Dynamin 2 is required for starvation-mediated breakdown of lipid droplets in hepatocytes by promoting vesiculation of autolysosomal tubules to release protolysosomes.
Abstract
Lipid droplets (LDs) are lipid storage organelles that in hepatocytes may be catabolized by autophagy for use as an energy source, but the membrane-trafficking machinery regulating such a process is poorly characterized. We hypothesized that the large GTPase Dynamin 2 (Dyn2), well known for its involvement in membrane deformation and cellular protein trafficking, could orchestrate autophagy-mediated LD breakdown. Accordingly, depletion or pharmacologic inhibition of Dyn2 led to a substantial accumulation of LDs in hepatocytes. Strikingly, the targeted disruption of Dyn2 induced a dramatic four- to fivefold increase in the size of autolysosomes. Chronic or acute Dyn2 inhibition combined with nutrient deprivation stimulated the excessive tubulation of these autolysosomal compartments. Importantly, Dyn2 associated with these tubules along their length, and the tubules vesiculated and fragmented in the presence of functional Dyn2. These findings provide new evidence for the participation of the autolysosome in LD metabolism and demonstrate a novel role for dynamin in the function and maturation of an autophagic compartment.
Introduction
Autophagy is a catabolic process by which cytoplasmic material is recycled in a lysosome-mediated manner for the purposes of energy production and survival in response to cell stress. An important energy reservoir common to most cells is the cytoplasmic lipid droplet (LD), a triglyceride- and cholesterol-rich organelle that sequesters esterified lipid into a readily accessible source of substrates for numerous physiological processes (Guo et al., 2009; Walther and Farese, 2012). Recently, hepatocytes have been shown to engage the autophagic machinery to support the breakdown of LDs under nutrient-limiting conditions (Singh et al., 2009). Also known as “lipophagy,” this mechanism appears to be of particular importance in hepatocytes, and is currently being investigated as a means for LD catabolism in other cell types such as adipocytes (Singh and Cuervo, 2012). Whereas the molecular intricacies of autophagic induction have been studied extensively, very little is known about the regulation of autophagy in the context of hepatic LD turnover.
During lipophagy, cytoplasmic LDs are engulfed by a growing limiting membrane that eventually encloses the entire LD, forming an autophagosome (Liu and Czaja, 2013). After the fusion of lysosomes with autophagosomes and the subsequent degradation of cargo in autolysosomes, new lysosomes are continually regenerated upon the extrusion of the autolysosomal membrane into LAMP1-positive tubular structures from which protolysosomes are then released. This process is known as autophagic lysosome reformation (ALR; Yu et al., 2010). The resulting tubular extensions from the autolysosomes are associated with clathrin, the AP2 adapter complex, and are enriched in the membrane phospholipid phosphatidylinositol-(4,5)-bisphosphate (PI(4,5)P2; Rong et al., 2012; Sridhar et al., 2013). These same components are also key constituents of endocytic invaginations of the plasma membrane targeted during clathrin-mediated endocytosis by the well-characterized membrane pinchase Dynamin 2 (Dyn2). Dyn2 is a 100-kD mechanoenzyme that uses GTP hydrolysis to provide the mechanical torque necessary to catalyze the scission of cellular membranes (Warnock et al., 1997). The similarity in components present at endocytic buds and ALR tubules therefore led us to examine whether there might also be a role in lipophagy for an important mediator of membrane trafficking such as Dyn2.
Numerous proteins populate the hepatocellular LD surface, having been detected in several proteomic screens to date (Fujimoto et al., 2004; Sato et al., 2006; Turró et al., 2006). Interestingly, a number of these identified proteins also have well-established roles in endocytic and membrane-trafficking pathways. Notable among these proteins are the caveolins and several members of the Rab family of small GTPases (Cermelli et al., 2006). The appearance of such proteins on LDs of many cell types suggests a critical interplay between membrane-trafficking events and lipid homeostasis, implicating a potentially important role for proteins such as Dyn2 in the maintenance of metabolic homeostasis.
In this study, we find that inhibition of Dyn2 by a variety of different methods prevents starvation-mediated breakdown of LDs in hepatocytes. This abrogation of lipophagy can be directly attributed to an enlarged and severely compromised autolysosomal compartment. These compartments frequently exhibit extensive membrane tubulation and, upon release of the Dyn2 perturbation, vesiculation resumes from the ALR tubules. We hypothesize that Dyn2 normally participates in the liberation of protolysosomes from autolysosomal tubules. These results suggest a novel, central, and direct role for this mechanoenzyme in the maintenance of lysosomal homeostasis and the autophagic clearance of hepatic lipid content.
Results
Knockdown of Dyn2 reduces starvation-mediated breakdown of LDs
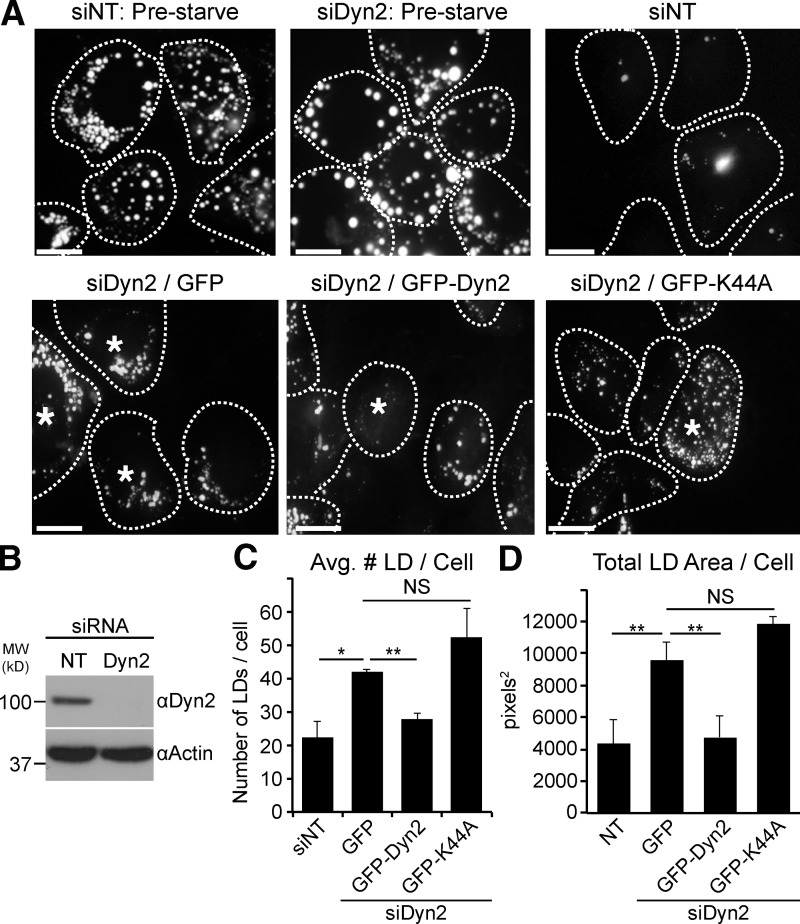
Nutrient limitation is known to promote hepatic LD breakdown for use of free fatty acids as a cellular energy source (Singh et al., 2009). To assess a potential role for Dyn2 in LD catabolism, we first applied a knockdown/re-expression approach in cultured Hep3B hepatocellular carcinoma cells. Cells were depleted of Dyn2 by siRNA treatment, lipid-loaded overnight with 150 µM oleic acid to promote LD formation, and then starved in low-serum media (containing 0.1% FBS) for 48 h. Lipid loading and LD breakdown was assessed using Oil Red O stain and the number and area of LDs were then quantified. The loading of cells transfected with nontargeting control siRNA (siNT) or siRNA targeting Dyn2 (siDyn2) was comparable before starvation (Fig. 1 A; Fig. S1, A–C). Whereas the 48-h starvation significantly reduced the LD content of siNT-treated cells, LD breakdown was greatly impacted by Dyn2 depletion. Knockdown of Dyn2 (knockdown efficiency >90%, shown in a representative blot in Fig. 1 B) resulted in an approximately twofold increase in both the average LD number per cell as well as average LD area per cell compared with control cells (Fig. 1, C and D, respectively). Importantly, re-expression of GFP-tagged wild-type Dyn2 (GFP-Dyn2) rescued this phenotype, whereas the GTPase-defective Dyn2-K44A mutant did not (Fig. 1, A, C, and D). In general, we observed that control cells reduced their lipid content under these experimental conditions by ∼60%, whereas Dyn2-depleted cells exhibited only a 10–30% reduction in LD area and number, respectively (Fig. S1 C). Taken together, these data suggest that an active Dyn2 enzyme plays an important role in LD breakdown.
