Abstract
The electronic linking of neuroscience information, including data embedded in the primary literature, would permit powerful queries and analyses driven by structured databases. This task would be facilitated by automated procedures which can identify biological concepts in journals. Here we apply an approach for automatically mapping formal identifiers of neuroanatomical regions to text found in journal abstracts, and apply it to a large body of abstracts from the Journal of Comparative Neurology (JCN). The analyses yield over one hundred thousand brain region mentions which we map to 8,225 brain region concepts in multiple organisms. Based on the analysis of a manually annotated corpus, we estimate mentions are mapped at 95% precision and 63% recall. Our results provide insights into the patterns of publication on brain regions and species of study in the Journal, but also point to important challenges in the standardization of neuroanatomical nomenclatures. We find that many terms in the formal terminologies never appear in a JCN abstract, while conversely, many terms authors use are not reflected in the terminologies. To improve the terminologies we deposited 136 unrecognized brain regions into the Neuroscience Lexicon (NeuroLex). The training data, terminologies, normalizations, evaluations and annotated journal abstracts are freely available at http://www.chibi.ubc.ca/WhiteText/.
Keywords: neuroinformatics, brain mapping, homology, brain reference system, brain atlases
Introduction
One of the challenges in neuroscience research is making the best use of the large bodies of existing knowledge and data. This challenge has been increasingly taken up under the broad rubric of “neuroinformatics”, which, among other things, is concerned with the formal encoding and combination of existing neuroscience knowledge, and the mining of that information to make new discoveries (French and Pavlidis, 2007; Koslow, 2005; Shepherd et al., 1998). Thus an aim has been to create “knowledgebases” that are formal enough to be usable for computation, but flexible enough to encompass a wide range of query situations. While there have been many efforts to create such knowledgebases with various scopes, the major bottleneck is simply assembling the existing information. The difficulty is that enormous amounts of information are available primarily through the biomedical literature, which is available only as “free text”, that is, natural human written language without formal structures that a computer can easily use. General-purpose systems such as Google may provide useful performance in document retrieval in response to simple queries, but as queries span more modalities and include data-driven aspects, the challenges mount and computational approaches that are “domain-aware” become important. In this paper we are concerned with a subset of this problem of how to turn documents like journal articles into “computable” knowledge. In this introduction we describe some of the broader goals and challenges that motivate our work.
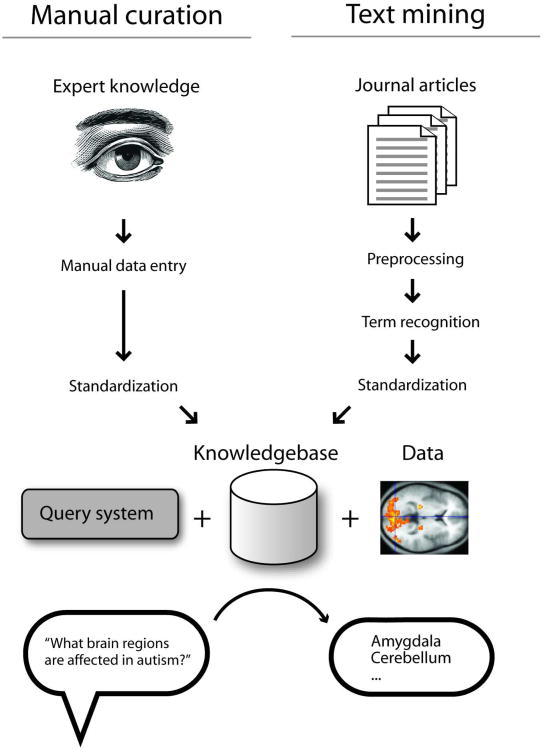
Figure 1 is a schematic expanding on the goals outlined above. The vision for this kind of system has been in circulation for some time, with a relatively recent exhortation to the field provided by David Van Essen in a 2007 editorial (Van Essen, 2007). Van Essen expressed the desire for a system that has the ability to automatically answer queries such as “which brain regions are affected by autism?” or, “which genes are implicated in autism and in which brain regions are they expressed?”. Answering such questions requires that the system contain the necessary expert knowledge in a way that is accessible to a query system, and ideally able to be integrated with large data sets from sources such as brain imaging, genetics, or genomics. As depicted in Figure 1, there are many pieces that must be assembled to accomplish this goal, but the first step is getting information into the system for processing. As shown near the top of Figure 1, there are two general ways that information in the biomedical literature can make its way into structured databases. First, at left in the figure, the information can be entered by a human expert. However, this is very expensive and time-consuming and is not free of other problems. It is therefore attractive to “mine” such information directly from the literature, with minimal or no human intervention. This text-mining approach is outlined in the right-hand branch of Figure 1.
Figure 1.
High-level overview of a hypothetical neuroscience query system, emphasizing the input of information. At top left, data entry by experts is depicted, while at top right, the approach taken in this paper, which is to use automated methods. Both methods require the “standardization” step, which is the focus of this paper. Once information is standardized and stored, appropriate methods can be used to combine it with data analysis to permit complex queries and data mining. See main text for more discussion. Image credits: Washington Irving/Wikipedia and Briar Press.
In Figure 1, a step common to both methods of data entry is “standardization”. By standardization (referred to as “normalization” or “resolution” in the text mining literature), we mean the mapping of a piece of text (a “mention”) to the concepts referred to by the text, in a formal way that can be used by computers. This addresses the difference between the concept of, for example, the substantia nigra pars compacta and the text “substantia nigra pars compacta”. In a computer system, we want all mentions of the concept “substantia nigra pars compacta” to be accessible in a consistent way. For example, the text “SNPC”, in the appropriate context, might refer to the same concept. If the computer system stores the information for occurrences of the text “SNPC” separately from that for the text “substantia nigra pars compacta”, queries accessing the latter will not successfully retrieve information linked to the former. Researchers in neuroinformatics have therefore designed ways to formalize the representation of neuroscience concepts. While this is a difficult task due to the scope and complexity of the field, fundamentally the solution is very simple: in the computer system, the concept of the substantia nigra pars compacta is represented by an agreed-upon code, which might be an identifier such as “birnlex_990” (Bug et al., 2008). This separates the meaning of the concept from its representation. This means that any piece of data need only be linked to the code “birnlex_990”, without worrying about the effects of spelling variation or abbreviations. When these codes are adopted widely by database developers, sophisticated queries become possible. This conversion of relatively informally-described data into formal, standardized codes is fundamental to the types of query systems envisioned by Van Essen. These types of approaches should also be seen as complementary to free-text based retrieval methods exemplified by Google.
