in PRKAR1α. The diverse signal transduction proteins that interact with R1α and cAMP-dependent PKA, as well as other PKA subunits, are candidates to be studied for somatic mutation in these tumors. Such a two-gene model of tumorigenesis is similar to that observed in murine neurofibromatosis models, which require null alleles in the NF1 tumor suppressor gene as well as p53 for tumorigenesis (24, 25).
Loss of the R1α regulatory subunit promotes cell proliferation and growth of benign tumors in multiple tissues. The cell biological consequences of the genetic abnormalities in R1α are likely mediated via changes in PKA activity. The PKA holoenzyme is a tetramer comprised of two catalytic subunits and two regulatory subunits. Genes for four regulatory subunits have been identified — R1α, R1β, R2α, and R2β (3, 18) — which differ in their tissue-specific expression patterns (3, 26). In its tetrameric form, PKA is inactive. Upon binding of cAMP to the regulatory subunits, the regulatory subunit dimer undergoes a conformational change and dissociates from the catalytic subunits; the free catalytic subunits are enzymatically active. Activity of cAMP-dependent PKA can either inhibit or stimulate cell proliferation depending on the cell type (18, 26–28). PKA activity in any given cell type is dependent not only upon the concentration of free catalytic subunit but also upon the cell type–specific ratio of regulatory subunits (3). R1:R2 is modulated not only at the gene expression level (18, 19, 29) but at the protein level by a wide array of different cell type–specific binding proteins for the regulatory subunits (A kinase–anchoring proteins, or AKAPs) (26–31). Changes in one regulatory subunit concentration may incompletely compensate for changes in another. For instance, knockout of R1β or R2β in mice leads to a partial increase in R1α, but overall PKA activity is still decreased (26, 29). McKnight and colleagues (26, 29) have hypothesized that maintenance of adequate R1α is a critical intracellular protective mechanism against unregulated catalytic subunit activity.
Hyperendocrine states, benign endocrine neoplasms (e.g., pituitary and thyroid adenomas), and the cutaneous spotty pigmentation of the skin seen in Carney complex are all likely related to increased PKA activity. Elevated PKA activity can be caused by increased free active catalytic subunit in the absence of the regulatory/inhibitory R1α regulatory subunit. For instance, constitutive activation of cAMP-dependent PKA activity produces unregulated growth of autonomous hyperfunctioning human thyrocytes, and mutations in Gsα, which yield elevated cAMP levels and elevated PKA activity, cause growth hormone–secreting pituitary adenomas and thyroid adenomas (32, 33). Thus, loss of the R1α regulatory subunit and increased PKA activity may result in thyroid adenomas as seen in individual I-1 in family YB. Similarly, PKA activity is required for tyrosine hydroxylase (TH) activity that contributes both to catecholamine synthesis and biosynthesis of melanin skin pigmentation (34). PKA deficiency decreases TH activity not only due to loss of TH phosphorylation but also because PKA is required for transcriptional regulation of TH gene expression (34).
In nonendocrine cell types, loss of PKA activity may be associated with increased cell proliferation (18, 26, 28). For instance, Svenningsen and Kanje (28) demonstrated that cAMP-dependent PKA inhibits Schwann cell proliferation, and pharmacologic inhibition of PKA promotes Schwann cell growth. Thus, benign schwannomas seen in Carney complex patients, including individual III-10 from family YA, may be a consequence of decreased Schwann cell PKA activity. In these cells, loss of PRKAR1α may result in decreased PKA activity through regulatory isoform switching, as in the atrial myxoma studied here, or through altered intracellular PKA binding and sequestration by Schwann cell–specific AKAPs that have not yet been identified.