Figure 1.
Dyn2 knockdown interferes with starvation-induced LD breakdown in Hep3B hepatocytes. Hep3B hepatocytes were treated with nontargeting control siRNA (siNT) or siRNA targeting Dyn2 (siDyn2), followed by re-expression of either vector alone, or GFP-tagged versions of Dyn2, loaded with 150 µM oleate overnight and starved for 48 h in medium containing 0.1% FBS. LDs were visualized by immunofluorescence using Oil Red O staining. Cell boundaries are outlined and those cells re-expressing GFP, GFP-wtDyn2, or GFP--K44A are denoted with asterisks. Bars, 20 µM. (B) Representative blot showing the efficiency of the Dyn2 knockdown in these cells. (C and D) Quantitation of the average LD number and area (in pixels2) per cell from three independent experiments. The data are represented as mean ± SE. *, P ≤ 0.05; **, P ≤ 0.01. NS, not significant.
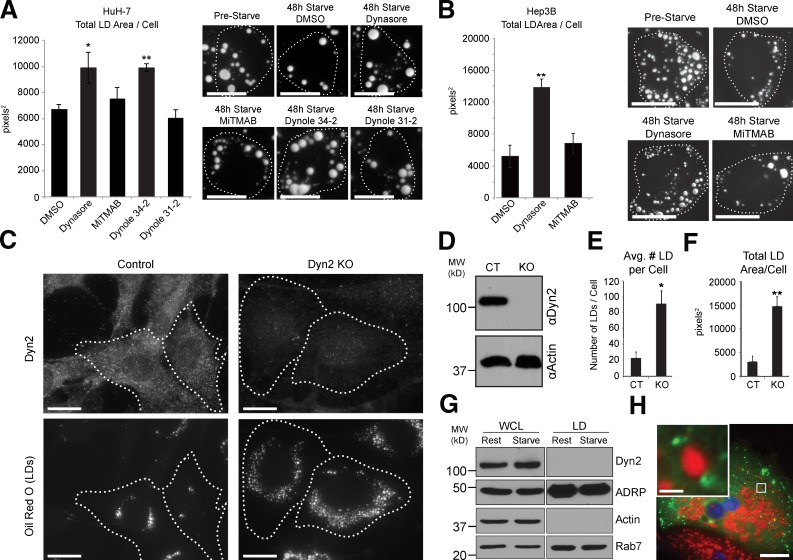
To further test these findings, pharmacological inhibitors of Dyn2 were also applied to HuH-7 and Hep3B hepatocytes that were first oleate-loaded overnight and subsequently starved in the presence of various Dyn2 inhibitors for 48 h. Interestingly, only the drugs interfering with the Dyn2 GTPase activity (Dynasore and Dynole 34-2) resulted in a reduction in LD breakdown in HuH-7 cells, whereas MiTMAB and the Dynole 31-2 negative control had no effect (Fig. 2 A). Similar results were obtained in Hep3B cells (Fig. 2 B). These data further support a role for Dyn2 activity in LD breakdown.
Figure 2.
Pharmacological and genetic inhibition of Dyn2 function also reduces starvation-induced LD breakdown. HuH-7 (A) and Hep3B (B) hepatocytes were loaded with 150 µM oleate overnight and starved for 48 h in medium containing 0.1% FBS in the presence of Dyn2 inhibitors or DMSO as indicated. Representative images of inhibitor-treated and control cells (stained with Oil Red O) are shown in A and B together with the quantitation of the average LD area per cell from three independent experiments. Pharmacological inhibitors used were: Dynasore (inhibits Dyn2 GTPase activity), MiTMAB (targets PH domain and interferes with membrane binding), Dynole 34-2 (allosteric GTPase inhibitor), and Dynole 31-2 (negative control for Dynole 34-2). Bars, 20 µM. (C) Representative images from control and Dyn2 knockout MEFs after an overnight lipid loading with 400 µM oleate for 17 h. Knockout of Dyn2 was induced by treatment with 2 µM 4-hydroxy-tamoxifen for 7 d and was confirmed by immunostaining of endogenous Dyn2 (top row) and by immunoblot (D). Bars, 20 µM. (E and F) Average LD number (E) and area (F) per cell from five independent experiments. All data are represented as mean ± SE. *, P ≤ 0.05; **, P ≤ 0.01. (G) Whole-cell lysates and LD fractions isolated from HuH-7 hepatocytes under resting or starved (2 h HBSS starvation) conditions. (H) Primary hepatocyte expressing Dyn2-GFP, showing an absence of colocalization with the LD surface (stained with Oil Red O). Bar, 20 µM. Inset shows magnification of boxed region (bar, 2 µM).
As an additional method to examine the role for Dyn2 in LD metabolism, we took advantage of the inducible Dyn2 knockout fibroblast cell line that was developed by Ferguson, de Camilli, and colleagues (Ferguson et al., 2009). In this cell line, Cre recombinase–mediated knockout of Dyn2 is induced by incubating cells with 4-hydoxy-tamoxifen for 5–7 d, monitored by immunofluorescence (Fig. 2 C, top row) and Western blotting (Fig. 2 D). Interestingly, when cells were loaded for 17 h with 400 µM oleate, a significant increase in both the average LD number and total LD area in the Dyn2-depleted cells was observed (Fig. 2 C, bottom row; and Fig. 2, E and F). In summary, three independent approaches (knockdown, pharmacological inhibition, and knockout) indicate a role for the large GTPase Dyn2 in hepatic LD metabolism.
As a result of our findings described above, we tested if Dyn2, based on its well-known mechanochemical ability to deform membranes, might participate directly in LD breakdown by promoting tubulation and/or scission of the LD lipid monolayer. This would result in an increased number of smaller vesicles and total surface area available to cytosolic lipases such as adipose tissue triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL). Inconsistent with this premise, however, Dyn2 did not co-purify with LD fractions isolated by density gradient centrifugation of HuH-7 cell lysates (Fig. 2 G). Further, it did not appear to associate directly with the LD surface, as expression of GFP-tagged Dyn2 in primary hepatocytes demonstrated localization to membranous compartments surrounding or adjacent to LDs, suggesting association with a proximal organelle (Fig. 2 H).
