Positioning of the mitotic spindle relative to the luminal domain in epithelial cells is determined by Par1b in a RhoA-mediated signaling pathway that involves the astral microtubule anchoring complex LGN–NuMA.
Abstract
Columnar epithelia establish their luminal domains and their mitotic spindles parallel to the basal surface and undergo symmetric cell divisions in which the cleavage furrow bisects the apical domain. Hepatocyte lumina interrupt the lateral domain of neighboring cells perpendicular to two basal domains and their cleavage furrow rarely bifurcates the luminal domains. We determine that the serine/threonine kinase Par1b defines lumen position in concert with the position of the astral microtubule anchoring complex LGN–NuMA to yield the distinct epithelial division phenotypes. Par1b signaling via the extracellular matrix (ECM) in polarizing cells determined RhoA/Rho-kinase activity at cell–cell contact sites. Columnar MDCK and Par1b-depleted hepatocytic HepG2 cells featured high RhoA activity that correlated with robust LGN–NuMA recruitment to the metaphase cortex, spindle alignment with the substratum, and columnar organization. Reduced RhoA activity at the metaphase cortex in HepG2 cells and Par1b-overexpressing MDCK cells correlated with a single or no LGN–NuMA crescent, tilted spindles, and the development of lateral lumen polarity.
Introduction
Symmetric cell divisions in nonstratified epithelial cells serve to generate equal daughters that both remain in the plane of the monolayer. In columnar epithelia this is accomplished by aligning the metaphase spindle parallel to the basal surface, resulting in a cleavage furrow perpendicular to the basal domain, which distributes luminal and basolateral surfaces in equal parts to both daughters. Thus, within their x-y-z cell space, the x-z orientation of the mitotic spindle determines whether apical and basolateral surface identities are maintained in both daughters (Reinsch and Karsenti, 1994). In multipolar hepatocytes, which organize their luminal domains perpendicular to their two basal domains, the x-y orientation of the mitotic spindle is equally important for a symmetric versus asymmetric outcome of the division (Fig. 1, Hepatocytic polarized) and hence for the maintenance of their polarized surface domain identities when hepatocytes proliferate during regeneration from injury. Because epithelial spindle positioning has been almost exclusively studied in columnar epithelial cells, little is known about the mechanisms for epithelial spindle orientation in the x-y plane. In cell lines which lack cell–cell adhesion junctions such as HeLa cells, cell–matrix signaling defines mitotic spindle orientation in both the x-z and x-y planes, but there is general consensus that cell–cell contacts provide the dominant signal for the stereotypic x-z orientation of metaphase spindles in polarized columnar epithelial cells such as kidney-derived MDCK cells (Théry et al., 2005, 2007; Toyoshima and Nishida, 2007; Toyoshima et al., 2007; den Elzen et al., 2009; Streuli, 2009). However, in the Drosophila follicle epithelium the integrin β-subunit is essential for spindle orientation and symmetric divisions, suggesting that dominant cell–ECM signaling processes for spindle alignment remain to be discovered in epithelial cells (Fernández-Miñán et al., 2007).
Figure 1.
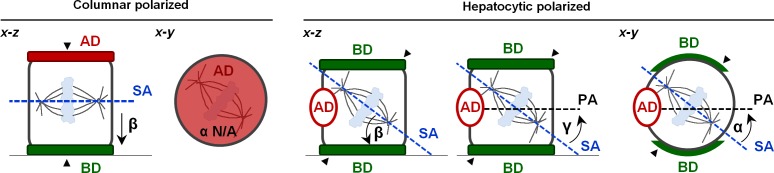
The β angle determines the symmetry of cell divisions in columnar cells, whereas α and β angles define hepatocytic cell divisions. Parameters that define spindle orientation in columnar (i.e., MDCK) or hepatocytic (i.e., WIF-B9, HepG2) metaphase cells. The β angle represents the angle between the spindle axis (SA) and the basal domain (BD) and defines division outcomes in both hepatocytic and columnar cells. The α angle measures the angle between the spindle axis (SA) and the apical–basolateral polarity axis (PA) in the x-y dimension and defines division outcome in hepatocytic cells, but is irrelevant for the inheritance of apical–basolateral domains in columnar cells. Similarly, the γ angle between the spindle axis (SA) and the apical–basolateral polarity axis (PA) in the x-z dimension is a predictor for the division outcome in hepatocytic cells. Because the cleavage furrow (black arrowheads) organizes perpendicular to the spindle pole, a β angle of 0° yields symmetric and a β angle of 90° asymmetric divisions in columnar cells. By contrast, small α angles favor asymmetric divisions in hepatocytic cells when the β and γ angles are also small. AD, apical domain.
We describe a novel cell–ECM signaling pathway that determines spindle orientation and promotes asymmetric divisions in hepatocyte-derived epithelial cells. It is mediated by the serine/threonine kinase and polarity determinant Par1b, which has been previously implicated in asymmetric cell divisions in the Caenorhabditis elegans zygote (Guo and Kemphues, 1995; Wu and Rose, 2007) and the Xenopus neuroectoderm (Tabler et al., 2010).
Results
Par1b determines mitotic spindle orientation in the x-y-z space of MDCK cells and hepatocyte WIF-B9 and HepG2 cells
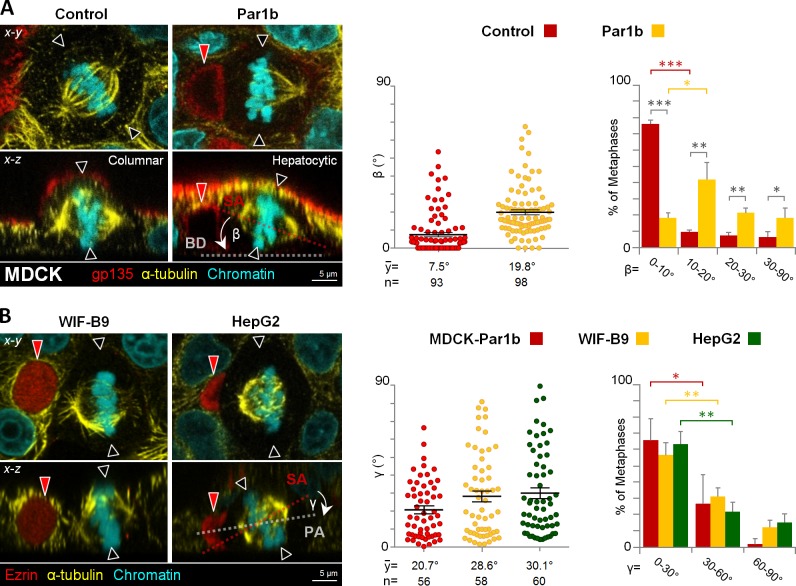
When cultured in 3D matrices, MDCK cells organize into hollow cysts in which the epithelial monolayer encloses a single luminal domain (O’Brien et al., 2002). We previously reported that overexpression of Par1b (MDCK-Par1b) resulted in cysts with multiple lumina (Cohen et al., 2011), a phenotype that can be caused by defects in mitotic spindle orientation with respect to the basal surface of columnar epithelial cells (Jaffe et al., 2008; Hao et al., 2010; Qin et al., 2010; Rodriguez-Fraticelli et al., 2010). Indeed, when grown as 2D monolayers, MDCK-Par1b metaphase spindles were “tilted,” i.e., they formed a mean β angle between the spindle axis and the substratum of 19.8 ± 1.4° while control cells aligned their metaphase spindles with the substratum, presenting a mean β angle of 7.5 ± 1.2° as previously reported (den Elzen et al., 2009; Fig. 2 A, see Fig. 1 for the definition of the parameters measured).
Figure 2.