As mentioned, the “codes” (or “identifiers”) that are used to formally represent information are themselves created by humans (with the assistance of computers). These identifiers are assembled into collections that are variously called ontologies, vocabularies, terminologies, or lexicons. Keeping with the example of neuroanatomical subdivisions of the brain (which is the focus of this paper), in these terminologies each brain region is listed along with its code. Importantly, these formalized terminologies provide much more power than simply offering a common encoding scheme. In particular, concepts can have relationships with one another. An important and commonly used type of relationship is “part of”. In this manner the terminologies contain the information that the ventral tegmental area (VTA) is “part of” the midbrain. Using these relationships, software can infer that a query for “midbrain” should include information that refers to the VTA, because the terminology encodes the fact that the VTA is part of the midbrain. In contrast, a purely text-based search for “midbrain” would not be guaranteed to retrieve information on the VTA. Of course these same general concepts apply to domains other than brain regions including drugs and diseases. As the common formal encodings are increasingly adopted by the field, the power of the knowledgebases becomes clearer. For example, the fact that the VTA contains dopaminergic neurons is formally encoded and could be discovered automatically even with a query for “monoaminergic neuron”, because the relationship between dopaminergic and monoaminergic neurons could be captured by the terminologies (in this case, a “is a” relationship instead of “part of”) (Gardner et al., 2008). Software can be used to automatically learn that tyrosine hydroxylase is one of the genes most specifically expressed in the VTA, using the formal brain region encodings in the Allen Brain Atlas (Lein et al., 2007). Additional genome-scale expression experiments that studied the VTA can be identified by links to the Gemma system (Lee et al., 2004). Similar integration approaches will reveal patterns of anatomical connectivity (Bota et al., 2005) or functional imaging results (Nielsen, 2003). A third use of formal encoding is to directly extract information from journal articles. For example, the co-occurrence of mentions of a brain region in articles might be used to infer a functional or structural connection between them. Similarly, associations between brain region mentions could be linked to other concepts found in text such as “addiction” and “Parkinson's” (Hayasaka et al., 2011; Kalar et al., 2011).
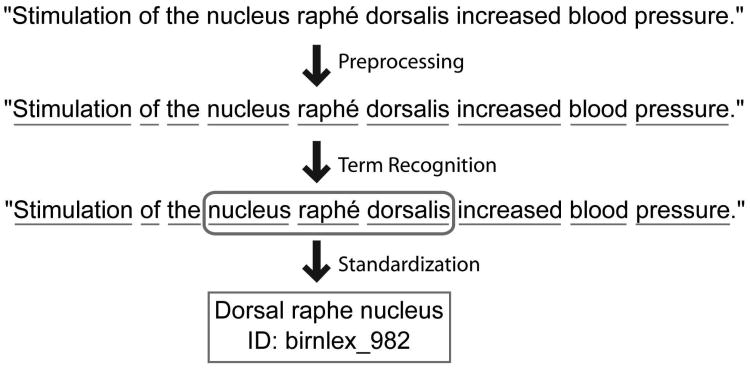
In this paper we are concerned with the right branch of Figure 1, which uses automated methods to extract information from journal articles. Our specific focus is the development of methods for automatically linking journal abstracts to formal brain region identifiers. In previous work, we presented high-performance methods for the first step needed to perform this task, which is identifying which parts of a document mention a brain region (French et al., 2009). In this paper we address the step of automatically standardizing text that is recognized as mentioning a brain region, by resolving them to formal identifiers in neuroanatomical databases. Doing this successfully will allow improved implementation of the scenarios we outline above as well as many others. The workflow which this paper contributes to is illustrated with an example in Figure 2.
Figure 2.
Example of the problem addressed in this paper. A sentence from an abstract is first pre-processed, for example to identify words or to expand abbreviations. Next, text that is predicted to refer to a brain region is identified (French et al., 2009). Finally, the text is converted into a standard identifier from a controlled terminology. The last step is the focus of the methods developed in this paper.
To our knowledge, very little work has explored automatically converting brain region mentions to database identifiers. The most relevant studies are by Srinivas at al., who extracted terms from atlases of thalamic anatomy and manually filtered them for neuroanatomical concepts (Srinivas et al., 2003; Srinivas et al., 2005). They then attempted to map the acronyms and terms across brain atlases for cat, primate, human and monkey, with the goal of assisting comparative analysis. Most other efforts in this area are focused on information retrieval tasks, in which a free-text query is given and used to search a database (Bowden and Dubach, 2002; Nielsen, 2003). The most advanced system is the Neuroscience information framework (NIF) which matches user queries to existing brain region terminologies to expand the input query with synonyms (Gardner et al., 2008). We note that none of these approaches are designed to work on complex documents such as journal articles. Other literature retrieval search engines attempt to match mentions to lists of regions but they do not use formal identifiers (Crasto et al., 2003; Muller et al., 2008). Recently, Yarkoni et al. developed methods to accomplish a related task, which is to automatically extract numerical activation coordinates from journal articles describing functional brain imaging studies (Yarkoni et al., 2011), and converting those coordinates to brain regions. There are also several general-purpose tools which extract biomedical concepts from the literature (Aronson and Lang, 2010; Jonquet et al., 2009). These tools discover terms from large biomedical terminologies, which include some brain region concepts.
There are several challenges to successfully converting text mentions of a brain region into a formal identifier. One is ambiguity; for example the hypothalamus, medulla and thalamus each have an “arcuate nucleus”. Another is that the terminology databases are incomplete, so authors may refer to brain regions which do not have a formal identifier. A related problem is that terminologies do not exist for some organisms, and even for those that do, the terminologies are obviously different for different nervous systems. We present several novel solutions to these problems and evaluate their effectiveness, yielding a method that provides a high level of accuracy in mapping text to brain region concepts. We apply our approach to the analysis of a large set of abstracts from the Journal of Comparative Neurology, providing information on the distributions of brain region mentions. Our results are a starting point for linking diverse neuroinformatics data sources to literature-based information on brain regions.
Methods
Benchmark data set
We used our previously described manually-curated data set (“corpus”) of brain region mentions in journal abstracts (French et al., 2009). Briefly, the corpus of 1377 abstracts consists of 1258 abstracts randomly chosen from the Journal of Comparative Neurology and 119 abstracts selected from other neuroscience journals. Before curation, we applied an abbreviation expansion algorithm to all abstracts (Schwartz and Hearst, 2003). All extracted mentions of an abbreviation short form were thus expanded to their long forms in a given abstract. The brain region mentions were then manually curated. The corpus provides no resolution of the brain region mentions into database identifiers.