Dyn2 inhibition dramatically alters the morphology and function of autophagic compartments
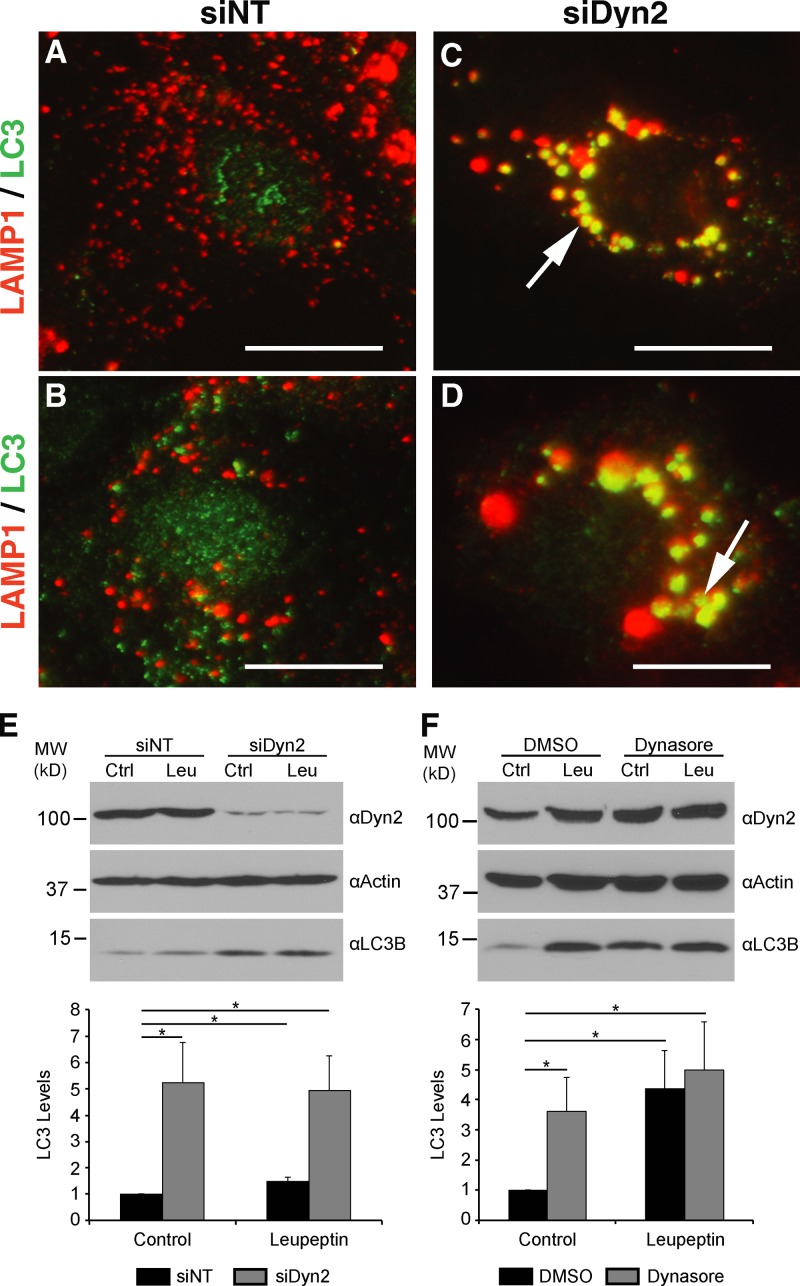
Hepatocytes are believed to use lysosomal components to drive an autophagy-based degradation of LDs (Singh et al., 2009; Dong and Czaja, 2011). As cells with impaired Dyn2 function exhibited a compromised breakdown of LDs under conditions of nutrient deprivation, we tested for indirect effects on lipolysis via alterations of lysosome function. Interestingly, Hep3B hepatocytes subjected to siRNA-mediated Dyn2 knockdown exhibited an exceptionally graphic redistribution of lysosomes into significantly larger membranous, juxtanuclear clusters, in contrast to control cells (Fig. 3, A–D; Fig. S2). These LAMP1-stained lysosomal aggregates also co-labeled with the autophagic marker LC3 and were similar to the persistent structures observed when autophagy is blocked at the autolysosomal stage (Rong et al., 2012). Consistent with an autophagic block in these cells is the observation that Dyn2 inhibition by siRNA knockdown or treatment with Dynasore in resting cells resulted in a four- to fivefold increase in levels of LC3-II, a classical marker of autophagosome biogenesis (Fig. 3, E and F, control lanes). To test if this LC3 increase represented an induction of the autophagic process in Dyn2-perturbed cells or, alternatively, a block in LC3 degradation by the autolysosome, the experiments were repeated in the presence of the lysosomal protease inhibitor leupeptin. In the case of siDyn2- or Dynasore-treated cells, this treatment resulted in no further significant increase in LC3 levels, indicative of a compromised autolysosomal compartment (Fig. 3, E and F, leupeptin lanes).
Figure 3.
Dyn2 inhibition leads to enlarged autolysosomal structures and prevents the autophagy of lipid droplets. Hep3B cells treated with either a nontargeting control siRNA (A and B, siNT) or an siRNA targeting human Dyn2 (C and D, siDyn2) were fixed and co-stained with antibodies specific for LAMP1 (red) and LC3 (green). After Dyn2 knockdown, a juxtanuclear aggregation and enlargement of the LAMP1-positive compartment is observed (C and D, arrows). Increased labeling of LC3 is also detectable after knockdown of Dyn2. (E) Western blotting of Hep3B lysates after a 3-d treatment with either the control or Dyn2-targeted siRNA and further treatment with or without 50 µM leupeptin. Densitometry-based analysis of six independent experiments is shown at the bottom of the figure. (F) Western blotting of Hep3B lysates after treatment for 2 h with DMSO or 80 µM Dynasore, in the presence or absence of 50 µM leupeptin. Quantitation of LC3-II levels relative to control are shown below the blots. The data are represented as mean ± SE; *, P ≤ 0.05.
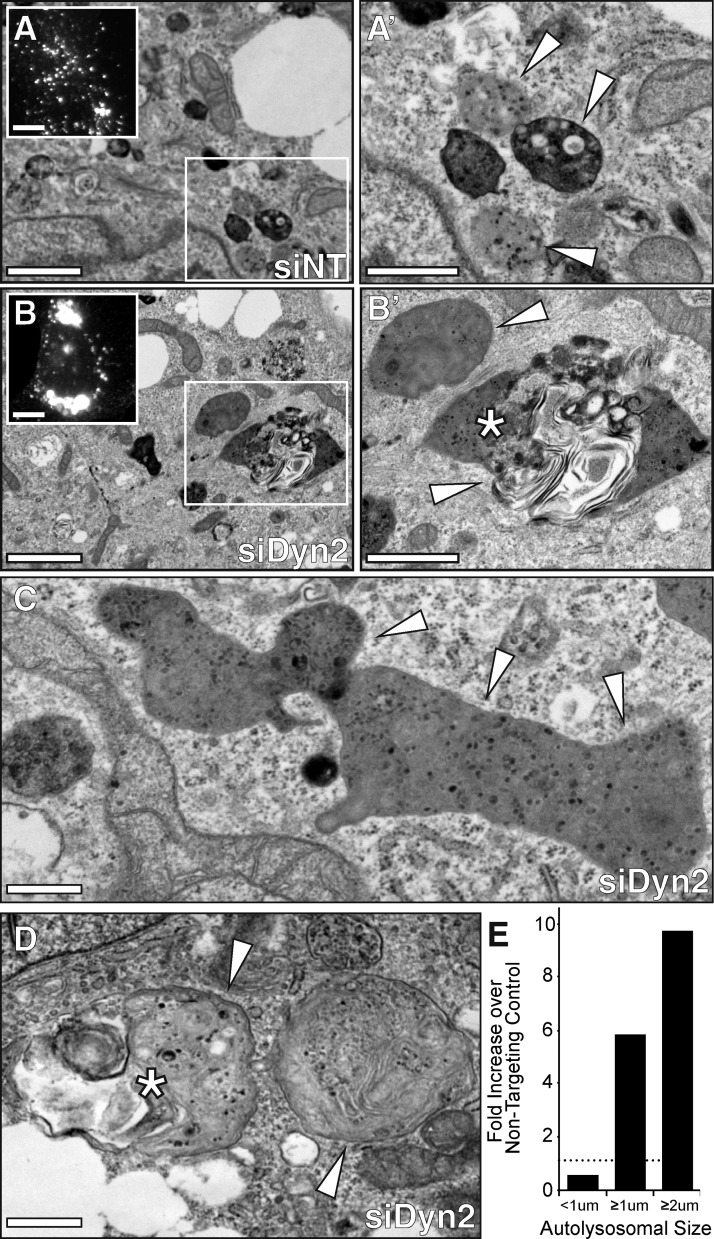
To better define the formation of the large LAMP1/LC3 organelles that form in hepatocytes with impaired Dyn2 function, Hep3B cells were loaded with 150 µM oleic acid to induce formation of LDs, treated with a nontargeting control siRNA or Dyn2-specific siRNA as above, then incubated for a further 24 h before fixation, embedding, and sectioning for transmission electron microscopy (TEM). As displayed in Fig. 4 A, control cells generally exhibited numerous small electron-dense lysosomes and autophagic structures of less than 1 µM in diameter (arrowheads). Dyn2 knockdown, however, induced the formation of much larger autophagosomes or autolysosomes (Fig. 4 B), in accordance with immunofluorescence images (insets), or those shown in Fig. 3. In the knockdown condition (Fig. 4, B–D), we observed a 10-fold increase (Fig. 4 E) in the number of large, dense, amorphous structures with diameters well in excess of 2 µM in size (4–5 times larger than those present in control cells). Many of these organelles contained large membranous voids reminiscent of engulfed LDs (asterisks). Together, these findings suggest that Dyn2 plays an essential role in LD breakdown via a participation in lysosomal dynamics that, in turn, contribute to the autophagic process.
Figure 4.