The cellular mechanism by which loss of PRKAR1α modifies PKA activity in the heart and produces cardiac myxomas remains uncertain. Cardiac myxomas arise from rare pluripotent subendocardial mesenchymal stem cells (2–3). These unusual cardiac cells have not been isolated or cultured (35), and the effects of PKA on their biology are unexplored. Rheumatologic symptoms in patients with cardiac myxomas are often attributed to elevated serum IL-6, whose synthesis is PKA-dependent (36). Normal cardiac structure and function require physiologic expression of the CREB transcription factor (37), which is at least partially regulated by PKA-dependent phosphorylation. Both the Cα catalytic subunit and the R1α regulatory subunit are expressed in the developing heart (16–18). Imaizumi-Scherrer et al. (17) concluded that there were low levels of R1α mRNA throughout the E16 mouse heart. However, close examination of their in situ hybridization data (17) suggests that there may be focal increase of R1α expression at the interatrial septum, where most cardiac myxomas, nonsyndromic and syndromic, arise.
We have previously observed (38) that lipomatous hypertrophy of the interatrial septum is common in young adults affected by Carney complex. Knockout mice homozygous-null for the R2β subunit exhibit a partial compensatory increase in R1α expression but an overall decrease in PKA activity, and their phenotype includes a loss of adipose tissue via increased lipolysis (39). In the atrial myxoma studied here, haploinsufficiency of R1α combined with tumorigenesis is associated with a reversal of the R1α:R2β ratio. These findings remain to be confirmed in other tumor samples as they become available. We hypothesize that haploinsufficiency of the R1α subunit in the heart of Carney complex patients may lead to decreased PKA activity with consequent partial compensatory increase in R2β expression and thereby induce adipose tissue growth in the interatrial septum. Furthermore, additional sporadic mutation of PRKAR1α and/or other genes in the cAMP-dependent signal transduction pathway may contribute to decreased PKA activity and altered PKA regulatory isoform expression which, in turn, ultimately leads to cardiac myxoma formation. In fact, the propensity of R2β-regulated PKA activity to induce cellular differentiation (18) may explain the wide variety of differentiated cell lineages (2, 3) observed within cardiac myxomas. In vitro and in vivo experimental models that probe PRKAR1α and other PKA regulatory subunit expression in the heart will elucidate PKA-dependent pathways for cardiac cell growth and differentiation. Manipulation of such PKA-dependent pathways has already prompted therapeutic modalities for malignancies (18) and ultimately may suggest novel therapies to prompt cardiac cell regeneration in the ischemic and cardiomyopathic heart.
Acknowledgments
We are grateful to family members and their physicians (including A. Merliss and A. Denio) for their participation in this study, and to B. Batog for technical assistance. PACs were provided by the Resource Center of the German Human Genome Project at the Max-Planck-Institut for Molecular Genetics. The authors appreciate discussion and comments from M. Gershengorn, C. Nathan, and B. Lerman. This work was supported by an American College of Cardiology/Merck Research Fellowship (to C.J. Vaughan), a National Heart, Lung, and Blood Institute (NHLBI) Minority Postdoctoral Fellowship (to C.J. Hatcher), an American Heart Association Student Scholarship in Cardiovascular Disease and Stroke (to J.M. Winter), a National Cancer Institute grant (CA78895 to C.C. Morton), and grants from the Wendy Will Case Cancer Fund, the American Heart Association (Grant-in-Aid), New York City Affiliate Inc., and NIH/NHLBI (R01 HL-61785) to C.T. Basson.
Footnotes
Mairead Casey and Carl J. Vaughan contributed equally to this work.
References
- 1.Reynen K. Cardiac myxomas. N Engl J Med. 1995;333:1610–1617. doi: 10.1056/NEJM199512143332407. [DOI] [PubMed] [Google Scholar]
- 2.Ferrans VJ, Roberts WC. Structural features of cardiac myxomas: histology, histochemistry, and electron microscopy. Hum Pathol. 1973;4:111–146. doi: 10.1016/s0046-8177(73)80051-6. [DOI] [PubMed] [Google Scholar]
- 3.Burke AP, Virmani R. Cardiac myxomas: a clinicopathologic study. Am J Clin Pathol. 1993;100:671–680. doi: 10.1093/ajcp/100.6.671. [DOI] [PubMed] [Google Scholar]
- 4.Kanda T, et al. Interleukin-6 and cardiac myxoma. Am J Cardiol. 1994;74:965–967. doi: 10.1016/0002-9149(94)90601-7. [DOI] [PubMed] [Google Scholar]
- 5.OMIM™: Online Mendelian Inheritance in Man. Center for Medical Genetics, Johns Hopkins University, Baltimore, Maryland, USA, and National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USA. http://www.ncbi.nlm.nih.gov/omim/.