Brain region identifiers
We assembled a large dictionary of brain regions with formal identifiers from multiple sources. We used NeuroNames (Bowden et al., 2007), NIFSTD/BIRNLex (Bug et al., 2008), Brede Database (Nielsen, 2003), the Brain Architecture Management System (BAMS) (Bota and Swanson, 2008) and the Allen Mouse Brain Reference Atlas (ABA) (Dong, 2007). All terms were converted to lowercase and are linked to the provided identifiers. Of the five sources only BAMS and ABA are true neuroanatomical atlases that provide direct links between brain region names and 3D volumes in a digital or print format. The Brede database provides similar spatial data with 3D coordinates for named regions of interest. We did not extract abbreviation terms because we expand abbreviations as described above.
The total number of concepts in these five source terminologies is 7,145, but it is clear that there is extensive redundancy among them (even after accounting for species-specificity of concepts). Unfortunately, because there are limited direct mappings of concepts across the terminologies, it is difficult to estimate how many different brain regions are represented in total. We arrived at a rough estimate of 1,000 different mammalian brain region concepts based on the sizes of four of the terminologies, and the fact that the much larger NeuroNames (at over 3,000 concepts) has an expanded concept of “brain region” that includes “ancillary” terms that tend not to be recognized as distinct concepts by the other terminologies. Technical details of the processing of each input terminology are given in the next paragraphs.
NeuroNames terms were extracted from all worksheets in the NeuroNames Ontology of Mammalian Neuroanatomy (NN2010) and Nomenclatures of Canonical Mouse and Rat Brain Atlases (NN2007) excel files (Bowden and Dubach, 2002). Classical, ancillary, Latin and synonym terms were added to the terminology. Further, terms from all four mouse and rat atlases were added to the terminology. Overall 9,188 unique terms were extracted to represent 3,238 Neuroname concepts.
The 2,391 NIFSTD terms were extracted from the 1,272 classes in the Anatomy subontology. Synonyms and the main labels were extracted for ontology classes that were regional parts of the eye, ear, brain, spine and ganglion of peripheral nervous system.
Terms from the Brede Database were extracted from the supplied worois.xml file. Terms were obtained from all name and variation XML tags. Hemispheric “left” and “right” prefixes were removed to be consistent with the rest of the terminology. In total 1,006 terms were extracted from Brede to represent 763 concepts.
For BAMS, we extracted terms from the primary terminology - Swanson-1998 (Swanson, 1999). This terminology allows linking to the rich connectivity information curated into BAMS. The version of the BAMS database we use contains 962 rat brain region terms and is accessible via bulk download (http://brancusi.usc.edu/bkms/xml/swanson-98.xml). Instead of parsing the original XML we used a converted semantic web version created by John Barkley (http://sw.neurocommons.org/2007/kb-sources/bams-from-swanson-98-4-23-07.owl).
Allen Brain Atlas terms were obtained from the OWL formatted version downloaded from the Allen Brain Atlas API documentation. Like the above sources, abbreviations were excluded from the extraction. In total 910 terms and concepts were extracted (no synonym information).
Resolvers
We employed five methods of resolving (or “converting”) textual mentions to region names in the ontologies and atlases. The most basic is the “Exact String Matching Resolver”. This resolver simply converts the mention to lower case and attempts to match all characters to a region name in the terminology. The next step is implemented in the “Bag of Words Resolver” which splits the mention strings into words (tokenization) and then looks for exact string matches for each of these words. This is a common information retrieval technique that matches the same text but ignores word order.
To remove lexical variation we again tokenized the phrases into words. We converted the words into a base form by using a stemmer. A stemmer normalizes words to their base form by removing common endings. For example “ventral striatopallidal parts of the basal ganglia” is stemmed to “ventr striatopallis part of th bas gangl”. We employed the Lovin's stemmer as implemented by Eibe Frank (Lovins, 1968). We created two additional resolvers analogous to the ones described above. After tokenizing and stemming, the first resolver will match the stemmed tokens to the stemmed terms in the terminology (Stem Resolver). The second will match them in any order (Bag of Stems Resolver). The Bag of Stems resolver is similar to the orderless gap-edit global string-matching algorithm used by Srinivas et al. (2005). In their implementation they allowed half of the stems to match for terms greater than two words in length (uninformative common words excluded). Our Bag of Stems method is slightly different with use of a different stemmer and a strict requirement of all words to match (our mentions might be modified to remove specific terms).
To compare these to an externally designed method we employed the Lexical OWL Ontology Matcher (LOOM). LOOM is a simple method for mapping across biomedical ontologies. While LOOM is not designed for matching free text mentions we found its approximate string matching technique to be of value. LOOM uses a string comparison function that requires an exact match for words longer than four characters and allows one character mismatch for longer strings (after removing spaces and parentheses). The LOOM authors show it provides comparable performance to more complicated tools for ontology mapping (Ghazvinian et al., 2009).
Modifiers
To improve the resolution process, we employed several techniques that work by modifying the text from the abstract. In total nine modifiers are employed (Table 1). Three of the modifiers remove important words from the text, and thus we consider them “lossy”. Each modifier is applied in the order presented in Table 1, and only to the mentions that have not been previously matched. The result of the modifier does not replace the original mention, but instead expands it, creating a set of alternative representations of the original mention. Each modifier is executed once except the Direction Remover which is run a second time at the end to extract more general regions from very specific mentions.
Table 1.
Modifier descriptions and examples.
| Name | Description | Lossy Example | |||
|---|---|---|---|---|---|
| Direction splitter | Splits conjunctions that use neuroanatomical directions | No | dorsal and posterior hypothalamic areas | ≫ | [dorsal hypothalamic areas, posterior hypothalamic areas] |
| Hemisphere stripper | Removes prefixes that specify hemisphere | No | contralateral inferior olivary | ≫ | inferior olivary |
| Bracketed text remover | Removes text that is enclosed by brackets | No | secondary somatosensory (sii) cortex | ≫ | secondary somatosensory cortex |
| “n.”expander | Expands “n.” to nucleus | No | n. ambiguus | ≫ | nucleus ambiguus |
| “of the” remover | Removes subdivision descriptors | No | medial portion of the entorhinal cortex | ≫ | medial entorhinal cortex |
| Region suffix remover | Removes “region” suffixes | No | posterior cingulate region | ≫ | posterior cingulate |
| Cyto prefix remover | Removes prefixes that mention cytoarchitectural descriptions | Yes | parvocellular red nucleus | ≫ | red nucleus |
| Direction remover | Removes neuroanatomical direction specifiers | Yes | caudal cuneate nucleus | ≫ | cuneate nucleus |
| “nucleus of the” remover | Removes nucleus specifiers | Yes | nucleus of the pontobulbar body | ≫ | pontobulbar body |
Example input and output mention strings are separated by “≫”. The Direction splitter modifier expands the single input string into two mentions. Modifiers that discard important words from the mention are classified as ‘Lossy’.
