The presence of members of an NPC subcomplex containing Nsp1p is required for NPC movement into a daughter yeast cell, allowing intact NPCs to bypass a filter that prevents movement of defective complexes.
Abstract
In the yeast Saccharomyces cerevisiae, organelles and macromolecular complexes are delivered from the mother to the emerging daughter during cell division, thereby ensuring progeny viability. Here, we have shown that during mitosis nuclear pore complexes (NPCs) in the mother nucleus are actively delivered through the bud neck and into the daughter cell concomitantly with the nuclear envelope. Furthermore, we show that NPC movement into the daughter cell requires members of an NPC subcomplex containing Nsp1p and its interacting partners. NPCs lacking these nucleoporins (Nups) were blocked from entry into the daughter by a putative barrier at the bud neck. This selection process could be observed within individual cells such that NPCs containing Nup82p (an Nsp1p-interacting Nup) were transferred to the daughter cells while functionally compromised NPCs lacking Nup82p were retained in the mother. This mechanism is proposed to facilitate the inheritance of functional NPCs by daughter cells.
Introduction
Molecular exchange across this nuclear envelope (NE) occurs through macromolecular assemblies termed nuclear pore complexes (NPCs). Yeast NPCs (∼60 MD) are composed of ∼30 distinct proteins termed nucleoporins (Nups; Aitchison and Rout, 2012), many of which are organized into subcomplexes (see Alber et al., 2007). These subcomplexes, which are present in multiple copies within each NPC, contribute to the distinct eightfold symmetry of the NPC core scaffold. Attached to the core are cytoplasmic filaments and a nuclear basket structure. Transport of most soluble proteins, protein complexes, and ribonucleoprotein particles across the NE occurs through the central channel of the NPC core. This channel is lined with Nups rich in FG repeats, which facilitate the movement of transport factors and their bound cargos through the NPC (Hoelz et al., 2011).
Once formed, yeast NPCs are assumed to remain intact throughout the life of the cell, and it is thought that at least a portion of those NPCs present in the mother cell are inherited by their daughters. Molecular inheritance in yeast has been widely studied, and various mechanisms facilitate the movement of cellular components between mother and daughter cells as the nascent bud emerges and grows throughout the cell cycle. Small molecules and most soluble proteins appear to passively diffuse from the mother to the daughter; however, the transmission of most organelles, including the cortical ER (Estrada et al., 2003), mitochondria (Itoh et al., 2004; Förtsch et al., 2011), vacuoles (Ishikawa et al., 2003), and peroxisomes (Fagarasanu et al., 2006), requires a facilitated process that uses microtubule-based motors or the actin/myosin network to distribute the organelle between the mother and daughter cells.
In many organisms, including Saccharomyces cerevisiae, cells undergo a closed mitosis, meaning the NE, including NPCs, remains intact and surrounds the separating daughter chromosomes until NE membrane fusion and fission events produce distinct mother and daughter nuclei late in M phase. These events are thought to produce two nuclei with a similar complement of NPCs. The processes that lead to the equal distribution of NPCs in mother and daughter nuclei, however, have been unclear, and distinctly different mechanisms have been proposed. It has been proposed that NPC movement along the NE and through the bud neck is too slow to account for the accumulation of daughter-specific NPCs (Shcheprova et al., 2008; Boettcher et al., 2012), and a model was put forward in which mother-specific NPCs are excluded from the daughter during anaphase and those populating the daughter nucleus are assembled de novo (Shcheprova et al., 2008). In contrast, a second study disputes this conclusion and provides evidence that NPCs from the mother are distributed to the daughter during anaphase (Khmelinskii et al., 2010).
In this study, we show that NPC segregation between mother and daughter is regulated during mitosis by the functional competence of each individual NPC. We present data that supports a model in which the accumulation of NPCs in the daughter nucleus during anaphase stems from their active delivery from the mother cell NE rather than by diffusion along the NE. Moreover, we show that transmission of existing NPCs from the mother to the daughter NE requires members of an NPC subcomplex containing Nsp1p. Specifically, we show that NPCs lacking members of the Nsp1p subcomplex are blocked at the bud neck as the NE enters the daughter cell while NPCs containing these Nups are allowed passage. Thus, NPC inheritance is shown to be a regulated process, capable of assessing the functional state of NPCs and ensuring that the emerging daughter receives a complement of functional NPCs required for its robust viability.
Results and discussion
Movement of NPCs from mother to daughter is an active process
We have investigated the movement of NPCs along the NE as the mother and daughter nuclei are formed during mitosis. Fluorescent protein-tagged Nup188p, an NPC core component, and Sur4p membrane protein (Kohlwein et al., 2001) were used to track the movements of NPCs and membrane proteins along the NE as the metaphase/anaphase nucleus passes through the bud neck and into the daughter cell. Imaging of Nup188-GFP and Sur4-mCherry within the NE revealed that both proteins accumulated in the daughter cell with similar kinetics, reaching a plateau ∼15 min after initial entry (Fig. 1, A and B). To assess the mechanism for the movement of these proteins into the emerging daughter cell NE, we examined their membrane diffusion characteristics using FRAP analysis. Cells in early anaphase were identified and NE regions extending into the daughter cell were photobleached. Signal recovery was monitored by collecting a series of z-stack images every 20 s. Sur4-GFP showed rapid recovery of the daughter-specific signal (Fig. 1, C and D; and Video 1), which is consistent with its rapid diffusion along the NE from the mother, through the bud neck, and into the daughter. In contrast, after bleaching of Nup188-GFP in the daughter NE, we observed little signal recovery over the time course of the experiment (Fig. 1, C and D). The minimal signal recovery detected likely arose by further movement of unbleached regions of NE into the daughter cells as they progressed into late anaphase, as little or no recovery was observed when the daughter NE was bleached in late anaphase (unpublished data). Furthermore, when both the mother and daughter cells were bleached no signal recovery was detected within the time course examined, which implies that GFP folded after bleaching is unlikely to account for the Nup188-GFP signal detected in the bleached regions (Fig. 1 C).