- 6.Carney JA, Gordon H, Carpenter PC, Shenoy BV, Go VL. The complex of myxomas, spotty pigmentation, and endocrine overactivity. Medicine. 1985;64:270–283. doi: 10.1097/00005792-198507000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Carney JA. Differences between nonfamilial and familial cardiac myxoma. Am J Surg Pathol. 1985;9:53–55. doi: 10.1097/00000478-198501000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Casey M, et al. Identification of a novel genetic locus for familial cardiac myxomas and Carney complex. Circulation. 1998;98:2560–2566. doi: 10.1161/01.cir.98.23.2560. [DOI] [PubMed] [Google Scholar]
- 9.Stratakis CA, et al. Carney complex, a familial multiple neoplasia and lentiginosis syndrome: analysis of 11 kindreds and linkage to the short arm of chromosome 2. J Clin Invest. 1996;97:699–705. doi: 10.1172/JCI118467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaughan CJ, et al. A t(2;19)(p13;p13.2) in a giant invasive cardiac lipoma from a patient with multiple lipomatosis. Genes Chromosomes Cancer. 2000;28:133–137. doi: 10.1002/(sici)1098-2264(200006)28:2<133::aid-gcc1>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 12.Benson DA, et al. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor SS, Buechler JA, Yonemoto W. CAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 14.DeMarco L, et al. Sporadic cardiac myxomas and tumors from patients with Carney complex are not associated with activating mutations of the Gsα gene. Hum Genet. 1996;98:185–188. doi: 10.1007/s004390050187. [DOI] [PubMed] [Google Scholar]
- 15.Boshart M, Weih F, Nichols M, Schutz G. The tissue-specific extinguisher locus TSE1 encodes a regulatory subunit of cAMP-dependent protein kinase. Cell. 1991;66:849–859. doi: 10.1016/0092-8674(91)90432-x. [DOI] [PubMed] [Google Scholar]
- 16.Jones KW, Shapero MH, Chevrette M, Fournier REK. Subtractive hybridization cloning of a tissue-specific extinguisher: TSE1 encodes a regulatory subunit of protein kinase A. Cell. 1991;66:861–872. doi: 10.1016/0092-8674(91)90433-y. [DOI] [PubMed] [Google Scholar]
- 17.Imaizumi-Scherrer T, Faust DM, Benichou JC, Hellio R, Weiss MC. Accumulation in fetal muscle and localization to the neuromuscular junction of cAMP-dependent protein kinase A regulatory and catalytic subunits R1α and Cα. J Cell Biol. 1996;134:1241–1254. doi: 10.1083/jcb.134.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho-Chung YS. Role of cyclic AMP receptor proteins in growth, differentiation, and suppression of malignancy: new approaches to therapy. Cancer Res. 1990;50:7093–7100. [PubMed] [Google Scholar]
- 19.Solberg R, et al. The human gene for the regulatory subunit R1α of cyclic adenosine 3’,5′-monophosphate-dependent protein kinase: two distinct promoters provide differential regulation of alternatively spliced messenger ribonucleic acids. Endocrinology. 1997;138:169–181. doi: 10.1210/endo.138.1.4864. [DOI] [PubMed] [Google Scholar]
- 20.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 21.Lem J, Chin AC, Thayer MJ, Leach RJ, Fournier REK. Coordinate regulation of two genes encoding gluconeogenic enzymes by the trans-dominant locus Tse-1. Proc Natl Acad Sci USA. 1988;85:7302–7306. doi: 10.1073/pnas.85.19.7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenne DE, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 23.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cichowski K, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 25.Vogel KS, et al. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amieux PS, et al. Compensatory regulation of RIα protein levels in protein kinase A mutant mice. J Biol Chem. 1997;272:3993–3998. doi: 10.1074/jbc.272.7.3993. [DOI] [PubMed] [Google Scholar]