Par1b promotes mitotic spindles that are oriented toward the lateral lumen and not aligned with the basal surface in MDCK cells, a characteristic of hepatocytic cells. (A) MDCK cells overexpressing Par1b under a doxycycline (dox)-dependent promoter (Par1b-MDCK-Tet-Off) were fixed and stained for the apical protein gp135, microtubules (α-tubulin), and chromatin with DAPI. The β angle was quantified (right panels). MDCK cells had significantly more cells with β angles in the 0–10° than in the 10–20° category, whereas the opposite was the case for MDCK-Par1b cells. (B) MDCK-Par1b, WIF-B9, and HepG2 cells were fixed and stained for α-tubulin, chromatin, and the apical domain (ezrin). The γ angle was quantified in cells with one spindle pole oriented toward the lateral lumen (right panels). In all three cell lines the percentage of cells with γ angles in the 0–30° category was significantly different from that of cells in the 30–60° category, indicating that the orientation was not random. Red arrowheads, BC-like lumina; black arrowheads, predicted cleave furrow plane. Error bars indicate ± SEM (dot graphics) and ± SD (bar graphics). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
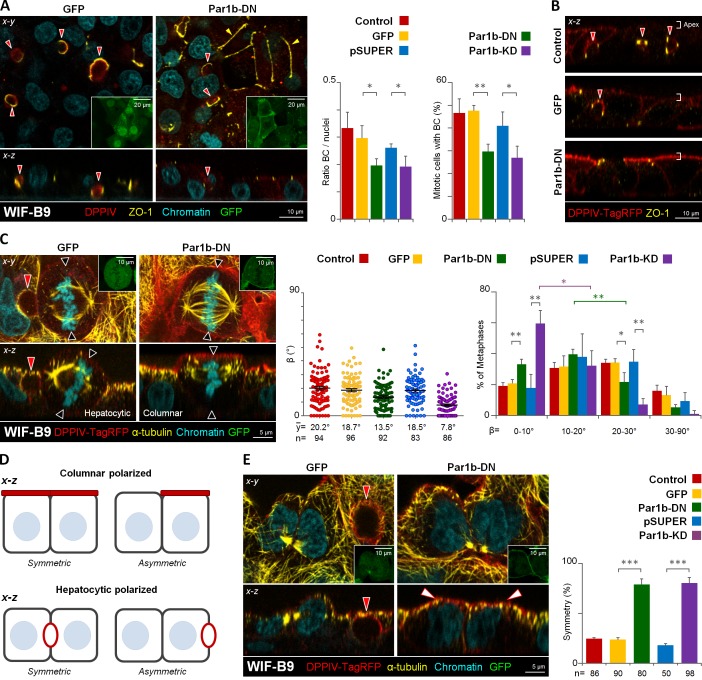
Par1b overexpression causes several hallmarks of hepatocyte polarity in MDCK cells, notably the organization of the apical domain as bile canalicular (BC)–like lumina that interrupt the lateral domains of neighboring cells (Cohen et al., 2004), similar to the polarized hepatocyte lines WIF-B9 and HepG2 (Ihrke et al., 1993; van IJzendoorn et al., 1997). Metaphase profiles in both of these hepatocytic lines exhibited a similar spindle tilt as those in MDCK-Par1b cells (control WIF-B9 cells in Fig. 3 C and control HepG2 cells in Fig. S1 B). Strikingly, in more than half of the x-z metaphase profiles one spindle pole faced the luminal domain in the three cell lines, which is manifested in a preferred γ angle between the spindle pole axis and apical–basal polarity axis of 20–30° (Fig. 2 B). Thus, the combined x-y-z spindle orientation predicted asymmetric divisions for MDCK-Par1b, WIF-B9, and HepG2 cells, which we indeed observed (unpublished data). Depletion of Par1b in WIF-B9 and HepG2 cells by siRNA (Fig. S1 A, Par1b-KD) or inhibition of Par1b activity by expression of dominant-negative Par1b in WIF-B9 cells (Par1b-DN; Cohen et al., 2004) had the opposite effect as Par1b overexpression in MDCK cells: it reduced the spindle tilt and caused alignment of metaphase spindles with the substratum (Fig. 3 C for WIF-B9 cells and Fig. S1 B for HepG2 cells). Concomitantly, Par1b-depleted WIF-B9 and HepG2 cells lost their hepatocytic polarity phenotype, as apparent from the significantly reduced number of lateral lumina (Fig. 3, A and B; Fig. S1 C and Video 1 for WIF-B9 cells, and Fig. S1 D for HepG2 cells). At the same time, we observed evidence for columnar polarity in Par1b-depleted WIF-B9 cultures: a chickenwire-like tight junction belt (Fig. 3 A, yellow arrowheads) and a luminal domain at the apex (Fig. 3 B) were detectable in areas of the WIF-B9 monolayer. Because in epithelial cells with columnar organization spindle alignment with the substratum results in symmetric divisions, it follows that Par1b depletion favors symmetric divisions in WIF-B9 cells, which we confirmed by analyzing cytokinesis profiles in fixed cells (Fig. 3, D and E).
Figure 3.
Par1b depletion in WIF-B9 cells promotes columnar polarity, x-z spindle alignment with the basal domain, and symmetric cell divisions. WIF-B9 cells (A) or DPPIV-TagRFP expressing WIF-B9 cells (B, C, and E) were transduced with adenoviruses expressing either GFP-tagged dominant-negative Par1b (Par1b-DN) or a shRNA-Par1b construct (Par1-KD) or their respective controls (GFP) or (pSUPER-GFP), then fixed and stained for the apical protein (DPPIV), Zonulae occludens 1 tight junctions (ZO-1; A and B), or microtubules (α-tubulin; C and E) and chromatin. GFP expression was monitored (A, C, and E; insets, left panels). The amount of BC-like lumina in interphase or mitotic cells (A; right panels), the β angle (C, right panels), and the distribution of the BC-like lumen or the apical domain into one or both daughters in cytokinetic cells (E, right) were quantified. The shift from larger to smaller β angles caused by Par1b inhibition was statistically significant for β angles in the 0–10° (59.5 ± 8.3%) category compared with the 10–20° (32.3 ± 9.6%) category (Par1b-KD) and between the 10–20° (39.7 ± 2.3) category and 20–30° (21.7 ± 5.6) category for Par1b-DN cells. (D) Modes of cell division encountered in cells with columnar and hepatocytic polarity (depicted in the x-z dimension). Red arrowheads, BC-like lumina; black arrowheads, predicted cleavage furrow plane; white arrowheads, apical domain; yellow arrowheads, chickenwire belt of tight junctions. Error bars indicate ± SEM (dot graphics) or + SD (bar graphics). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
We wondered whether the position of the lateral luminal domain, which interrupts most of the cell–cell-contacting surfaces in MDCK-Par1b and in polarized HepG2 and WIF-B9 cells (see Fig. 2, A and B), causes the Par1b-dependent spindle tilt in x-z. Indeed, metaphase spindles that oriented toward the luminal domain in MDCK-Par1b, WIF-B9, and HepG2 cells showed the largest β angle (Fig. 4, A and B; “apical domain oriented”[AD-O]). More precisely, we determined an inverse correlation between the β angles and the α angles, which indicate how much the nearest spindle pole faces away from the lumen in the x-y dimension (Fig. 4 B). However, even spindles that completely avoid a lumen-containing surface (i.e., α = 90°; Fig. 4 A, “basal domain oriented” [BD-O]), as well as mitotic spindles in MDCK-Par1b cells without lateral luminal domain (Fig. 4 A; BC-Ø), showed a significantly larger β angle than the corresponding control MDCK cells with columnar phenotype. Therefore, although Par1b coordinates spindle orientation with the formation of a lateral luminal domain in the majority of the cells, the kinase can inhibit the alignment of the epithelial mitotic spindle with the substratum independently of the presence of a lateral luminal domain.
Figure 4.
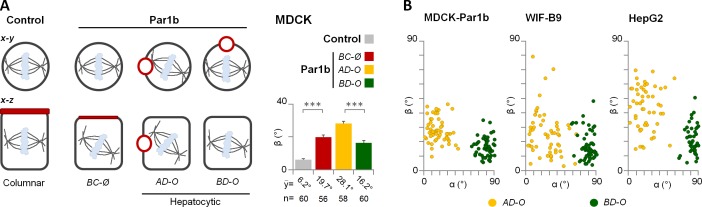
Lateral lumen architecture contributes to but is not the sole reason for the mitotic spindle tilt. (A) Control and MDCK-Par1b cells were fixed and stained for gp135, α-tubulin, and chromatin (not depicted). The β angle was calculated in polarized control cells (gray bar) and cells expressing Par1b without defined lumen (“Non polarized”; BC-Ø, absence of lateral lumen) or with lateral lumen (AD-O and BD-O). In AD-O cells one spindle pole faced the lumen, whereas in BD-O cells none of the spindle poles oriented toward the lumen. Error bars indicate ± SD. ***, P ≤ 0.001. (B) Correlation between β and α angles in MDCK-Par1b (n = 114), WIF-B9 (n = 115), and HepG2 (n = 98) cells with hepatocyte polarity.
Par1b regulates spindle orientation via ECM signaling that involves astral microtubule anchoring via the LGN–NuMA complex
Par1b, which is localized to the tight junctions, lateral and basal surfaces in interphase (Böhm et al., 1997; Cohen et al., 2004, 2011; Suzuki et al., 2004), and mitotic (Fig. S5, A and C) MDCK cells, is implicated in both E-cadherin–mediated cell–cell adhesion and in cell–ECM signaling (Böhm et al., 1997; Elbert et al., 2006; Saadat et al., 2007; Masuda-Hirata et al., 2009; Cohen et al., 2011). Spindle orientation along the x-z axis in polarized epithelial cells is thought to be controlled by the former. Indeed, mitotic spindles are “tilted” upon E-cadherin depletion in MDCK cells (den Elzen et al., 2009), but were insensitive to the inhibition of ECM-mediated signaling pathways that operate in HeLa cells to align metaphase spindles with the substratum (Toyoshima and Nishida, 2007). However, because Par1b overexpression strengthens rather than weakens E-cadherin–mediated adhesion (Elbert et al., 2006), we hypothesized that Par1b might function in yet uncharacterized ECM signaling pathway(s) that determine spindle orientation in polarized epithelial cells.
