Abstract
Purpose
To describe the relationships of selected candidate genes to the prevalence of early age-related macular degeneration (AMD) in a cohort of whites, blacks, Hispanics, and Chinese Americans.
Design
Cross-sectional study.
Methods
Setting
Multicenter study.
Study Population
2456 persons aged 45–84 years with genotype information and fundus photographs.
Procedures
Twelve of 2862 single nucleotide polymorphisms (SNPs) from 11 of 233 candidate genes for cardiovascular disease were selected for analysis based on screening with marginal unadjusted P value <0.001 within 1 or more racial/ethnic groups. Logistic regression models tested for association in case-control samples.
Main Outcome Measure
Prevalence of early AMD.
Results
Early AMD was present in 4.0% of the cohort and varied from 2.4% in blacks to 6.0% in whites. The odds ratio increased from 2.3 for one to 10.0 for four risk alleles in a joint effect analysis of Age-Related Maculopathy Susceptibility 2 rs10490924 and Complement Factor H Y402H (P for trend=4.2×10−7). Frequencies of each SNP varied among the racial/ethnic groups. Adjusting for age and other factors, few statistically significant associations of the 12 SNPs with AMD were consistent across all groups. In a multivariate model, most candidate genes did not attenuate the comparatively higher odds of AMD in whites. The higher frequency of risk alleles for several SNPs in Chinese Americans may partially explain their AMD frequency approaching that of whites.
Conclusions
The relationships of 11 candidate genes to early AMD varied among 4 racial/ethnic groups, and partially explained the observed variations in early AMD prevalence among them.
Introduction
Age-related macular degeneration (AMD) is an important cause of visual impairment in older persons in the United States1,2 and is more prevalent in whites compared with blacks and Hispanics.3–15 Such differences may be due to variation in environmental exposures (e.g., smoking, physical activity, diet), variation in the frequencies of protective and deleterious genetic alleles for risk of AMD among the different racial/ethnic groups, or both. In the Multi-Ethnic Study of Atherosclerosis (MESA), we previously observed that racial/ethnic differences in the prevalence of AMD could not be explained by smoking status, levels of inflammatory factors, cardiovascular disease (CVD), or by the Complement Factor H (CFH) Y402H polymorphism, a major AMD risk allele.16–18
Additional genes for AMD susceptibility have been identified by association studies in whites,19,20 but there is less information regarding the frequencies of protective and risk alleles in non-Caucasian populations of the United States.21–28 In this report, we examine the relationships of candidate genes and AMD in a cohort of whites, blacks, Hispanics, and Chinese Americans participating in the MESA. A candidate gene study across all of MESA afforded the opportunity to test both the association of AMD with AMD candidate genes as well as genes selected for association with CVD and cardiovascular phenotypes including cholesterol metabolism, innate immunity, and atherosclerosis. We aimed to examine the association of these candidate genes with AMD in four racial/ethnic groups (whites, blacks, Hispanics, and Chinese Americans) in the MESA and to test the hypothesis that differences in the frequencies of protective and risk alleles in these candidate genes may account, at least in part, for our previous observations of differences in the frequency of AMD signs among the 4 racial/ethnic groups represented in the MESA.
Methods
Study Sample
The MESA is a prospective cohort study of men and women aged 45–85 years without a history of clinical CVD living in 6 United States communities.29 The study objectives of the MESA are to identify risk factors for subclinical CVD, progression of subclinical CVD, and transition from subclinical to clinical CVD. Selection of the study population has been reported in detail elsewhere.29 At the first examination carried out between July 17, 2000 and August 29, 2002, there were 6814 participants: 1086 from Baltimore, Maryland; 1164 from Chicago, Illinois; 1077 from Forsyth County, North Carolina; 1319 from Los Angeles County, California; 1102 from New York, New York; and 1066 from St. Paul, Minnesota. Tenets of the Declaration of Helsinki were followed, the work was compliant with the Health Insurance Portability and Accountability Act, and institutional review board approval was granted at each study site prior to examining any participants. Written informed consent was obtained from every participant before examination.
Retinal Photography and Grading of Age-Related Macular Degeneration
Fundus photography using a 45° 6.3 megapixel digital nonmydriatic camera was performed at each site at the second examination conducted between September 9, 2002 and February 7, 2004 (immediately after the baseline examination) using a standardized protocol.13,30 Two photographic fields were taken of each eye, the first centered on the optic disc (Early Treatment Diabetic Retinopathy Study field 1) and the second centered on the fovea (Early Treatment Diabetic Retinopathy Study field 2).31 Images were obtained from 6176 participants.
Grading of digital images and quality control are described in detaile elsewhere.32,33 Each image was graded twice (a preliminary and a detail grade) using a modification of the Wisconsin Age-Related Maculopathy Grading Scheme.33 Of all participants photographed, 5887 (95.3%) had at least 1 eye that could be evaluated for AMD (right eye only, n=211; left eye only, n=200; both eyes, n=5476) and were included in the analyses for the purposes of this report. There were no statistically significant differences in gradability for AMD among the 4 racial/ethnic groups (data not shown).
Definitions of Age-Related Macular Degeneration Endpoints
Among the AMD features evaluated were drusen size, type, and area, increased retinal pigment, retinal pigment epithelial (RPE) depigmentation, pure geographic atrophy, and signs of exudative macular degeneration (subretinal hemorrhage, subretinal fibrous scar, RPE detachment, and/or serous detachment of the sensory retina or laser or photodynamic treatment scar for neovascular AMD). Soft distinct drusen were defined by size (between 63 and 300 μm in diameter) and appearance (sharp margins and a round nodular appearance with a uniform density). When 2 eyes of a participant were discrepant for the severity of a lesion, the grade assigned for the participant was that of the more severely involved eye. For example, in assigning the prevalence of soft drusen, if soft drusen were present in 1 eye but not in the other eye, the participant was considered to have soft drusen. When drusen or signs of AMD could not be graded in an eye, the participant was assigned a grade based on severity of the lesion in the other eye.
Assessment of Risk Factors
Participants underwent an interview and assessment of cardiovascular risk factors during the course of the study.29,34 Variables for this analysis were based on data collected at the baseline examination when most of the systemic hematological tests were conducted. Resting blood pressure was measured 3 times with participants in the seated position using the Dinamap model Pro 100 automated oscillometric sphygmomanometer (Critikon, Tampa, FL). The average of the last 2 measurements was used in analyses. Hypertension was defined as systolic blood pressure ≥ 140 mmHg and/or diastolic blood pressure ≥ 90 mmHg and/or current use of antihypertensive medications. Height and weight were measured with participants wearing light clothing and no shoes; body mass index (BMI) was calculated as the weight in kg divided by the square of height in m. A detailed questionnaire was used to obtain information about past medical history such as hypertension, cigarette smoking, alcohol consumption and medication use, including antihypertensive and hypoglycemic agents and lipid-lowering agents.
C-Reactive Protein
Fasting (≥8 hours) venous blood samples were drawn from participants and aliquots were prepared for central analysis and storage at the University of Vermont and the University of Minnesota.29 A standardized protocol was used to measure serum high-sensitivity C-reactive protein.34
Genotyping and the Multi-Ethnic Study of Atherosclerosis Candidate Gene Study
DNA was extracted from whole blood and quantified using standard methods (DNA isolation by Puregene kits, Gentra Systems, Minneapolis, MN; PicoGreen kits, Molecular Probes, Inc., Eugene, OR).