Figure 1.
NPCs are actively delivered to the daughter NE during mitosis. (A) Anaphase progression of cells producing Nup188-GFP and Sur4-mCherry was followed using epifluorescence microscopy. Images were acquired at 4-min intervals. 0 min was defined as the moment the fluorescing protein entered the daughter. Bar, 5 µm. (B) The daughter-specific signal intensity of the indicated fluorescent protein was quantified at each time point (n = 13). The fluorescence intensities within the daughter cell during anaphase were normalized as follows: signal levels in the daughter cell before entry of the NE (t < 0 min) were set at 0, and those after entry into telophase (t ≥ 20 min) at 1. Error bars represent standard deviation. (C) Anaphase cells producing Nup188-GFP or Sur4-GFP were analyzed by FRAP. Daughter (D) or both daughter and mother (D+M) cell signals were bleached and fluorescence recovery was monitored using confocal microscopy at 20-s intervals. Each image was derived from a summed projection of a z-stack series. Bar, 5 µm. (D) Plots of integrated fluorescent intensities of Nup188-GFP and Sur4-GFP in the daughter cell (n = 8–10) at various time points after photobleaching. Minimal Nup188-GFP recovery gave a straight-line plot (circles), whereas Sur4-GFP recovered with relaxation kinetics (squares) and a rate constant of 1.09 ± 0.03 × 10−2 s−1. Error bars represent standard deviation.
The lack of Nup188-GFP signal recovery in the daughter NE is consistent with a model in which minimal NPC diffusion occurs along the mitotic NE. In line with this conclusion, FRAP analyses of regions of the mother NEs in anaphase cells also showed little diffusion of Nup188-GFP (Fig. S1 A). Similarly, results were also obtained when interphase NEs were analyzed (Fig. S1, B and C; and Fig. S2 D). Together with previous reports (Khmelinskii et al., 2010), our results lead us to conclude that the movement of NPCs from the mother cell into the daughter cell NE did not occur by diffusion but rather is an active process.
An Nsp1p subcomplex is required for NPC delivery to the daughter cell
To explore the molecular basis for NPC movement from mother to daughter cell, we have investigated whether distinct structural features of the NPCs were required. For these experiments, the transfer of NPCs from mother to daughter cell during mitosis was monitored in nup mutant strains. Viable nup null strains lacking NUP170, NUP157, NUP60, MLP1, MLP2, POM34, or POM152 were examined, and several essential NUP genes, including NSP1, NUP82, NUP57, and NUP192, or a double mutant lacking the paralogs NUP170 and NUP157, were assessed by using a regulatable MET3 promoter. These Nups are present at multiple positions within the NPC. Mutants lacking each of the nonessential Nups revealed no change in the inheritance of NPCs, as revealed using Nup188-GFP or Ndc1-GFP. Similarly, depletion of Nup192p or Nup170p in the absence of Nup157p (PMET3-NUP170 nup157Δ), although required for NPC assembly, showed no alteration in NPC movement from mother to daughter cell (unpublished data and Fig. S1 C). Strikingly, however, after depletion of Nsp1p (PMET3−NSP1; +Met), NPCs were retained in the mother NE and largely absent from the daughter NE as the cell progressed through mitosis (Figs. 2 and S1 D). Quantification of Nup188-GFP fluorescence intensity in Nsp1p-depleted telophase cells showed a daughter-to-mother ratio (IDaughter/IMother) of ∼0.25 (Fig. 2 C; +Met 4 h) as compared with ∼0.65 for cells expressing NSP1. In contrast, Sur4-mCherry entered the daughter cells in a time frame similar to wild-type (WT) cells as assayed by time-lapse microscopy or FRAP analysis (Fig. 2 B and not depicted). Like Nsp1p, depletion of Nup57p or Nup82p, two Nups that physically interact with Nsp1p, also exhibited a reduced Nup188-GFP IDaughter/IMother ratio, albeit to a lesser degree (∼0.4; Fig. S1 E). These observations lead us to conclude that the Nsp1p subcomplex plays an essential role in NPC inheritance. Consistent with this conclusion, Nsp1p depletion leads to the mislocalization and increased cytoplasmic levels of complex partners such as Nup49p and Nup82p, but not other Nups examined, including Nup188p, Nup60p, and Ndc1p (Fig. 2 D).
Figure 2.