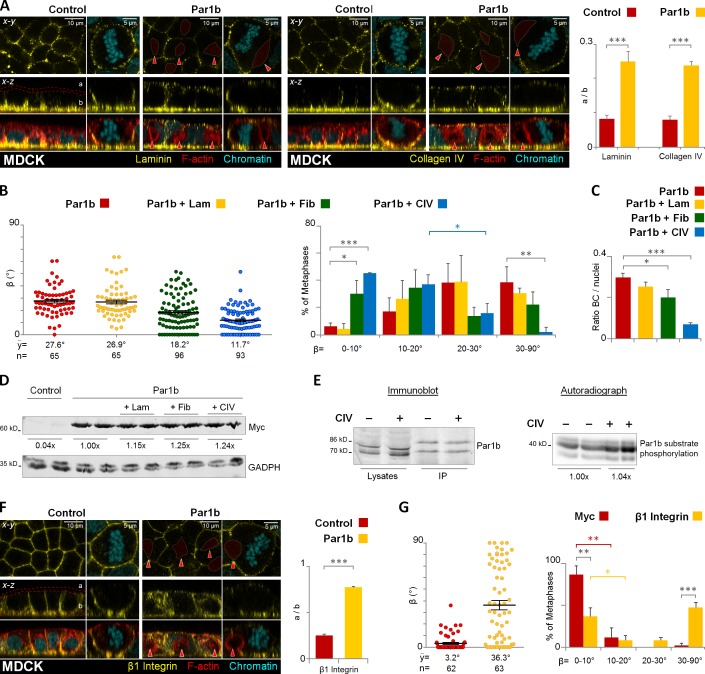
The two main components of the MDCK basal lamina, laminin and collagen IV, showed a mislocalization from the substrate-contacting surface (“b” in Fig. 5 A) to the cell apex (“a” in Fig. 5 A), but ECM components were absent from the lateral lumina (red arrowheads) in interphase and mitotic cells (Fig. 5 A). In further support of a role for Par1b upstream of ECM signaling we had observed that Par1b expression in MDCK cells diminished paxillin-positive focal contacts and adhesions (Cohen et al., 2011), whereas Par1b-DN-GFP expression in WIF-B9 cells yielded larger and more numerous paxillin-positive clusters (Fig. S2, A and B, respectively). Plating MDCK-Par1b cells on collagen IV or fibronectin- but not laminin-coated surfaces reestablished the x-z spindle alignment with the basal domain that was lost in Par1b-MDCK cells grown on uncoated coverslips (Fig. 5 B). Collagen IV also rescued the columnar polarity phenotype (Fig. 5 C), giving rise to cells with apical rather than lateral luminal domains. Likewise, HepG2 and WIF-B9 cells plated on collagen IV or fibronectin (but not laminin) aligned their spindle more with the basal domain (Fig. S3, A and C, respectively) and had fewer cells organizing with lateral lumen polarity (Fig. S3, B and D, respectively). Columnar polarity in HepG2 cells plated on collagen IV was apparent by the relocalization of PKCζ/λ from lateral lumina to the cell apex (Fig. S3 E). In addition, when grown as 3D cultures in gelifying Matrigel that supplied exogenous collagen IV, WIF-B9 cells aligned their metaphase spindles parallel to the lumen, with the cleavage furrow plane crossing a central lumen, consistent with the mitotic spindle orientation expected for cells with ascinar, i.e., columnar polarity (Fig. S3 F; and Video 2, Matrigel). Fewer symmetric divisions were observed in 3D WIF-B9 cultures grown in collagen I, which is not a component of the epithelial basement membrane (Fig. S3 F; and Video 2, Collagen I).
Figure 5.
Par1b regulates lumen polarity and mitotic spindle orientation in MDCK cells via ECM signaling. Control and MDCK cells overexpressing Par1b (A–E) were fixed and stained for F-actin with phalloidin-TRITC, chromatin, and either laminin, collagen IV (A), or integrin β1 (F). The distribution of the ECM proteins or the integrin β1 in the apex (a) versus base (b) in 2D cultures monolayers was quantified (A and F, right panels, respectively). MDCK-Par1b (B and C) were plated on either uncoated (Control), laminin (+Lam), fibronectin (+ Fib), or collagen IV (+ CIV) matrices. The cells were fixed and stained for gp135, α-tubulin, and chromatin (not depicted). The β angle (B) and the amount of BC-like lumina in interphase cells (C) were quantified. The percentage of (+CIV) Par1b cells with β angles between 10–20° (37.1 ± 7.1%) differed significantly from that in the 20–30° category (15.8 ± 7.3%). (D) Recombinant Par1b expression levels when MDCK-Par1b cells were plated on either plastic, laminin (+ Lam), fibronectin (+ Fib), or collagen IV (+ CIV). (E) Par1b activity in MDCK cells plated on either plastic (−) or CIV (+). Immunoblot: endogenous Par1b levels (lysates), and the immunoprecipitated fraction used for in vitro kinase assays (IP). The double band corresponds to two splice variants of Par1b. Audioradiograph: phosphorylation of a standard substrate in an IP-kinase assay. (G) MDCK cells were incubated 4 h in vivo with myc (control) or integrin β1 antibodies. Cells were fixed stained as in B. The β angle was quantified. Red arrowheads, BC-like lumina. Error bars indicate ± SEM (dot graphics) or + SD (bar graphics). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Our data thus indicate that Par1b promotes hepatocyte lumen polarity as well as hepatocyte x-z spindle orientation and asymmetric cell divisions by reducing the asymmetry in cell–ECM signals generated by the polarized deposition of a basal lamina, which is indeed absent in hepatocytes in vivo (Martinez-Hernandez and Amenta, 1993). We ruled out that the exogenous ECM altered Par1b levels or activity (Fig. 5, D and E). Furthermore, two findings let us suggest that the ECM signals are at least in part transmitted via the integrin β1. We determined that (1) β1 integrin labeling showed a similar relative decrease at the substrate-contacting surface as collagen IV/laminin in MDCK-Par1b cells (Fig. 5 F), and that (2) the function-blocking β1 integrin antibody AIIB2 caused a metaphase spindle tilt when added to the basal surface of control MDCK cells (Fig. 5 G).
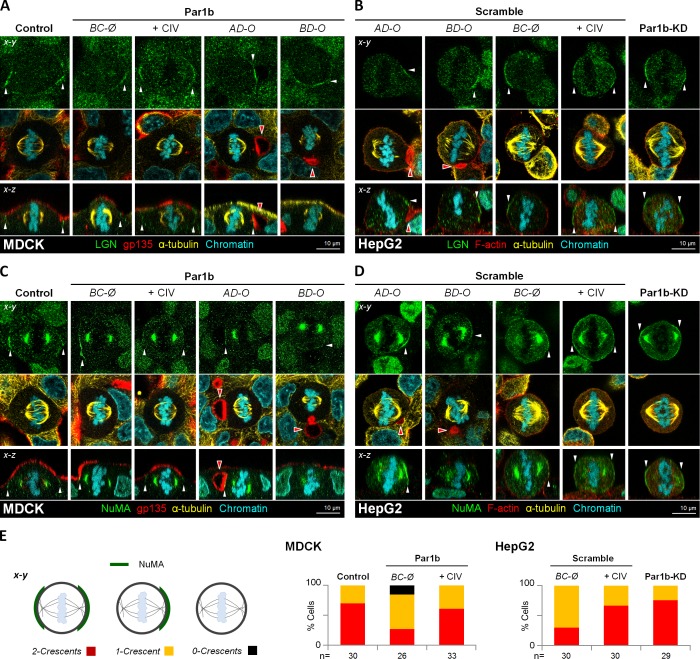
To elucidate how cell matrix/integrin signaling contributes to spindle alignment, we focused on the evolutionary conserved mitotic protein complex of myristoylated Gαi/leucine-glycine-asparagine repeat protein (LGN) and nuclear mitotic apparatus protein (NuMA) that has been proposed to serve as spatial cue for the cortical attachment of astral microtubules to dynein. Depletion of NuMA or LGN, inhibition of Gαi GTP-loading, or a uniform cortical distribution of the complex do not generally interfere with the progression of mitosis, per se, but all cause spindle orientation defects in epithelial cells (Hao et al., 2010; Woodard et al., 2010; Zheng et al., 2010; Peyre et al., 2011; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012). In agreement with previous reports, we observed that Gαi was restricted to the lateral domain (Fig. S4 A; x-y views, Control) and that LGN and NuMA formed two crescents parallel to the basal surface below the tight junctions in mitotic MDCK cells, which aligned with the two astral microtubule fans (Fig. 6, A and C, x-y views; and E, Control). The apical domain was devoid of the spindle-anchoring complex (Fig. 6, A and C; and Fig. S4 A, x-z views, Control). Similarly, during metaphase in polarized HepG2 and MDCK-Par1b cells, LGN and NuMA became restricted to a cortical region flanking the luminal domains (Fig. 6, A–D, AD-O and BD-O; and Fig. S4 C) where Gαi was also present (Fig. S4, A and B; AD-O and BD-O). Unlike LGN–NuMA, however, which were excluded from the MDCK-Par1b and HepG2 lumina (see Fig. S4 C for NuMA in HepG2 cells), Gαi also localized to the lateral lumina in these cells. We observed that metaphase profiles, in which the astral microtubules faced the lumen almost head on, had a tightly restricted sub-luminal belt of LGN–NuMA (AD-O panels), whereas those with larger α angles (BD-O panels) featured a broader LGN–NuMA distribution along the cell–cell contacting domain. Because one spindle pole always faced the sub-luminal NuMA patch, this landmark likely determines the preferential orientation of the spindle toward the lumen in x-y. With no second anchoring cue opposite the luminal domain along the luminal-basolateral polarity axis, the sub-luminal LGN–NuMA patch likely also accounts for the spindle tilt in x-z. In agreement with this interpretation, MDCK-Par1b and HepG2 or WIF-B9 cells (not depicted) without luminal domains likewise showed predominantly tilted metaphase spindles (see Fig. 4 A, BC-Ø) that correlated with a single LGN–NuMA patch (Fig. 6, A–E; BC-Ø) and reduced cortical Gαi staining (Fig. S4, A and B; BC-Ø). As expected from the lower β angles in MDCK-Par1b and HepG2 cells plated on collagen IV (see Fig. 5 B and Fig. S3 A), these cells favored the two-crescent LGN–NuMA phenotype (Fig. 6, A–E; +CIV) accompanied by recovery of Gαi staining (Fig. S4, A and B; +CIV). Likewise, Par1b depletion promoted a homogeneous cortical Gαi staining and a two-crescent LGN–NuMA phenotpye in HepG2 cells (Fig. S4 B; and Fig. 6, B, D, and E, Par1b-KD). Thus, Par1b-dependent ECM signaling determines mitotic spindle orientation by affecting the cortical recruitment of the LGN–NuMA complex.