Dyn2 knockdown results in the formation of enlarged autophagic structures. (A–D) Transmission electron micrographs (TEMs) of oleate-loaded Hep3B hepatocytes treated with nontargeting control (siNT) or Dyn2 (siDyn2) siRNA for 72 h. Bars: (A and B) 2 µm; (A′ and B′) 1 µm; (C and D) 0.5 µm. Insets in A and B show fluorescent micrographs of LAMP1-stained cells (bars, 10 µM). Control cells (A) contain an abundance of small (<1 µm) electron-dense lysosomes and autolysosomes (arrowheads). Under conditions in which Dyn2 expression is suppressed (B–D), far fewer small lysosomes are observed. Instead, the cells are populated by larger autolysosomes (≥1 µm) with aberrant morphologies and containing putative LDs (*). (E) Quantitative measure of autolysosomal size from cells in which Dyn2 was knocked down as compared with control cells. Values represent the fold-increase in the number of autophagic structures of a given diameter (measured in microns) observed over siNT-treated control cells. Data were obtained from a single experiment with n = 6 cells examined by electron microscopy from each condition.
Dyn2 inhibition significantly impairs autophagic lysosomal reformation
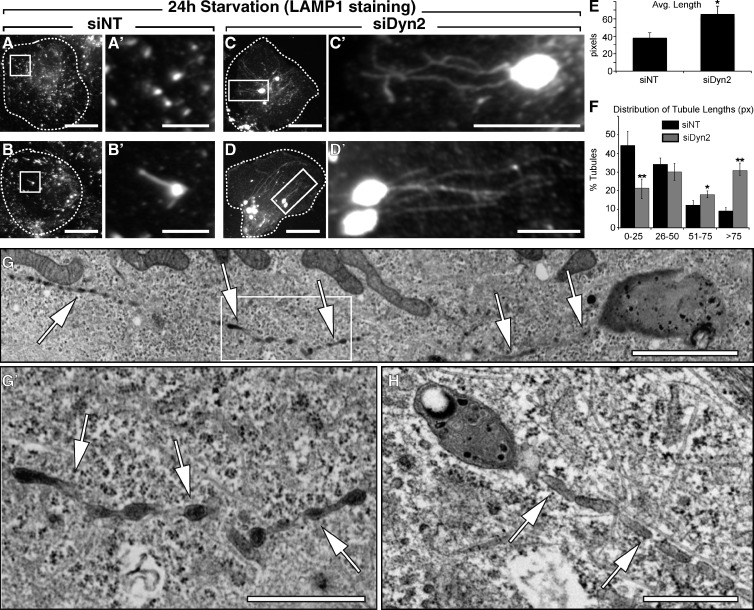
To pursue the concept that Dyn2 inhibition might affect lysosomal function and hepatocyte breakdown of LDs, cells were stimulated to metabolize stored lipids by being subjected to a 24-h period of nutrient deprivation/starvation in the context of Dyn2 inhibition. Cells were then fixed and lysosomal compartments were visualized by immunostaining with antibodies to LAMP1. Under starvation conditions, untreated cells would be expected to display autolysosomal compartments with small tubular protrusions that represent the de novo formation of nascent lysosomes used for autophagy (Yu et al., 2010). Depletion of clathrin or PI(4,5)P2 prevents the budding of these protrusions (Rong et al., 2012), resulting in the formation of aberrantly long tubules accompanied by a cessation of autophagy. We also observed the formation of short tubules emanating from LAMP1-positive compartments as a result of acute starvation (Fig. 5, A and B). Most graphic was the formation of remarkably long LAMP1-positive tubules in starved cells also subjected to Dyn2 siRNA treatment (Fig. 5, C and D). Quantification revealed that under these conditions, reformation tubules from those cells treated with Dyn2 siRNA were, on average, ∼60% longer (Fig. 5 E), with significant increases in the proportion of longer tubules compared with those found in control cells (Fig. 5 F). Indeed, some of these aberrant tubules exceeded 30 µM in length. Ultrastructural viewing by TEM revealed that these electron-dense tubules emanate from autolysosomes, and possess numerous varicosities at regularly spaced intervals along their length (Fig. 5, G and H), suggestive of membrane constrictions with defects in the final scission event. To confirm that these electron-dense structures that generated the extensive tubules were components of the late endocytic pathway, we pretreated cells with the fluid phase marker HRP, which is chased exclusively to late endosomes and lysosomes, before fixation and embedding for EM. As shown in thick EM sections (Fig. S3), tubules staining positive for HRP clearly originate from large autolysosomes in cells treated with Dyn2 siRNA.
Figure 5.
Dyn2 knockdown disrupts autophagic lysosomal reformation (ALR) and lysosomal tubule scission. Hep3B cells treated with either a nontargeting control siRNA (A and B, siNT) or an siRNA targeting human Dyn2 (C and D, siDyn2) were fixed and stained with an antibody specific for LAMP1 after a 24-h starvation period in media containing 0.1% FBS. Control cells displayed normal lysosomal compartments, with LAMP1-stained structures dispersed throughout the cytosol, some containing short reformation tubules (B′). After Dyn2 knockdown, many cells contained enlarged autolysosomes with a noticeable increase in persistent and lengthy reformation tubules emanating from LAMP1-positive compartments (C′ and D′). Bars: (A–D) 20 µM; (A′ and B′) 5 µM; (C′ and D′) 10 µM. (E and F) Measurements of the length (E) and distribution (F) of LAMP1-stained ALR tubules in control (siNT) and siDyn2-treated cells. Data represent the mean derived from measurements of tubules in a minimum of 40 cells for each experimental group over 6 independent experiments. Error bars represent SE; *, P ≤ 0.05; **, P ≤ 0.01. (G) TEM of siDyn2-treated Hep3B hepatocyte exhibiting enlarged autolysosomal structures with extensive tubulation (arrows). Bar, 2 µm. (G′) Inset showing enlargement of autolysosomal tubule (arrows). Bar, 1 µm. (H) Tubulating autolysosome with engulfed lipid droplet. Bar, 0.5 µm.
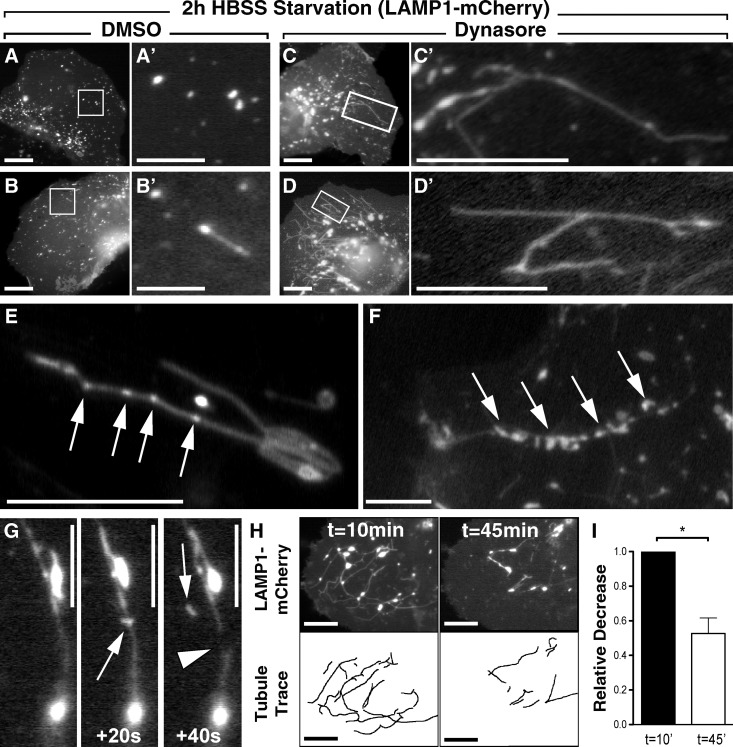
Based on the experimentation described above using siRNAs to reduce Dyn2 levels, it appears that this enzyme participates in LD breakdown indirectly by supporting lysosomal dynamics and biogenesis. A caveat of these findings is the duration (24–72 h) required for an siRNA knockdown, potentially resulting in either indirect or off-target effects. To circumvent this issue, we again used the pharmacological inhibitor Dynasore to provide a rapid, acute, and reversible block of Dyn2 function and assessed the consequences on lysosomal function. Hep3B hepatocytes expressing LAMP1-mCherry were plated onto glass-bottomed imaging dishes, starved in HBSS for 2 h, and treated with either DMSO or 40 µM Dynasore for 30 min. Still images (Fig. 6, A–D) from movies of live cells (Videos 1 and 2) show that, as expected, short-term starvation in HBSS with DMSO resulted in the appearance of punctate LAMP1-positive compartments (Fig. 6 A) that occasionally formed short tubular extensions (Fig. 6 B; Video 1). In contrast, the cells incubated in the presence of Dynasore for just 30 min exhibited numerous dynamic tubules emanating from enlarged LAMP1-positive compartments (Fig. 6, C and D; Video 2), similar to those observed with siRNA-mediated inhibition of Dyn2 expression (Fig. 5, C and D). These tubules were dynamic yet persistent, and importantly, upon drug washout with full-serum media, developed small varicosities or buds along their lengths (Fig. 6, E and F, arrows). Some tubules were actually observed to undergo scission events, resulting in the release of nascent vesicles or putative protolysosomes (Fig. 6 G; Videos 3 and 4). This vesiculation process was particularly exciting as it suggested that drug-induced autolysosomal hypertubulation is a readily reversible phenomenon once Dyn2 function has been restored. Comparative analysis of movie stills taken at the beginning and end of a 45-min drug washout period showed a nearly 55% reduction in total LAMP1-positive tubule content (Fig. 6, H and I), reflecting the fragmentation process. Together, these findings demonstrate an essential role for Dyn2 activity in autolysosomal tubule disassembly and the maintenance of lysosomal homeostasis.
