Abstract

The system specific chaperone DmsD plays a role in the maturation of the catalytic subunit of dimethyl sulfoxide (DMSO) reductase, DmsA. Pre-DmsA contains a 45-amino acid twin-arginine leader peptide that is important for targeting and translocation of folded and cofactor-loaded DmsA by the twin-arginine translocase. DmsD has previously been shown to interact with the complete twin-arginine leader peptide of DmsA. In this study, isothermal titration calorimetry was used to investigate the thermodynamics of binding between synthetic peptides composed of different portions of the DmsA leader peptide and DmsD. Only those peptides that included the complete and contiguous hydrophobic region of the DmsA leader sequence were able to bind DmsD with a 1:1 stoichiometry. Each of the peptides that were able to bind DmsD also showed some α-helical structure as indicated by circular dichroism spectroscopy. Differential scanning calorimetry revealed that DmsD gained very little thermal stability upon binding any of the DmsA leader peptides tested. Together, these results suggest that a portion of the hydrophobic region of the DmsA leader peptide determines the specificity of binding and may produce helical properties upon binding to DmsD. Overall, this study demonstrates that the recognition of the DmsA twin-arginine leader sequence by the DmsD chaperone shows unexpected rules and confirms further that the biochemistry of the interaction of the chaperone with their leaders demonstrates differences in their molecular interactions.
Prokaryotic preproteins with an N-terminal twin-arginine (RR) motif-containing leader peptide are translocated in a fully folded state by the twin-arginine translocation (Tat) system.1−4 Many Tat preproteins are components of multisubunit respiratory enzymes that acquire cofactors, fold, and associate with partner proteins in the cytoplasm, prior to the translocation event. A system specific chaperone, also termed a redox enzyme maturation protein (REMP), is believed to play a role in assessing the folded and cofactor-loaded state of the Tat preprotein prior to translocation.5,6 While the details of REMP activity and function are yet to be defined, it is known that each REMP interacts specifically with its Tat preprotein substrate prior to translocation7 and deletion of the REMP significantly reduces the activity of the final holoenzyme.8,9
Understanding the specificity and affinity of the interaction between each REMP and Tat substrate pair has been the focus of studies over the past several years.7,10−19 Most REMPs bind to only their Tat substrate RR-leader peptide in vitro; however, some cross interaction does occur.7 Additionally, these chaperones have been shown to interact with different portions of the Tat preprotein, either the RR-leader peptide, the mature protein, or the entire preprotein (composed of the RR-leader peptide and mature protein).7,18 Binding affinities between REMPs and their leader peptides vary depending on the Tat substrate construct and technique used but consistently range from nanomolar to micromolar dissociation constants (Kd).7,11−13,19
The Escherichia coli REMP DmsD binds to the DmsA preprotein, the catalytic subunit of the dimethyl sulfoxide (DMSO) reductase molybdoenzyme (DmsABC), through an interaction with the DmsA twin-arginine leader peptide (DmsAL)8 and not the mature portion of the DmsA protein.7,18 DmsD has some cross specificity and was shown to interact with the RR-leader peptides of DmsA homologues, YnfE and YnfF,7 as well as pre-TorA (the catalytic subunit of trimethylamine N-oxide reductase).8 The DmsAL peptide consists of 45 amino acids and, like other RR-leader peptides, is composed of four regions:20 the amino-terminal N-region (residues 1–14), the twin-arginine -SRRGLVK- motif (residues 15–21), the hydrophobic H-region (residues 22–39), and the carboxy-terminal C-region (residues 40–45) with residues 43–45 composing the cleavage recognition site (Figure 1). The interaction between DmsD and the 43 N-terminal amino acids of the DmsA RR-leader peptide (DmsAL1–43) has been characterized with a variety of in vitro techniques in which the leader peptide was fused to the N-terminus of either glutathione S-transferase (GST) or streptavidin binding peptide (SBP).7,8,13,15,21 The Kd values between DmsD and the GST fusion (DmsAL1–43::GST) and the SBP fusion (DmsAL1–43::SBP) were found to be in the same range (0.22 and 0.06 μM, respectively).7,13
Figure 1.

Sequence comparison of twin-arginine leader peptides capable of binding DmsD. The amino-terminal N-region (light gray), including the twin-arginine (RR) motif, the hydrophobic H-region (black), and the C-region (dark gray) are highlighted. Twin-arginine leader peptide sequences capable of binding DmsD are aligned, and the consensus is shown below with identical (asterisks) and similar (colons) residues indicated.
In this study, the region of the DmsAL peptide capable of interacting with the DmsD chaperone is identified, and residues of DmsAL that allow specificity of binding are highlighted. Toward this end, peptides composed of various portions of the DmsAL peptide were synthesized and isothermal titration calorimetry (ITC) was used to characterize the affinity and thermodynamics of the interaction with DmsD. Circular dichroism (CD) spectroscopy was used to investigate the secondary structure of the DmsAL peptides in aqueous solution as well as a more hydrophobic environment of 50% trifluoroethanol (TFE) to determine if the propensity to form the helical structure of the peptide plays a role in binding. Differential scanning calorimetry (DSC) was used to examine the thermostability of DmsD alone and in complex with each of the DmsAL peptides to examine if DmsD undergoes a conformational change upon binding the DmsAL peptide.