Figure 6.
Par1b alters cortical LGN–NuMA distribution in metaphase. Control, Par1b-expressing (A and C) or control (Scramble) and Par1b-depleted (Par1b-KD) HepG2 cells (B and D) plated on uncoated or collagen IV–coated (+CIV) coverslips were fixed and stained for LGN (A and B) or NuMA (C and D), gp135 or F-actin (MDCK and HepG2, respectively), α-tubulin, and chromatin. The cortical pattern (E, left: 2 crescents, red; 1 Crescent, yellow; or 0 crescents, black) of the NuMA distribution in cells was plotted (E, right panels). See the schematics in Fig. 4 A for the nomenclature. Red arrowheads, BC-like lumina; white arrowheads, cortical distribution of NuMA (A and B) or LGN (C and D).
Par1b governs LGN–NuMA recruitment via ECM-dependent RhoA signaling
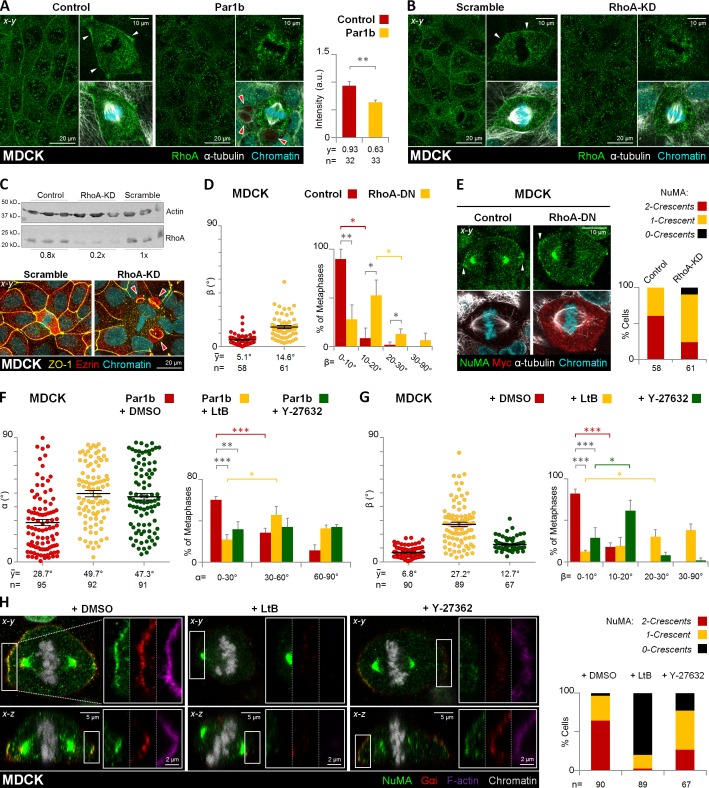
We noticed that cortical F-actin thickness and retraction fibers were reduced in mitotic MDCK-Par1b cells compared with the control cells, whereas both actin structures increased upon Par1b-DN-GFP expression in WIF-B9 cells (Fig. S2, C–F). In addition, we detected a robust ring of RhoA at the lateral cortex of metaphase MDCK cells (Fig. 7 A, Control), which was reduced upon Par1b overexpression (Fig. 7 A, Par1b), and restored when MDCK-Par1b cells were plated on collagen IV (not depicted). RhoA depletion using a published siRNA (Wakayama et al., 2011; Fig. 7 C, immunoblot) likewise abolished the cortical RhoA signal (Fig. 7 B), indicating that the antibody staining is specific.
Figure 7.
RhoA/RhoK inhibition alters mitotic spindle orientation and cortical LGN–NuMA distribution in metaphase. Control, Par1b-expressing (A) or control (Scramble) and RhoA-depleted (B, RhoA-KD) MDCK cells were fixed and stained for RhoA, α-tubulin, and chromatin. The mean fluorescence intensity of the cortical RhoA was quantified (A, right). (C) Endogenous RhoA levels in control, scrambled, and RhoA-KD MDCK cells (top) and immunostaining of scrambled and RhoA-KD cells for ZO-1 and ezrin (bottom). Note the presence of lateral lumina in the RhoA-depleted cells (red arrowheads). Control and RhoA-DN expressing MDCK cells (D and E), were fixed and stained for NuMA, myc, α-tubulin, and chromatin. Par1b (F) and control MDCK cells (G and H) were treated with DMSO (0.001% vol/vol), microfilament-depolymerizing agent, latrunculin B (LtB, 2 µM/45 min), or Rho-kinase inhibitor Y-27632 (50 µM/45 min). Cells were fixed and stained for NuMA, Gαi, F-actin, and chromatin (H). The α (F) and β angle (G) were quantified. The percentage of cells with β angles in the 10–20° category for Y-27632 or in the 20–30° category for LtB was significantly higher (61.4 ± 12.6% and 30.3 ± 8.5%, respectively) than that of cells with β angles in the 0–10° category (29.0 ± 12.2% and 12.3 ± 1.7%, respectively). The cortical pattern (2 crescents, red; 1 crescent, yellow; or 0 crescents, black) of the NuMA distribution in cells was plotted (E and H, right panels). Red arrowheads, BC-like lumina; white arrowheads, cortical distribution of RhoA (A and B) or NuMA (E). Error bars indicate ± SEM (dot graphics) or + SD (bar graphics).*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
We therefore hypothesized that the connection between Par1b-dependent cell–matrix/integrin signaling and cortical LGN–NuMA recruitment might be a RhoA-dependent organization of the actin cytoskeleton at the mitotic cell cortex. Microfilaments have been implicated in mitotic spindle orientation (Busson et al., 1998; Kaushik et al., 2003; Maddox and Burridge, 2003; Théry and Bornens, 2008; Kunda and Baum, 2009). Most relevantly, microfilament disruption has been shown to abolish cortical LGN in mitotic HeLa cells (Kaushik et al., 2003; Kaji et al., 2008), whereas a different HeLa cell study reported an x-z spindle tilt upon pharmacological inhibition of the RhoA effector Rho-kinase (Heng et al., 2012).
We determined that like microfilament disruption with latrunculin B (LtB), inducible expression of a dominant-negative form of RhoA (RhoA-N19) and pharmacological Rho-kinase (RhoK) inhibition with Y-27632 all caused a spindle tilt in MDCK cells, yielding β angles comparable to those caused by Par1b overexpression (Fig. 7, D and G). Inhibition of RhoK, and LtB treatment also prevented the stereotypical orientation of the spindle toward the lateral lumen in MDCK-Par1b cells (Fig. 7 F), resulting in a random distribution of α angles that is expected to cause more divisions with a cleavage furrow bisecting the luminal domain.
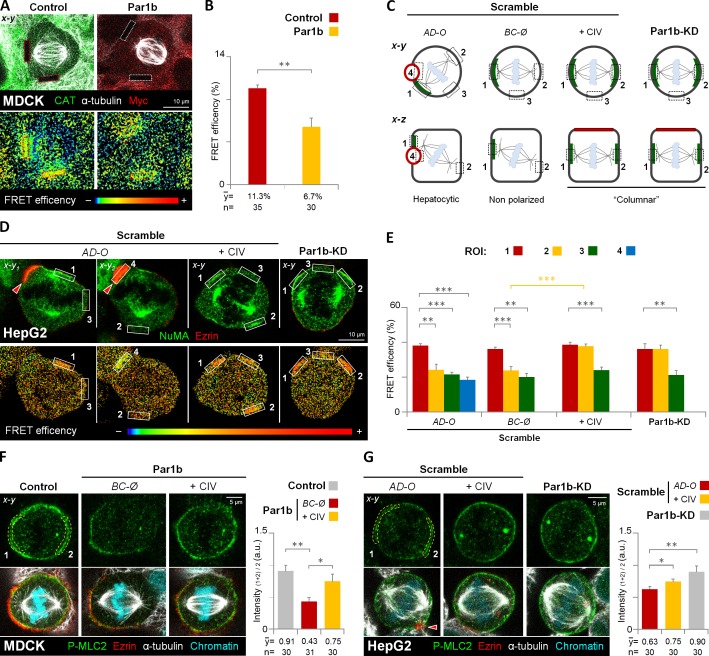
Importantly, RhoA-N19 and RhoK inhibition both mimicked the Par1b-mediated loss of cortical Gαi and LGN–NuMA metaphase crescents (Fig. 7, E and H). This prompted us to investigate a possible Par1b dependence of RhoA activity at sites of spindle attachment in mitotic MDCK and HepG2 cells with a FRET-based RhoA biosensor (Pertz et al., 2006). We compared the FRET efficiency at the lateral cortex in metaphase between control MDCK cells and MDCK-Par1b cells without a lateral lumen. We chose regions of interest (ROIs) at x-z positions where the mitotic spindles and astral MTs were in focus (Fig. 8 A). As expected from their compromised LGN–NuMA recruitment, MDCK-Par1b cells had significantly lower RhoA activity at the metaphase cortex than the controls (Fig. 8 A, bottom, and B). In agreement with this finding, MDCK-Par1b cells also had comparably less phosphorylated myosin light chain (P-MLC2), a RhoK-substrate, than controls (Fig. 8 F, BC-Ø). Of note, exogenous collagen IV restored the P-MLC2 staining in these cells to control levels (Fig. 8 F, +CIV). We further investigated the spatial relationship between RhoA activity and NuMA recruitment in HepG2 cells. We found that high RhoA activity always coincided with the presence of a NuMA crescent (Fig. 8 D). Thus, in polarized cells (Fig. 8, C–E; AD-O), FRET efficiency was high only adjacent to the luminal domain (Fig. 8, D and E; ROI #1), but low at the luminal domain itself, (Fig. 8, D and E; ROI #4) at the astral MT anchoring site away from the luminal domain (Fig. 8, D and E; ROI #2) and at a cortical site perpendicular to the spindle pole axis (Fig. 8, D and E; ROI #3). Cortical P-MLC2 labeling was discontinuous with the highest signal present adjacent to the lateral lumina (Fig. 8 G; AD-O). Non-polarized HepG2 cells featuring a single NuMA crescent (Fig. 8 C; BC-Ø) had high RhoA activity only at the spindle attachment point that coincided with their NuMA patch (Fig. 8 E), whereas collagen IV (Fig. 8 C; +CIV) or Par1b depletion (Fig. 8 C; Par1b-KD) resulted in higher FRET efficiency at both cortical regions where astral MTs attached (i.e., ROIs #1 and #2; Fig. 8, D and E; +CIV and Par1b-KD, respectively). These cells also feature a more robust staining of P-MLC2 at the circumferential cortex (Fig. 8 G, +CIV and Par1b-KD, respectively). Taken together, our data suggest that Par1b-mediated ECM signaling governs LGN–NuMA recruitment by regulating cortical RhoA and RhoK activity at the mitotic cortex.