Figure 6.
Acute inhibition of Dyn2 reversibly disrupts autophagic lysosomal reformation (ALR) and lysosomal tubule scission. (A–D) Still frames from time-lapse movies of Hep3B cells expressing LAMP1-mCherry. Cells were starved for 2 h in HBSS and subsequently treated for 30 min with either DMSO (A and B) or 40 µM Dynasore (C and D), which induced extensive tubulation of LAMP1-positive compartments. Bars (A–D): 20 µM; (A′–B′) 2 µM; (C′–D′) 10 µM. (E–G) To demonstrate the reversibility of this tubulation, Dynasore-treated cells were washed extensively with drug-free media containing 10% FBS and monitored by time-lapse microscopy for 45 min. Frequently, after drug washout, LAMP1-positive tubules exhibited noticeable varicosities (E and F, arrows; bars, 10 µM) along their length. These sites are suggestive of areas of scission and resumed budding of nascent protolysosomes from the reformation tubules (G; bars, 10 µM). (H) Tubules from cells undergoing drug washout were quantified by tracing their lengths at the beginning and end of these movies. Still frames from a representative movie show tubule content at t = 10 and 45 min after drug washout. Bars, 20 µM. (I) Analysis of five independent movies showed an average decrease in total tubulation of ∼50% after drug washout. Data represent the average relative change in total tubule length between the first and last frames of the time-lapse movies. Error bars represent SE; *, P < 0.05.
Dyn2 localizes to the autolysosomal compartment and participates in ALR
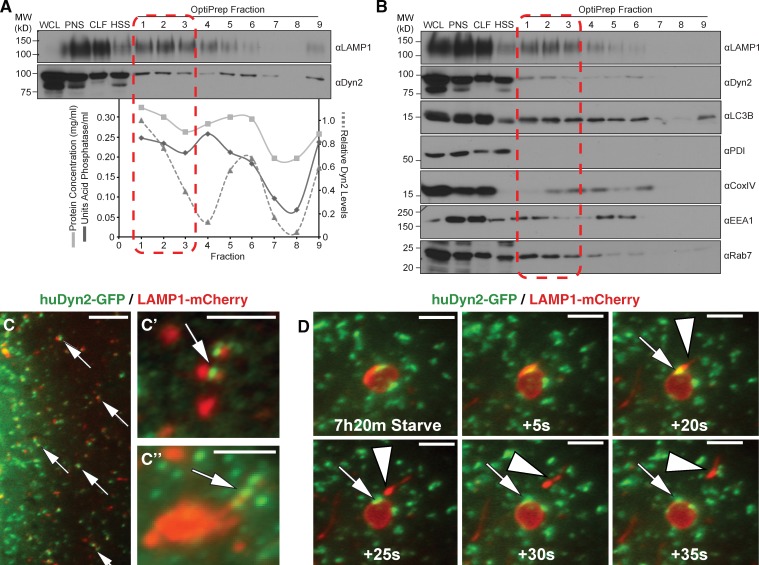
With the observation that Dyn2 has a direct role in the generation and disassembly of tubulated autolysosomes, it became important to determine whether Dyn2 is physically localized to the site of action. To this end, Hep3B hepatocytes were subjected to a subcellular fractionation approach to enrich for intact lysosomes. After a 2-h HBSS starvation and treatment with 40 µM Dynasore for 30 min, the same conditions used to induce autolysosomal tubulation in Fig. 6, C and D, autolysosomes were enriched by ultracentrifugation of a crude lysosomal fraction through an iodixanol gradient. Subsequent assays for acid phosphatase activity and immunoblotting analysis of gathered fractions with an antibody against LAMP1 revealed specific fractions (1–3) that were enriched in lysosomal content (Fig. 7 A). These same fractions also contained significant levels of Dyn2, suggestive of an intimate association with the lysosomal compartment. Additional Western blotting of these fractions for a variety of subcellular components (Fig. 7 B) suggests that Dyn2 is enriched in the lysosome fractions compared with those containing markers of other organelles such as mitochondria (COXIV) or the endoplasmic reticulum (PDI). Although these fractionation experiments can only be supportive of a Dyn2 association with lysosomes, as cross-contamination of cell components by these methods is an issue, they are consistent with both the functional findings described above and the fluorescence imaging observations described below.
Figure 7.
Dyn2 associates with autolysosomal tubules. (A and B) Subcellular density gradient fractionation of Hep3B hepatocytes starved for 2 h in HBSS and treated with 40 µM Dynasore, to induce tubule formation, as in Fig. 5. Cells were lysed (WCL), and the post-nuclear supernatant (PNS) was pelleted by centrifugation to produce a crude lysosomal fraction (CLF) and high-speed supernatant (HSS). The CLF was subsequently loaded onto an 8–27% discontinuous iodixanol (OptiPrep) gradient for separation by ultracentrifugation. Nine fractions were collected from the top of the gradient and blotted for Dyn2 and the lysosomal resident protein, LAMP1. Lysosomal acid phosphatase activity roughly correlates with LAMP1 protein levels in each fraction. Levels of Dyn2 are highest in fraction 2, corresponding with both the peak levels of LAMP1 and lysosomal activity. (A) The data shown are from a single representative experiment out of three repeats. (B) Blotting for subcellular components shows that Dyn2 is specific for these same fractions, unlike other organelle markers such as EEA1 (early endosomes) and COXIV (mitochondria). (C and D) Dyn2 localizes to the surface of LAMP1-positive compartments. Fluorescence imaging of Hep3B hepatocytes transfected with Dyn2-GFP and LAMP1-mCherry under resting (C and C′) or starvation (D) conditions. (C′′) Dyn2-GFP localizing to LAMP1-labeled lysosome structures under starvation conditions. Arrows indicate regions of protein colocalization. Dyn2 (arrows) is present at the site of scission of LAMP1-positive tubules (arrowhead) from large autolysosomal structures. Bars: (C) 5 µM; (C′, C′′, and D) 3 µM.
Using fluorescence live-cell imaging (Fig. 7, C and C′; Videos 5 and 6) and immunostaining (Fig. 7 C′′), we examined the distribution of cytoplasmic Dyn2 in relationship to LAMP1-positive compartments. In Hep3B hepatocytes, we observed a significant amount of colocalization between Dyn2-GFP– and LAMP1-mCherry–positive compartments (Fig. 7, C and C′, arrows; Video 5). Immunostaining of endogenous Dyn2 (Fig. 7 C′′) revealed Dyn2-positive puncta along the length of a LAMP1-positive tubule, reminiscent of the puncta appearing after Dynasore washout in Fig. 6 E. A 7.5-h serum withdrawal also results in the formation of ALR tubules in these cells, consistent with the timeframes observed by Yu et al. (2010). As shown in time-lapse stills (Fig. 7 D), Dyn2 can be resolved at the base of nascent LAMP1-positive tubules that are being released from enlarged autolysosomes (Video 6). These morphological observations support a central role for Dyn2 as a direct participant in the reformation of nascent lysosomes from tubular extensions of autolysosomes in the autophagic process.