Materials and Methods
Protein and Peptide Preparation for Isothermal Titration Calorimetry
The purification and storage procedures for recombinant E. coli DmsD (His6T7::DmsD construct13) have been described elsewhere.15 Briefly, the protein was purified with Ni affinity chromatography and eluted using two imidazole concentrations to separate monomeric and dimeric DmsD protein populations that were assessed via size exclusion chromatography and fast performance liquid chromatography. Prior to ITC experiments, purified monomeric DmsD was thawed and exchanged into ITC buffer [25 mM Tris-HCl (pH 8.0) and 100 mM NaCl] via the application to a 5 mL HiTrap column and the collection of eluted fractions. The protein concentration was determined from absorption at 280 nm [extinction coefficient of 72085 M–1 cm–1, determined using the online ExPASy tool ProtParam (available at http://web.expasy.org/protparam/)].23 DmsD protein was diluted to 25 μM with ITC buffer, prepared in 10 mL batches to allow for triplicate titrations, and stored at 4 °C prior to titration experiments. For each individual titration, 3 mL of DmsD was degassed with a thermovac at 28 °C for 10 min while being stirred. The ITC sample cell was loaded with 1.5 mL of degassed DmsD sample, and the sample remaining in the syringe after fill was used to confirm the protein concentration with a Bradford assay and absorbance at 280 nm.
DmsAL peptides were chemically synthesized and purified to >95% purity by GenScript, as determined by high-performance liquid chromatography. 1H nuclear magnetic resonance (NMR) spectroscopy of selected peptides, compared to an internal standard [4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS)], was used to determine that 70% of the lyophilized peptide mass corresponded to peptide mass, and this conversion was used to set the final peptide concentration in solution. Peptides were weighed on an analytical balance and dissolved in distilled water to a concentration of 1 mM. All peptides were completely soluble in water, and no precipitate was visible after centrifugation at 10000 rpm for 10 min. Peptide samples were then diluted to a concentration of 0.5 mM with 2× ITC buffer and stored at 4 °C prior to titration. For each titration, 0.5 mL of the DmsAL peptide sample was degassed and equilibrated to 28 °C for 10 min with a thermovac. The ITC syringe was loaded with degassed and temperature-equilibrated peptide.
Isothermal Titration Calorimetry
ITC experiments were conducted with a MicroCal VP-ITC microcalorimeter at 303.15 K (30 °C). Thirty 10 μL injections of 500 μM DmsAL peptide were added to 25 μM DmsD protein while the mixture was being stirred at 300 rpm. The reference power applied was 10 (units), and an initial delay and spacing of 300 s was used. DmsD titration experiments were performed in triplicate. The heat of dilution was determined by injection of each peptide ligand into ITC buffer, and the resulting values were subtracted from the individual heats of reaction to obtain the observed heat of binding. Origin 7.0 was used for data fitting with a single-binding site model. The mean and standard error of the mean of binding constants and corresponding thermodynamics of binding are reported. Buffer ionization enthalpies24 were used to determine if protonation or deprotonation occurred during formation of the complex.
Secondary Structure Prediction of DmsAL Peptides
Two algorithms were used to predict the secondary structure of DmsAL peptide sequences shown in Figure 4: PSIPRED (version 3.3) available at http://bioinf.cs.ucl.ac.uk/psipred/(25,26) and AGADIR available at http://agadir.crg.es/agadir.jsp.27,28
Figure 4.
Helical content of DmsA leader peptides. Far-UV circular dichroism spectra were collected for DmsAL peptides in water (dashed line) and 50% TFE (solid line) for DmsAL2–20 (A), DmsAL15–33 (B), DmsAL15–41 (C), DmsAL17–41 (D), DmsAL21–41 (E), and DmsAL15–41 KK (F). The helical content (percent) of each peptide was approximated using Dichroweb analysis and is shown for each peptide (in water → in 50% TFE) (A–F). (G) Helical content predictions were determined for DmsAL peptide sequences using AGADIR and PSIPRED. Regions of the peptide predicted to be helical by the PSIPRED algorithm are underlined. nd, not determined.
Circular Dichroism Spectroscopy
Circular dichroism spectroscopy experiments were conducted with a Jasco (J-810) spectrometer in 0.1 nm path length quartz cells. Peptide solutions (1 mg/mL) were prepared in water and diluted to 0.1 mg/mL for analysis in water (pH ∼5.0), ITC buffer (pH 8.0), or 50% (v/v) TFE. For each sample, six scans were collected at a rate of 20 nm/min from 260 to 170 nm with a 0.1 nm bandwidth and averaged. Peptide samples in water (pH 5) and ITC buffer (pH 8) were identical with the exception of the higher absorbance below 200 nm for the ITC buffer; therefore, only the water spectra were included. The DICHROWEB server available at http://dichroweb.cryst.bbk.ac.uk/html/home.shtml(29,30) was subsequently used to approximate the helical content of each of the peptides in aqueous and TFE environments. Helical content values (percent) were reported as the mean ± the standard error of the mean (SEM) of three different predictive algorithms (SELCON3, CONTIN, and CDSSTR).
For the CD spectra of DmsD in the presence of each of the DmsAL peptides, the DmsD concentration was 25 μM and the peptide concentration was 50 μM. Six scans were collected at a rate of 20 nm/min from 260 to 180 nm with a 0.5 nm bandwidth and averaged.
Differential Scanning Calorimetry
The heat of denaturation of DmsD in the absence and presence of DmsAL peptides was determined on a VP-DSC microcalorimeter (MicroCal Inc.) at a scan rate of 40 °C/h. Samples were degassed using a thermovac (MicroCal Inc.) for 10 min at room temperature prior to loading. Buffer [25 mM Tris-HCl and 100 mM NaCl (pH 8)] scans were performed to generate isotherms to subtract from protein scans. The isotherms were normalized to the concentration of protein used as determined by A280. Prior to each protein scan, five buffer scans were run to equilibrate the system and the fifth scan was used as a reference to subtract from the protein scan. Typically, by the second or third scan, the system was equilibrated. Protein samples were scanned only once because the denaturation was found to be irreversible and a second scan did not produce a transition peak over the temperature range. DmsD (25 μM) was mixed with 50 μM DmsA leader peptide and incubated for 1 h at 4 °C prior to being loaded into the thermovac.