Figure 8.
Par1b-mediated ECM signaling inhibits RhoA activity in metaphase cells. (A and B) MDCK cells expressing a RhoA FRET biosensor were transduced with adenoviruses expressing either CAT (Control) or Par1b-myc (Par1b). Cells were fixed and stained with α-tubulin and either CAT or myc antibodies (A, top). The FRET efficiency from acceptor to donor species in the cell cortex (A, dashed rectangles in bottom panel) was quantified (B). (C) Schematic x-y and x-z views of the regions of interest (ROIs) for RhoA activity measurements in HepG2 cells. The ROIs correspond to the cortical area adjacent to the left (1) and right (2) centrosome, a cortical region perpendicular to the spindle pole axis (3), and the lateral lumina membrane (4). Green labeling represents the NuMA localization. (D and E) Control (Scramble) and Par1b-depleted HepG2 cells (Par1b-KD) expressing the RhoA FRET biosensor were fixed and stained with NuMA and ezrin antibodies (D, top). The FRET efficiency (D, bottom) from acceptor to donor species was quantified (E, n = 30 cells for either AD-O, BC-Ø, +CIV, and Par1b-KD). (F and G) Control, Par1b-expressing (F) or control (Scramble) and Par1b-depleted HepG2 cells (G) were fixed and stained for the phosphorylated form of myosin light chain 2 (P-MLC2), ezrin, α-tubulin, and chromatin. The mean fluorescence intensity of the cortical P-MLC2 was quantified (right panels). Red arrowheads, BC-like lumina. a.u., arbitrary unit. Error bars indicate ± SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Discussion
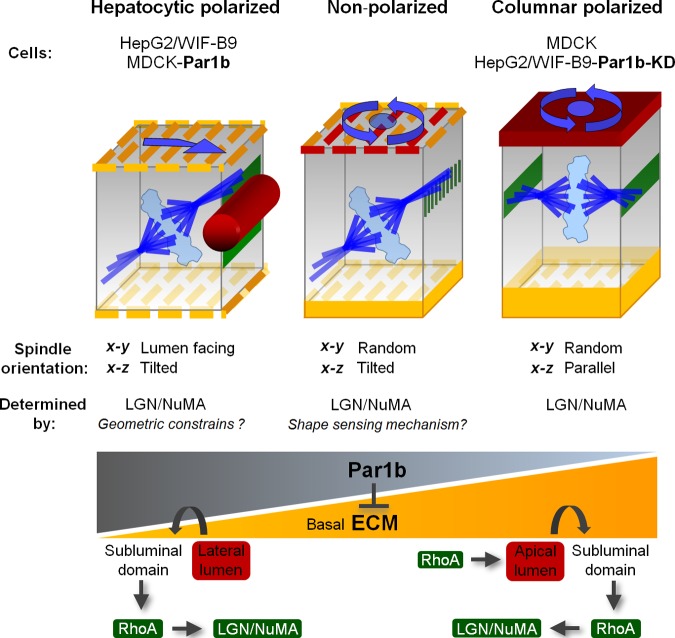
We propose that the epithelial architecture of columnar and hepatocytic epithelial cells defines their distinct mitotic spindle orientation phenotypes and modes of cell division: columnar epithelial cells, such as MDCK cells, establish their luminal domains at the apex; apical–junctional and cell–cell adhesion complexes organize in a “chickenwire” pattern, and the spindle-anchoring complex LGN–NuMA forms crescents parallel to the apical surface. This results in mitotic spindle alignment with the basal domain and in symmetric cell divisions (Fig. 9, Columnar polarized). Polarized hepatocytic HepG2 and WIF-B9 cells have lumina interrupting the lateral domain of neighboring cells, and accordingly, their lumen-flanking crescents of LGN–NuMA beneath the apical–junctional complexes are organized perpendicular to the two basal domains that interact with ECM proteins. They serve as astral microtubule attachment sites just as in the columnar cells; however, only one of the spindle poles anchors there. Geometric constraints, i.e., the spindle’s inability to curl around the lateral lumen, likely preclude that both spindle poles attach at the sub-luminal cortex. So, one set of astral microtubules attaches at the cortex opposite the lumen. This predominant spindle orientation yields asymmetric divisions (Fig.9, Hepatocytic polarized). We found that in multipolar hepatocytes, which face two or more BCs at opposite cell surfaces, two sub-luminal LGN–NuMA patches serve as spindle-anchoring domains (unpublished data). This will cause BC domains to be distributed to both daughters without the cleavage furrow bisecting a lumen, which is in agreement with observations made in histological sections of mitotic profiles in regenerating livers (Bartles and Hubbard, 1986; Stamatoglou et al., 1992). Increasing Par1b levels converts columnar MDCK cells into cells with hepatocyte lumen polarity, with hepatocyte LGN–NuMA distribution, spindle orientation, and asymmetric cell divisions, whereas depletion or inhibition of Par1b in HepG2 and WIF-B9 cells promotes columnar cell polarity and spindle organization. A comparison of Par1b localization (Fig. S5, A and B) and kinase levels and activity (Fig. S5, D and E) between MDCK, WIF-B9, and HepG2 cells did not reveal differences in subcellular distribution or higher Par1b activity in the hepatocyte-derived lines, suggesting that the cell lines differ in the availability of Par1b substrates relevant for the distinct polarity phenotypes.
Figure 9.
Model. Par1b inhibits an ECM–RhoA–RhoK pathway to promote hepatocytic lumen polarity and to regulate mitotic spindle orientation. Par1b-mediated inhibition of ECM-RhoA signaling in nonpolarized cells causes single or no cortical LGN–NuMA crescents in metaphase, leaving shape-sensing mechanisms to dictate spindle orientation (Grill and Hyman, 2005). RhoA inhibition also promotes polarization of these cells with hepatocytic polarity where the highest RhoA activity in metaphase presents adjacent to the lateral lumen and results in a robust cue for the recruitment of the LGN–NuMA complex to the subluminal domain and to a stereotypic spindle orientation shaped by the combination of the NuMA cue and the geometric constraints of the cell. The asymmetric deposition of a strong basal lamina as observed at lower Par1b signaling levels is associated with higher RhoA activity, which favors polarization with apical luminal domains (columnar polarity). In columnar epithelia, high cortical RhoA activity in metaphase is present along the sub-apical adhesion belt parallel to the basal surface, allowing Gαi (not depicted) and LGN–NuMA recruitment. A Ran-GTP gradient emanating from the chromosomes prevents LGN–NuMA from attaching to cortical regions closest to the metaphase plate (Kiyomitsu and Cheeseman, 2012), resulting in two LGN–NuMA crescents at two opposite membrane domains that align the spindle with the substratum. See Discussion for further explanations.