Species identification
We employed LINNAEUS, a species name identification system for biomedical literature for extracting species mentions from the corpus (Gerner et al., 2010). LINNAEUS provided an open and accurate tool for quantifying species mentions with accuracies above 90%. We used the default configuration properties to tag the abstracts for NCBI species identifiers. Of the 209 species found we manually deemed 44 to be not relevant (list available at supplement website). These primarily included mentions of reagents for tract tracing (“horseradish”, “phaseous vulgaris”, “pseudorabies virus”). We noted some false positives, including brain regions that were tagged as species (“n. superficialis”, “n. ambiguus”).
Data model
To capture the relations between abstracts, mentions, terms and ontology concepts we employed a resource description framework (RDF) model (W3C, 2004). For NIFSTD, BAMS, ABA and Brede concepts we link to the original identifiers for future integration. The full RDF dataset is available on our supplement website at http://www.chibi.ubc.ca/WhiteText/.
Evaluation
By automatically testing for exact string matches we were able to review the complete set of mention-to-region pairings. The exact string matches were automatically accepted while the remaining pairings were manually evaluated. Each mention-to-concept pairing was marked as “accept”, “reject” or “specific-to-general” (a “part-of” relationship). A specific-to-general marking applies to mentions where the region was mapped to an enclosing region (e.g. “nucleus deiters dorsalis” mapped to “nucleus of deiters”). This applies to many cases, as several of our modifiers discard information. Resolutions of ambiguous terms were accepted only if they matched a majority of the contexts. For example, all mappings of “arcuate nucleus” were rejected because the abstracts in which they occur are not consistently referencing the arcuate nucleus of the thalamus, medulla or hypothalamus. To reduce redundant evaluations, pairings were grouped when the main text label for the matched region is the same across ontologies. The abstracts in which the mention occurred were used to judge the context and correctness of the resolution. Resolutions were accepted across species unless it was a specific parcellation scheme for a species, for example - “area 10a of Vogts”.
Resolution coverage represents the proportion of mentions that have been mapped to at least one brain region concept. This proportion of mapped mentions is dominated by frequently occurring terms like “cortex”. To control for mention popularity we provide two additional measures of coverage. The first ignores the number of times a mention occurs and treats each unique mention equally (rare mentions are given equal weight as common terms). The second ignores repeat mentions of a mention within an abstract and weights each mention by the number of abstracts it appears in.
Resolution accuracy was measured by dividing the number of accepted concept to mention links by all total mention-to-concept resolutions made. We also take into account frequency of the mention by multiplying the concept to mention links by number of abstracts the mention appears in. We considered specific-to-general mappings to be an accepted resolution while also measuring their frequency individually.
Although the species name recognizer we chose has been previously evaluated we compared it to a subset of our abstracts that we previously annotated with species. Because the annotated tags were entered in free text we employed LINNAEUS to convert them to NCBI taxonomy identifiers. These converted identifiers were then compared to those extracted from the abstracts automatically.
Results
Figure 2 shows an overview of the system we developed, starting from journal abstract to mapped concept. In developing the approach, we examined the properties of the input terminologies, and carefully evaluated the quality of the mappings we obtained, as described in the next sections. In the final section we describe the application of the process to a large set of JCN abstracts and present findings on the patterns of brain region concept usage.
Summary of the terminologies
We first established the basic properties of the target terminologies (or “lexicons”) we used for mapping. It is important that these terminologies encompass the range of concepts used in the literature. In total we extracted 11,909 terms from five terminologies. These terms represent a total of an estimated 1,000 different mammalian brain regions (see Methods). On average a concept in the aggregated terminologies had 1.6 terms or labels (for example representing synonyms; note that we must distinguish between “concepts” and their textual representation as “terms”). While we estimate that concept overlaps among the terminologies are high, term overlap across terminologies was remarkably low, with terms being linked to just 1.3 of the five terminologies on average, with 79.8% of the terms appearing in only one terminology. Across the ontologies the highest amount of overlap was between ABA and NeuroNames with 62.7% of the ABA terms appearing in the much larger NeuroNames set. In addition 53.5% of the NIFSTD terms appear in NeuroNames. This is expected because NIFSTD was originally based on NeuroNames (Bug et al., 2008). Although the NeuroNames curators have imported some of the ABA and BAMS terminology, it is not complete. While some “singleton” terms are minor variants of terms found in other terminologies (e.g., raphé vs. raphe), the terminologies contain many apparently obscure or rarely-used terms such as “area 22 of mauss 1908”.
Standardization
We ran our resolvers on an annotated set of 17,585 brain region mentions (see Methods). These initial results showed that 47.1% of the 17,585 could be resolved to a formal identifier in the terminologies (Table 2). However, many of brain regions mentions are repeated: there are 5,941 unique brain region mentions in the set of 17,585. When viewed this way, 18.8% of unique mentions are resolved. As shown in Table 2, mentions that occur more often are better resolved. These measures of “coverage” do not address whether the mappings are correct. To assess accuracy, we first considered mappings that exactly match the text in the abstract to the term in the terminologies to be correct; this accounts for 41.1% of mapped mentions (7,228). The more advanced methods that allow partial matches are shown in Table 2 (detailed results at supplement website). We found the Simple Mapping Matcher performed the worst with 3.6% of unique mention mappings rejected as incorrect. Overall the combined set of resolvers result in 4.3% of unique mentions being rejected and 52.9% of mentions failing to map.
Table 2.
Mention coverage and rejection rates across resolvers.
| Coverage | Rejection rates | |||||
|---|---|---|---|---|---|---|
| Resolver | Mentions | Abstract-Mentions | Unique Mentions | Mentions | Abstract-mentions | Unique Mentions |
| Exact String Match | 41.1% | 36.0% | 14.3% | 0.0% | 0.0% | 0.0% |
| Bag of Words | 42.1% | 37.1% | 15.8% | 0.2% | 0.2% | 1.0% |
| Stem | 45.1% | 39.4% | 16.2% | 0.5% | 0.5% | 1.0% |
| Bag of Stems | 46.4% | 40.8% | 18.0% | 0.7% | 0.7% | 1.9% |
| LOOM Matcher | 41.1% | 35.8% | 14.3% | 2.5% | 2.5% | 3.6% |
| All | 47.1% | 41.6% | 18.8% | 3.1% | 3.2% | 4.3% |
Coverage is provided at three different levels to quantify repeated mentions. For the “Unique Mentions” and “Reject Unique” columns the number of times a mention occurs is ignored (rare terms are given equal weight as common terms). The “Abstract-Mentions” and “Reject Abs-Mention pairs” statistics ignores the number of times a mention occurs in an abstract. The “Mentions” and “Reject Frequency” columns weight each unique mention by the number of times it occurs in the corpus.