Results
The Entire Contiguous Hydrophobic Region of the DmsA Twin-Arginine Leader Peptide Is Required for DmsD To Bind
To determine the region of the DmsAL peptide important for binding to DmsD, various portions of the 45-amino acid DmsAL peptide were synthesized and assayed for binding to DmsD using ITC (Figure 2). Our previous research demonstrated that the complete DmsAL peptide (DmsAL1–43), which included the twin-arginine motif (15SRRGLVK21) and all three regions (N-terminal, hydrophobic, and C-terminal), bound to DmsD with a Kd of 0.2 μM when it was fused to the N-terminus of GST.13 Here, the ITC results indicate that DmsD could not bind to the DmsA RR motif alone (DmsAL15–21) or if the entire N-region was also present (DmsAL2–20) (Table 1). DmsAL peptides that included the RR motif in addition to portions of the H-region (DmsAL15–27 and DmsAL15–33) also did not bind DmsD (Table 1 and Figure 2). Only when the amino acids of the entire H-region were added to the RR motif (DmsAL15–41) could binding be observed by ITC (Kd = 1.7 μM) (Table 1 and Figure 2). Leaving the H-region intact, but removing amino acids from the RR motif at the N-terminus of the peptide, caused binding to DmsD to become progressively weaker. Specifically, removing the N-terminal serine (S15) and the first arginine (R16) reduced the affinity [Kd = 2.1 μM (DmsAL17–41)], and removing the entire RR motif reduced the affinity further [Kd = 3.9 μM (DmsAL21–41)], corresponding to 1.2- and 2.3-fold reductions in the affinity for DmsAL17–41 and DmsAL21–41, respectively, relative to DmsAL15–41. When the arginines of the tightest binding peptide (DmsAL15–41) were changed to lysines (DmsAL15–41 KK), the affinity was reduced to 2.9 μM, a 1.7-fold reduction in affinity from that of the native peptide sequence (Table 1). Overall, these results indicate that the entire H-region of the DmsAL peptide must be present to allow DmsD to bind and the RR motif contributes marginal additional affinity.
Figure 2.
Representative ITC thermograms of DmsA leader peptides added to DmsD. DmsAL peptides containing residues 15–33 (A), 15–41 (B), 17–41 (C), or 21–41 (D) were added to DmsD protein, and isothermal titration calorimetry thermograms are shown. Experiments were performed in triplicate; binding was observed, and the results were tabulated.
Table 1. Evaluation of Regions of the DmsA Twin-Arginine Leader Peptide for Binding to DmsD.

DmsAL1–43 fused to the N-terminus of GST.13
NB, no binding could be detected by ITC under the conditions utilized.
Mixture of two peptides as indicated to give the DmsAL peptide region for evaluation. H-Region residues are shown in bold. If the complete and contiguous H-region is present, the residues are underlined.
To further explore the H-region of DmsAL in the binding of DmsD, a peptide mixture of DmsAL15–33 and DmsAL34–45 was assayed for binding. Individually, each of these peptides showed no binding (Table 1); however, the peptide mixture provides all the amino acids of the DmsAL peptide necessary for binding (residues 15–45), and a single polypeptide composed of similar amino acids (DmsAL15–41) was able to bind tightly (Kd = 1.7 μM). However, this peptide mixture showed no binding to DmsD and provided further proof that the entire and contiguous H-region of the DmsAL peptide is necessary to allow binding to DmsD. Only DmsAL peptides containing the complete and contiguous H-region (residues 22–39) showed binding to DmsD (Table 1).
Interaction between DmsD and DmsA RR-Leader Peptides Is Promoted by a Favorable Enthalpy
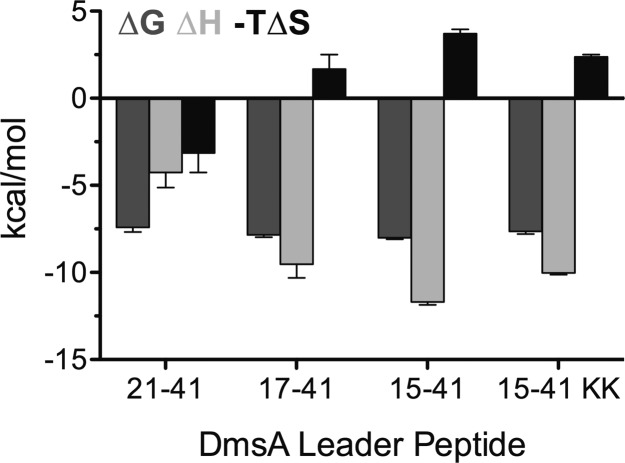
To understand the nature of the binding between each of the DmsAL peptides and DmsD, the thermodynamic contributions were analyzed for all peptides that were capable of binding. All DmsAL peptides that bound to DmsD had a 1:1 stoichiometry (Table 2). Overall, each of the four DmsAL peptides assayed for binding to DmsD exhibited similar free energies (minimum of −7.4 kcal/mol for DmsAL21–41 and maximum of −8.0 kcal/mol for DmsAL15–41); however, the contributions from enthalpy and entropy differed substantially (Table 2 and Figure 3). When “entropy” is discussed, the values refer to the entire “–TΔS” term, so that the equivalent units of kilocalories per mole may be compared with enthalpy. The three native DmsA RR-leader peptides, which showed binding to DmsD, have similar association constants, ranging from 2.6 to 6.0 × 105 M–1, and as such, there is a difference of only 0.6 kcal/mol in the overall apparent free energies (ΔG) (Table 2). Considering the DmsAL peptides, the most favorable binding reaction occurred between DmsD and DmsAL15–41 (ΔG = −8.0 kcal/mol); this reaction showed the largest favorable enthalpy (−11.7 kcal/mol) combined with the most unfavorable entropy (3.7 kcal/mol) (Table 2 and Figure 3). When the first two amino acids of the RR motif were removed (DmsAL17–41), the entropy became more favorable (difference of 2.0 kcal/mol) at the cost of enthalpy (2.2 kcal/mol), and the overall free energy became 0.2 kcal/mol less favorable because of a net loss of enthalpy (Table 2 and Figure 3). Removal of the entire RR motif, leaving only the H-region of the DmsAL peptide (DmsAL21–41), caused the enthalpy to be reduced to only −4.3 kcal/mol (a further loss of 5.2 kcal/mol), while the entropy again became even more favorable (by 4.8 kcal/mol); as a result, more than one-third of the overall free energy was from entropy (−3.1 kcal/mol). Changing the arginines to lysines in the RR motif reduced the overall free energy by only 0.4 kcal/mol because of a loss of enthalpy (1.7 kcal/mol), which outweighed the increase in favorable entropy (1.3 kcal/mol) when compared to that of the twin-arginine peptide (Table 2 and Figure 3).