Although Par1b itself is present below the apical–junctional complexes in mitotic cells where the spindle-anchoring complexes are localized (Fig. S5, A and B), we suggest that the kinase does not act directly at the sites of spindle anchoring as observed for astral microtubule attachment at the posterior cortex of the C. elegans zygote (Wu and Rose, 2007). Instead, key to both the hepatocyte lumen polarity and spindle orientation phenotype is that Par1b antagonizes a robust and asymmetric basal lamina assembly, focal adhesion formation, and cell–ECM signaling pathway(s) that activate RhoA at the lateral cortex in polarizing cells. We previously reported that Par1b inhibits RhoA activity in contact-naive MDCK cells and that pharmacological inhibition of RhoK during MDCK polarization triggered by Ca2+-dependent cell–cell contact formation promotes hepatocytic lumen polarity (Cohen et al., 2007). In the current study we extended this finding by showing that siRNA-mediated RhoA depletion also induced lateral lumen polarity in MDCK cells (Fig. 7 C, bottom panels). RhoA inhibition had also been associated with lumen organization in HepG2 cells (Herrema et al., 2006). Here we show that Par1b controls cortical RhoA activity at the metaphase cortex, and that NuMA recruitment coincides with areas of high RhoA activity. Par1b overexpression in MDCK cells reduces both RhoA activity and LGN–NuMA at the lateral cortex in metaphase, whereas Par1b depletion in HepG2 cells increases cortical RhoA activity as well as NuMA recruitment to the region of astral microtubule attachment. Where a single NuMA crescent is present it always correlates with a patch of high RhoA activity, while the sister spindle pole faces a cortical area with low RhoA activity. Because direct RhoA and RhoK inhibition in MDCK cells also prevented Gαi, LGN, and NuMA recruitment, we propose that RhoK is essential for the recruitment of the spindle-anchoring complex. In our model (Fig. 9), fully polarized columnar and hepatocyte cells possess high sub-luminal mitotic RhoA activity, resulting in stereotypic LGN–NuMA recruitment, whereas in nonpolarized cells Par1b-mediated RhoA signaling levels determine both their trajectory toward either columnar (high RhoA) or hepatocyte (low RhoA) polarity and their LGN–NuMA recruitment pertinent to mitotic spindle orientation. The reduced cortical RhoA activity in MDCK-Par1b cells that failed to establish lateral lumina compared with control MDCK cells might result from compromised apical–junctional complex formation because tight junction proteins such as cingulin have been linked to the recruitment of RhoA-GEFs to the subapical region (Terry et al., 2011; Citi et al., 2012). Thus, although tilted mitotic spindles are observed in hepatocytic-polarized and in nonpolarized cells, the molecular reasons are likely different in the two cases: In the polarized cells, fully established cell–cell junctions result in high RhoA activity at the cell–cell contacting domains, providing robust astral MT-anchoring cues flanking the lumen, but the lateral lumen architecture forces one spindle pole to orient toward the basal cortex that is devoid of RhoA activity (except when the cell–cell contacting area is large compared with the luminal area and a spindle orientation away from the lateral lumen is observed, i.e., the BD-O, orientation in Fig. 4 A). The nonpolarized cells, by contrast, fail to align the spindle with the substratum because incomplete cell–cell junction formation prevents robust RhoA activation necessary for the reliable recruitment of spindle-anchoring complexes to two opposite faces of the lateral cortex. The nonrandom β angles in both scenarios might be the result of cell shape–sensing mechanisms for mitotic spindle orientation (Grill and Hyman, 2005) that operate in the absence of dominant cortical cues.
How RhoA/RhoK contribute to LGN–NuMA recruitment remains to be established. That Gαi is removed from the cortex upon RhoK inhibition along with LGN and NuMA makes it likely that the kinase acts upstream of Gαi membrane anchoring and unlikely that Gαi functions as coactivator of RhoA, as was shown for other Gα subunits (Chikumi et al., 2002).
Exogenous collagen IV overcomes Par1b-mediated RhoA inhibition, presumably by promoting the clustering of integrins, among them, β1 integrins, which we found to be important for spindle orientation in MDCK cells. Accordingly, exogenous collagen IV at the substrate-contacting surface reverted both the hepatocytic lumen polarity phenotype and the hepatocytic spindle alignment and division phenotypes in MDCK-Par1b cells to columnar characteristics of control MDCK cells. Likewise, plating WIF-B9 and HepG2 cells on a collagen IV matrix was sufficient to abolish their lateral lumina and to cause mitotic spindle alignment with the substratum. In an unbiased screen for Par1b substrates in MDCK cells we identified many putative direct and indirect regulators of the ECM and of RhoA activity (unpublished data). We therefore consider it likely that Par1b orchestrates RhoA activity downstream of the ECM via multiple substrates.
In summary, a picture emerges in which the inhibition of cell–ECM signaling pathways is central to Par1b’s role as a regulator of multiple key features that distinguish columnar and hepatocytic polarity phenotypes.
Materials and methods
Cell lines and culture
WIF-B9 cells (provided by A. Hubbard, John Hopkins University School of Medicine, Baltimore, MD) were grown in modified F-12 Coon’s modification medium (F6636; Sigma-Aldrich) supplemented with 10−6 M hypoxanthine, 4 × 10−8 M aminopterin, 1.6 × 10−6 M thymidine, 5% (vol/vol) FBS (100-106; Gemini), 1% Glutamax (Invitrogen), 0.5 µg/ml amphotericin, and 10 mM Hepes. For culture maintenance, cells were seeded in Falcon plastic dishes (BD) at 10 × 103 cells/cm2 and cultivated up to 4 d before replating. For experiments, differentiated cultures (10–12 d) were plated on water-prewashed German glass coverslips (EMS), and MatTek or CELLview (Geiner Bio-One) chambers at 15 × 104 cells/cm2. HepG2 (provided by A. Wolkoff, Albert Einstein College of Medicine, Bronx, NY) and MDCK-Tet-Off cells (provided by K. Mostov, University of California, San Francisco, San Francisco, CA) were grown in DMEM without phenol red (17-205; Corning) supplemented with 10% FBS (S11050; Atlanta Biologicals) and 2 mM l-glutamine. 3D cyst-like cultures of WIF-B9 cells were obtained with cells growing in a medium containing 1.2 mg/ml rat collagen I or pure Matrigel. In brief, cells were plated at 2.5 × 104 cells/cm2 in 150 µl gel solution on 0.4-µm pore-size polyester Costar Transwells, prewarmed at 37°C. Media was refreshed every other day for the first 48 h and every day thereafter. Cultures were analyzed after 6 d. Stable MDCK cell lines expressing RhoA ECFP/Citrine-YFP FRET biosensor (RhoA-BS; Pertz et al., 2006) were generated from MDCK-Tet-Off cells and Par1b-GFP clones from parental MDCK-II cells, respectively. The RhoA-N19 (RhoA-DN) MDCK-Tet-Off cell line was provided by Jou and Nelson (1998). Recombinant Par1b, RhoA-BS, and RhoA-DN expression in MDCK-Tet-Off cells was repressed by 200 ng/ml doxycycline and induced for 2–3 d after the doxycycline washout. Cells expressing DPPIV-TagRFP, GFP, Par1b-DN-GFP, pSUPER-GFP, or shRNA-Par1b-GFP (Par1b-KD-GFP), chloramphenicol acyl transferase (CAT), and Par1b were obtained by adenovirus transduction in Opti-MEM (Invitrogen) for 1 h with 1–10 plaque-forming units/cell and 8–12 h expression at 37°C in 12–24 h fresh seeded cells. siRNA duplexes targeting the same sequence as the shRNA construct (Cohen et al., 2004) were transfected (for Par1b-KD) via Lipofectamine RNAiMAX (12.5 pM/cm2) into HepG2 cells (40 × 103 cells/cm2) or electroporated (5 µg of RhoA-KD) into MDCK cells (1.5 × 105 cells/cm2). For the ECM experiments, cells were seeded at the same confluency on uncoated, laminin-, fibronectin- or collagen IV–coated (50 µg/cm2 each) glass coverslips or 0.4-µm pore-size polycarbonate Costar Transwells. Cells were maintained at 37°C in a 5% CO2 (MDCK, HepG2) or 7% (WIF-B9) humidified atmosphere.
Antibodies, plasmids, and reagents
The following antibodies were used in this study: α-tubulin (ab6160; Abcam), collagen IV (ab6586; Abcam), DPPIV (MCA1317T or MCA924R; AbD Serotec), ezrin (ab41672, Abcam; or clone p81, NICHD Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), β1 integrin (clone AIIB2; NICHD Developmental Studies Hybridoma Bank; Werb et al., 1989), Gαi (sc-13533; Santa Cruz Biotechnology, Inc.), gp135 (clone 3F21D8, mouse monoclonal provided by G. Ojakian, SUNY Downstate Medical Center, Brooklyn, NY), GADPH (ab9484; Abcam), laminin (L 9393; Sigma-Aldrich), Myc (ab9106, Abcam; or sc-40, Santa Cruz Biotechnology, Inc.), NuMA (provided by D. Cleveland, University of California, San Diego, La Jolla, CA; or ab36999, Abcam), CAT (ab50151; Abcam), LGN (provided by Q. Du, Medical College of Georgia, Augusta, GA), Par1b (rabbit polyclonal; provided by B. Schaar, Stanford University, Stanford, CA; Schaar et al., 2004; or ab77641, Abcam or the previously described Par1 rabbit polyclonal raised against the 310 C-terminal amino acid of mouse EMK2; Cohen et al., 2004), paxillin (610568; BD), phospho-myosin light chain 2 Thr18/Ser19 (P-MLC2, #3674; Cell Signaling Technology), PKCζ (sc-216; Santa Cruz Biotechnology, Inc.), RhoA (sc-418, Santa Cruz Biotechnology, Inc.; or ab86297, Abcam) and ZO-1 (clone R40.76, EMD Millipore; or 33-9100, Invitrogen). The RhoA single-chain biosensor pBabe-sin-tet-CMV-puro retroviral vector for Tet-On/Off system was as in Pertz et al. (2006). Scrambled/Par1b siRNA (targeting sequence: 5′-GAGGTAGCTGTGAAGATCA-3′; Cohen et al., 2004) and RhoA siRNA (targeting sequence: 5′-GCAGGTAGAGTTGGCTTTG-3′; Wakayama et al., 2011) was custom synthesized by Thermo Fisher Scientific. Laminin, fibronectin, collagen IV, collagen I, and growth factor–reduced Matrigel were from BD. Y-27632 was from Enzo Life Sciences. Phalloidin-TRITC and latrunculin B (LtB) were from Sigma-Aldrich. DAPI and DRAQ5 were from Invitrogen.