The details of the MESA Candidate Gene Study have been published previously.35,36 In brief, a subgroup of 2880 MESA subjects were selected from all subjects who participated in the third MESA examination, supplemented by random selection from the remaining cohort to give approximately equal numbers across the four MESA ethnic/racial groups and between sexes. A complete dataset of 2456 participants with AMD grading and gene polymorphisms was available for this study.
Candidate genes were selected by the MESA Genetics Committee after considering suggestions by all MESA investigators. For each candidate gene, single nucleotide polymorphisms (SNPs) were selected for compatibility with the Illumina GoldenGate technology37–39 as determined by the Assay Design Tool (TechSupport, Illumina, San Diego, CA) and ability to tag Caucasian and Yoruban haplotypes in the HapMap Release 2 dataset using “Tagger.”40–42 In total, 2862 SNPs from 233 genes were completed by the Illumina genotyping service. An additional 199 previously identified ancestry-informative markers selected from HapMap data for four racial/ethnic groups were also genotyped and used for estimating principal component variables. Details of the genotyping quality have been published elsewhere.35,36
Analyses of Association
All statistical analyses were performed with SAS version 9.1 (SAS Institute, Cary, NC) and PLINK version 1.07.43 Logistic regression was used to estimate the odds ratio (OR) and 95% confidence interval (95% CI) for risk factors and AMD as a discrete variable (early AMD vs. none). SNP association tests were performed by two-stage design: initial screening of 2862 SNPs and follow-up analysis of 12 SNPs identified by screening. Although only 12 SNPs were selected for further analysis in detail, a Bonferroni corrected P value adjusted for 2862 SNPs (1.7×10−5) was considered statistically significant for the SNP-association. P values before and after Bonferroni correction are presented in the Results.
Screen
We first performed a screen of all 2862 SNPs that passed quality control to find those SNPs associated with AMD. An arbitrary cut-point with an unadjusted marginal P value of less than 0.001 in 1 or more racial/ethnic groups was used as an initial approach to identify the best SNP associations in the data and to identify those SNPs for a combined analysis.44 For multiple SNPs identified in strong linkage disequilibrium (r2>0.8) in the same region, we used the most significant SNP (the lead SNP) to represent the region. Based on this screening analysis, a total of 12 SNPs from 11 genes were selected for a more complete analysis of the association with AMD.
Association of individual single nucleotide polymorphisms
The association of the 12 SNPs with AMD was evaluated by logistic regression initially within each racial/ethnic group separately, and then combined by meta-analysis using the inverse variance method. Age, sex, study site, 2 principal component variables, and smoking status were selected as covariates. The 2 most statistically significant principal component variables representing global continental ancestry were obtained using the ancestry-informative markers.
Association of multiple single nucleotide polymorphisms
In order to test the association of multiple SNPs together, we performed the association analyses by combining the significant SNPs from the single gene analyses into a single model. First, the joint additive effect was tested for 2 interesting SNPs (Age-Related Maculopathy Susceptibility 2 [ARMS2] rs10490924, the most significant SNP associated with early AMD in our data; and Complement Factor H [CFH] Y402H, the most reported SNP associated with AMD in the literature). To test the association of combinations of these two SNPs, we grouped the combination by the number of risk alleles. For example, 2 risk alleles in ARMS2 (AA) and 2 risk alleles in CFH (CC) were grouped as the highest risk, and 2 protective alleles each in ARMS2 (CC) and CFH (TT) were grouped as the lowest risk. For the additive effect (i.e., using a combination of significant SNPs together), we performed the association test by logistic regression among all combinations of groups with AMD.
Second, to identify the interaction effect among all tested SNPs, the multiplicative term of genotypes between 2 significant SNPs was included and evaluated in the model as an interaction term. A total of 144 SNP-SNP pair interactions by 3 genotype groups, i.e., 9 genotype combinations for each SNP-SNP pair interaction, were performed. Due to limited sample size, association tests for interactions and the combination of two SNPs were performed by using all samples together and adjusted for principal component variables. Logistic regression was performed to evaluate the significance level of the interaction by including both the main effects of SNPs and the interaction in the model. The Bonferroni corrected P value adjusted for 144 tests (3.0×10−4) was considered statistically significant.
In addition, we conducted a logistic regression analysis to identify whether these SNPs contributed to the observed racial/ethnic differences of early AMD. Using whites as the reference group, we first performed the tests to check the overall AMD difference between whites and each of the other 3 racial/ethnic groups after controlling for age, sex, study site, and smoking status. We then conditioned on a particular SNP and re-ran the model to compare the difference between 2 racial/ethnic groups. If parameters of the model (e.g., P value and OR) changed after conditioning on a particular SNP, then this particular SNP was considered to contribute to the variation in genetic susceptibilities between the two tested racial/ethnic groups. For example, if the overall significance between whites and blacks existed in the original model but became less significant after conditioning on a specific SNP, we concluded that this SNP might contribute to the overall racial difference in AMD between whites and blacks. Since more than 1 SNP was independently associated with the differences between whites and Chinese Americans, a combined analysis of all the SNPs was further tested in an independent model with only these 2 racial/ethnic groups.
Results
Complete genotyping and gradable images were available for 2456 MESA subjects. Age, sex, and smoking status for these subjects together and by racial/ethnic group are shown in Table 1. The cohort under analysis consisted of 634 (25.8%) whites, 630 (25.7%) Chinese Americans, 593 (24.1%) blacks, and 599 (24.4%) Hispanics. Early AMD was present in 4.0%, late AMD in 0.5%, large drusen in 9.8%, soft drusen in 16.3%, increased retinal pigment in 1.8%, RPE depigmentation in 0.9%, neovascular AMD in 0.3%, and pure geographic atrophy in 0.2% of the cohort. These features were most frequent in whites and least frequent in blacks (Table 1). The distributions of the specific SNPs in the 11 candidate AMD genes in each racial/ethnic group and the whole cohort are also presented in Table 1. The frequency of the effect variant (in parentheses) was lower in Chinese Americans for CFH (C), PPARG (A), and CAPN5 rs11237081 (G) SNPs compared to the other racial/ethnic groups, while the frequency of MRPL10 rs3209 (G) and ABCA4 rs548122 (C) SNPs was lower and frequency of the SOD3 rs2284659 (A) SNP was higher in blacks compared to the other racial/ethnic groups.
Table 1.