Table 2. Thermodynamics of Binding of the DmsA Leader Peptide to DmsDa.
| DmsAL peptide | n | ΔG (kcal/mol) | ΔH (kcal/mol) | –TΔS (kcal/mol) |
|---|---|---|---|---|
| DmsAL21–41 | 1.14 ± 0.09 | –7.4 ± 0.2 | –4.3 ± 0.9 | –3.1 ± 1.1 |
| DmsAL17–41 | 1.04 ± 0.04 | –7.8 ± 0.1 | –9.5 ± 0.8 | 1.7 ± 0.8 |
| DmsAL15–41 | 1.04 ± 0.03 | –8.0 ± 0.1 | –11.7 ± 0.2 | 3.7 ± 0.2 |
| DmsAL15–41 KK | 1.09 ± 0.04 | –7.6 ± 0.1 | –10.0 ± 0.2 | 2.4 ± 0.2 |
Thermodynamic parameters were determined from ITC experiments in which each peptide was titrated into a DmsD protein solution at 30 °C and pH 8.0. The mean of triplicate experiments is presented with the corresponding SEM. Values of the Gibbs free energy (ΔG), enthalpy (ΔH), entropy (−TΔS), and stoichiometry of binding (n) are listed.
Figure 3.
Thermodynamics of binding of DmsD to various regions of DmsA twin-arginine leader peptides. Isothermal titration calorimetry experiments were conducted at 30 °C and pH 8.0. Mean values of free energy (ΔG), enthalpy (ΔH), and entropy (−TΔS) of binding with the corresponding SEM of triplicate titrations are reported.
In some cases, buffer ionization can contribute to the apparent enthalpy. To determine the buffer-independent enthalpy of binding (ΔHbind) and the net number of exchanged protons (between the bulk solution and the complex), ITC experiments were repeated for the four DmsAL peptides capable of binding to DmsD. These experiments were performed under the same conditions, except in phosphate buffer, which has an ionization enthalpy (0.7 kcal/mol) very different from that of Tris (11.3 kcal/mol) at 30 °C.24 The free energy and apparent enthalpy determined in phosphate buffer did not change from that determined in Tris buffer; therefore, the apparent enthalpy values were considered to be equivalent to the buffer-independent binding enthalpy. Additionally, by plotting the apparent enthalpy of binding versus the ionization enthalpy of each buffer and seeing a slope of approximately zero, we determined that upon binding the various DmsAL peptides at pH 8.0, the DmsD::DmsAL complexes do not have a net gain or loss of protons for any of the peptides (Figure 1 of the Supporting Information).
DmsA Leader Peptides Able To Bind DmsD Are Unstructured in Aqueous Solution but Can Acquire Helicity in a Hydrophobic Environment
Having determined the portion of the DmsAL peptide capable of binding to DmsD, we investigated the potential of each peptide to form secondary structure. We wanted to determine if the secondary structure of the DmsAL peptide played a role in binding. Does the sequence of the DmsAL peptide influence its ability to adopt a helical structure? To answer these questions, we used two methods. CD spectroscopy was performed on DmsAL peptides in aqueous solution and in a 50% TFE solution (a solvent known to influence secondary structure and mimic that of a hydrophobic environment),31,32 and secondary structure predictions were performed on DmsAL peptide sequences (Figure 4).
All DmsAL peptides were found to contain minimal helical content (3–10%) in aqueous solution (Figure 4A–F). However, in a 50% TFE solution, the peptides with the complete H-region adopted increased amounts of helical content (24% for DmsAL15–41, 21% for DmsAL17–41, and 15% for DmsAL21–41) (Figure 4C–E). If the H-region was incomplete, no helical content was found regardless of the environment (DmsAL15–33; 3% in water and 5% in 50% TFE) (Figure 4B). Two secondary structure prediction algorithms were used for the DmsAL peptides and showed a range of helical content. AGADIR27,28 predicts the secondary structure of peptides, while PSIPRED25,26 is used to predict the secondary structure within protein sequences. If the H-region is not intact as in DmsAL15–33, then no helical structure was predicted to occur by either algorithm (Figure 4G). However, if the complete H-region was present, PSIPRED predicted between 33 and 48% helical content while AGADIR predicted almost no helical content regardless of the sequence. The results of the CD experiments with the DmsAL peptides in TFE supported what was found in the PSIPRED predictions.
Because only DmsAL peptides containing the intact contiguous H-region were able to bind as well as form helical structure, it is possible that a portion of the DmsAL peptide adopts a helical structure upon binding to DmsD. To further investigate this question, we performed CD spectroscopy on DmsD alone as well as in complex with each of the peptides that were investigated above. If the peptide was shown to have helical content in TFE, then there was also an increased helical content in the complex with DmsD (Figure 5B–D and Table 3). Importantly, the complex of DmsD with the DmsAL peptide containing an incomplete H-region (DmsAL15–33) showed almost no increase in helical content over that of DmsD alone (Figure 5A and Table 3).
Figure 5.
DmsA leader peptides shown to bind and form helical structure in a hydrophobic environment increase the helicity of the complex with DmsD. Far-UV CD spectra were collected for 50 μM DmsAL peptide (dashed line), 25 μM DmsD (dotted line), and 25 μM DmsD in complex with 50 μM DmsAL peptide with the 50 μM DmsAL peptide spectra subtracted (solid line): DmsAL15–33 (A), DmsAL15–41 (B), DmsAL17–41 (C), and DmsAL21–41 (D).