The importance of lysosomal reformation in cellular lipid catabolism was further tested by inhibiting clathrin, an established regulator of ALR (Rong et al., 2012). As with the inhibition of Dyn2, we observed a nearly complete block in lipid droplet breakdown when Hep3B hepatocytes were treated with clathrin siRNA (Fig. S4). Like clathrin, Dyn2 therefore plays a vital role in ALR and consequently regulates the autophagic breakdown of LDs in the hepatocyte.
Discussion
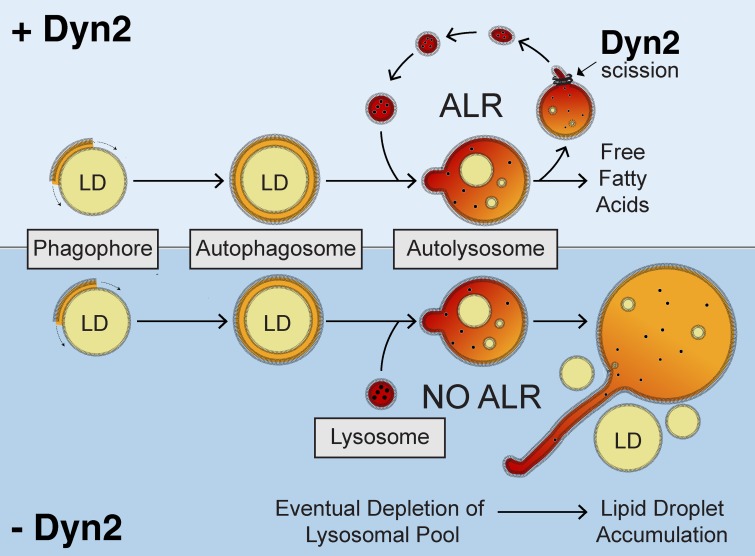
In this study, we have identified an important new role for Dyn2 in the autophagic degradation of hepatic LDs. Using a combination of genetic and pharmacological approaches that inhibit the function of Dyn2 (Figs. 1 and 2), we demonstrate that these perturbations alter the dynamics of the autolysosomal compartment (Figs. 3 and 4), leading to excessively long lysosomal tubules (Figs. 5–7). In hepatocytes, continued regeneration of the lysosomal pool is vital for the regulated mobilization of lipid stores in times of energetic deficit. We therefore propose a model (Fig. 8) that reflects the central importance of Dyn2 in hepatic lysosome function as it relates to lipophagy, based on the observable defects arising from Dyn2 depletion or inhibition during nutrient deprivation. Under these conditions, the ongoing liberation and formation of nascent lysosomes from the autophagosomal/autolysosomal compartment is compromised, resulting in enlarged LAMP1-positive organelles, rendering the cell unable to further catabolize LDs by lipophagy.
Figure 8.
Model for a role of Dyn2 in autophagic breakdown of lipid droplets in hepatocytes. After initiation of starvation, lipid droplets are enclosed by the double-membrane isolation membrane (phagophore), which engulfs the droplet, forming an autophagosome. Subsequent fusion with lysosomes results in the release of hydrolytic enzymes and lysosomal lipases into the autolysosomal compartment, which degrade the lipid droplets within. During the process of autophagic lysosomal reformation (top), nascent lysosomes are generated from membrane tubule extensions of this autolysosomal compartment. Dyn2 mediates the scission of these tubular structures. Depletion of Dyn2 by siRNA knockdown or inhibition of Dyn2 activity by pharmacological reagents (bottom) prevents tubular scission, eventually resulting in depleted lysosomal pools within the cell. Continued fusion of remaining lysosomes causes enlargement of the autolysosomal structures. A complete decrease in the recycled lysosomal population will prevent autophagic-mediated breakdown of hepatic lipid droplets.
Dyn2 as a mediator of late endosomal traffic
Dyn2 has been implicated in vesicle budding from the late endosome via a direct interaction with the endocytic adaptor CIN85, where this complex appears to mediate the scission of EGFR-containing vesicles from the MVB to the lysosome for subsequent degradation (Schroeder et al., 2010). Key to the recruitment of Dyn2 to the cellular membrane is a recognition of regions with high levels of membrane curvature (Roux et al., 2010), clathrin-coated structures (Taylor et al., 2012), and the membrane phosphoinositide PI(4,5)P2 (Achiriloaie et al., 1999; Zoncu et al., 2007). Interestingly, reformation tubules generated from autolysosomes have been shown to possess all three of these characteristics (Rong et al., 2012). Therefore, it is consistent that Dyn2 is also recruited to and functions on this late endomembrane platform.
Similar to the requirement of Dyn2-mediated GTP hydrolysis at the plasma membrane to promote scission during endocytosis (Warnock et al., 1997), we demonstrate that the same holds true in the context of the autolysosomal reformation tubule. Indeed, pharmacological inhibition of Dyn2 enzymatic activity significantly hindered LD breakdown in cultured hepatocytes (Fig. 2). The use of various pharmacological inhibitors of dynamin has the advantage of providing insights into the molecular mechanism of Dyn2 interactions with the autolysosomal membrane. It is surprising to us that MiTMAB, which targets the pleckstrin homology domain of Dyn2 and inhibits membrane binding, does not seem to affect LD catabolism. Our finding that Dynole 34-2 (another pharmacological inhibitor of Dyn2 GTPase function), like Dynasore, does have an effect seems to suggest that Dyn2 nucleotide hydrolysis is paramount in lysosome reformation, and that membrane binding might be dispensable. An alternative explanation for why no effect was seen with MiTMAB may be due to the low concentration (2 µM) used during the 48-h period of drug treatment for this particular experiment. This low concentration was used to avoid toxicity issues in the hepatocyte cell lines tested (concentrations >4 µM were not tolerated for these experiments). Future analysis with other members of the MiTMAB series of inhibitors, such as OcTMAB, or PH domain mutants of Dyn2, may provide valuable insights into the molecular mechanism of Dyn2 interactions with the autolysosomal membrane. Knockdown of Dyn2 in a non-hepatocyte cell line (Fig. 2 C) also demonstrated aberrant lipid accumulation in the absence of starvation, likely reflecting defects in basal LD turnover.
Importantly, the use of acute pharmacological inhibition complements the siRNA-based observations and reinforces the hypothesis that the observed autolysosomal tubulation is a direct consequence of inhibiting Dyn2 activity and not a response to inhibiting the scission of clathrin-coated pits from the plasma membrane over a 1–3-d period during siRNA knockdown or expression of mutant Dyn2 constructs. Indeed, LAMP1-positive tubule formation can be observed after just 30 min of treatment with relatively low concentrations of the drug. Of equal importance is the near-complete reversibility of the tubulation phenotype that can be observed as soon as 30 min after media replacement. Interestingly, the evenly spaced puncta that form along the length of the lysosomal tubules (Fig. 6 E) during the drug washout are consistent with the regularly spaced varicosities we observed by electron microscopy (Fig. 5, G and H), and suggest that these sites represent points of membrane constriction and scission not unlike those observed at the cell surface by a variety of different groups. Further, the localization of GFP-Dyn2 to these structures both biochemically and morphologically (Fig. 7) provides an important correlation between this novel distribution and function. Finally, thick sections of siDyn2-treated cells that were first allowed to endocytose HRP for several hours to label late endocytic compartments verified that these specific tubules are emanating from an endomembrane and not the PM (Fig. S3).
How Dyn2 is conditionally recruited to the autolysosomal membrane during nutrient starvation to participate in ALR remains unclear and will require further investigation. Under normal growth conditions, the formation of and budding from ALR tubules occurs rapidly. Very few instances of LAMP1-positive compartments with persistent tubule outgrowths are evident at steady-state; in these circumstances, Dyn2 may only be transiently associated with the tubules. Such short-lived interactions might provide an explanation for previous difficulties in the identification and localization of scission machinery at the autolysosomal compartment (Sridhar et al., 2013). The finding that clathrin and the AP2 adaptor complex are physically located on the ALR tubules, however, suggests that their Dyn2-mediated scission is very similar to, if not an extension of, events occurring at the PM.