Characteristics of the Multi-Ethnic Study of Atherosclerosis Cohort with Genotype Information and Gradable Early Age-related Macular Degeneration
| Characteristic | % or Mean±SD
|
||||
|---|---|---|---|---|---|
| All (N=2456) | White (N=634) | Chinese (N=630) | Black (N=593) | Hispanic (N=599) | |
|
|
|||||
| Age, years | 60.9±10.0 | 61.0±10.3 | 61.5±10.1 | 60.6±9.5 | 60.3±9.8 |
| Sex, male | 47.2 | 48.0 | 49.1 | 45.4 | 46.2 |
| Current smoker | 13.6 | 15.1 | 5.6 | 18.7 | 15.4 |
| Early AMD | 4.0 | 6.0 | 3.8 | 2.4 | 3.8 |
| Large drusen | 9.8 | 9.9 | 12.9 | 6.2 | 10.0 |
| Soft drusen | 16.3 | 14.0 | 23.3 | 11.3 | 16.4 |
| Increased retinal pigment | 1.8 | 3.9 | 1.3 | 0.7 | 1.0 |
| RPE depigmentation | 0.9 | 1.7 | 1.3 | 0.3 | 0.3 |
| Late AMD | 0.5 | 0.5 | 1.1 | 0.2 | 0.0 |
| Pure GA | 0.2 | 0.3 | 0.2 | 0.2 | 0.0 |
| Neovascular AMD | 0.3 | 0.2 | 1.0 | 0.0 | 0.0 |
| Risk Alleles | |||||
| CFH Y402H | |||||
| CT genotype | 33.3 | 45.6 | 9.8 | 45.1 | 33.1 |
| CC genotype | 8.1 | 12.8 | 0.3 | 12.0 | 7.2 |
| ARMS2 rs10490924 | |||||
| AC genotype | 39.7 | 36.0 | 49.7 | 36.6 | 37.8 |
| CC genotype | 51.8 | 60.1 | 30.0 | 57.5 | 57.2 |
| PPARG rs2972164 | |||||
| AG genotype | 34.8 | 49.4 | 10.9 | 38.2 | 40.7 |
| AA genotype | 24.7 | 26.0 | 0.3 | 53.7 | 20.0 |
| PLEKHA1 rs4311997 | |||||
| AG genotype | 47.4 | 51.3 | 44.4 | 45.7 | 47.5 |
| AA genotype | 28.0 | 23.1 | 43.5 | 14.9 | 31.9 |
| PLEKHA1 rs2421018 | |||||
| AG genotype | 38.0 | 48.0 | 31.7 | 31.5 | 39.8 |
| AA genotype | 54.2 | 36.3 | 64.5 | 64.5 | 53.7 |
| MRPL10 rs3209 | |||||
| AG genotype | 25.0 | 47.2 | 2.7 | 20.4 | 29.4 |
| AA genotype | 70.4 | 41.0 | 97.3 | 78.6 | 65.3 |
| ABCA4 rs1237081 | |||||
| CG genotype | 48.0 | 53.1 | 49.9 | 42.1 | 46.5 |
| CC genotype | 19.9 | 14.9 | 31.9 | 8.8 | 25.3 |
| CAPN5 rs 11237081 | |||||
| CG genotype | 38.8 | 49.1 | 29.8 | 35.5 | 39.3 |
| GG genotype | 11.5 | 23.9 | 4.2 | 4.0 | 12.4 |
| IL1B rs1143629 | |||||
| AG genotype | 48.8 | 45.5 | 51.8 | 51.0 | 46.7 |
| AA genotype | 31.8 | 43.7 | 26.8 | 28.9 | 27.4 |
| FOXO1 rs12583418 | |||||
| AG genotype | 42.5 | 39.6 | 38.3 | 49.2 | 42.7 |
| AA genotype | 28.2 | 11.4 | 53.2 | 29.8 | 21.8 |
| SOD3 rs2284659 | |||||
| AC genotype | 41.2 | 46.9 | 42.7 | 28.9 | 45.9 |
| AA genotype | 38.5 | 13.0 | 44.8 | 67.1 | 30.7 |
| MTR rs3754255 | |||||
| AG genotype | 50.6 | 52.1 | 46.4 | 52.9 | 51.2 |
| GG genotype | 30.0 | 32.3 | 28.6 | 29.4 | 29.6 |
Abbreviations: AMD, age-related macular degeneration; GA, geographic atrophy; RPE, retinal pigment epithelium; SD, standard deviation.
Associations
Associations of the selected SNPs with early AMD in the initial screen are presented in Table 2. Statistically significant associations of early AMD with PLEKHA1 rs4311197, MRPL10, ABCA4, and CAPN5 were found in whites only, PLEKHA1 rs2421018 in whites and Chinese Americans only, CFH in whites and Hispanics only, and IL1B rs114629 in blacks only (Table 2). However, after controlling for multiple tests by Bonferroni correction (1.7×10−5), only the association of ARMS2 remained significant in the combined sample. All SNPs met Hardy-Weinberg equilibrium in each racial/ethnic group (all P≥0.01; data not shown).
Table 2.
Prevalence of Early Age-Related Macular Degeneration by Gene Polymorphism, Genotype, and Racial/Ethnic Group
| Gene and Genotype | All | White | Chinese | Black | Hispanic | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| N | % | N | % | N | % | N | % | N | % | |
|
|
||||||||||
| CFH Y402H | ||||||||||
| TT | 1434 | 3.2 | 262 | 3.8 | 560 | 3.9 | 254 | 2.8 | 358 | 2.0 |
| CT | 814 | 4.9 | 288 | 7.3 | 61 | 3.3 | 267 | 1.5 | 198 | 6.6 |
| CC | 197 | 6.6 | 81 | 8.6 | 2 | 0.0 | 71 | 4.3 | 43 | 7.0 |
| OR (C) | 1.61 | 1.60 | 0.48 | 1.03 | 2.59 | |||||
| P valuea | 0.007 | 0.07 | 0.35 | 0.94 | 0.003 | |||||
| ARMS2 rs10490924 | ||||||||||
| AA | 200 | 10.0 | 25 | 16.0 | 110 | 6.4 | 35 | 2.9 | 30 | 26.7 |
| AC | 939 | 3.9 | 227 | 7.1 | 270 | 3.7 | 216 | 3.2 | 226 | 1.8 |
| CC | 1224 | 3.0 | 379 | 4.8 | 163 | 1.2 | 340 | 1.8 | 342 | 3.2 |
| OR (A) | 2.12 | 1.77 | 2.46 | 1.61 | 2.84 | |||||
| P valuea | 1.1×10−5 b | 0.05 | 0.01 | 0.26 | 0.002 | |||||
| PPARG rs2972164 | ||||||||||
| AA | 604 | 5.3 | 164 | 6.7 | 2 | 0.0 | 318 | 2.8 | 120 | 10.0 |
| AG | 850 | 4.4 | 312 | 6.7 | 68 | 2.9 | 226 | 2.2 | 244 | 3.7 |
| GG | 991 | 3.0 | 155 | 3.9 | 553 | 4.0 | 48 | 0.0 | 235 | 0.9 |