Table 3. Molar Ellipticities of DmsD Alone and in Complex with DmsAL Peptides.
| θ225 (mdeg)a |
|||
|---|---|---|---|
| DmsAL peptide | DmsD | complex | difference |
| DmsAL15–33 | –76.7 | –76.8 | –0.1 |
| DmsAL15–41 | –75.5 | –81.5 | –6.0 |
| DmsAL17–41 | –76.8 | –83.6 | –7.2 |
| DmsAL21–41 | –75.4 | –78.7 | –3.3 |
The raw machine units (θ) at 225 nm (millidegrees) recorded from circular dichroism experiments with DmsD alone or in complex with DmsAL peptides are listed.
The Conformational Change of DmsD Is Small upon Binding of the DmsA Leader Peptide
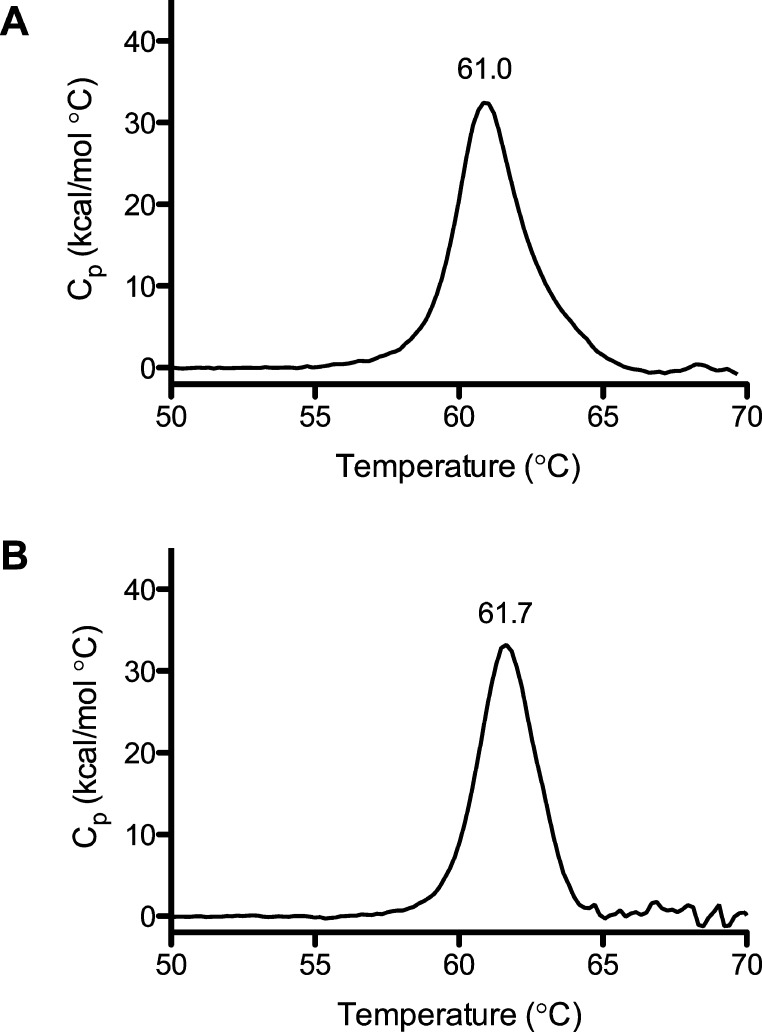
E. coli DmsD is a globular single-domain protein consisting of 11 α-helices.33,34 Differential scanning calorimetry was used to investigate the thermal stability of DmsD in the absence and presence of various DmsAL peptides. The melting temperature (Tm) of DmsD alone was determined to be just below 61 °C (Figure 6A and Table 4). When peptides shown not to bind (DmsAL15–33 and DmsAL34–45) were added, the Tm showed no change (Table 4). However, when DmsAL peptides that did bind were added to DmsD, the melting temperature increased to 61.5 °C (DmsAL21–41), 61.8 °C (DmsAL17–41), 61.7 °C (DmsAL15–41), or 61.9 °C (DmsAL15–41 KK). These small increases (within 1 °C) in the thermal stability of DmsD in the presence of bound peptides suggest a very minimal conformational change in DmsD upon binding of the DmsAL peptide.
Figure 6.
Melting temperature of DmsD that increases in the presence of DmsAL peptides shown to bind. Differential scanning calorimetry (DSC) of DmsD alone (A) yielded a melting temperature of 61.0 °C, while in the presence of DmsAL15–41 (B), the melting temperature increased to 61.7 °C. The DSC results of DmsD in the presence of other DmsAL peptides are listed in Table 4.
Table 4. Melting Temperatures of DmsD–DmsAL Complexesa.
| DmsAL peptide | Tm (°C) |
|---|---|
| DmsD alone | 60.9 ± 0.2 |
| DmsD–DmsAL15–41 | 61.7 |
| DmsD–DmsAL17–41 | 61.8 |
| DmsD–DmsAL21–41 | 61.5 |
| DmsD–DmsAL15–33–DmsAL34–45 | 60.8 |
| DmsD–DmsAL15–41 KK | 61.9 |
Differential scanning calorimetry was used to determine the temperature of melting (Tm) for DmsD (25 μM), and its mean and standard error of the mean of triplicate experiments is reported. The Tm values of single trials of 2:1 complexes of DmsAL and DmsD (50 and 25 μM, respectively) are also reported.