Tuning and final evaluation
While the accuracy of the mappings obtained in these experiments was high, the coverage was lower than desired, with too many mentions left unmapped to formal identifiers. After reviewing the results, we decided to modify our process and the input terminologies. The first change was the addition of previously undocumented synonyms to the terminologies. We were able to make synonym-to-region links for 42 of the 122 top unmatched mentions that occurred more than 9 times in the corpus. Examples include “cortical”, “thalamic” and “si” (the full table is provided on the supplement website). Although we sought to remove acronyms and abbreviations, several occur in the list. These are primarily terms that the automatic abbreviation expander failed to resolve or a long form was not provided by the author. The rate of these errors is roughly 5%, and is similar to the tested accuracy of the abbreviation expander (Schwartz and Hearst, 2003). The addition of these synonyms produces a 7.7 percentage point increase in mention coverage (Table 3). We list these values separately because they reflect post-hoc additions to the process, but they present true increases in coverage expected for a production system.
Table 3. Incremental improvements from several additional methods.
| Added modifier | Added Mentions | Added Mappings | Percent accepted | Specific-to-general Mappings | Matched Mentions |
|---|---|---|---|---|---|
| Baseline | 8280 | 2963 | 95.7% | 0.8% | 47.1% |
| Synonym additions | 1346 | 91 | 97.8% | 0.0% | 54.7% |
| Direction splitting editor | 35 | 109 | 86.2% | 7.3% | 54.9% |
| Hemisphere stripper | 131 | 265 | 100.0% | 0.0% | 55.7% |
| Bracketed text remover | 29 | 73 | 90.4% | 2.7% | 55.8% |
| Converts n. to nucleus | 5 | 14 | 100.0% | 0.0% | 55.9% |
| Remover of “of the” type phrases | 27 | 67 | 85.1% | 10.4% | 56.0% |
| Region suffix remover | 36 | 56 | 100.0% | 0.0% | 56.2% |
| Cytoarchitecture prefix remover | 37 | 76 | 97.4% | 96.1% | 56.4% |
| Direction prefix and suffix remover | 1092 | 2204 | 95.8% | 94.4% | 62.7% |
| Remover of “nucleus of the” phrase | 21 | 72 | 100.0% | 100.0% | 62.8% |
| Direction prefix and suffix remover | 125 | 205 | 84.9% | 84.9% | 63.5% |
We implemented further improvements using what we call “modifiers” (Table 1). Modifiers make slight changes (“edits”) to the original text and, as a last resort, remove qualifying terms such as “dorsal”. The last resort “lossy” modifiers would result in mapping “dorsal hippocampus” to “hippocampus”, what we call a “specific-to-general” mapping. When combined, the modifiers raise the coverage of abstract-mention pairs to 58.4% and 35.9% of unique mentions with 39.4% specific-to-general mappings. As Table 3 shows, each modifier provided a modest contribution to the coverage while maintaining accuracy. Finally, the lossy modifiers (designed to discard qualifiers) created primarily general-to-specific mappings for the mentions that failed to match after applying the preceding modifiers.
To gain insight into remaining problem areas, we manually examined a random subset of 100 unmatched mentions. Fourteen of the 100 can be explained by annotation errors in our corpus, including text spans that missed the first character of a term and annotations of tracts. This result was expected given previously measured rates of annotator agreement (French et al., 2009). Another 25 refer to regions in species not represented by the terminologies. The remaining unmatched mentions can be categorized as unique variants, very specific mentions and ambiguous mentions (a complete list is available on the supplement website). Thus most failures to map are due to gaps in the terminologies.
Species-specific evaluation
The evaluation above suggests that the quality of resolution would depend on the organism used in the study, as the terminologies are species-specific and many taxa lack terminologies. To filter for species of study we ran LINNAEUS to identify species mentions in the abstracts (Gerner et al., 2010). We compared the automatically tagged species information to a subset of 396 abstracts with manually annotated species information. LINNAEUS was able to recall 97.4% of the annotated species mentions that could be mapped to a specific species. Precision could not be fully evaluated because many mentions of species are too general and refer to a genus or other taxonomic level. LINNAEUS does not extract these terms and as a result terms like “Macaque monkey”, “pigeon” and “squirrel monkey” could not be extracted (but were still annotated manually for the subset). Overall, LINNAEUS identified species terms in 88% of abstracts. Co-occurrence of species within abstracts is relatively low; the most common pair of species is rat and human which occur together in 30 abstracts.
As predicted, the coverage of mentions and specific-to-general matches varied greatly across species. Table 4 presents the results for a selected set of top occurring species, the full table is available on the supplement website. Species that lacked terminologies resolved less well and specific-to-general mappings occurred much more often. The top occurring species benefited from terminologies of their species. To determine the accuracy of the commonly studied species targeted by our terminologies we combined the mentions that co-occur with rat, mouse, human, rhesus monkey and macaca fasicularis mentions. Coverage of unique mentions in this group increases by 7 percentage points, and specific-to-general mappings are reduced to 33.7% from 39.4% on all mentions. Accepted mappings slightly increased from 95.1% to 96.6%.
Table 4.
Resolution of species linked mentions.
| Species | Species terms | Mentions | Unique Coverage | Mention Coverage | Rejected Mappings | Specific-to-general Mappings |
|---|---|---|---|---|---|---|
| Cat | cats, kitten, cat, Cat, kittens | 1001 | 42.5% | 60.6% | 3.0% | 24.3% |
| Rabbit | rabbit, rabbits | 200 | 60.0% | 73.3% | 4.0% | 16.5% |
| Pigeon | Columba livia | 157 | 40.1% | 45.0% | 1.1% | 19.6% |
| Clawed frog | clawed frog, Xenopus laevis, African clawed frog, X. Laevis | 107 | 57.0% | 66.8% | 8.1% | 37.1% |
| Rat | rat, rats, Norway rat, Sprague-Dawley rats, Wistar rats, Sprague-Dawley rat | 2434 | 44.6% | 67.7% | 3.2% | 31.9% |
| Mouse | mice, mouse, murine, transgenic mice | 396 | 55.8% | 75.7% | 2.6% | 13.6% |
| Human | patient, patients, human, infant, children, humans, infants, people, participants, man | 409 | 57.7% | 73.5% | 4.2% | 11.6% |
| Rhesus Monkey | rhesus monkey, rhesus monkeys, Macaca mulatta | 406 | 49.5% | 63.6% | 2.5% | 29.6% |
| Macaca f. | macaca fascicularis, cynomolgus monkey, cynomolgus monkeys | 143 | 64.3% | 67.9% | 5.9% | 17.5% |
| Macaca f., Rhesus, Human, Mouse and Rat | 3061 | 42.9% | 68.6% | 3.4% | 33.7% | |
| All | 5941 | 35.9% | 63.5% | 4.9% | 39.4% |
The “Species terms” column lists all recognized terms for a given species. Coverage is provided at two levels by counting mention frequency (“Mention Coverage”) and ignoring it (“Unique Coverage”).