| OR (A) | 1.69 | 1.14 | 0.58 | 1.89 | 4.35 | |||||
| P valuea | 0.004 | 0.60 | 0.48 | 0.21 | 1.0×10−5 c | |||||
| PLEKHA1 rs4311997d | ||||||||||
| AA | 661 | 5.9 | 146 | 10.3 | 236 | 5.1 | 88 | 1.1 | 191 | 5.8 |
| AG | 1119 | 3.2 | 324 | 4.6 | 241 | 2.1 | 270 | 3.0 | 284 | 2.8 |
| GG | 583 | 3.3 | 161 | 5.0 | 66 | 3.0 | 233 | 2.2 | 123 | 3.3 |
| OR (A) | 1.54 | 1.89 | 1.75 | 0.80 | 1.61 | |||||
| P valuea | 0.01 | 0.02 | 0.16 | 0.58 | 0.14 | |||||
| PLEKHA1 rs2421018e | ||||||||||
| AA | 1281 | 4.8 | 229 | 9.6 | 350 | 4.9 | 381 | 2.1 | 321 | 4.4 |
| AG | 899 | 2.7 | 303 | 4.0 | 172 | 1.2 | 186 | 2.7 | 238 | 2.1 |
| GG | 183 | 4.9 | 99 | 4.0 | 21 | 0.0 | 24 | 4.2 | 39 | 13.0 |
| OR (A) | 1.49 | 2.22 | 5.00 | 0.76 | 0.97 | |||||
| P valuea | 0.04 | 0.007 | 0.03 | 0.56 | 0.93 | |||||
| MRPL10 rs3209 | ||||||||||
| AA | 1721 | 4.4 | 259 | 9.7 | 606 | 3.8 | 465 | 2.8 | 391 | 3.6 |
| AG | 612 | 3.6 | 298 | 4.0 | 17 | 5.9 | 121 | 0.8 | 176 | 4.6 |
| GG | 112 | 1.8 | 74 | 1.4 | 0 | 0.0 | 6 | 0.0 | 32 | 3.1 |
| OR (A) | 1.69 | 2.86 | 0.47 | 4.35 | 0.93 | |||||
| P valuea | 0.02 | 0.001 | 0.51 | 0.15 | 0.85 | |||||
| ABCA4 rs548122 | ||||||||||
| CC | 470 | 6.4 | 94 | 13.8 | 173 | 4.6 | 52 | 3.9 | 151 | 4.6 |
| CG | 1133 | 3.5 | 335 | 5.4 | 271 | 3.3 | 249 | 2.8 | 278 | 2.2 |
| GG | 760 | 3.2 | 202 | 3.5 | 99 | 2.0 | 290 | 1.7 | 169 | 5.9 |
| OR (C) | 1.48 | 2.37 | 1.59 | 1.49 | 0.76 | |||||
| P valuea | 0.02 | 0.002 | 0.20 | 0.34 | 0.36 | |||||
| CAPN5 rs11237081 | ||||||||||
| CC | 1175 | 2.9 | 170 | 2.9 | 358 | 3.4 | 358 | 2.0 | 289 | 3.5 |
| CG | 917 | 4.3 | 310 | 7.1 | 162 | 3.1 | 210 | 1.9 | 235 | 3.4 |
| GG | 271 | 7.8 | 151 | 7.3 | 23 | 8.7 | 23 | 13.0 | 74 | 6.8 |
| OR (G) | 1.50 | 1.68 | 1.31 | 1.84 | 1.20 | |||||
| P valuea | 0.01 | 0.04 | 0.51 | 0.16 | 0.58 | |||||
| IL1B rs1143629 | ||||||||||
| AA | 778 | 5.8 | 276 | 8.3 | 167 | 3.6 | 171 | 4.7 | 164 | 4.9 |
| AG | 1192 | 3.6 | 287 | 3.8 | 323 | 5.0 | 302 | 2.0 | 280 | 3.6 |
| GG | 475 | 2.3 | 68 | 5.9 | 133 | 1.5 | 119 | 0.0 | 155 | 3.2 |
| OR (A) | 1.52 | 1.59 | 1.35 | 3.57 | 1.16 | |||||
| P valuea | 0.01 | 0.11 | 0.35 | 0.009 | 0.61 | |||||
| FOXO1 rs12583418 | ||||||||||
| GG | 691 | 4.6 | 309 | 4.9 | 46 | 2.2 | 124 | 6.5 | 212 | 3.8 |
| AG | 1003 | 3.9 | 250 | 7.6 | 207 | 2.4 | 291 | 2.1 | 255 | 3.5 |
| AA | 666 | 3.5 | 72 | 5.6 | 288 | 4.5 | 176 | 0.0 | 130 | 4.6 |
| OR (G) | 1.03 | 0.90 | 0.62 | 3.91 | 0.94 | |||||
| P valuea | 0.85 | 0.67 | 0.26 | 0.004 | 0.82 | |||||
| SOD3 rs2284659 | ||||||||||
| CC | 495 | 5.1 | 253 | 6.3 | 78 | 0.0 | 24 | 0.0 | 140 | 6.4 |
| AC | 1008 | 3.4 | 296 | 5.1 | 266 | 2.3 | 171 | 2.3 | 275 | 3.3 |
| AA | 942 | 4.3 | 82 | 8.5 | 279 | 6.5 | 397 | 2.5 | 184 | 2.7 |
| OR (A) | 1.12 | 0.94 | 4.17 | 2.13 | 0.66 | |||||
| P valuea | 0.52 | 0.83 | 0.002 | 0.23 | 0.18 | |||||
| MTR rs3754255 | ||||||||||
| AA | 475 | 3.0 | 98 | 5.1 | 156 | 4.5 | 105 | 1.9 | 116 | 0.0 |
| AG | 1237 | 3.6 | 329 | 5.8 | 289 | 3.1 | 313 | 1.6 | 306 | 3.6 |
| GG | 733 | 5.6 | 204 | 6.9 | 178 | 4.5 | 174 | 4.0 | 177 | 6.8 |
| OR (G) | 1.49 | 1.23 | 1.09 | 1.82 | 2.94 | |||||
| P valuea | 0.01 | 0.43 | 0.79 | 0.15 | 0.003 | |||||
% = percent with early age-related macular degeneration; N = number with the polymorphism; OR, odds ratio.
Uncorrected P values adjusted for age, sex, study site, and ancestry-informative markers, but not Bonferroni corrected.
Statistically significant after Bonferroni correction (P=0.031)
Statistically significant after Bonferroni correction (P=0.029)
R2=0.3 with ARMS2
R2=0.6 with ARMS2
Association of two single nucleotide polymorphisms with early age-related macular degeneration
Joint effect
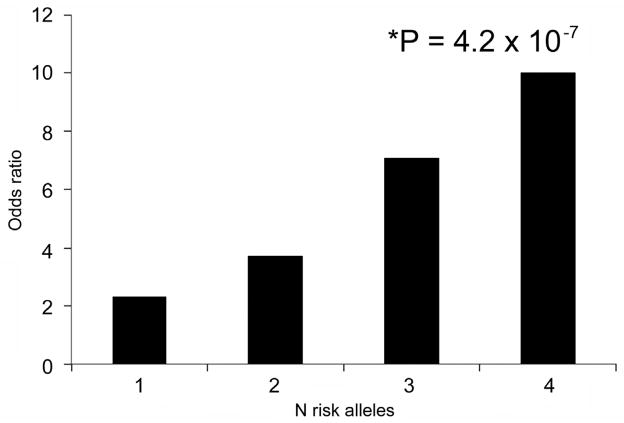
We first evaluated the joint effect of two SNPs in our data, ARMS2 and CFH. We did this for all possible genotypes in the following manner. Using ARMS2 (CC) and CFH (TT) as the reference group (4 protective alleles together, OR=1), we estimated the ORs by number of risk alleles (Figure 1). The ORs increased from 2.3 for one, 3.7 for two, and 7.1 for three to 10.0 for four risk allele combinations (P for trend=4.2×10−7, Figure 1). The results were similar when adjusted for smoking (data not shown). Thus, the joint effect on risk of AMD increased significantly with the increasing number of risk alleles at these 2 loci.
Figure 1.
The joint effect between CFH Y402H and ARMS2 rs10490924 and odds ratio of age-related macular degeneration.