Discussion
We have determined that the complete and contiguous H-region of the DmsAL peptide is recognized by DmsD. Previous research has shown that DmsD can interact with four RR-leader peptides (or preproteins): DmsA, YnfE, YnfF, and TorA.7,8 Aside from the RR motif, the H-region within these leader peptides is the most similar and identical (Figure 1) and as such is the likely determinant of specificity. DmsAL21–41 was the smallest peptide shown to bind DmsD and is almost exclusively composed of the H-region. Binding of this peptide was driven by favorable entropy and enthalpy, suggesting that hydrophobic interactions play an important role. A previous study showed that the kinetics of dissociation (off rate) had the greatest ability to differentiate binding of DmsD to the complete DmsA leader peptide (koff = 2.0 ms–1) or the YnfE leader peptide (koff = 127 ms–1), with a 60 fold-greater propensity to dissociate even though the rate of association (kon) for the YnfE peptide was 6-fold higher than for the DmsA peptide.7 The variance of the residues within the H-region of each of these leader sequences may cause the subtle differences in the on and off rates toward DmsD but still allow binding to occur.
The display of the RR-leader peptide off the parent substrate or a carrier protein likely plays a role in the determined affinity for the REMP. Because the substrates of most RR-leader peptides are labile, likely due to the complex cofactors that must be incorporated, most studies investigating the interaction employ fusion proteins at the N- or C-terminus of the leader peptide. When a variety of RR-leader peptide fusion proteins were used in combination with far-Western blot assays, it was shown that the fusion partner of the leader peptide was critical for some interactions between REMPs and their leader peptide.7 Identical DmsAL peptide constructs have been fused to either GST or SBP. When placed at the C-terminus of GST (GST::DmsAL1–43), no interaction with DmsD was observed, while at the N-terminus (DmsAL1–43::GST), a tight interaction was observed (Kd = 0.2 μM).13 Subsequently, the Kd of a DmsAL1–43::SBP construct with DmsD was determined to be 0.06 μM.7 The increased affinity of the DmsAL1–43::SBP fusion compared to that of the DmsAL1–43::GST fusion is likely due to the resulting peptide display. To overcome the effects of the fusion partner on the determined affinity, we chose to synthesize various portions of the leader peptide to investigate the interaction. Our results suggest that some of the peptides may be behaving slightly differently in isolation and within a fusion or the larger native construct; however, clear thermodynamic trends were observed.
DmsD and TorD are often considered to behave in an almost identical manner with regard to binding and maturation of their respective substrates DmsA and TorA. In a study performed with the REMP TorD and various TorA leader peptides, some results similar to those described here were found. The TorA leader peptide that included the entire H-region as well as the RR motif bound to TorD with a Kd of 1.8 μM.11 The affinity is almost identical to that shown here for DmsD and DmsAL15–41 (Kd = 1.7 μM). However, the same study showed that TorD was able to bind to a TorA leader peptide devoid of half of the H-region, while here, DmsD had no interaction with the DmsA leader peptide unless the entire H-region was present. Additionally, TorD is able to bind to the mature portion of the TorA protein, while DmsD binds pre-DmsA only if the DmsA leader peptide is present.7,18 Thus, while the REMPs DmsD and TorD both bind to the H-region of their RR-leader peptide, the overall recognition and interactions with their substrates are different.
Recently, green fluorescent protein (GFP) fusions to chimeras of various regions of DmsA and TorA leader peptides were used to investigate the specificity of binding of DmsD and TorD.35 The H-region of DmsA and TorA determined binding to either DmsD or TorD; however, how the H-region was defined within that study eludes to the importance of the 38PFSR41 portion of the DmsAL for binding to DmsD, as omission of this sequence did not allow binding to occur. Further evidence that the H-region is important for REMP binding was seen when three different leucines (L27, L31, and L32) within the H-region of the TorA leader peptide were changed to a polar amino acid (glutamine); this abolished binding to TorD in vivo.16 This same study showed that when L21 in the TorA leader was changed to glutamine, binding to TorD was not affected. However, if the equivalent residue within the DmsA leader (L28) is changed to a valine, binding to DmsD was abolished (data not shown). This further supports the idea that subtle differences within the leader peptide sequences are able to distinguish binding to either TorD or DmsD.
As portions of the twin-arginine motif were added to the N-terminus of the H-region of DmsAL peptides, the entropy of binding became more unfavorable, such that the interaction was driven exclusively by enthalpy accompanied by an entropic cost. Overall, the presence of the RR motif was not critical to the interaction between DmsAL and DmsD and contributed little additional affinity. A similar result was also shown for binding of TorD to the TorA leader peptide.11 However, the nitrate reductase REMP, NarJ, behaves differently in that it is able to bind to the 15 amino-terminal residues of its substrate NarG with more than 10-fold tighter affinity (Kd ∼ 0.1 μM) than either DmsD or TorD.12,19 The 15 N-terminal residues of NarG contain a remnant RR motif,6,36 and nitrate reductase has been shown to be Tat-dependent;37 however, the enzyme complex (NarGHI) remains on the cytoplasmic side of the membrane. It is known that TatC binds specifically to the RR motif of Tat-dependent substrates.38 This binding then recruits TatB and TatA, which allows translocation of the RR-leader substrate protein. It is possible that each REMP binds specifically to the H-region of its leader peptide while still allowing TatC to bind to the RR motif. Because NarJ binds more tightly to the N-region of NarG and remains on the cytoplasmic side of the membrane, the TatC protein likely plays a different role with respect to the atypical vestige RR-leader peptide of NarG.
Depending on the region of the twin-arginine leader peptide that is present, a structural component may also play a role in binding to the REMP. The effect that the peptide sequence alone may have within the bound structure is difficult to determine in the absence of a structure of the complex itself. Two E. coli RR-leader peptides (HiPIP and SufI) with no known system specific chaperone (REMP) have been studied, and both peptides were shown to have almost no secondary structure in aqueous solution.39,40 Our results show that DmsAL peptides are also unstructured in aqueous solution, but the ability to form helical structure was influenced by the sequence that was present as well as the environment in which it was placed. The helical tendency of the peptide was dependent on the portion of the peptide sequence that was present, specifically, the entire hydrophobic region. Only those DmsAL peptides that contained a complete and contiguous H-region became helical when they were examined in a more hydrophobic environment, like that which might be found at the binding site of DmsD. A putative interaction site in DmsD, determined through mutagenesis screening, presented a hydrophobic cleft.15 The intrinsic propensity of these peptides to form helical structure correlated with the DmsAL peptides that were also able to bind to DmsD. It is possible that RR-leader peptides of Tat protein substrates adopt secondary structure within a portion of their sequence upon binding, which further defines the specificity of binding among the REMPs.