Depletion of Nsp1p causes asymmetric NPC segregation. (A) PMET3-HA-NSP1 cells were incubated with methionine for the indicated times to repress NSP1 expression. Cell lysates were analyzed by Western blotting using anti-HA and anti-Gsp1p (load control) antibodies. (B) PMET3-NSP1 NUP188-GFP SUR4-mCherry cells were grown in the absence (−Met) or presence (+Met, 3 h) of methionine. Early anaphase cells were identified, and images were acquired every 2 min as they progressed through mitosis using an epifluorescence microscope. Arrowheads indicate the position of the bud neck. Arrows highlight accumulated Nup188-GFP signal close to the bud neck in Nsp1p-depleted cells. Bar, 5 µm. (C) Cells expressing NUP188-GFP in WT and the indicated PMET3-NUP backgrounds were grown as described in A. Before (+Met 0 h) and after (+Met 4 h) Nup depletion, telophase cells (n = 23–33 per condition) were imaged using a confocal microscope, and the daughter-to-mother ratio (Idaughter/Imother) of the Nup188-GFP signal was determined. Error bars express standard deviation. (D) Cells expressing the indicated Nup-GFP fusions in the PMET3-NSP1 background were analyzed by epifluorescence microscopy before (−Met) and after depletion of Nsp1p (+Met, 6 h). Arrowheads identify mother (blue) and daughter (red) nuclei. Bar, 5 µm.
NPCs distribution in NSP1-depleted cells was also examined by electron microscopy. Thin sections of cells showing a defined NE were examined (Fig. 3). The number of NPCs per micrometer of NE was determined for multiple sections, and the distribution of NPCs per micrometer per section were graphed as a percentile rank plot. PMET3-NSP1 and a control strain, PMET3-NUP170 nup157Δ, were examined. Depletion of Nup170p in the nup157Δ background inhibits NPC assembly, but existing NPCs distribute equally between mother and daughter NEs, leading to twofold decreases in NPC numbers with each cell division (Makio et al., 2009; Fig. 3 B). Thus, the overall number of NPCs observed per NE section decreases after Nup170p depletion, and variations in NPC densities between sections conform to a Poisson distribution before and after Nup170p depletion. In contrast, while cells producing Nsp1p showed linear NPC densities approximating a Poisson distribution, the distributions of NPC densities among sections of cells depleted of Nsp1p (4 h) revealed increased numbers of sections containing relatively higher (>0.6 NPCs/µm) and lower (<0.2 NPCs/µm) NPC densities (Fig. 3 B). These data are consistent with our conclusion that Nsp1p depletion leads to an asymmetric distribution of NPCs between mother and daughter cells.
Figure 3.
Nsp1p depletion creates two cell populations with distinct NPC linear densities. (A) Shown is an electron micrograph of PMET3-NSP1 cells after depletion of Nsp1p (+Met 4 h). The positions of NPCs are visible as gaps (≤100 nm across) within the NE (arrowheads). Bar, 2 µm. (B) PMET3-NSP1 cells and PMET3-NUP170 nup157Δ cells were incubated with methionine for the indicated times and subjected to the EM analysis. Transmission EM images containing sections through the nucleus were selected and the linear density of NPCs for each nuclear section was calculated (NPCs per micrometer of the NE). For each time point, the number of NPCs per micrometer in individual sections (x axis) is plotted versus its percentile rank within the dataset of sections examined (n; black points). The gray line in each graph represents a cumulative Poisson distribution estimated on an assumption that NPCs were randomly distributed throughout the NE. Also plotted is the deviation of the observed NPC density (black) from the theoretical distribution for each rank (gray).
The reduced levels of NPCs seen in daughter cells upon Nsp1p depletion were also accompanied by asymmetry in the size of nuclei, with the mother nucleus appearing distinctly larger than the daughter nucleus (Figs. 2 and S2). This size difference was not caused by a detectable defect in chromosome segregation. FACS analysis revealed that Nsp1p depletion did not alter 1C and 2C DNA content (Fig. S2 B), and bulk chromatin, as visualized with mCherry-tagged histone H2B, appeared to segregate normally (Fig. S2, A and C).
