Large, insoluble aggregates of a yeast prion protein are required for the prion phenotype, but soluble oligomers contain all the information necessary to transmit the prion conformation.
Abstract
Amyloidogenic proteins aggregate through a self-templating mechanism that likely involves oligomeric or prefibrillar intermediates. For disease-associated amyloidogenic proteins, such intermediates have been suggested to be the primary cause of cellular toxicity. However, isolation and characterization of these oligomeric intermediates has proven difficult, sparking controversy over their biological relevance in disease pathology. Here, we describe an oligomeric species of a yeast prion protein in cells that is sufficient for prion transmission and infectivity. These oligomers differ from the classic prion aggregates in that they are soluble and less resistant to SDS. We found that large, SDS-resistant aggregates were required for the prion phenotype but that soluble, more SDS-sensitive oligomers contained all the information necessary to transmit the prion conformation. Thus, we identified distinct functional requirements of two types of prion species for this endogenous epigenetic element. Furthermore, the nontoxic, self-replicating amyloid conformers of yeast prion proteins have again provided valuable insight into the mechanisms of amyloid formation and propagation in cells.
Introduction
The amyloid hypothesis proposes that large, protease-resistant amyloid fibers underlie the toxicity associated with several neurodegenerative diseases (Caughey and Lansbury, 2003; Chiti and Dobson, 2006). A definitive link between the amyloid aggregate and toxicity and neurodegeneration has not been established (Haass and Selkoe, 2007). A recent alternative proposal posits that an intermediate in the amyloid pathway is the primary toxic agent, while the large, insoluble aggregates may sequester oligomers and perhaps aid in cell survival (Kirkitadze et al., 2002). Soluble oligomers of several amyloidogenic proteins including amyloid-β, huntingtin, α-synuclein, and PrP have been detected both from analysis of amyloid-forming recombinant proteins and in cell and mouse models (Lasmézas et al., 1997; Conway et al., 1998; Tzaban et al., 2002; Sánchez et al., 2003; Silveira et al., 2005; Lesné et al., 2006; Sajnani et al., 2012). These oligomers, characterized as putative intermediates in amyloid formation, encompass a variety of sizes and structures that cause toxicity when introduced into disease models (Klyubin et al., 2005; Silveira et al., 2005; Sajnani et al., 2012). Isolation of these dynamic, soluble oligomers has remained largely elusive and, as such, investigation of their role in amyloid formation has proven challenging.
The yeast prion protein Sup35 forms self-perpetuating amyloid conformers that are transmissible and infectious (Patino et al., 1996; Paushkin et al., 1996; Serio et al., 2000). To propagate the [PSI+] prion, Sup35 aggregates must undergo remodeling by the chaperone Hsp104, which facilitates monomer addition by severing amyloid structures to generate transmissible species, or propagons (Chernoff et al., 1995; Kryndushkin et al., 2003; Shorter and Lindquist, 2006; Satpute-Krishnan et al., 2007). Until now, in vivo studies have primarily reported on the role of Hsp104 in aggregate fragmentation (Ness et al., 2002; Satpute-Krishnan et al., 2007). Interestingly, Hsp104 has also been shown to catalyze amyloid formation in vitro, specifically impacting the formation of amyloid oligomers (Shorter and Lindquist, 2004). Moreover, recent in vitro evidence identified Sup35 oligomers as intermediates during amyloid formation under some conditions (Ohhashi et al., 2010). Here, we identify soluble, more SDS-sensitive oligomers of Sup35 as prion propagons and show that Hsp104 plays a role in their maintenance.