Analysis of all Journal of Comparative Neurology abstracts
We ran our final method on 12,557 JCN abstracts that are not already in our corpus (covering 1975 to January 2011). This required first running the abbreviation expander, then the brain region mention detector as described previously (French et al., 2009), followed by the tuned normalization process described above. In total we found 142,178 brain region mentions. Of these 95,895 were resolved to a concept in a terminology, representing 7,923 unique region mentions and 57,185 unique abstract-region pairs (on average 4.6 per abstract; 86% of abstracts having at least one). The resolution results resemble those from the manually annotated abstracts with 67.5% of mentions resolved and 27.4% of unique mentions matched to a terminology entry. For the subset of commonly studied species that cover the terminologies coverage reaches 71.6% of mentions and 32.3% of unique terms. The slight increase in mention coverage and decrease in unique coverage is expected from a larger corpus size generating a larger set of rare terms. Table 5 presents the top 25 most frequently occurring NIFSTD concepts. The types of unmatched mentions are similar to those found previously with many broad terms that are not explicitly in the terminologies and several insect brain regions such as “mushroom bodies”.
Table 5.
Top 25 most frequent brain region concepts in the Journal of Comparative Neurology.
| Region | Frequency |
|---|---|
| Retina | 7341 |
| Cerebral cortex | 5578 |
| Spinal cord | 3915 |
| Thalamus | 2290 |
| Hippocampus | 2098 |
| Cerebellum | 1953 |
| Hypothalamus | 1800 |
| Olfactory bulb | 1551 |
| Brainstem | 1512 |
| Superior colliculus | 1457 |
| Neostriatum | 1343 |
| Amygdala | 1312 |
| Midbrain tectum | 1109 |
| Midbrain | 1093 |
| Forebrain | 962 |
| Solitary nucleus | 819 |
| Locus ceruleus | 769 |
| Substantia nigra | 764 |
| Cochlea | 762 |
| Entorhinal cortex | 712 |
| Lateral geniculate body | 705 |
| Dentate gyrus | 684 |
| Central gray substance of midbrain | 662 |
| Telencephalon | 660 |
| Cochlear nuclear complex | 651 |
Regions are limited to the NIFSTD terminology with frequency determined from the full JCN corpus. Both the manually curated and automatically predicted brain region spans were used to generate the table.
We examined the extent to which terms in the terminologies are used. We found that 44.1% of the 7,145 available concepts are used at least once. Viewed another way, over 55% of the concepts (and 77% of terms) in the terminologies do not appear to be used in any JCN abstract. These results suggest that many of the concepts (and terms) in the terminologies are rarely used by working scientists.
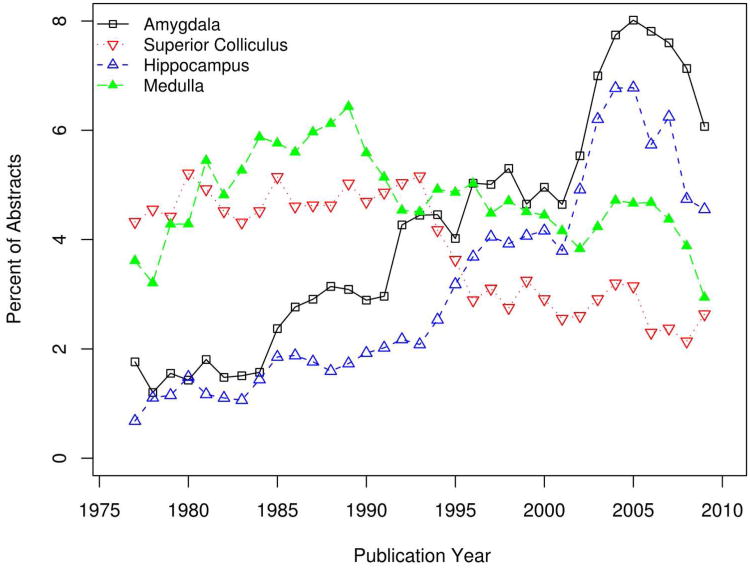
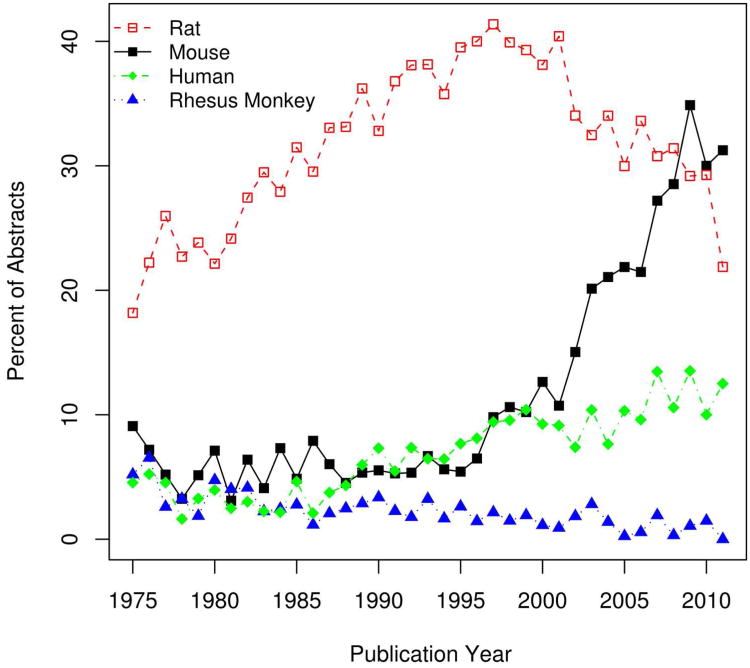
Because our analysis includes information on species and publication date as well as brain region use, the final data set allows interesting temporal analyses of the JCN. We first asked whether there is a tendency for more recent articles to use more narrowly defined brain regions. By comparing the publication year with the proportion of specific-to-general mappings in the training set we observe a slight but non-significant positive trend (Spearman correlation 0.18; p-value = 0.31). Our analysis is also able to reveal trends in the “popularity” of brain regions over the years. For example, we found that there was an abrupt dip in the mentions of “superior colliculus” in the early 1990s, while the hippocampus and amygdala enjoyed rising mentions until recently (Figure 3). A similar analysis of species of study shows that mentions of mouse and humans are increasing, while rat and Rhesus monkey mentions are fading (Figure 4).