NPCs lacking components of the Nsp1p complex are selectively retained in mother cells
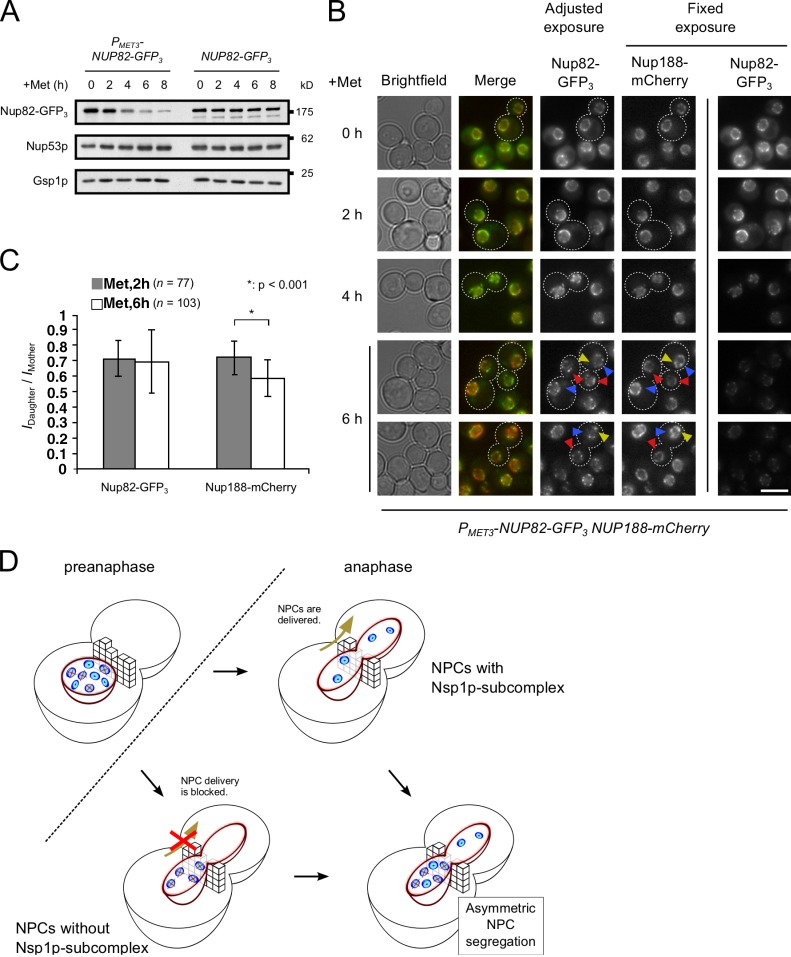
NPCs depleted of Nsp1p, while showing slow diffusion characteristics similar to WT NPCs (Fig. S2 D), accumulated in mother NEs during mitosis, often appearing to concentrate where the NE extends through the bud neck and into the daughter cell (Fig. 2 B, arrows). These results imply that Nsp1p-depleted NPCs encountered a barrier at or near the bud neck that restricts their movement into the daughter NE. To further assess whether the loss of the Nsp1p subcomplex was directly affecting NPC movement into the daughter cell, we examined whether, in a single cell, NPCs containing or lacking components of the Nsp1p complex could be identified, and if NPCs containing this complex are selectively transferred to the daughter cells. For these experiments, we used the PMET3-NUP82 strain. Loss of Nup82p proceeds at a slower rate than Nsp1p depletion (compare Fig. 2 A to Fig. 4 A), allowing a larger time window for detection and analysis of NPCs that contained or lacked Nup82p. Repression of NUP82-GFP3 expression led to a decrease in NE-associated signal, and by 6 h after repression, the number of Nup82-GFP3 foci was decreased and foci were sparsely distributed along the NE (Fig. 4 B). Importantly, the Nup82-GFP3 containing NPCs appeared equally distributed between the mother and daughter nuclei. This was in contrast to the total pool of NPCs detected with Nup188-mCherry, which exhibited an asymmetric distribution pattern and a reduced Nup188-GFP IDaughter/IMother ratio (Fig. 4, B and C). Mother cells contained many NPCs visible with Nup188-mCherry but lacking a detectable Nup82-GFP3 signal. These results are consistent with our model in which NPCs containing a functional Nsp1p subcomplex can equally distribute between mother and daughter cells during mitosis while NPCs functionally compromised by the loss of this complex are detected and selectively retained in the mother cell (Fig. 4 D).
Figure 4.
Nup82p-depleted NPCs are selectively excluded from the daughter NE during mitosis. (A) Western analysis using anti-GFP, anti-Nup53p, or anti-Gsp1p (load control) antibodies of PMET3-NUP82-GFP3 and NUP82-GFP3 cell lysates after growth in methionine-containing media for the indicated times. (B) A PMET3-NUP82-GFP3 NUP188-mCherry strain was grown for the indicated times in medium containing methionine to repress NUP82-GFP3 expression. Nup82-GFP3 and Nup188-mCherry were visualized using an epifluorescence microscope. Selected telophase cells are outlined. Two sets of Nup82-GFP3 images are shown. Fixed exposure times reveal decreasing intensities of Nup82-GFP3 across the time course, whereas Nup188-mCherry levels are unaffected. Adjusted exposures allow enhanced visualization of Nup82-GFP3 foci and reveal their symmetrical distribution between mother (blue arrowheads) and daughter (red arrowheads) NEs. In contrast, Nup188-mCherry reveals greater numbers of NPCs retained in the mother, including those with no detectable Nup82-GFP3 (yellow arrowheads). Bar, 5 µm. (C) Cells grown as described in B were examined by confocal fluorescence microscopy at 2 and 6 h after methionine addition. Z stacks of telophase cells were used to determine the Idaughter/Imother signal intensity ratios of Nup82-GFP3 and Nup188-mCherry. Error bars represent standard deviation. (D) A schematic model based on data presented in B and C is shown. NPCs depleted of Nup82p and compromised for Nsp1p subcomplex function are crossed with a red X. A diffusion barrier (brick wall) is proposed to prevent these damaged NPCs from entering the daughter cell. In contrast, NPCs containing a functional Nsp1p subcomplex can traverse the barrier and enter the daughter. These two events result in asymmetric NPC segregation during mitosis.