A high-resolution structure of any of the DmsD/TorD/NarJ family of REMPs bound to their twin-arginine leader peptide has not been determined. However, the NMR solution structure of NapD, the REMP for the periplasmic nitrate reductase NapA, which has a fold very different from that of the DmsD/TorD/NarJ family, has been determined [Protein Data Bank (PDB) entry 2JSX]14 along with a second structure in complex with the NapAL1–35 peptide (PDB entry 2PQR, unpublished observations). In the NapD–NapA complex structure, NapA residues 5–22, encompassing the twin-arginine motif and the H-region, form an α-helix and bind within a hydrophobic pocket of NapD. Additionally, the NapD protein shows very little structural change upon binding the NapA RR-leader peptide.14
Here, DmsD was shown to have a melting temperature of 61 °C using DSC. In the presence of DmsAL peptides capable of binding, the Tm of DmsD showed only a minor increase, up to 1 °C, which suggests very little conformational change in DmsD upon binding. Further, the DmsAL peptide was shown to bind over the surface of DmsD in a recent study.41 Overall, the results suggest that there are very few conformational changes occurring in DmsD upon binding of the DmsAL peptide.
Conclusions
The redox enzyme maturation protein DmsD binds to the H-region of the DmsA twin-arginine leader peptide sequence, and this interaction is promoted by favorable entropy and enthalpy. While the conformation of DmsD does not appear to change upon binding, a portion of the H-region of the DmsA RR-leader peptide may form a helix upon binding to a hydrophobic pocket of DmsD.
Acknowledgments
We acknowledge Dr. Evan Haney for technical assistance with NMR spectroscopy.
Glossary
Abbreviations
- CD
circular dichroism
- DmsAL
DmsA leader
- DMSO
dimethyl sulfoxide
- DSC
differential scanning calorimetry
- ITC
isothermal titration calorimetry
- Kd
dissociation constant
- REMP
redox enzyme maturation protein
- RR
twin-arginine
- Tat
twin-arginine translocase
- TFE
trifluoroethanol.
Supporting Information Available
Effect of the enthalpy of ionization (Figure S1). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Operating grant to R.J.T. from the Canadian Institutes of Health Research and Canada Graduate Scholarship to T.M.L.W. from the Natural Sciences and Engineering Research Council of Canada.
Supplementary Material
References
- Weiner J. H.; Bilous P. T.; Shaw G. M.; Lubitz S. P.; Frost L.; Thomas G. H.; Cole J. A.; Turner R. J. (1998) A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93, 93–101. [DOI] [PubMed] [Google Scholar]
- Sargent F.; Bogsch E.; Stanley N.; Wexler M.; Robinson C.; Berks B.; Palmer T. (1998) Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17, 3640–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks B. C.; Sargent F.; Palmer T. (2000) The Tat protein export pathway. Mol. Microbiol. 35, 260–274. [DOI] [PubMed] [Google Scholar]
- Palmer T.; Berks B. C. (2012) The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10, 483–496. [DOI] [PubMed] [Google Scholar]
- Jack R. L.; Buchanan G.; Dubini A.; Hatzixanthis K.; Palmer T.; Sargent F. (2004) Coordinating assembly and export of complex bacterial proteins. EMBO J. 23, 3962–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R. J.; Papish A. L.; Sargent F. (2004) Sequence analysis of bacterial redox enzyme maturation proteins (REMPs). Can. J. Microbiol. 50, 225–238. [DOI] [PubMed] [Google Scholar]
- Chan C. S.; Chang L.; Rommens K. L.; Turner R. J. (2009) Differential interactions between Tat-specific redox enzyme peptides and their chaperones. J. Bacteriol. 191, 2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oresnik I. J.; Ladner C. L.; Turner R. J. (2001) Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 40, 323–331. [DOI] [PubMed] [Google Scholar]
- Ray N.; Oates J.; Turner R. J.; Robinson C. (2003) DmsD is required for the biogenesis of DMSO reductase in Escherichia coli but not for the interaction of the DmsA signal peptide with the Tat apparatus. FEBS Lett. 534, 156–160. [DOI] [PubMed] [Google Scholar]
- Tranier S.; Mortier-Barriere I.; Ilbert M.; Birck C.; Iobbi-Nivol C.; Mejean V.; Samama J. P. (2002) Characterization and multiple molecular forms of TorD from Shewanella massilia, the putative chaperone of the molybdoenzyme TorA. Protein Sci. 11, 2148–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzixanthis K.; Clarke T. A.; Oubrie A.; Richardson D. J.; Turner R. J.; Sargent F. (2005) Signal peptide-chaperone interactions on the twin-arginine protein transport pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 8460–8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S.; Howell J. M.; Workentine M. L.; Turner R. J. (2006) Twin-arginine translocase may have a role in the chaperone function of NarJ from Escherichia coli. Biochem. Biophys. Res. Commun. 343, 244–251. [DOI] [PubMed] [Google Scholar]
- Winstone T. L.; Workentine M. L.; Sarfo K. J.; Binding A. J.; Haslam B. D.; Turner R. J. (2006) Physical nature of signal peptide binding to DmsD. Arch. Biochem. Biophys. 455, 89–97. [DOI] [PubMed] [Google Scholar]
- Maillard J.; Spronk C.; Buchanan G.; Lyall V.; Richardson D. J.; Palmer T.; Vuister G. W.; Sargent F. (2007) Structural diversity in twin-arginine signal peptide-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 15641–15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S.; Winstone T. M. L.; Chang L.; Stevens C. M.; Workentine M. L.; Li H.; Wei Y.; Ondrechen M. J.; Paetzel M.; Turner R. J. (2008) Identification of residues in DmsD for twin-arginine leader peptide binding, defined through random and bioinformatics-directed mutagenesis. Biochemistry 47, 2749–2459. [DOI] [PubMed] [Google Scholar]