Figure 3.
Trends in the proportion of yearly abstracts mentioning amygdala (black square), superior colliculus (red triangle), hippocampus (blue triangle) and medulla (green triangle). Proportion values are smoothed by averaging the previous, current and following years.
Figure 4.
Changes in the proportion of yearly abstracts mentioning rat (red square), mouse (black circle), people (green diamonds) and Rhesus monkey (blue triangle) over time. Only 32 abstracts are considered for the 2011 year. Rat, mouse and human are significantly increasing over time (p < 0.05). Abstracts mentioning Rhesus monkey are significantly declining (p < 0.001).
Discussion
Our contribution in this paper is the development and thorough evaluation of a process for mapping specific brain region concepts to free text in journal abstracts. We achieved a high degree of coverage (64.5%) and precision (95.1%), and yield a data set of value for additional analyses. Our research has also identified many challenges and current limitations that need to be addressed to most effectively use information on brain regions in the neuroscience literature.
The primary problem we encountered was with the terminologies, which are not well-standardized and also, apparently, incomplete. The terminologies we used have surprisingly little overlap, despite some of them having common target organisms. This reflects the extensive variation in how neuroanatomical concepts are expressed in natural language, but the lack of harmonization across terminologies is striking.
We observed that authors often write about regions that are beyond the granularity of the terminologies (for example, by adding a modifier such as “mediolateral” to a recognized term). While we presented a “lossy” mapping method that handles this problem, it is likely that some of these fine-grained terms should be added to the terminologies. To this end, we have contributed 136 new brain region concepts to NeuroLex (Larson et al., 2010). We selected the regions by filtering our results for specific-to-general mappings to an existing NITSFD brain region concept. We then selected terms that are co-mentioned with rhesus monkey, macaca fasicularis, rat, mouse or human in at least two separate abstracts, assuming that repeated use in the literature is evidence of utility. This automatically generated list of 152 region terms was reduced to 136 after manual adjustments for synonyms and conjunctions. Although this is a small first step, formalization of these mention-to-concept pairings would reduce the specific-to-general mapping rate in our study by 2.5 percentage points. Further, these 136 terms occur over 2,400 times in the complete set of JCN abstracts. Because NeuroLex is presented in a wiki format, the community can review and edit these additions (http://neurolex.org). Another potential avenue for improving terminologies is the International Neuroinformatics Coordinating Facility (INCF) Program on Ontologies of Neural Structures (PONS) which seeks to establish formalized terminologies for neuroanatomy (http://www.incf.org/core/programs/pons).
By inspecting unmatched mentions, we were able to tune the system and improve the coverage from 47.1% to 63.5%. Other attempts at tuning resulted in very modest improvements in comparison. Our analysis of one hundred unmatched mentions suggests more advanced methods employing contextual information could be used to resolve ambiguous and co-referenced mentions. Context information has already been applied to cross-species mapping and may be adaptable to brain region mapping outside of atlases (Srinivas et al., 2005).
In addition to pointing out gaps in the existing terminologies, our results also point to a mismatch in the other direction, in that the terminologies contain numerous terms that do not appear in any JCN abstracts. For example, the rodent term “Perireunensis nucleus” never appears in any PubMed abstract; a wider web search turns up just a single mention in the accessible literature (Jacobsson et al., 2010). An overall picture emerges of terminologies that are incomplete while simultaneously full of terms which may not be actually used in practice. Our results may thus aid the developers of terminologies and highlights the need for more work in this area.
Improvements to the terminologies would increase the coverage of our methods, that is, the number of textual mentions of brain regions which can be mapped to an identifier.
In contrast to coverage, there is little improvement that can be realized in the precision of our method. If our method is able to map text in an abstract to a formal identifier, the mapping is almost always valid. We believe this is due in part to our conservative approach which relies on strict matches and avoids the ambiguity of abbreviations. The best single method appears to be the Bag of Stems Resolver, which nearly reaches the coverage of all the methods combined, while maintaining a very low 1.9% unique term rejection rate. This agrees with previous work that tested a similar resolver for cross species mapping of thalamic atlases (Srinivas et al., 2005). The LOOM Simple Mapping Matcher, designed for a different task (ontology mapping) performs worse than any other method. Our motivation for testing it is that it provides mappings our other resolvers cannot. Unfortunately, we find that by allowing one letter mismatches, it leads to frequent serious errors such as conflating “central” and “ventral”. Interestingly, past work in the ontology mapping domain has placed LOOM on par with other more advanced methods (Ghazvinian et al., 2009). Overall, our results suggest that more complicated methods will not yield substantial improvements.
Our analysis broken down by species confirmed our expectation that the organism of study is a key piece of information for high-quality mapping. As expected, brain region mentions from amphibians, insects and fish had increased rejection and more specific-to-general mappings. Mammalian species such as rabbit and cat performed at levels close to the average. Rat, the most common species of study in the corpus, had an above average coverage but also a high amount of specific-to-general mappings (31.9%). In comparison, mouse abstracts had only 13.6% specific-to-general mappings while achieving the highest coverage (75.7%). This may reflect that the larger rat brain is commonly used for study of detailed rodent neuroanatomy that extends beyond the standard atlases. In addition the human abstracts have results similar to mouse with high coverage of mentions and few specific-to-general mappings compared to Rhesus monkey abstracts. Approximately half of the mentions are linked to species matching the target terminologies. These mentions from commonly studied species are accurately normalized, with low rejected (3.4%) and specific-to-general mappings (33.7%).
We applied the methods to an unseen set of automatically tagged region mentions from the remaining JCN abstracts. The results mirror those from within the manually annotated corpus and suggest the methods could readily be extended to larger scales. Our method appears to scale well, with over 1,000 brain region concepts appearing in the extended corpus but not the original annotated set.
To increase the value of the data set to the neuroscience community, our results have been incorporated into the NeuroLex database, where work is in progress to display, for example, time-trends of brain region mentions in the JCN alongside other information on each region (Anita Bandrowksi, personal communication). We also provide a bulk version of the data suitable for third-party analyses on our website (http://www.chibi.ubc.ca/WhiteText/). As mentioned in the introduction, having brain regions mapped to abstracts is only one step in making full use of the information embedded in the literature. Future work will focus on the linking of brain region mentions to each other and to other concepts such as drugs and diseases. Our eventual goal is to provide computationally rich linkages of brain regions to diverse neuroinformatics resources.