The role of the Nsp1p subcomplex in NPC inheritance is distinct from its transport function
Mutations in members of the Nsp1p subcomplexes give rise to defects in nuclear transport (Bailer et al., 2001). Thus, we examined whether conditions that lead to asymmetric NPC distribution also altered transport. Steady-state localizations of protein reporters imported by four importins were examined. By 4 h after repression, asymmetric NPC distribution was visible; however, little or no change was detected in import mediated by the Kap95p–Kap60p complex, Kap104p, or Kap123p (Fig. 5). In contrast, Kap121p-mediated import was inhibited, as was mRNA export, beginning as early as 2 h after NSP1 repression. To explore these relationships, we examined NPC distributions in mutants with known defects in Kap121p-mediated import (kap121-34) and mRNA export (dbp5-1 and gle1-4). No detectable defects were seen in NPC inheritance under conditions that restrict the functions of these mutant alleles (Fig. 5 C). We conclude that Kap121p-mediated import and mRNA export are not required for NPC inheritance, and that the role of the Nsp1p subcomplex in the latter process is likely separate from its function in transport.
Figure 5.
Analysis of nuclear transport in Nsp1p-depleted cells. (A) PMET3-NSP1 cells expressing the indicated NLS reporters were incubated with methionine (0–4 h) to repress NSP1 expression. Reporter location was analyzed by epifluorescence microscopy. (B) Poly-A mRNA was visualized under similar conditions using FISH analysis. (C) The specified temperature-sensitive mutants were grown at room temperature and then shifted to 37°C for the indicated times. Nup188-GFP localization was analyzed by epifluorescence microscopy. Bars, 5 µm.
Barriers to NPC inheritance and quality control
Our data lead us to conclude that the movement of NPCs from mother to daughter cell during mitosis is an active process. Our analysis and previous studies (Shcheprova et al., 2008) support the existence of a barrier at the bud neck that restricts the passage of NPCs, and we have shown that the Nsp1p subcomplex is required for NPCs to traverse the bud neck (Fig. 2). The force necessary for NPCs to traverse the bud neck may arise from various sources. NPCs could “piggyback” on structural elements of the NE as it is pulled into the daughter cell or engage factors in the cytoplasm and/or nucleoplasm that facilitate their movement through the bud neck. Cytoplasmic motor proteins represented possible candidates because they appear to play a role in NPC’s mobility in the interphase NE (Steinberg et al., 2012). However, NPCs are effectively delivered to daughter cells in a variety of microtubule- and actin-based motor mutants (see Fig. S3), which suggests that cytoplasmic motors may not play a role. An alternative idea is that intranuclear factors contribute to NPC inheritance. NPCs interact with transcriptionally active and inactive chromatin (Dieppois and Stutz, 2010). Tethering NPCs to chromatin would allow the forces applied to chromatin by the spindle during anaphase to assist in transmission of NPCs to the daughter cells. Intriguingly, during anaphase in Schizosaccharomyces japonicus, NPCs are concentrated in the poles of the fusiform-shaped nucleus in close proximity to the condensed chromosomes (Aoki et al., 2011). Moreover, NPCs appear uncoupled from chromatin in Nsp1p-depleted cells, as chromosome segregation occurs normally in these cells while NPCs are retained in the mother cell (Fig. S2). Detailed testing of these hypotheses will await greater understanding of the molecular basis for the interactions of NPCs with chromatin.
The retention of Nsp1p-depleted NPCs in the mother cell during mitosis led us to hypothesize that a bud neck–associated structure or structures forms a barrier preventing compromised NPCs from entering the daughter cell. We examined the consequences of mutations in individual genes encoding various bud neck proteins as well as those proteins deemed to be potentially involved in barrier function (Fig. S3). Among those examined were mutations in BUB6 and SHS1, two genes encoding proteins proposed to restrict of the movement of integral membrane proteins through the bud neck (Shcheprova et al., 2008). However, none of the mutations appeared to affect the NPC barrier, perhaps indicating that multiple factors contribute to the barrier or that an alternative mechanism influences NPC movement.
Importantly, we have uncovered a process for selectively retaining functionally compromised NPCs in the mother and inhibiting their transmission to the daughter cell. As not all nup mutants cause NPC retention in the mother, this quality control mechanism may sense specific functional defects linked to the Nsp1p subcomplex. We propose that this mechanism is functionally analogous to that performed by other cellular systems that promote retention of toxic materials in the mother cell. For example, as yeast cells age, they accumulate deleterious molecules such as protein aggregates (Aguilaniu et al., 2003) and nonsegregating DNA elements (e.g., extrachromosomal ribosomal DNA circles; Sinclair and Guarente, 1997), and their retention in the mother protects the daughter cell from their aging events (Shcheprova et al., 2008; Liu et al., 2010). We suggest that this cellular rejuvenation process extends to NPC inheritance. Such a process was previously suggested, but it was proposed that all the mother’s NPCs are prevented from entering the daughter cell (Shcheprova et al., 2008). However, NPCs examined in this study were likely functionally compromised by GFP tagging of the Nsp1p-interacting protein Nup49p, leading to NPC retention in the mother (Chadrin et al., 2010; Fig. S1 E). Instead, we propose that NPCs compromised for Nsp1p subcomplex function are selectively retained in the mother cells, and only functional NPCs are capable of passing to the daughter cell. This phenomenon was visible in cells during Nup82p depletion where NPCs lacking visible Nup82p were retained in the mother while those containing Nup82p were distributed evenly between the mother and daughter cells (Fig. 4).