Immunoblot, immunoprecipitation, and kinase assay
Par1b was immunoprecipitated from confluent cell monolayers by harvesting the cells in RIPA lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.25% deoxycholate, and 1% NP-40, with protease and phosphatase inhibitors). Debris was cleared by centrifugation at 13,500 rpm for 10 min. Lysates were standardized for their protein concentration, which was determined using a Bradford assay. The lysates were precleared on pansorbin cells and then incubated with a rabbit polyclonal Par1b antibody. Subsequently, the lysates were placed onto Protein A–coupled agarose at 4°C for 90 min. The agarose was washed with cold buffer solutions of: three times RIPA buffer, once with high salt RIPA (RIPA with 500 mM NaCl), and once with 20 mM Tris, pH 7.5, and 150 mM NaCl. The agarose beads were then frozen in 40 mM Hepes, 5 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100, and 20% glycerol for subsequent use in either standardizing Par1b levels by Western blot or for in vitro kinase assays. For in vitro kinase assays, fractions of the immuno-isolated Par1b were mixed with a recombinant GST C-terminal fragment of human IRSp53 (aa 319–521) in a kinase buffer (40 mM Hepes, 5 mM MgCl, 2 mM EGTA, 0.1% Triton X-100, 2.5 uCi of ATP-[γ]-P32, and 0.5 mM ATP, with protease and phosphatase inhibitors) for 30 min at 30°C. Sample buffer was added directly to the mixture and the samples were run on a 9% polyacrylamide gel that was stained with Coomassie blue, dried, and exposed to a phosphor screen. The phosphor screen and polyacrylamide gel were imaged on a molecular imager (FLA-9000; Fujifilm). Quantitation of the images was done using MultiGauge software (Fujifilm). For quantitative immunoblotting, nitrocellulose blots were probed with primary antibodies and with Dylight 680–conjugated secondary antibodies, and fluorescence was scanned using a molecular imager (FLA-9000; Fujifilm) and quantified using MultiGauge software.
Immunofluorescence and confocal microscopy
For immunofluorescence experiments, cells were fixed in 4% paraformaldehyde (PFA) in PBS buffer for 20 min at room temperature or 4°C (PKCζ and Par1b antibodies). Cells were permeabilized in 0.2% Triton X-100 for 10 min before antibodies incubation. For RhoA staining, cells were fixed in 10% trichloroacetic acid in distilled water for 15 min on ice (Yonemura et al., 2004). Laminin, collagen IV, and β1 integrin antibodies were incubated on living cells at 4°C for 1 h on both sides of the polycarbonate Transwells before fixation in PFA. F(ab’)2 fragments secondary antibodies coupled to DyLight 488, Rhodamine Red-X, or DyLight 649 were used. Chromatin was stained with DAPI or DRAQ5. Microscopy was performed in a confocal microscope (TCS SP5; Leica) equipped with a motorized x-y-z stage for multiple position finding and with an 8,000-Hz resonant scanner. Fixed cells were imaged using an HCX PL APO 63×/1.4–0.60 NA oil λBL CS objective on glass coverslips mounted in nonhardening, glycerol-based aqueous mounting medium to avoid sample compression artifacts. Confocal (pinhole, 1 AU; pixel size, 80.02 nm) x-y-z or x-z-y stacks were taken from the monolayer. Live-cell imaging was performed using an HCX PL APO 40×/1.25–0.75 NA oil CS objective on MatTek or CELLview chambers at 37°C in a CO2-enriched atmosphere in growth medium without phenol red. x-y-z-t stack frames (pinhole, 2–3 AU; pixel size, 100.1–252.8 nm) were recorded. Image x-y-z and x-y-z-t stacks were processed with LAS AF 2.3.1 and ImageJ 1.46 software (National Institutes of Health, Bethesda, MD). For 3D modeling, x-y-z stacks of 30–40 µm (2-µm/section) were processed in IMOD 4.3.7 software (Boulder Laboratory for 3-Dimensional Electron Microscopy of Cells, Boulder, CO).
FRET analysis
Föster resonance energy transfer (FRET) of RhoA ECFP/citrine-YFP was measured in stable cell lines expressing the biosensor constitutively (HepG2) or under a doxycycline-dependent promoter (MDCK). FRET efficiency was calculated with the acceptor-bleaching FRET module (FRET-AB) of the LAS AF 2.3.1 software. For the setup an HCX PL APO 40×/1.25–0.75 NA oil CS objective was used to obtain pre- and post-bleaching confocal (pinhole, 1 AU; pixel size, 75.8 nm; line average: 4) x-y-z sections. For the bleaching, ROIs at the cell cortex (1–3) and in the lateral lumen (4; see Fig. 8 C) were exposed to the 488 argon laser (main power; 50% with 514-nm laser line; 100%, for 30 passes). The efficiency of FRET was evaluated only in ROIs with at least 75% of reduced fluorescence intensity in the acceptor species.
Quantification and statistical analysis
The α, β, and γ angles were estimated with the ImageJ angle tool in x-z-y confocal sections. Alternatively, the β angle was measured relative to the x-y-z sections by tag β = a/b, where “a” and “b” are the distances between the two centrosomes in z and x-y maximal projection, respectively. The ratio of BC/nuclei and the percentage of mitotic cells with BC were obtained from 10–20 random x-y fields of ∼150 µm2. The distribution of the ECM proteins or the integrin β1 in 2D cultures was measured as the mean fluorescence intensity ratio (a/b) between the apex (a) and base (b) of the monolayer ROIs from 10–20 random x-z maximum projections of 10 confocal sections from 10-µm stacks using ImageJ. The F-actin or the RhoA and the P-MLC2 fluorescence were measured as the thickness or the mean intensity of ROIs, respectively, at lateral lumen-free regions of the cell cortex from x-y confocal sections using ImageJ. For statistical computation and estimation of significance, we used Prism version 6.00 (GraphPad Software) for Windows. Graphs represent mean (y) + SD or ± SEM, as indicated, where (n) represents the number of cells analyzed. Statistical significance was determined using unpaired t test for three independent experiments.
Online supplemental material
Fig. S1 shows that Par1b depletion in WIF-B9 and HepG2 cells inhibits lateral lumen polarity and promotes spindle alignment with the basal domain. Fig. S2 shows that Par1b overexpression in MDCK cells inhibits and Par1b depletion in WIF-B9 cells promotes focal adhesions and cortical F-actin. Fig. S3 shows that collagen IV promotes columnar polarity in HepG2 and WIF-B9 cells. Fig. S4 shows that NuMA is excluded from the lateral lumina of HepG2 cells, whereas Gαi is present at the lateral and luminal domains in MDCK-Par1b and HepG2 cells. Fig. S5 shows that the global Par1b activity in HepG2 and WIF-B9 cells is lower than that in MDCK cells while Par1b localization during mitosis is similar in the three cell lines. Video 1 shows that Par1b depletion in WIF-B9 inhibits lateral lumen polarity. Video 2 shows the metaphase plate orientation with respect to the lumen in WIF-B9 cells growing in cyst-like structures generated in Matrigel or collagen I, predicting symmetric or asymmetric cell divisions, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201303013/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201303013.dv.
Supplementary Material
Acknowledgments
The authors are grateful to Q. Du and D. Cleveland for their gifts of LGN and NuMA antibodies, respectively.
This work was supported by RO1 NIDDK R01KD064842-07 to A. Müsch and NIH GM093121 to L. Hodgson.