Results and discussion
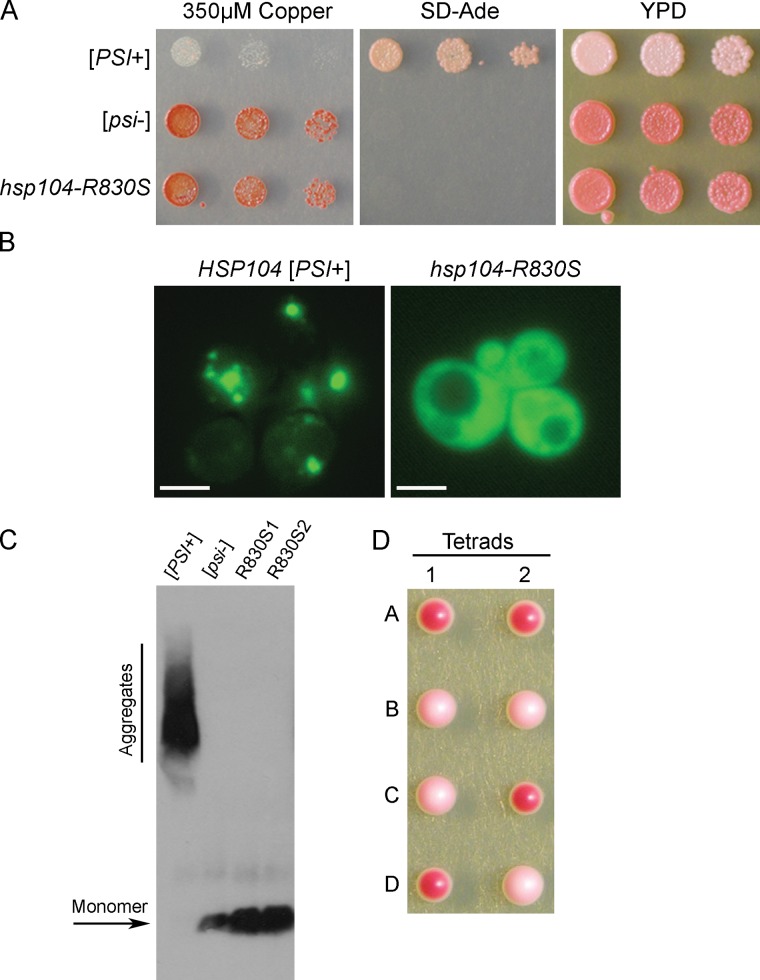
We performed a screen to identify cellular changes that rescued cells from toxic overexpression of SUP35 in [PSI+] cells (Vishveshwara et al., 2009). As this toxicity is dependent on [PSI+], we expected to uncover factors that affected Sup35 aggregation. One mutation identified, hsp104-R830S in the C-terminal domain of Hsp104, caused [PSI+] cells to appear phenotypically [psi−] by a readout of translation termination efficiency of a reporter that is indicative of the functional, soluble state of Sup35 (Fig. 1 A). Fluorescent imaging showed that Sup35 in hsp104-R830S cells appeared diffuse (Fig. 1 B), and by semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE), only monomeric Sup35 was detected in hsp104-R830S cells (Fig. 1 C). Strikingly, mating hsp104-R830S cells that appeared [psi−] to wild-type HSP104 [psi−] cells produced many diploids that were phenotypically [PSI+], as seen by increased nonsense suppression (Fig. S1). Furthermore, sporulation of these diploids produced two [PSI+] and two [psi−] haploid progeny (Fig. 1 D), thereby indicating that prion-competent propagons had been maintained in hsp104-R830S cells that resulted in cryptic [PSI+] (phenotypically undetectable). Indeed, transformation of hsp104-R830S cell lysates into wild-type [psi−] cells demonstrated that these propagons were infectious and produced [PSI+] cells. Out of 1,154 cells transformed, 45 were [PSI+] (3.9% infectivity). Control transformations of [psi−] lysates into [psi−] cells resulted in only red colonies. Of the thousands of red colonies resulting from these control transformations, over 400 were further analyzed and confirmed to be [psi−]. Thus, the infectious propagon in hsp104-R830S cells is unable to cause observable nonsense suppression, but reestablishes and maintains the [PSI+] phenotype in the presence of wild-type HSP104.
Figure 1.
hsp104-R830S propagates cryptic [PSI+]. (A) [PSI+], [psi−], and hsp104-R830S cells were spotted on media containing CuSO4 to induce toxic overexpression of Sup35, media lacking adenine (SD-Ade) to assess nonsense suppression of the premature stop codon in ade1-14, and rich media (YPD). (B) HSP104 [PSI+] and hsp104-R830S cells containing pSup35NM-GFP were imaged by fluorescence microscopy. Bar, 10 µm. (C) Western blot of an SDS-containing agarose gel (SDD-AGE) shows Sup35 aggregate status in lysates of indicated strains. This blot is one representative of three individual experiments. (D) An example of two tetrads where mating hsp104-R830S to [psi−] cells, both containing the ade1-14 mutation, resulted in tetrads with two red (efficient translation termination, hsp104-R830S) and two light pink (increased nonsense suppression, [PSI+] HSP104) haploids.