Interaction
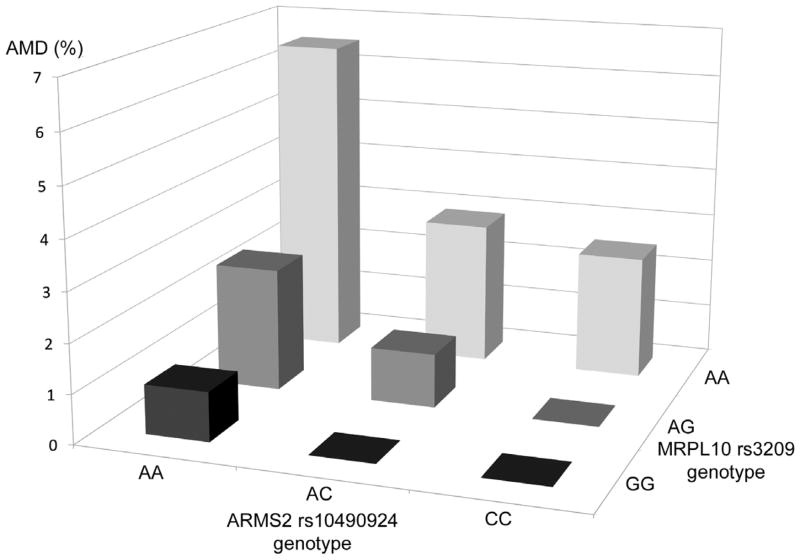
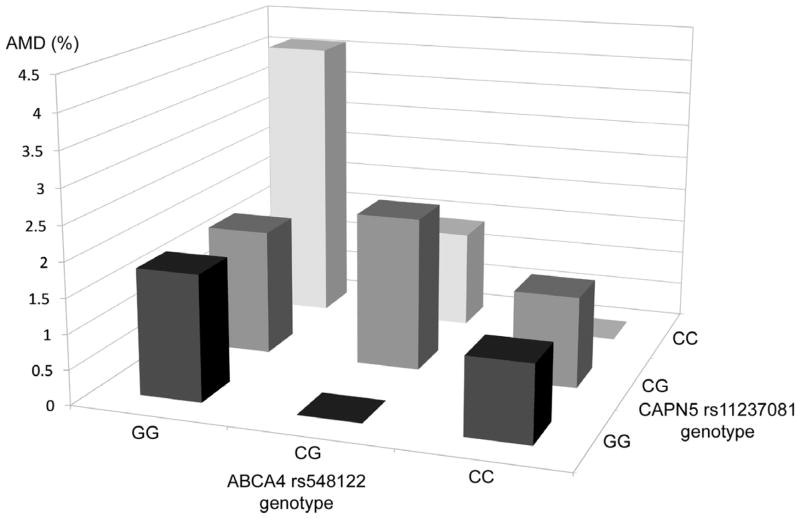
We next examined all SNP-SNP interactions of 12 analyzed SNPs, using the prevalence of early AMD in each genotype group. Thus, with a total of 9 genotype groups in 1 SNP-SNP pair present, the differences of the prevalence among 9 groups were tested for significant interactions. Two statistically significant interactions (ARMS2 and MRPL10 [P=0.003] and CAPN5 and ABCA4 [P=0.002]) were found between 144 SNP-SNP pairs from 12 significant candidate SNPs for early AMD, after adjusting for age, sex, smoking status and principal components using all samples (Figure 2). No interactions remained significant after adjustment for multiple testing by the Bonferroni method. Athough strong associations were present for the single and joint effects of ARMS2 and CFH SNPs, no interactions were identified for these 2 SNPs with AMD (P=0.19).
Figure 2.
Interactions between genes of interest and prevalence of age-related macular degeneration. These interactions were no longer significant after correction for multiple hypothesis testing. Upper. ARMS2 rs10490924 and MRPL10 rs3209; Lower. CAPN5 rs11237081 and ABCA4 rs548122
Effect of candidate gene single nucleotide polymorphisms on racial/ethnic variation of prevalence of early age-related macular degeneration
When controlling for age, sex, study site, and smoking status, blacks, Chinese Americans, and Hispanics had lower ORs for early AMD than whites (Table 3). However, the association was statistically significant only when the comparison was made between whites and blacks (P=0.004).
Table 3.
Prevalence of Early Age-related Macular Degeneration Comparing Whites With Other Racial/Ethnic Groups While Adjusting for Genotypes and Other Factors in the Multi-Ethnic Study of Atherosclerosis
| Adjusted for | White | Chinese | Black | Hispanic | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| OR (95% CI) | OR (95% CI) | Pb | OR (95% CI) | Pb | OR (95% CI) | Pb | |
|
|
|||||||
| Age, sex, and study site | 1.00 (ref) | 0.61 (0.36, 1.05) | 0.08 | 0.39 (0.20, 0.74) | 0.004 | 0.66 (0.38, 1.14) | 0.14 |
| a | 1.00 (ref) | 0.67 (0.38, 1.17) | 0.16 | 0.39 (0.20, 0.74) | 0.004 | 0.68 (0.39, 1.19) | 0.18 |
| a and CFH Y402H genotype | 1.00 (ref) | 0.85 (0.45, 1.61) | 0.62 | 0.39 (0.22, 0.73) | 0.004 | 0.80 (0.45, 1.43) | 0.45 |
| a and ARMS2 rs10490924 genotype | 1.00 (ref) | 0.45 (0.24, 0.85) | 0.01 | 0.38 (0.20, 0.73) | 0.004 | 0.66 (0.38, 1.16) | 0.15 |
| a and PPARG rs2972164 genotype | 1.00 (ref) | 0.76 (0.38, 1.55) | 0.46 | 0.36 (0.18, 0.69) | 0.002 | 0.80 (0.45, 1.43) | 0.46 |
| a and PLEKHA1 rs4311997 genotype | 1.00 (ref) | 0.50 (0.27, 0.93) | 0.03 | 0.42 (0.22, 0.80) | 0.009 | 0.63 (0.36, 1.10) | 0.10 |
| a and PLEKHA1 rs2421018 genotype | 1.00 (ref) | 0.44 (0.24, 0.82) | 0.01 | 0.32 (0.16, 0.63) | <0.001 | 0.61 (0.35, 1.07) | 0.09 |
| a and MRPL10 rs3209 genotype | 1.00 (ref) | 0.38 (0.21, 0.71) | 0.002 | 0.25 (0.12, 0.48) | <0.001 | 0.55 (0.31, 0.98) | 0.04 |
| a and ABCA4 rs548122 genotype | 1.00 (ref) | 0.49 (0.26, 0.91) | 0.02 | 0.45 (0.23, 0.86) | 0.02 | 0.63 (0.36, 1.11) | 0.11 |
| a and CAPN5 rs11237081 genotype | 1.00 (ref) | 0.78 (0.41, 1.47) | 0.44 | 0.54 (0.27, 1.07) | 0.08 | 0.76 (0.43, 1.35) | 0.35 |
| a and IL1B rs1143629 genotype | 1.00 (ref) | 0.74 (0.42, 1.30) | 0.29 | 0.44 (0.23, 0.85) | 0.01 | 0.75 (0.42, 1.32) | 0.32 |
| a and FOXO1 rs12583418 genotype | 1.00 (ref) | 0.51 (0.26, 1.01) | 0.05 | 0.44 (0.23, 0.86) | 0.02 | 0.66 (0.38, 1.16) | 0.15 |
| a and SOD3 rs2284659 genotype | 1.00 (ref) | 0.52 (0.28, 0.95) | 0.03 | 0.35 (0.17, 0.75) | 0.006 | 0.71 (0.40, 1.25) | 0.24 |
| a and MTR rs3754255 genotype | 1.00 (ref) | 0.68 (0.39, 1.20) | 0.18 | 0.39 (0.21, 0.75) | 0.005 | 0.71 (0.41, 1.24) | 0.23 |
CI, confidence interval; OR, odds ratio; P, P value.
Age, sex, study site, and smoking status.
P value for difference in prevalence of early AMD prevalence between this racial/ethnic group and whites.
We then examined the effect of each of the 12 specific candidate SNPs on the association of race/ethnicity with early AMD by conditioning, 1 at a time, on each candidate SNP while controlling for age, sex, study site, and smoking status (Table 3). In blacks, the P value for CAPN5 changed from significant at baseline (P=0.004) to non-significant in the model conditioned on CAPN5 (P=0.08), suggesting that adjusting for CAPN5 attenuates the OR of early AMD in blacks when compared to whites and that this SNP contributed to the variation in genetic susceptibilities of early AMD between blacks and whites. The OR for early AMD in blacks compared to whites increased from 0.39 to 0.54 (before and after conditioning on CAPN5) but did not change significantly when adjusting for any of the other SNPs (Table 3).