We envisage that an NPC quality-control mechanism would detect NPC damage arising from environmental stress factors and aging. For example, previous studies have shown that oxidative stress conditions cause NPC damage that leads to increased NPC permeability and reduced nucleocytoplasmic transport. Importantly, these stress conditions lead to significant reductions in cellular levels of various Nups, including Nsp1p and Nup159p (Mason et al., 2005). These results establish the Nsp1p subcomplex among the targets of the stress response. Based on our results, the cellular response to these stress-induced NPC changes is to retain these compromised NPCs in the mother cell as a means of promoting the survival of the daughter.
Materials and methods
Yeast strains, media, and plasmids
Yeast strains used in this study are shown in Table S1. NLS reporter plasmids (cNLS, the SV-40 large T antigen NLS; rg-NLS, Nab2 [200–249 aa]; rpl25-NLS, Rpl25 [1–45 aa]; and pho4-NLS, Pho4 [140–166 aa], fused with tandem GFP genes) used in this study have been previously described (Makio et al., 2009). Introducing the PMET3 promoter or coding sequences for GFP and mCherry at specific genomic loci was performed as described previously (Makio et al., 2009), using the plasmids pTM1046 (Makio et al., 2009), pFA6a-GFP-HISMX (Longtine et al., 1998), and pmCherry-NAT (a gift from R. Rachubinski, University of Alberta, Edmonton, Canada) as templates. All strains were grown in YPD (1% yeast extract, 2% Bacto Peptone, and 2% glucose) or synthetic media containing 0.17% yeast nitrogen base (without amino acids and ammonium sulfate), 0.5% ammonium sulfate, 2% glucose, and appropriate amino acid supplements. Cultures were incubated at 30°C. Repression of genes under the control of PMET3 was initiated by the addition of methionine into the medium to a final concentration of 200 µg/ml.
Western blotting
Cells were grown in synthetic minimal medium lacking methionine, and PMET3 regulated gene repression initiated by the addition of 200 µg/ml methionine. 1 ml of cells was harvested from cultures having a cell density of OD600 ∼1.0, and the resulting cell pellet was resuspended in 100 µl of SDS-PAGE sample buffer, followed by brief sonication and heat denaturation. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Proteins were detected by Western blotting using the anti-Gsp1p (Makhnevych et al., 2003), anti-Nup53p (Makhnevych et al., 2003), anti-GFP (Lusk et al., 2002), and anti-HA (F7; Santa Cruz Biotechnology, Inc.) antibodies.
Fluorescence microscopy
The locations of GFP and mCherry fusion proteins in live cells were visualized with either epifluorescence or confocal microscopy. Epifluorescence images were obtained using a microscope (Axiovert AX10) equipped with a Plan-Apochromat 100×/1.40 NA oil immersion objective lens and a charge coupled device (CCD) camera (AxioCam MRm), controlled by AxioVision software (all from Carl Zeiss). Confocal images were visualized using a microscope (Axiovert 200M) equipped with a confocal scanning system (LSM 510 META) and a Plan-Apochromat 63×/1.4 NA oil objective lens (all from Carl Zeiss), controlled by Zen software. Images were acquired at room temperature.
Cells, grown in liquid culture, were placed directly on a slide, or on a slide with an agarose pad (1.8% agarose containing supplemented minimal medium) for time-lapse experiments, covered with a coverslip, and examined using the indicated microscopy technique.
FRAP experiments with z-stack acquisition were performed using confocal fluorescence microscopy. Initially, fluorescing fusion proteins in early anaphase cells were bleached either in the daughter only or in both daughter and mother, using 20 iterations of full intensity 488 nm laser light. Fluorescence recovery was then followed by acquiring a z stack (30 sections spaced 0.16 µm apart with a scanning speed of 1.25 pixels/µs, 3.7 s total acquisition time) every 5, 10, or 20 s as indicated. Images from each time point (t) were quantified using ImageJ software to determine: the integral signal intensity within the daughter (N), the integral signal intensity within a neighboring cell as a control (C), and the integral signal intensity of the corresponding background (BN and BC, respectively). Fluorescence recovery (I) was then calculated as I(t) = (N(t) − BN(t))/(C(t) − BC(t)). The amount of GFP fusion protein having diffused from mother to daughter, at each time point, was estimated by calculating IDiffusion(t) = ID(t) − ID+M(t). These values were then plotted and subjected to a nonlinear least square fitting with first-order kinetics: IDiffusion(t) = Ae−Bt + C, where A, B, and C were fitting parameters used by Gnuplot software.
FRAP experiments with single focal plane acquisition were performed using a confocal microscope. Bleaching of fluorescent fusion proteins in the regions of interest was accomplished by 20 iterations of full intensity 488 nm laser light. Fluorescence recovery was followed by acquiring images every 250 ms. Images from each time point (t) were quantified using ImageJ software as described in the previous paragraph.