- 27.Indolfi C, et al. Activation of cAMP-PKA signaling in vivo inhibits smooth muscle proliferation induced by vascular injury. Nat Med. 1997;3:775–779. doi: 10.1038/nm0797-775. [DOI] [PubMed] [Google Scholar]
- 28.Svenningsen AF, Kanje M. Regulation of schwann cell proliferation in cultured segments of adult rat sciatic nerve. J Neurosci Res. 1998;52:530–537. doi: 10.1002/(SICI)1097-4547(19980601)52:5<530::AID-JNR5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 29.Burton KA, et al. Type II regulatory subunits are not required for the anchoring-dependent modulation of Ca2+ channel activity by cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1997;94:11067–11072. doi: 10.1073/pnas.94.20.11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miki K, Eddy EM. Single amino acids determine specificity of binding of protein kinase A regulatory subunits by protein kinase A anchoring proteins. J Biol Chem. 1999;274:29057–29062. doi: 10.1074/jbc.274.41.29057. [DOI] [PubMed] [Google Scholar]
- 31.Kapiloff MS, Schillace RV, Westphal AM, Scott JD. mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J Cell Sci. 1999;112:2725–2736. doi: 10.1242/jcs.112.16.2725. [DOI] [PubMed] [Google Scholar]
- 32.Duprez L, et al. TSH receptor mutations and thyroid disease. Trends Endocrinol Metab. 1998;9:133–140. doi: 10.1016/s1043-2760(98)00036-8. [DOI] [PubMed] [Google Scholar]
- 33.Harris PE. Gs protein mutations and the pathogenesis and function of pituitary tumors. Metabolism. 1996;45:120–122. doi: 10.1016/s0026-0495(96)90104-3. [DOI] [PubMed] [Google Scholar]
- 34.Kim KS, et al. A dual role for the cAMP-dependent protein kinase in tyrosine hydroxylase gene expression. Proc Natl Acad Sci USA. 1993;90:3471–3475. doi: 10.1073/pnas.90.8.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams RS. Cell cycle control in the terminally differentiated myocyte: a platform for myocardial repair. Cardiol Clin. 1998;16:739–754. doi: 10.1016/s0733-8651(05)70048-5. [DOI] [PubMed] [Google Scholar]
- 36.Murakami S, et al. Adenosine regulates the production of interleukin-6 by human gingival fibroblasts via cyclic AMP/protein kinase A pathway. J Periodontal Res. 2000;35:93–101. doi: 10.1034/j.1600-0765.2000.035002093.x. [DOI] [PubMed] [Google Scholar]
- 37.Fentzke RC, et al. Dilated cardiomyopathy in transgenic mice expressing a dominant-negative CREB transcription factor in the heart. J Clin Invest. 1998;101:2415–2426. doi: 10.1172/JCI2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein MM, Casey M, Carney JA, Basson CT. Molecular genetic diagnosis of the familial myxoma syndrome (Carney complex) Am J Med Genet. 1999;86:62–65. doi: 10.1002/(sici)1096-8628(19990903)86:1<62::aid-ajmg12>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 39.Planas JV, Cummings DE, Idzerda RL, McKnight GS. Mutation of the RIIβ subunit of protein kinase A differentially affects lipolysis but not gene induction in white adipose tissue. J Biol Chem. 1999;274:36281–36287. doi: 10.1074/jbc.274.51.36281. [DOI] [PubMed] [Google Scholar]