In contrast, when comparing Chinese Americans to whites, the P values for the OR of AMD by SNP for ARMS2, PLEKHA1 (rs4311997 and rs2421018), MRPL10, ABCA4, and SOD3 changed from non-significant (P=0.08) at baseline to significant, conditioning on each SNP independently (Table 3, “Chinese” column). The OR for early AMD in Chinese Americans compared to whites decreased from 0.61 at baseline to 0.45 for ARMS2 (P=0.01), 0.50 for PLEKHA1 rs4311997 (P=0.03), 0.44 for PLEKHA1 rs2421018 (P=0.01), 0.38 for MRPL10 (P=0.002), 0.49 for ABCA4 (P=0.02), and 0.52 for SOD3 (P=0.03). This decrease also suggested that these SNPs independently contributed to the variation in genetic susceptibilities of early AMD between Chinese Americans and whites.
As all 6 SNPs were independently associated with the racial/ethnic difference in prevalence of early AMD in Chinese Americans versus whites, we performed a combined analysis by grouping all 6 SNPs in 1 model. When combined, these 6 SNPs were associated with an even greater decrease in OR for AMD; the ORs decreased from 0.67 at baseline to 0.14 in the combined analysis (P<0.0001).
Discussion
This study provided a unique opportunity to examine the relationships of candidate genes to early AMD in whites, blacks, Chinese Americans, and Hispanics participating in a large multi-ethnic cohort study in the United States. After controlling for age, sex, ancestry-informative markers, study site, and smoking status, we found that associations of several candidate genes with early AMD varied among the racial/ethnic groups. However, differences in genotype frequencies did not explain the higher prevalence of early AMD in whites compared to other racial/ethnic groups, with the possible exception of Chinese American and white differences, in which the higher frequency of risk alleles for a number of SNPs in Chinese Americans may in part be responsible for the disease frequency in Chinese Americans approaching that of whites. Allele frequencies of most SNPs, except for rs3754255 in MTR, were statistically significant between Chinese Americans and whites after adjusting for multiple testing (data not shown).
We have previously reported on the racial/ethnic differences in the frequency of early AMD in the MESA (whites 5.4%, Chinese Americans 4.6%, Hispanics 4.2%, and blacks 2.4%) and in the 2005–2008 NHANES (whites 7.3%, Mexican Americans 5.1%, and blacks 2.4%).13,14 The differences in whites and blacks are well known and were statistically significant in both studies. In the MESA, reasons for these differences between whites and blacks were not explained by potential known risk factors of AMD such as cigarette smoking, history of alcohol drinking, BMI, inflammatory factors, hypertension, diabetes status, or markers of subclinical CVD.16–18 We also showed that whites, blacks, and Hispanics who were all homozygous for the CFH Y402H CC variant genotype had the highest frequencies of early AMD compared with those with the CFH Y402H TT wild genotype and that the CFH Y402H CC variant genotype distributions varied among the racial/ethnic groups but did not explain the higher early AMD in non-Hispanic whites compared to blacks. The current analyses show that despite differences in distributions of other protective (e.g., MRPL10) and deleterious genotypes (e.g., ARMS2), these SNPs only partially explained the differences in AMD frequencies among the different racial/ethnic groups. However, it should be noted that we have moderate power to identify the differences for SNPs with moderate effect size, i.e., OR less than 2, given our current sample sizes. At the 1.7×10−5 significance level (after Bonferroni correction), we tested the power under the additive genetic model. For genetic relative risk greater than 2.5 (most reported ORs of CFH and ARMS2 genes are greater than 3), we had enough power (>0.80) to identify the association with disease allele frequency greater than 0.20.
In the MESA, despite the distributions of CFH Y402H being similar in blacks and whites, there was a higher frequency of AMD in whites compared to blacks. This differential in prevalence in whites and blacks is consistent with the findings of other studies.21,22,28 It has been attributed, in part, to a higher frequency of protective genes for AMD in blacks compared to whites.17,21,22,28 Our current analyses show that while there are some differences in candidate gene distributions (Table 2) and main effects of these genes (Table 3) on AMD prevalence between blacks and whites, only CAPN5 appears to contribute to the differences in frequency of early AMD between blacks and whites. The relation of CAPN5 attenuated the OR from 0.39 to 0.54 (in reference to whites at OR=1.00), so this gene could very well account for some of the difference in genetic susceptibility to AMD between whites and blacks. However, the ORs altered by this and other SNPs remain within the 95% CI of the baseline OR, suggesting that these results may lie within an expected range of variation. Because of this, the reader should exercise caution with inferences regarding candidate gene SNPs explaining racial/ethnic differences that resulted from these analyses.
The strong relationship of the ARMS2 variant genotype to AMD in Chinese Americans in the MESA and the infrequency of CFH variants are consistent with other studies of Han Chinese living in China.23–26 Our study describes associations of a number of additional candidate genes (e.g., PLEKHA1 rs2421018, MRPL10, ABCA4, SOD3) not previously reported to be associated with AMD in Chinese Americans that need to be replicated in other cohorts. Each of these 6 identified SNPs independently contribute to the difference in genetic susceptibilities of early AMD between Chinese Americans and whites. The combined effect of all 6 SNPs resulted in a more significant difference, further supporting evidence of an underlying genetic difference between the 2 racial/ethnic groups. Furthermore, as can be seen in Table 2, in all 6 of these SNPs, the risk allele is more frequent in Chinese Americans than in whites. Thus, the higher frequency of these risk alleles in Chinese Americans may have resulted in the trend toward a disease frequency similar to whites when adjusted for other risk factors in this dataset. Thus, Chinese Americans would more likely have a lower frequency of AMD if these SNPs were present in frequencies similar to the white population.
There are too few other studies of Hispanic persons to compare differences in the distributions and main effects of candidate genes on AMD between Hispanics and whites, though our data suggests that MRPL10 did contribute to the differences in frequency of early AMD between Hispanics and whites. Of interest, MRPL10 is the only common candidate gene between Chinese Americans and Hispanics when compared to whites and has a similar direction of effect (i.e., the odds of early AMD decrease in both Chinese Americans and Hispanics but increase in blacks). It is also of interest that the 6 SNPs that differ in frequency between Chinese Americans and whites are intermediate in frequency in Hispanics, thus supporting prior evidence of a common ancestral lineage in Hispanics (Amerindian genes) and East Asians.45
AMD is a complex disease with multiple genetic variants involved in its etiology. The biological interactions of gene-gene or gene-environment were not clear. Several studies of the joint effect between CFH and ARMS2 were reported, but the interaction results were conflicting.46–48 Consistent with other reports, we identified the strongest joint risk of CFH and ARMS2, but the formal interaction analysis was not significant. However, the moderate sample size and multiple testing issues limited our power for identifying interaction effects.