- Buchanan G.; Maillard J.; Nabuurs S. B.; Richardson D. J.; Palmer T.; Sargent F. (2008) Features of a twin-arginine signal peptide required for recognition by a Tat proofreading chaperone. FEBS Lett. 582, 3979–3984. [DOI] [PubMed] [Google Scholar]
- Guymer D.; Maillard J.; Sargent F. (2009) A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 191, 519–528. [DOI] [PubMed] [Google Scholar]
- Chan C. S.; Chang L.; Winstone T. M.; Turner R. J. (2010) Comparing system-specific chaperone interactions with their Tat dependent redox enzyme substrates. FEBS Lett. 584, 4553–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian S.; Lafitte D.; Vergnes A.; Pimentel C.; Sebban-Kreuzer C.; Toci R.; Claude J. B.; Guerlesquin F.; Magalon A. (2010) Basis of recognition between the NarJ chaperone and the N-terminus of the NarG subunit from Escherichia coli nitrate reductase. FEBS J. 277, 1886–1895. [DOI] [PubMed] [Google Scholar]
- Berks B. C. (1996) A common export pathway for proteins binding complex redox cofactors?. Mol. Microbiol. 22, 393–404. [DOI] [PubMed] [Google Scholar]
- Sarfo K. J.; Winstone T. L.; Papish A. L.; Howell J. M.; Kadir H.; Vogel H. J.; Turner R. J. (2004) Folding forms of Escherichia coli DmsD, a twin-arginine leader binding protein. Biochem. Biophys. Res. Commun. 315, 397–403. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., and Bairoch A. (2005) Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook (Walker J. M., Ed.) pp 571–607, Humana Press, Totowa, NJ. [Google Scholar]
- Goldberg R. N.; Kishore N.; Lennen R. M. (2002) Thermodynamic quantities for the ionization reactions of buffers. J. Phys. Chem. Ref. Data 31, 231–370. [Google Scholar]
- Bryson K.; McGuffin L. J.; Marsden R. L.; Ward J. J.; Sodhi J. S.; Jones D. T. (2005) Protein structure prediction servers at University College London. Nucleic Acids Res. 33, W36–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D. W.; Ward S. M.; Lobley A. E.; Nugent T. C.; Bryson K.; Jones D. T. (2010) Protein annotation and modelling servers at University College London. Nucleic Acids Res. 38, W563–W568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix E.; Viguera A. R.; Serrano L. (1998) Elucidating the folding problem of α-helices: Local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 284, 173–191. [DOI] [PubMed] [Google Scholar]
- Munoz V.; Serrano L. (1997) Development of the multiple sequence approximation within the AGADIR model of α-helix formation: Comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers 41, 495–509. [DOI] [PubMed] [Google Scholar]
- Whitmore L.; Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L.; Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 89, 392–400. [DOI] [PubMed] [Google Scholar]
- Buck M. (1998) Trifluoroethanol and colleagues: Cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31, 297–355. [DOI] [PubMed] [Google Scholar]
- Kentsis A.; Sosnick T. R. (1998) Trifluoroethanol promotes helix formation by destabilizing backbone exposure: Desolvation rather than native hydrogen bonding defines the kinetic pathway of dimeric coiled coil folding. Biochemistry 37, 14613–14622. [DOI] [PubMed] [Google Scholar]
- Stevens C. M.; Winstone T. M.; Turner R. J.; Paetzel M. (2009) Structural analysis of a monomeric form of the twin-arginine leader peptide binding chaperone Escherichia coli DmsD. J. Mol. Biol. 389, 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S. K.; Clemons W. M. Jr. (2009) Structure of the twin-arginine signal-binding protein DmsD from Escherichia coli. Acta Crystallogr. F65, 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugham A.; Bakayan A.; Voller P.; Grosveld J.; Lill H.; Bollen Y. J. (2012) The hydrophobic core of twin-arginine signal sequences orchestrates specific binding to Tat-pathway related chaperones. PLoS One 7, e34159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ize B.; Coulthurst S. J.; Hatzixanthis K.; Caldelari I.; Buchanan G.; Barclay E. C.; Richardson D. J.; Palmer T.; Sargent F. (2009) Remnant signal peptides on non-exported enzymes: Implications for the evolution of prokaryotic respiratory chains. Microbiology 155, 3992–4004. [DOI] [PubMed] [Google Scholar]
- Li H.; Turner R. J. (2009) In vivo associations of Escherichia coli NarJ with a peptide of the first 50 residues of nitrate reductase catalytic subunit NarG. Can. J. Microbiol. 55, 179–188. [DOI] [PubMed] [Google Scholar]
- Alami M.; Luke I.; Deitermann S.; Eisner G.; Koch H. G.; Brunner J.; Muller M. (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946. [DOI] [PubMed] [Google Scholar]
- Kipping M.; Lilie H.; Lindenstrauss U.; Andreesen J. R.; Griesinger C.; Carlomagno T.; Bruser T. (2003) Structural studies on a twin-arginine signal sequence. FEBS Lett. 550, 18–22. [DOI] [PubMed] [Google Scholar]
- San Miguel M.; Marrington R.; Rodger P. M.; Rodger A.; Robinson C. (2003) An Escherichia coli twin-arginine signal peptide switches between helical and unstructured conformations depending on the hydrophobicity of the environment. Eur. J. Biochem. 270, 3345–3352. [DOI] [PubMed] [Google Scholar]
- Stevens C. M.; Okon M.; McIntosh L. P.; Paetzel M. (2013) 1H, 13C and 15N resonance assignments and peptide binding site chemical shift perturbation mapping for the Escherichia coli redox enzyme chaperone DmsD. Biomol. NMR Assignments 7, 193–197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.