Acknowledgments
We thank the providers of the neuroanatomical atlases for making the ontologies available and accessible online. In particular we thank Maryanne Martone, Stephen Larson and Anita Bandrowski for facilitating our additions to NeuroLex. We thank Amir Ghazvinian for kindly providing the source code of the LOOM simple mapping matcher. We are grateful to Dr. Claudia Krebs, the anonymous reviewers and the editor for their constructive criticism which led to many improvements to the manuscript.
This work was supported in part by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (371348). Further support was provided by a National Institutes of Health grant GM076990 to PP, the Canadian Foundation for Innovation (Leaders Opportunities Fund), the Michael Smith Foundation for Health Research (Career Investigator award to PP), and the Canadian Institutes of Health Research (New Investigator Salary Award to PP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Literature cited
- Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Dong HW, Swanson LW. Brain architecture management system. Neuroinformatics. 2005;3(1):15–48. doi: 10.1385/NI:3:1:015. [DOI] [PubMed] [Google Scholar]
- Bota M, Swanson LW. BAMS Neuroanatomical Ontology: Design and Implementation. Frontiers in neuroinformatics. 2008;2:2. doi: 10.3389/neuro.11.002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden DM, Dubach M, Park J. Methods in molecular biology. Vol. 401. Clifton, NJ: 2007. Creating neuroscience ontologies; pp. 67–87. [DOI] [PubMed] [Google Scholar]
- Bowden DM, Dubach MF. BrainInfo. An Online Interactive Brain Atlas and Nomenclature. In: K R, editor. Neuroscience Databases. Dusseldorf: Kluwer Academic Press; 2002. pp. 259–274. [Google Scholar]
- Bug WJ, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird AR, Larson SD, Rubin D, Shepherd GM, Turner JA, Martone ME. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. 2008;6(3):175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasto CJ, Marenco LN, Migliore M, Mao B, Nadkarni PM, Miller P, Shepherd GM. Text mining neuroscience journal articles to populate neuroscience databases. Neuroinformatics. 2003;1(3):215–237. doi: 10.1385/NI:1:3:215. [DOI] [PubMed] [Google Scholar]
- Dong HW. The Allen Atlas: A Digital Brain Atlas of C57BL/6J Male Mouse. Hoboken, NJ: Wiley; 2007. [Google Scholar]
- French L, Lane S, Xu L, Pavlidis P. Automated recognition of brain region mentions in neuroscience literature. Frontiers in neuroinformatics. 2009;3:29. doi: 10.3389/neuro.11.029.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Pavlidis P. Informatics in neuroscience. Briefings in bioinformatics. 2007;8(6):446–456. doi: 10.1093/bib/bbm047. [DOI] [PubMed] [Google Scholar]
- Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, Goldberg DH, Grafstein B, Grethe JS, Gupta A, Halavi M, Kennedy DN, Marenco L, Martone ME, Miller PL, Muller HM, Robert A, Shepherd GM, Sternberg PW, Van Essen DC, Williams RW. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6(3):149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner M, Nenadic G, Bergman CM. LINNAEUS: a species name identification system for biomedical literature. BMC bioinformatics. 2010;11:85. doi: 10.1186/1471-2105-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazvinian A, Noy NF, Musen MA. Creating mappings for ontologies in biomedicine: simple methods work. AMIA Annual Symposium proceedings / AMIA Symposium. 2009;2009:198–202. [PMC free article] [PubMed] [Google Scholar]
- Hayasaka S, Hugenschmidt CE, Laurienti PJ. A network of genes, genetic disorders, and brain areas. PloS one. 2011;6(6):e20907. doi: 10.1371/journal.pone.0020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson JA, Stephansson O, Fredriksson R. C6ORF192 forms a unique evolutionary branch among solute carriers (SLC16, SLC17, and SLC18) and is abundantly expressed in several brain regions. J Mol Neurosci. 2010;41(2):230–242. doi: 10.1007/s12031-009-9222-7. [DOI] [PubMed] [Google Scholar]
- Jonquet C, Shah NH, Musen MA. The Open Biomedical Annotator. San Francisco, CA: 2009. pp. 56–60. [PMC free article] [PubMed] [Google Scholar]
- Kalar D, Sabb F, Parker D, Bilder R, Poldrack RA. PubBrain: A literature-based visualization tool for neuroscience. Review 2011 [Google Scholar]
- Koslow SH. Discovery and integrative neuroscience. Clin EEG Neurosci. 2005;36(2):55–63. doi: 10.1177/155005940503600204. [DOI] [PubMed] [Google Scholar]
- Larson S, Iman F, Bakker R, Pham L, Martone M. Neuroinformatics. Vol. 2010. Kobe, Japan: 2010. A multi-scale parts list for the brain: community-based ontology curation for neuroinformatics with NeuroLex.org. [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome research. 2004;14(6):1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lovins JB. Development of a Stemming Algorithm. Mechanical translation and computational linguistics. 1968;(11):22–31. [Google Scholar]
- Muller HM, Rangarajan A, Teal TK, Sternberg PW. Textpresso for neuroscience: searching the full text of thousands of neuroscience research papers. Neuroinformatics. 2008;6(3):195–204. doi: 10.1007/s12021-008-9031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen FA. The Brede database: a small database for functional neuroimaging. 9th International Conference on Functional Mapping of the Human Brain; New York, NY. 2003. [Google Scholar]
- Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. Pacific Symposium on Biocomputing. 2003:451–462. [PubMed] [Google Scholar]
- Shepherd GM, Mirsky JS, Healy MD, Singer MS, Skoufos E, Hines MS, Nadkarni PM, Miller PL. The Human Brain Project: neuroinformatics tools for integrating, searching and modeling multidisciplinary neuroscience data. Trends in neurosciences. 1998;21(11):460–468. doi: 10.1016/s0166-2236(98)01300-9. [DOI] [PubMed] [Google Scholar]
- Srinivas PR, Gusfield D, Mason O, Gertz M, Hogarth M, Stone J, Jones EG, Gorin FA. Neuroanatomical term generation and comparison between two terminologies. Neuroinformatics. 2003;1(2):177–192. doi: 10.1007/s12021-003-0004-z. [DOI] [PubMed] [Google Scholar]
- Srinivas PR, Wei SH, Cristianini N, Jones EG, Gorin FA. Comparison of vector space model methodologies to reconcile cross-species neuroanatomical concepts. Neuroinformatics. 2005;3(2):115–131. doi: 10.1385/NI:3:2:115. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Brain Maps: Structure of the Rat Brain. Elsevier; 1999. p. 268. [Google Scholar]
- Van Essen D. Neuroinformatics - What's in It for You? Neuroscience Quarterly 2007 [Google Scholar]
- Manola F, Miller E, editors. W3C. RDF Primer. 2004. [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nature methods. 2011;8(8):665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]