Cellular metabolism and Dyn2 function
Although numerous Dyn2 mutations have been linked to several diseases including centronuclear myopathy (CNM) or Charcot-Marie-Tooth (CMT) disease, no human Dyn2 mutations have been shown to directly result in hepatic lipid accumulation. A recent publication has demonstrated a role for a particular Dyn2-CNM mutation in autophagy in newborn mice, which resulted in a significant accumulation of hepatic glycogen content (Durieux et al., 2012). It is now known that autophagic glycogenolysis is especially critical to glucose homeostasis in newborn animals (Kotoulas et al., 2006). Chemical inhibition of Dyn2 by Dynasore was recently shown to impair cholesterol trafficking in HeLa cells and macrophages (Girard et al., 2011). In that study, treatment of cells with Dynasore resulted in the formation of enlarged LAMP1-positive lysosomal structures (as we have also observed here) and was shown to prevent the normal efflux of free cholesterol from endolysosomal compartments to the ER. It is also worth noting that hepatocytes in which Dyn2 function is compromised also appear to possess a greater number of lipid-containing autolysosomes (Fig. 4, B–D; and Fig. S3 C), as if the LDs have progressively accumulated in these defective structures over time.
Although this study provides additional support for a role of the autophagosome and autolysosome in LD breakdown, these findings do not discount the likely and additive contributions of cytosolic lipases to hepatocellular LD breakdown. ATGL and HSL, enzymes of central importance to adipose tissue biology, are also expressed in the liver (Reid et al., 2008). It will be important to define how these cytosolic lipases work synergistically with the membranous autophagic pathways to mediate lipid metabolism in the hepatocyte. Defects in lysosomal homeostasis may also be manifest in perturbations of alternative types of autophagy such as mitophagy or pexophagy. Further studies will be required to analyze whether the effects of Dyn2 inhibition are specific to lipophagy or more global in nature.
Materials and methods
Cell culture and reagents
The HuH-7 cell line was a gift from the laboratory of G. Gores (Mayo Clinic, Rochester, MN). The Hep3B2.1-7 (Hep3B) cell line was obtained from ATCC (HB-8064). The conditional dynamin knockout cell line (Dnm1LoxP/LoxP;Dnm2LoxP/LoxP;Cre-ER+/0) was a gift from the laboratory of P. De Camilli (Yale University, New Haven, CT). In brief, this dynamin-depletion fibroblast cell line was isolated from conditional Dyn1/2 knockout mice that were generated through the use of a cre/loxP recombination strategy. Conditional knockout was achieved through culturing the cells in media containing 2 µM 4-hydroxy-tamoxifen for at least 5 d before experimentation. This treatment activated a Cre recombinase-estrogen receptor transgene, knocking out Dyn1 and Dyn2 (Ferguson et al., 2009). All cells were maintained in DMEM or MEM containing Earle’s salts and l-glutamine supplemented with 1 mM sodium pyruvate, nonessential amino acids, and 0.075% (wt/vol) sodium bicarbonate (Corning). Media for all cell lines contained 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin (Life Technologies), and cells were grown at 37°C in 5% CO2. Cells were grown on acid-washed coverslips for fluorescence microscopy and in plastic tissue culture dishes for biochemical analyses.
The rabbit polyclonal antibody raised against a C-terminal peptide representing residues 761–785 of mammalian Dyn2 has been described previously (Henley et al., 1998). The mouse monoclonal LAMP-1 (H4A3) antibody was from Santa Cruz Biotechnology, Inc. (sc-20011.), the LC3 and LC3B antibodies were from Novus Biologicals (NB100-2220 and NB600-1384, respectively), the antibody against ADRP was from LifeSpan Biosciences, Inc. (LS-B3121), the antibody against EEA1 was from BD (610457), the antibodies against PDI and COXIV were from Cell Signaling Technology (2446 and 4844, respectively), and the antibodies recognizing actin and Rab7 were from Sigma-Aldrich (A2066 and R8779, respectively). Secondary antibodies used for immunofluorescence were conjugated to Alexa Fluor 350, 488, or 594 (Invitrogen) and cells were mounted for microscopy in ProLong Antifade reagent (Invitrogen). Secondary antibodies for Western blot analysis were conjugated to horseradish peroxidase (HRP; Invitrogen). HRP-conjugated reagents for Western blot analysis were detected using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific). The Dynasore and MiTMAB inhibitory compounds were from EMD Millipore and the Dynole series of compounds were from Abcam.
Constructs, siRNA, and transfection
The expression constructs encoding full-length wild-type rat Dyn2 (cloned into pEGFP-N1; Takara Bio Inc.) or the dominant-negative Dyn2K44A mutant (generated by PCR mutagenesis from the wild-type construct) have been described previously (Jones et al., 1998; Cao et al., 2000). Constructs encoding for full-length wild-type human Dyn2, also cloned into pEGFP-N1 were a gift from D. Billadeau (Mayo Clinic, Rochester, MN) and have been described previously (Schroeder et al., 2010). Constructs encoding the dominant-negative Dyn2K44A protein were generated using wild-type Dyn2 plasmid as template and the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) according to the manufacturer’s directions. The WT and dominant-negative insert were subsequently subcloned into the pEGFP-N1 and pDsRed-N1 vectors (Takara Bio Inc.). The constructs encoding LAMP-1 were obtained through amplification from a rat brain cDNA library and cloned into the pEGFP-N1 and pmCherry-N1 vector (Takara Bio Inc.). All DNA constructs were verified by sequence analysis (Mayo Molecular Biology Core, Rochester, MN) and were analyzed using DNASTAR SeqMan analysis software (DNASTAR). Plasmid DNAs were purified using the QIAGEN Plasmid Purification kit.
Lipofectamine Plus reagent (Invitrogen) was used for transfection according to the manufacturer’s instructions. The siRNA oligos targeting Dyn2 and clathrin heavy chain were purchased from Thermo Fisher Scientific and cells were transfected with the RNAiMAX reagent (Invitrogen) according to the manufacturer’s instructions. For re-expression studies, cells were first treated with siRNA oligos for 24 h and then subsequently transfected with the proper expression construct.
Fluorescence microscopy
Cells were fixed and processed for immunofluorescence using standard procedures as described previously (Cao et al., 1998). In brief, cells were rinsed in PBS, fixed in 3% formaldehyde, and permeabilized in 0.1% Triton X-100 for 2 min. Coverslips were rinsed thoroughly with PBS between treatments. Fixed samples were blocked in buffer containing 5% goat serum and serially incubated in the appropriate primary and secondary antibodies at 37°C in blocking buffer. Images were acquired using epifluorescence microscopes (Axio Observer and Axiovert 200; Carl Zeiss) fitted with a 63× Carl Zeiss Plan Apochromat oil objective (1.4 NA) at room temperature. Digital images were captured with cameras (Orca II-ERG and Orca II; Hamamatsu Photonics) using iVision software (BioVision Technologies). Lipid droplet number and size were measured using the auto-segmentation tool within the iVision software. ALR tubule lengths were measured with iVision software through manual segmentation. All images and figures were assembled using Adobe Photoshop and Illustrator CS5 software (Adobe Systems). Movies were converted from 320 × 240-pixel TIF image sequences in Adobe Photoshop CS5 using H.264 encoding.
Time-lapse microscopy
Hep3B hepatocytes transfected with LAMP1-mCherry as described above were plated on glass-bottomed imaging dishes (MatTek Corporation) and starved for 2 h in HBSS + Ca2+/Mg2+ and 10 mM Hepes, pH 7.4. Dynasore was then added to 40 µM for 30 min to induce tubulation. For drug washout experiments, full-serum media (MEM + 10% FBS + 10 mM Hepes) was then used to extensively wash the cells. The cells were then immediately imaged using the above fluorescence microscopy setup on a heated stage at 37°C (Temp Controller 2000-2; PeCon GmbH) to find a cell exhibiting significant tubulation. This cell was then followed by acquiring images every 20 s with a 400-ms acquisition time.