For quantification of GFP- and mCherry-tagged Nup and histone segregation between daughter and mother during mitosis, 3D confocal fluorescence microscopy was used. Images of telophase cells were acquired as a z stack using a 63× objective lens and a piezoelectric actuator that allows for rapid image collection (Fagarasanu et al., 2005). Stacks of 30 optical sections spaced 0.16 µm apart with a scanning speed of 0.625 pixels/µs (7.5 s of total acquisition time) were collected. The integral signal intensities in the daughter and the mother sides of the dividing cell (SD and SM, respectively) and the corresponding background signals (BD and BM, respectively) were quantified using ImageJ software. The ratio of signal intensities (IDaughter/IMother) was calculated as: IDaughter/IMother = (SD − BD)/(SM − BM).
FISH analysis was performed as described previously (Gorsch et al., 1995; Makio et al., 2009). Cells were fixed with 4% formaldehyde in 0.1 M potassium phosphate, pH 6.5, and cell walls were removed by treatment with zymolyase 100T (ICN) in 1.2 M sorbitol. The samples were adhered to the Teflon-coated microscope slides (Cel-Line; Thermo Fisher Scientific) pretreated with 0.1% polylysine solution (Sigma-Aldrich). Cells were permeabilized with 0.1% IGEPAL CA-630 in 0.1 M potassium phosphate, pH 6.5, and then incubated with prehybridization solution (50% formamide, 4× SSC, 0.02% polyvinyl pyrrolidone, 0.02% bovine serum albumin, 0.02% Ficoll-400, 125 µg/ml tRNA, and 500 µg/ml denatured salmon testes DNA) for 1 h at 37°C. The samples were probed by hybridization with 500 ng/ml Texas red–conjugated oligo dT50 in the prehybridization buffer for 16 h at 37°C. After serial washes with 2×SSC, 1×SSC, and 0.5×SSC, the slides were mounted in DAPI-Fluoromount G (Southern Biotech) and covered by coverslips. The samples were examined using an epifluorescence microscope.
Electron microscopy
Transmission EM analysis of yeast cells, after potassium permanganate fixation, was performed as described previously (Makio et al., 2009). Cells were grown in synthetic minimal media lacking methionine, followed by the addition of 200 µg/ml methionine, and incubated for the indicated times. For fixation, cells were suspended in buffer (2% glutaraldehyde, 2% formaldehyde, and 100 mM sodium phosphate, pH 6.5) and incubated on ice for 1 h. The samples were then successively treated with 3% potassium permanganate for 30 min and 1% sodium periodate for 15 min, followed by 1% ammonium chloride for 10 min, all at room temperature. Samples were then dehydrated with a series of ethanol washes (50%, 70%, 95%, and 100% vol/vol) and two propylene oxide washes, and then embedded in epon. Thin sections were placed on copper grids and examined using an electron microscope (410; Phillips). The number of NPCs and the length/circumference of the NE in each slice were determined manually using ImageJ software.
Theoretical estimates of random NPC distribution (see Fig. 3) were determined as follows. Cumulative probability distribution of random events is well represented by a cumulative Poisson distribution function (CPD). Given the number of occurrences k and the expected number of events Λ, the cumulative probability distribution function P of a stochastic variable X is represented by:
where floor(k) gives the largest integral number equal to or less than the value k and Poisson(i; Λ) represents a Poisson distribution function of the number of occurrences i with the expected number of events Λ. Given the assumption that nuclear pores are distributed randomly and λ is the mean pore density over all nuclear slices, the cumulative distribution of NPC density (x) for each slice, F(x; λ), is estimated as:
where y denotes the normalized rank between 0 and 1. Because 0.1 (NPCs/µm) was used as a unit of interval for the analysis, the variables and the parameters were scaled accordingly, such as k = 10x and Λ = 10λ.
From the definition in the previous paragraph, the function F(x; λ) represents a step function of x. In Fig. 3 B, vertical lines are introduced to connect between the vertical gaps of F(x; λ) at the units of x. These vertical lines represent the inverted relation of the function F, providing the estimated NPC density x for the given rank y. The difference between the observed NPC density and the estimated one was calculated for each rank, and plotted as deviations along the y axis.
FACS analysis
FACS analysis of the yeast strains was performed as described previously (Ptak et al., 2009). Cells were grown in synthetic minimal media lacking methionine, then incubated with 200 µg/ml methionine for the indicated times. DNA was stained with propidium iodide and detected with a FACScan flow cytometer (BD).
Online supplemental material
Fig. S1 shows that NPCs exhibit limited movement in the NE and that their transfer to daughter cells is dependent on the Nsp1p subcomplex. Fig. S2 shows the pattern of chromosome segregation upon Nsp1p depletion. Fig. S3 shows that asymmetry of NPC segregation in the absence of Nsp1p is not altered by mutants lacking bud neck–associated proteins. Table S1 is a strain list. Video 1 shows rapid recovery of Sur4-GFP signal in the daughter cell after photobleaching. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304047/DC1.
Supplementary Material
Acknowledgments
We would like to thank Dr. Christopher Ptak for critical reading of the manuscript, Honey Chan and Dr. Nasser Tahbaz for assistance in the EM experiments, and members of Wozniak laboratory for helpful discussions.
Support for this work was provided to R.W. Wozniak by the Canadian Institutes of Health Research (MOP 36519 and MOP 106502), Alberta Innovates Health Solutions, Howard Hughes Medical Institute, and the Li Ka Shing Institute of Virology at the University of Alberta. T. Makio was partially supported by a postdoctoral fellowship from the Uehara Memorial Foundation.