Although our study has strengths, including objective grading of AMD and a large multi-ethnic cohort, caution must be exercised when interpreting the findings for several reasons. First, we examined many candidate genes which had been identified in previous studies as associated with AMD. This may have led to chance findings or type I error, particularly for associations that have not previously been reported in nonwhite racial/ethnic groups, as is the case with SNPs in minority groups. Second, the power to detect some associations may have been limited by the infrequency of AMD lesions. There are differences in the frequency of the candidate genes, and there is a suggestion of differences in the strength of the association for the same risk alleles, but the data do suggest that several genes contribute to the differences in frequency of AMD relationships among the different racial/ethnic groups. Many of the covariates were only measured in a sample of the population, further reducing the study power. Third, not all protective variants of candidate SNPs (e.g., CFH-related proteins 1 and 3, factor B/C2, C3 and factor 1) were included in the analyses.49 Fourth, the period cross-sectional nature of the study (retinal examination 2 years after the non-genetic risk factors were measured) also may have limited our ability to detect associations with factors that may manifest subsequently as pathologic changes over time. Finally, we cannot determine whether there were biases caused by nonparticipation or selective mortality that affected the relationships.
In summary, our study confirms the importance of candidate genes such as CFH and ARMS2 in a large multi-ethnic cohort in the United States, and shows that they and other candidate genes differ in both their distributions and their associations with early AMD among different racial/ethnic groups. The relative distributions of the gene variants appear to contribute to both the lower prevalence of AMD in certain racial/ethnic groups (e.g., blacks), and to a higher prevalence of AMD in others (e.g., Chinese Americans). Thus, genetic susceptibility appears to partially explain variations in the prevalence of AMD among the four racial/ethnic groups in the MESA cohort.
Acknowledgments
Financial Support: The Multi-Ethnic Study of Atherosclerosis (MESA) was supported by contracts N01-HC-95159 through N01-HC-95165 and N01-HC-95169 from the National Heart, Lung, and Blood Institute. MESA Family was conducted and supported in collaboration with MESA investigators by grants and contracts R01-HL071051, R01-HL071205, R01-HL071250, R01-HL071251, R01-HL071252, R01-HL071258, R01-HL071259, and NIH Intramural Research Award ZIAEY000403. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. Additional support was provided by NIH grant HL69979 (Klein R and Wong TY) and in part by the Cedars-Sinai Board of Governors’ Chair in Medical Genetics (Rotter JI). None of the above named sponsors or funding organizations had any role in the design or conduct of this research. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Heart, Lung, and Blood Institute, the National Eye Institute, or the National Institutes of Health.
Biography

Dr. Ronald Klein is a Professor of Ophthalmology and Visual Sciences at the University of Wisconsin School of Medicine and Public Health, as well as co-principal investigator of two large, long-running population-based cohort studies, the Beaver Dam Eye Study and the Wisconsin Epidemiologic Study of Diabetic Retinopathy. His primary research interests are the epidemiology of age-related eye diseases, especially age-related macular degeneration, and hypertensive and diabetic retinopathy.
Footnotes
Each of the coauthors has seen and agrees with each of the changes made to this manuscript in the revision and to the way his or her name is listed.
As of July 1, 2013, the following authors are affiliated with the Institute for Translational Genomics and Population Sciences, LA BioMed, Harbor-UCLA, Torrance, California, USA: Xiaohui Li, Kent D. Taylor, Jerome I. Rotter.
Financial Disclosures: The authors have no proprietary or commercial interest in any materials discussed in this article.
Contributions of Authors:
Design of the study (RK); conduct of the study (RK); collection of data (RK, MFC, JIR); analysis of data (XL, JZK, KDT); interpretation of the data (JIR, XL, JZK, BEKK, RK); manuscript preparation (RK, BEKK, JIR); manuscript review (XL, JZK, BEKK, MFC, TYW, KDT, JIR); manuscript approval (XL, JZK, BEKK, MFC, TYW, KDT, JIR).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Congdon N, O’Colmain B, Klaver CC, et al. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004;122(4):477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 2.Klein R, Wang Q, Klein BE, Moss SE, Meuer SM. The relationship of age-related maculopathy, cataract, and glaucoma to visual acuity. Invest Ophthalmol Vis Sci. 1995;36(1):182–191. [PubMed] [Google Scholar]
- 3.Klein BE, Klein R. Cataracts and macular degeneration in older Americans. Arch Ophthalmol. 1982;100(4):571–573. doi: 10.1001/archopht.1982.01030030573002. [DOI] [PubMed] [Google Scholar]
- 4.Klein R, Clegg L, Cooper LS, et al. Prevalence of age-related maculopathy in the Atherosclerosis Risk in Communities Study. Arch Ophthalmol. 1999;117(9):1203–1210. doi: 10.1001/archopht.117.9.1203. [DOI] [PubMed] [Google Scholar]
- 5.Klein R, Klein BE, Marino EK, et al. Early age-related maculopathy in the Cardiovascular Health Study. Ophthalmology. 2003;110(1):25–33. doi: 10.1016/s0161-6420(02)01565-8. [DOI] [PubMed] [Google Scholar]
- 6.Bressler SB, Munoz B, Solomon SD, West SK. Racial differences in the prevalence of age-related macular degeneration: the Salisbury Eye Evaluation (SEE) Project. Arch Ophthalmol. 2008;126(2):241–245. doi: 10.1001/archophthalmol.2007.53. [DOI] [PubMed] [Google Scholar]
- 7.Schachat AP, Hyman L, Leske MC, Connell AM, Wu SY. Features of age-related macular degeneration in a black population. The Barbados Eye Study Group. Arch Ophthalmol. 1995;113(6):728–735. doi: 10.1001/archopht.1995.01100060054032. [DOI] [PubMed] [Google Scholar]
- 8.Leske MC, Wu SY, Hennis A, et al. Nine-year incidence of age-related macular degeneration in the Barbados Eye Studies. Ophthalmology. 2006;113(1):29–35. doi: 10.1016/j.ophtha.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Friedman DS, Katz J, Bressler NM, Rahmani B, Tielsch JM. Racial differences in the prevalence of age-related macular degeneration: the Baltimore Eye Survey. Ophthalmology. 1999;106(6):1049–1055. doi: 10.1016/S0161-6420(99)90267-1. [DOI] [PubMed] [Google Scholar]
- 10.Cruickshanks KJ, Hamman RF, Klein R, Nondahl DM, Shetterly SM. The prevalence of age-related maculopathy by geographic region and ethnicity. The Colorado-Wisconsin Study of Age-Related Maculopathy. Arch Ophthalmol. 1997;115(2):242–250. doi: 10.1001/archopht.1997.01100150244015. [DOI] [PubMed] [Google Scholar]
- 11.Varma R, Fraser-Bell S, Tan S, Klein R, Azen SP. Prevalence of age-related macular degeneration in Latinos: the Los Angeles Latino Eye Study. Ophthalmology. 2004;111(7):1288–1297. doi: 10.1016/j.ophtha.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 12.Munoz B, Klein R, Rodriguez J, Snyder R, West SK. Prevalence of age-related macular degeneration in a population-based sample of Hispanic people in Arizona: Proyecto VER. Arch Ophthalmol. 2005;123(11):1575–1580. doi: 10.1001/archopht.123.11.1575. [DOI] [PubMed] [Google Scholar]
- 13.Klein R, Klein BE, Knudtson MD, et al. Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the Multi-Ethnic Study of Atherosclerosis. Ophthalmology. 2006;113(3):373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Klein R, Chou CF, Klein BE, Zhang X, Meuer SM, Saaddine JB. Prevalence of age-related macular degeneration in the US population. Arch Ophthalmol. 2011;129(1):75–80. doi: 10.1001/archophthalmol.2010.318. [DOI] [PubMed] [Google Scholar]