Lysosomal enrichment
∼2 × 108 Hep3B hepatocytes were grown to a confluence of 90% in 150-mm dishes. To induce tubulation, cells were first starved in HBSS + Ca2+/Mg2+ for 2 h before treatment with 40 µM Dynasore for 30 min. Cells were then washed in dPBS, resuspended in lysis buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 10 mM NaF, and protease inhibitors), and homogenized by 20 strokes in a Dounce homogenizer on ice. The lysate was centrifuged at 1,000 g for 10 min to pellet nuclei and unbroken cells. The post-nuclear supernatant was then collected and centrifuged at 20,000 g to generate a pelleted crude lysosomal fraction (CLF). The CLF was adjusted to 19% iodixanol and layered within an 8–27% iodixanol gradient. The gradient was then centrifuged at 150,000 g in a rotor (SW55Ti; Beckman Coulter) for 4 h at 4°C. 400-µl fractions were collected from the top of the gradient. Aliquots were resuspended in 2xSDS sample loading buffer for loading onto a 10% polyacrylamide gel in preparation for Western blotting. 40 µl of each fraction was used to measure acid phosphatase activity using the Acid Phosphatase Acitivity kit (Sigma-Aldrich) according to the manufacturer’s protocol.
Statistical analysis
All statistical tests were performed with the two-tailed Student’s t test.
Online supplemental material
Fig. S1 shows that Dyn2 inhibition has no effect on lipid loading. Fig. S2 shows that LAMP1-positive structures are enlarged after Dyn2 inhibition. Fig. S3 confirms the endocytic nature of the tubules observed by EM. Fig. S4 demonstrates that clathrin knockdown has a similar phenotype to Dyn2 inhibition. Video 1 shows fluorescently labeled lysosomes in a control Hep3B hepatocyte. Video 2 shows the same lysosomes from under conditions of Dyn2 inhibition by Dynasore. Video 3 shows the effect of Dynasore washout. Video 4 shows an autolysosomal tubule scission event after drug washout. Video 5 shows colocalization of Dyn2 with LAMP1-positive structures. Video 6 shows a scission event mediated by Dyn2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201306140/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201306140.dv.
Supplementary Material
Acknowledgments
The authors would like to thank members of the McNiven laboratory, especially Gina Razidlo and Zhipeng Li, for helpful discussion and critical reading of the manuscript.
This study was supported by grants 5R37DK044650 (to M.A. McNiven), 5R01AA020735 (to M.A. McNiven and C.A. Casey), and T32DK007352 (to R.J. Schulze); National Institutes of Health Challenge Grant AA19032 (M.A. McNiven and C.A. Casey); the Robert and Arlene Kogod Center on Aging; and the Optical Morphology Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology (NIDDK P30DK084567).
The authors declare that they have no conflict of interest.
Footnotes
Abbreviations used in this paper:
- ALR
- autophagic lysosomal reformation
- ATGL
- adipose triglyceride lipase
- CMT
- Charcot-Marie-Tooth
- CNM
- centronuclear myopathy
- HSL
- hormone-sensitive lipase
- LD
- lipid droplet
- PI(4,5)P2
- phosphatidylinositol-(4,5)-bisphosphate
- TEM
- Transmission electron microscopy
References
- Achiriloaie M., Barylko B., Albanesi J.P. 1999. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol. Cell. Biol. 19:1410–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Garcia F., McNiven M.A. 1998. Differential distribution of dynamin isoforms in mammalian cells. Mol. Biol. Cell. 9:2595–2609 10.1091/mbc.9.9.2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Thompson H.M., Krueger E.W., McNiven M.A. 2000. Disruption of Golgi structure and function in mammalian cells expressing a mutant dynamin. J. Cell Sci. 113:1993–2002 [DOI] [PubMed] [Google Scholar]
- Cermelli S., Guo Y., Gross S.P., Welte M.A. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16:1783–1795 10.1016/j.cub.2006.07.062 [DOI] [PubMed] [Google Scholar]
- Dong H., Czaja M.J. 2011. Regulation of lipid droplets by autophagy. Trends Endocrinol. Metab. 22:234–240 10.1016/j.tem.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux A.C., Vassilopoulos S., Lainé J., Fraysse B., Briñas L., Prudhon B., Castells J., Freyssenet D., Bonne G., Guicheney P., Bitoun M. 2012. A centronuclear myopathy—dynamin 2 mutation impairs autophagy in mice. Traffic. 13:869–879 10.1111/j.1600-0854.2012.01348.x [DOI] [PubMed] [Google Scholar]
- Ferguson S.M., Raimondi A., Paradise S., Shen H., Mesaki K., Ferguson A., Destaing O., Ko G., Takasaki J., Cremona O., et al. 2009. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev. Cell. 17:811–822 10.1016/j.devcel.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 1644:47–59 10.1016/j.bbamcr.2003.10.018 [DOI] [PubMed] [Google Scholar]
- Girard E., Paul J.L., Fournier N., Beaune P., Johannes L., Lamaze C., Védie B. 2011. The dynamin chemical inhibitor dynasore impairs cholesterol trafficking and sterol-sensitive genes transcription in human HeLa cells and macrophages. PLoS ONE. 6:e29042 10.1371/journal.pone.0029042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cordes K.R., Farese R.V.J., Jr, Walther T.C. 2009. Lipid droplets at a glance. J. Cell Sci. 122:749–752 10.1242/jcs.037630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley J.R., Krueger E.W., Oswald B.J., McNiven M.A. 1998. Dynamin-mediated internalization of caveolae. J. Cell Biol. 141:85–99 10.1083/jcb.141.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.M., Howell K.E., Henley J.R., Cao H., McNiven M.A. 1998. Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science. 279:573–577 10.1126/science.279.5350.573 [DOI] [PubMed] [Google Scholar]
- Kotoulas O.B., Kalamidas S.A., Kondomerkos D.J. 2006. Glycogen autophagy in glucose homeostasis. Pathol. Res. Pract. 202:631–638 10.1016/j.prp.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Liu K., Czaja M.J. 2013. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 20:3–11 10.1038/cdd.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B.N., Ables G.P., Otlivanchik O.A., Schoiswohl G., Zechner R., Blaner W.S., Goldberg I.J., Schwabe R.F., Chua S.C.J., Jr, Huang L.S. 2008. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. J. Biol. Chem. 283:13087–13099 10.1074/jbc.M800533200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y., Liu M., Ma L., Du W., Zhang H., Tian Y., Cao Z., Li Y., Ren H., Zhang C., et al. 2012. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 14:924–934 10.1038/ncb2557 [DOI] [PubMed] [Google Scholar]
- Roux A., Koster G., Lenz M., Sorre B., Manneville J.B., Nassoy P., Bassereau P. 2010. Membrane curvature controls dynamin polymerization. Proc. Natl. Acad. Sci. USA. 107:4141–4146 10.1073/pnas.0913734107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. 2006. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J. Biochem. 139:921–930 10.1093/jb/mvj104 [DOI] [PubMed] [Google Scholar]
- Schroeder B., Weller S.G., Chen J., Billadeau D., McNiven M.A. 2010. A Dyn2-CIN85 complex mediates degradative traffic of the EGFR by regulation of late endosomal budding. EMBO J. 29:3039–3053 10.1038/emboj.2010.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Cuervo A.M. 2012. Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012:282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S., Patel B., Aphkhazava D., Macian F., Santambrogio L., Shields D., Cuervo A.M. 2013. The lipid kinase PI4KIIIβ preserves lysosomal identity. EMBO J. 32:324–339 10.1038/emboj.2012.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.J., Lampe M., Merrifield C.J. 2012. A feedback loop between dynamin and actin recruitment during clathrin-mediated endocytosis. PLoS Biol. 10:e1001302 10.1371/journal.pbio.1001302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turró S., Ingelmo-Torres M., Estanyol J.M., Tebar F., Fernández M.A., Albor C.V., Gaus K., Grewal T., Enrich C., Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 7:1254–1269 10.1111/j.1600-0854.2006.00465.x [DOI] [PubMed] [Google Scholar]
- Walther T.C., Farese R.V.J., Jr 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock D.E., Baba T., Schmid S.L. 1997. Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol. Biol. Cell. 8:2553–2562 10.1091/mbc.8.12.2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 465:942–946 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Perera R.M., Sebastian R., Nakatsu F., Chen H., Balla T., Ayala G., Toomre D., De Camilli P.V. 2007. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 104:3793–3798 10.1073/pnas.0611733104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.