Footnotes
Abbreviations used in this paper:
- AD-O
- apical domain oriented
- BC
- bile canalicular
- BD-O
- basal domain oriented
- LGN
- leucine-glycine-asparagine repeat protein
- NuMA
- nuclear mitotic apparatus protein
- ROI
- region of interest
References
- Bartles J.R., Hubbard A.L. 1986. Preservation of hepatocyte plasma membrane domains during cell division in situ in regenerating rat liver. Dev. Biol. 118:286–295 10.1016/0012-1606(86)90095-3 [DOI] [PubMed] [Google Scholar]
- Böhm H., Brinkmann V., Drab M., Henske A., Kurzchalia T.V. 1997. Mammalian homologues of C. elegans PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Curr. Biol. 7:603–606 10.1016/S0960-9822(06)00260-0 [DOI] [PubMed] [Google Scholar]
- Busson S., Dujardin D., Moreau A., Dompierre J., De Mey J.R. 1998. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 8:541–544 10.1016/S0960-9822(98)70208-8 [DOI] [PubMed] [Google Scholar]
- Chikumi H., Vázquez-Prado J., Servitja J.M., Miyazaki H., Gutkind J.S. 2002. Potent activation of RhoA by Galpha q and Gq-coupled receptors. J. Biol. Chem. 277:27130–27134 10.1074/jbc.M204715200 [DOI] [PubMed] [Google Scholar]
- Citi S., Pulimeno P., Paschoud S. 2012. Cingulin, paracingulin, and PLEKHA7: signaling and cytoskeletal adaptors at the apical junctional complex. Ann. N. Y. Acad. Sci. 1257:125–132 10.1111/j.1749-6632.2012.06506.x [DOI] [PubMed] [Google Scholar]
- Cohen D., Brennwald P.J., Rodriguez-Boulan E., Müsch A. 2004. Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J. Cell Biol. 164:717–727 10.1083/jcb.200308104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Tian Y., Musch A. 2007. Par1b promotes hepatic-type lumen polarity in Madin Darby canine kidney cells via myosin II- and E-cadherin-dependent signaling. Mol. Biol. Cell. 18:2203–2215 10.1091/mbc.E07-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Fernandez D., Lázaro-Diéguez F., Müsch A. 2011. The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling. J. Cell Biol. 192:525–540 10.1083/jcb.201007002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Elzen N., Buttery C.V., Maddugoda M.P., Ren G., Yap A.S. 2009. Cadherin adhesion receptors orient the mitotic spindle during symmetric cell division in mammalian epithelia. Mol. Biol. Cell. 20:3740–3750 10.1091/mbc.E09-01-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert M., Cohen D., Müsch A. 2006. PAR1b promotes cell-cell adhesion and inhibits dishevelled-mediated transformation of Madin-Darby canine kidney cells. Mol. Biol. Cell. 17:3345–3355 10.1091/mbc.E06-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Miñán A., Martín-Bermudo M.D., González-Reyes A. 2007. Integrin signaling regulates spindle orientation in Drosophila to preserve the follicular-epithelium monolayer. Curr. Biol. 17:683–688 10.1016/j.cub.2007.02.052 [DOI] [PubMed] [Google Scholar]
- Grill S.W., Hyman A.A. 2005. Spindle positioning by cortical pulling forces. Dev. Cell. 8:461–465 10.1016/j.devcel.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K.J. 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 81:611–620 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Hao Y., Du Q., Chen X., Zheng Z., Balsbaugh J.L., Maitra S., Shabanowitz J., Hunt D.F., Macara I.G. 2010. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. 20:1809–1818 10.1016/j.cub.2010.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng Y.W., Lim H.H., Mina T., Utomo P., Zhong S., Lim C.T., Koh C.G. 2012. TPPP acts downstream of RhoA-ROCK-LIMK2 to regulate astral microtubule organization and spindle orientation. J. Cell Sci. 125:1579–1590 10.1242/jcs.096818 [DOI] [PubMed] [Google Scholar]
- Herrema H., Czajkowska D., Théard D., van der Wouden J.M., Kalicharan D., Zolghadr B., Hoekstra D., van Ijzendoorn S.C. 2006. Rho kinase, myosin-II, and p42/44 MAPK control extracellular matrix-mediated apical bile canalicular lumen morphogenesis in HepG2 cells. Mol. Biol. Cell. 17:3291–3303 10.1091/mbc.E06-01-0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrke G., Neufeld E.B., Meads T., Shanks M.R., Cassio D., Laurent M., Schroer T.A., Pagano R.E., Hubbard A.L. 1993. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J. Cell Biol. 123:1761–1775 10.1083/jcb.123.6.1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.B., Kaji N., Durgan J., Hall A. 2008. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 183:625–633 10.1083/jcb.200807121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou T.S., Nelson W.J. 1998. Effects of regulated expression of mutant RhoA and Rac1 small GTPases on the development of epithelial (MDCK) cell polarity. J. Cell Biol. 142:85–100 10.1083/jcb.142.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji N., Muramoto A., Mizuno K. 2008. LIM kinase-mediated cofilin phosphorylation during mitosis is required for precise spindle positioning. J. Biol. Chem. 283:4983–4992 10.1074/jbc.M708644200 [DOI] [PubMed] [Google Scholar]
- Kaushik R., Yu F., Chia W., Yang X., Bahri S. 2003. Subcellular localization of LGN during mitosis: evidence for its cortical localization in mitotic cell culture systems and its requirement for normal cell cycle progression. Mol. Biol. Cell. 14:3144–3155 10.1091/mbc.E03-04-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Cheeseman I.M. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C., Gönczy P. 2012. Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199:97–110 10.1083/jcb.201203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Baum B. 2009. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 19:174–179 10.1016/j.tcb.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Maddox A.S., Burridge K. 2003. RhoA is required for cortical retraction and rigidity during mitotic cell rounding. J. Cell Biol. 160:255–265 10.1083/jcb.200207130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez A., Amenta P.S. 1993. The hepatic extracellular matrix. I. Components and distribution in normal liver. Virchows Arch. A Pathol. Anat. Histopathol. 423:1–11 10.1007/BF01606425 [DOI] [PubMed] [Google Scholar]
- Masuda-Hirata M., Suzuki A., Amano Y., Yamashita K., Ide M., Yamanaka T., Sakai M., Imamura M., Ohno S. 2009. Intracellular polarity protein PAR-1 regulates extracellular laminin assembly by regulating the dystroglycan complex. Genes Cells. 14:835–850 10.1111/j.1365-2443.2009.01315.x [DOI] [PubMed] [Google Scholar]
- O’Brien L.E., Zegers M.M., Mostov K.E. 2002. Opinion: Building epithelial architecture: insights from three-dimensional culture models. Nat. Rev. Mol. Cell Biol. 3:531–537 10.1038/nrm859 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., Hahn K.M. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Peyre E., Jaouen F., Saadaoui M., Haren L., Merdes A., Durbec P., Morin X. 2011. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J. Cell Biol. 193:141–154 10.1083/jcb.201101039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Meisen W.H., Hao Y., Macara I.G. 2010. Tuba, a Cdc42 GEF, is required for polarized spindle orientation during epithelial cyst formation. J. Cell Biol. 189:661–669 10.1083/jcb.201002097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsch S., Karsenti E. 1994. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J. Cell Biol. 126:1509–1526 10.1083/jcb.126.6.1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli A.E., Vergarajauregui S., Eastburn D.J., Datta A., Alonso M.A., Mostov K., Martín-Belmonte F. 2010. The Cdc42 GEF Intersectin 2 controls mitotic spindle orientation to form the lumen during epithelial morphogenesis. J. Cell Biol. 189:725–738 10.1083/jcb.201002047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadat I., Higashi H., Obuse C., Umeda M., Murata-Kamiya N., Saito Y., Lu H., Ohnishi N., Azuma T., Suzuki A., et al. 2007. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 447:330–333 10.1038/nature05765 [DOI] [PubMed] [Google Scholar]
- Schaar B.T., Kinoshita K., McConnell S.K. 2004. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 41:203–213 10.1016/S0896-6273(03)00843-2 [DOI] [PubMed] [Google Scholar]
- Stamatoglou S.C., Enrich C., Manson M.M., Hughes R.C. 1992. Temporal changes in the expression and distribution of adhesion molecules during liver development and regeneration. J. Cell Biol. 116:1507–1515 10.1083/jcb.116.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli C.H. 2009. Integrins and cell-fate determination. J. Cell Sci. 122:171–177 10.1242/jcs.018945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Hirata M., Kamimura K., Maniwa R., Yamanaka T., Mizuno K., Kishikawa M., Hirose H., Amano Y., Izumi N., et al. 2004. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14:1425–1435 10.1016/j.cub.2004.08.021 [DOI] [PubMed] [Google Scholar]
- Tabler J.M., Yamanaka H., Green J.B. 2010. PAR-1 promotes primary neurogenesis and asymmetric cell divisions via control of spindle orientation. Development. 137:2501–2505 10.1242/dev.049833 [DOI] [PubMed] [Google Scholar]
- Terry S.J., Zihni C., Elbediwy A., Vitiello E., Leefa Chong San I.V., Balda M.S., Matter K. 2011. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat. Cell Biol. 13:159–166 10.1038/ncb2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Bornens M. 2008. Get round and stiff for mitosis. HFSP J. 2:65–71 10.2976/1.2895661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Racine V., Pépin A., Piel M., Chen Y., Sibarita J.B., Bornens M. 2005. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 7:947–953 10.1038/ncb1307 [DOI] [PubMed] [Google Scholar]
- Théry M., Jiménez-Dalmaroni A., Racine V., Bornens M., Jülicher F. 2007. Experimental and theoretical study of mitotic spindle orientation. Nature. 447:493–496 10.1038/nature05786 [DOI] [PubMed] [Google Scholar]
- Toyoshima F., Nishida E. 2007. Spindle orientation in animal cell mitosis: roles of integrin in the control of spindle axis. J. Cell. Physiol. 213:407–411 10.1002/jcp.21227 [DOI] [PubMed] [Google Scholar]
- Toyoshima F., Matsumura S., Morimoto H., Mitsushima M., Nishida E. 2007. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev. Cell. 13:796–811 10.1016/j.devcel.2007.10.014 [DOI] [PubMed] [Google Scholar]
- van IJzendoorn S.C., Zegers M.M., Kok J.W., Hoekstra D. 1997. Segregation of glucosylceramide and sphingomyelin occurs in the apical to basolateral transcytotic route in HepG2 cells. J. Cell Biol. 137:347–357 10.1083/jcb.137.2.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama Y., Miura K., Sabe H., Mochizuki N. 2011. EphrinA1-EphA2 signal induces compaction and polarization of Madin-Darby canine kidney cells by inactivating Ezrin through negative regulation of RhoA. J. Biol. Chem. 286:44243–44253 10.1074/jbc.M111.267047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werb Z., Tremble P.M., Behrendtsen O., Crowley E., Damsky C.H. 1989. Signal transduction through the fibronectin receptor induces collagenase and stromelysin gene expression. J. Cell Biol. 109:877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard G.E., Huang N.N., Cho H., Miki T., Tall G.G., Kehrl J.H. 2010. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30:3519–3530 10.1128/MCB.00394-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.C., Rose L.S. 2007. PAR-3 and PAR-1 inhibit LET-99 localization to generate a cortical band important for spindle positioning in Caenorhabditis elegans embryos. Mol. Biol. Cell. 18:4470–4482 10.1091/mbc.E07-02-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Hirao-Minakuchi K., Nishimura Y. 2004. Rho localization in cells and tissues. Exp. Cell Res. 295:300–314 10.1016/j.yexcr.2004.01.005 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Zhu H., Wan Q., Liu J., Xiao Z., Siderovski D.P., Du Q. 2010. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J. Cell Biol. 189:275–288 10.1083/jcb.200910021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.