- 15.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379(9827):1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 16.Klein R, Klein BE, Knudtson MD, et al. Subclinical atherosclerotic cardiovascular disease and early age-related macular degeneration in a multiracial cohort: the Multiethnic Study of Atherosclerosis. Arch Ophthalmol. 2007;125(4):534–543. doi: 10.1001/archopht.125.4.534. [DOI] [PubMed] [Google Scholar]
- 17.Klein R, Knudtson MD, Klein BE, et al. Inflammation, complement factor H, and age-related macular degeneration: the Multi-Ethnic Study of Atherosclerosis. Ophthalmology. 2008;115(10):1742–1749. doi: 10.1016/j.ophtha.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein R, Knudtson MD, Klein BE, Wong TY, Cotch MF, Barr G. Emphysema, airflow limitation, and early age-related macular degeneration. Arch Ophthalmol. 2010;128(4):472–477. doi: 10.1001/archophthalmol.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montezuma SR, Sobrin L, Seddon JM. Review of genetics in age related macular degeneration. Semin Ophthalmol. 2007;22(4):229–240. doi: 10.1080/08820530701745140. [DOI] [PubMed] [Google Scholar]
- 20.Kokotas H, Grigoriadou M, Petersen MB. Age-related macular degeneration: genetic and clinical findings. Clin Chem Lab Med. 2011;49(4):601–616. doi: 10.1515/CCLM.2011.091. [DOI] [PubMed] [Google Scholar]
- 21.Hageman GS, Hancox LS, Taiber AJ, et al. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization ethnic distribution and evolutionary implications. Ann Med. 2006;38(8):592–604. [PMC free article] [PubMed] [Google Scholar]
- 22.Grassi MA, Fingert JH, Scheetz TE, et al. Ethnic variation in AMD-associated complement factor H polymorphism p. Tyr402His. Hum Mutat. 2006;27(9):921–925. doi: 10.1002/humu.20359. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Zhao P, Tang S, et al. Association study of complement factor H, C2, CFB, and C3 and age-related macular degeneration in a Han Chinese population. Retina. 2010;30(8):1177–1184. doi: 10.1097/IAE.0b013e3181cea676. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y, Li Y, Xu L, et al. Complement factor H polymorphism in age-related maculopathy in the Chinese population: the Beijing Eye Study. Retina. 2010;30(3):443–446. doi: 10.1097/IAE.0b013e3181c2e086. [DOI] [PubMed] [Google Scholar]
- 25.Jiang H, Qu Y, Dang G, et al. Analyses of single nucleotide polymorphisms and haplotype linkage of LOC387715 and the HTRA1 gene in exudative age-related macular degeneration in a Chinese cohort. Retina. 2009;29(7):974–979. doi: 10.1097/IAE.0b013e3181a3b90e. [DOI] [PubMed] [Google Scholar]
- 26.Tsai YY, Lin JM, Wan L, et al. Interleukin gene polymorphisms in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49(2):693–698. doi: 10.1167/iovs.07-0125. [DOI] [PubMed] [Google Scholar]
- 27.Thakkinstian A, Bowe S, McEvoy M, Smith W, Attia J. Association between apolipoprotein E polymorphisms and age-related macular degeneration: A HuGE review and meta-analysis. Am J Epidemiol. 2006;164(9):813–822. doi: 10.1093/aje/kwj279. [DOI] [PubMed] [Google Scholar]
- 28.Nonyane BA, Nitsch D, Whittaker JC, et al. An ecological correlation study of late age-related macular degeneration and the complement factor H Y402H polymorphism. Invest Ophthalmol Vis Sci. 2010;51(5):2393–2402. doi: 10.1167/iovs.09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bild DE, Bluemke DA, Burke GL, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 30.Wong TY, Klein R, Islam FM, et al. Diabetic retinopathy in a multi-ethnic cohort in the United States. Am J Ophthalmol. 2006;141(3):446–455. doi: 10.1016/j.ajo.2005.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Early Treatment Diabetic Retinopathy Study Research Group. Grading diabetic retinopathy from stereoscopic color fundus photographs--an extension of the modified Airlie House classification ETDRS report number 10. Ophthalmology. 1991;98(5 Suppl):786–806. [PubMed] [Google Scholar]
- 32.Klein R, Meuer SM, Moss SE, Klein BE, Neider MW, Reinke J. Detection of age-related macular degeneration using a nonmydriatic digital camera and a standard film fundus camera. Arch Ophthalmol. 2004;122(11):1642–1646. doi: 10.1001/archopht.122.11.1642. [DOI] [PubMed] [Google Scholar]
- 33.Klein R, Davis MD, Magli YL, Segal P, Klein BE, Hubbard L. The Wisconsin Age-Related Maculopathy Grading System. Ophthalmology. 1991;98(7):1128–1134. doi: 10.1016/s0161-6420(91)32186-9. [DOI] [PubMed] [Google Scholar]
- 34.MESA Coordinating Center. Multi-Ethnic Study of Atherosclerosis Field Center Manual of Operations. Seattle: University of Washington; 2001. [Google Scholar]
- 35.Wassel CL, Pankow JS, Rasmussen-Torvik LJ, et al. Associations of SNPs in ADIPOQ and subclinical cardiovascular disease in the multi-ethnic study of atherosclerosis (MESA) Obesity (Silver Spring) 2011;19(4):840–847. doi: 10.1038/oby.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bielinski SJ, Pankow JS, Li N, et al. ICAM1 and VCAM1 polymorphisms, coronary artery calcium, and circulating levels of soluble ICAM-1: the multi-ethnic study of atherosclerosis (MESA) Atherosclerosis. 2008;201(2):339–344. doi: 10.1016/j.atherosclerosis.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunderson KL, Kruglyak S, Graige MS, et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14(5):870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan JB, Gunderson KL, Bibikova M, et al. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]
- 39.Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat Rev Genet. 2006;7(8):632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 40.de Bakker PI, Saxena R, Graham RR. Variation in the human genome and risk to common disease. pharmacogenomics; Keystone Symposium on ‘Human Genome Sequence Variation and the Inherited Basis of Common Diseases’; January 8–13; Breckenridge, Colorado, USA. 2004. pp. 157–161. [DOI] [PubMed] [Google Scholar]
- 41.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 42.The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haritunians T, Taylor KD, Targan SR, et al. Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis. 2010;16(11):1830–1840. doi: 10.1002/ibd.21293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokunaga K, Ohashi J, Bannai M, Juji T. Genetic link between Asians and native Americans: evidence from HLA genes and haplotypes. Hum Immunol. 2001;62(9):1001–1008. doi: 10.1016/s0198-8859(01)00301-9. [DOI] [PubMed] [Google Scholar]
- 46.Conley YP, Jakobsdottir J, Mah T, et al. CFH, ELOVL4, PLEKHA1 and LOC387715 genes and susceptibility to age-related maculopathy: AREDS and CHS cohorts and meta-analyses. Hum Mol Genet. 2006;15(21):3206–3218. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]
- 47.Francis PJ, Zhang H, Dewan A, Hoh J, Klein ML. Joint effects of polymorphisms in the HTRA1, LOC387715/ARMS2 and CFH genes on AMD in a Caucasian population. Mol Vis. 2008;14:1395–1400. [PMC free article] [PubMed] [Google Scholar]
- 48.Seitsonen SP, Onkamo P, Peng G, et al. Multifactor effects and evidence of potential interaction between complement factor H Y402H and LOC387715 A69S in age-related macular degeneration. PLoS One. 2008;3(12):e3833. doi: 10.1371/journal.pone.0003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charbel Issa P, Chong NV, Scholl HP. The significance of the complement system for the pathogenesis of age-related macular degeneration - current evidence and translation into clinical application. Graefes Arch Clin Exp Ophthalmol. 2011;249(2):163–174. doi: 10.1007/s00417-010-1568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]