Footnotes
Abbreviations used in this paper:
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- WT
- wild type
References
- Aguilaniu H., Gustafsson L., Rigoulet M., Nyström T. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 299:1751–1753 10.1126/science.1080418 [DOI] [PubMed] [Google Scholar]
- Aitchison J.D., Rout M.P. 2012. The yeast nuclear pore complex and transport through it. Genetics. 190:855–883 10.1534/genetics.111.127803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007. The molecular architecture of the nuclear pore complex. Nature. 450:695–701 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Aoki K., Hayashi H., Furuya K., Sato M., Takagi T., Osumi M., Kimura A., Niki H. 2011. Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes Cells. 16:911–926 10.1111/j.1365-2443.2011.01540.x [DOI] [PubMed] [Google Scholar]
- Bailer S.M., Balduf C., Hurt E. 2001. The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport. Mol. Cell. Biol. 21:7944–7955 10.1128/MCB.21.23.7944-7955.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher B., Marquez-Lago T.T., Bayer M., Weiss E.L., Barral Y. 2012. Nuclear envelope morphology constrains diffusion and promotes asymmetric protein segregation in closed mitosis. J. Cell Biol. 197:921–937 10.1083/jcb.201112117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadrin A., Hess B., San Roman M., Gatti X., Lombard B., Loew D., Barral Y., Palancade B., Doye V. 2010. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 189:795–811 10.1083/jcb.200910043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G., Stutz F. 2010. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J. Cell Sci. 123:1989–1999 10.1242/jcs.053694 [DOI] [PubMed] [Google Scholar]
- Estrada P., Kim J., Coleman J., Walker L., Dunn B., Takizawa P., Novick P., Ferro-Novick S. 2003. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 163:1255–1266 10.1083/jcb.200304030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu M., Fagarasanu A., Tam Y.Y., Aitchison J.D., Rachubinski R.A. 2005. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 169:765–775 10.1083/jcb.200503083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A., Fagarasanu M., Eitzen G.A., Aitchison J.D., Rachubinski R.A. 2006. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell. 10:587–600 10.1016/j.devcel.2006.04.012 [DOI] [PubMed] [Google Scholar]
- Förtsch J., Hummel E., Krist M., Westermann B. 2011. The myosin-related motor protein Myo2 is an essential mediator of bud-directed mitochondrial movement in yeast. J. Cell Biol. 194:473–488 10.1083/jcb.201012088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsch L.C., Dockendorff T.C., Cole C.N. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129:939–955 10.1083/jcb.129.4.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A., Debler E.W., Blobel G. 2011. The structure of the nuclear pore complex. Annu. Rev. Biochem. 80:613–643 10.1146/annurev-biochem-060109-151030 [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Catlett N.L., Novak J.L., Tang F., Nau J.J., Weisman L.S. 2003. Identification of an organelle-specific myosin V receptor. J. Cell Biol. 160:887–897 10.1083/jcb.200210139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Toh-E A., Matsui Y. 2004. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 23:2520–2530 10.1038/sj.emboj.7600271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Keller P.J., Lorenz H., Schiebel E., Knop M. 2010. Segregation of yeast nuclear pores. Nature. 466:E1 10.1038/nature09255 [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D., Eder S., Oh C.S., Martin C.E., Gable K., Bacikova D., Dunn T. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:109–125 10.1128/MCB.21.1.109-125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Larsson L., Caballero A., Hao X., Oling D., Grantham J., Nyström T. 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 140:257–267 10.1016/j.cell.2009.12.031 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lusk C.P., Makhnevych T., Marelli M., Aitchison J.D., Wozniak R.W. 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159:267–278 10.1083/jcb.200203079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T., Lusk C.P., Anderson A.M., Aitchison J.D., Wozniak R.W. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 115:813–823 10.1016/S0092-8674(03)00986-3 [DOI] [PubMed] [Google Scholar]
- Makio T., Stanton L.H., Lin C.C., Goldfarb D.S., Weis K., Wozniak R.W. 2009. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 185:459–473 10.1083/jcb.200810029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D.A., Shulga N., Undavai S., Ferrando-May E., Rexach M.F., Goldfarb D.S. 2005. Increased nuclear envelope permeability and Pep4p-dependent degradation of nucleoporins during hydrogen peroxide-induced cell death. FEMS Yeast Res. 5:1237–1251 10.1016/j.femsyr.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Ptak C., Anderson A.M., Scott R.J., Van de Vosse D., Rogers R.S., Sydorskyy Y., Aitchison J.D., Wozniak R.W. 2009. A role for the karyopherin Kap123p in microtubule stability. Traffic. 10:1619–1634 10.1111/j.1600-0854.2009.00978.x [DOI] [PubMed] [Google Scholar]
- Shcheprova Z., Baldi S., Frei S.B., Gonnet G., Barral Y. 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature. 454:728–734 [DOI] [PubMed] [Google Scholar]
- Sinclair D.A., Guarente L. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 91:1033–1042 10.1016/S0092-8674(00)80493-6 [DOI] [PubMed] [Google Scholar]
- Steinberg G., Schuster M., Theisen U., Kilaru S., Forge A., Martin-Urdiroz M. 2012. Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J. Cell Biol. 198:343–355 10.1083/jcb.201201087 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.