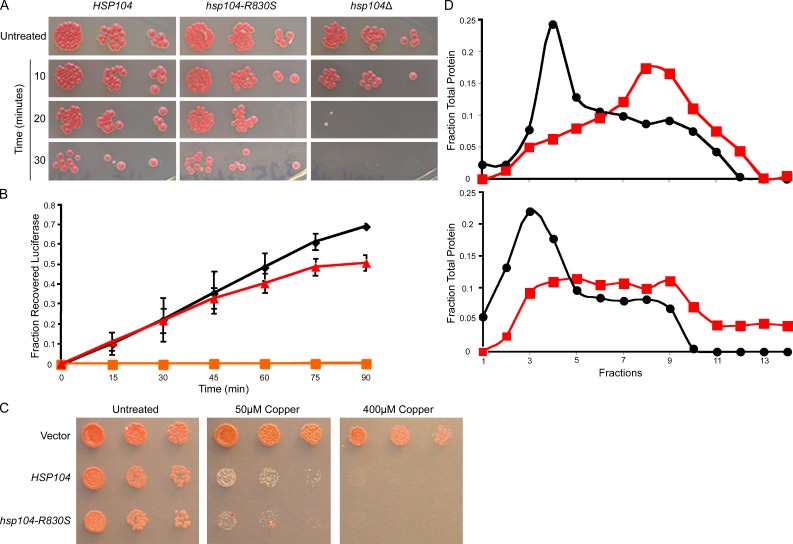
Hsp104 normally functions to disaggregate nonprion aggregates and promote cellular recovery from stress (Glover and Lindquist, 1998). Therefore, we tested the activity of the Hsp104-R830S mutant with other known substrates. Interestingly, the thermotolerance of hsp104-R830S cells resembled that of wild-type cells (Fig. 2 A). Moreover, hsp104-R830S cells efficiently resolubilized heat-aggregated luciferase (Fig. 2 B). Hsp104 threads substrates through a central channel as a mechanism of disaggregation (Tessarz et al., 2008). The Hsp104 variant, HAP, has been used to investigate threading activity by coupling Hsp104 to the ClpP protease so that threaded substrates are degraded, resulting in decreased viability (Tessarz et al., 2008). We created the HAP-R830S variant and found that the mutant maintained threading activity (Fig. 2 C). Hsp104 is an AAA+ ATPase. Mutations that inhibit ATP hydrolysis or hexamerization typically prevent [PSI+] propagation (Glover and Lum, 2009). We purified Hsp104-R830S and measured a reduced initial rate of ATP hydrolysis of 0.1659 ± 0.0308 nmol Pi·µg−1·min−1, as compared with the initial rate of wild-type Hsp104 of 0.2975 ± 0.412 nmol Pi·µg−1·min−1, which is comparable to previously published data (Tkach and Glover, 2004). We next analyzed the distribution of Hsp104-R830S, relative to wild-type Hsp104, using glycerol gradients and size-exclusion chromatography. These analyses revealed that Hsp104-R830S forms unstable hexamers (Fig. 2 D and Fig. S2). Together, these data reveal that the reduced activity of Hsp104-R830S impairs the maintenance of the [PSI+] phenotype.
Figure 2.
Analysis of Hsp104-R830S activity reveals the mechanism of altered activity. (A) Growth on YPD plates of the strains heat-shocked at 50°C for times indicated. Untreated cells were spotted before heat shock. (B) HSP104 (black), hsp104-R830S (red), and hsp104Δ (orange) cells expressing heat-aggregatable luciferase were heat-shocked at 44°C while blocking new protein synthesis. Luminescence during recovery at 30°C was plotted as a fraction of luminescence before heat shock. Error bars represent the SD. (C) hsp104Δ cells carrying pRS315CUP1-ClpP, covered by the indicated plasmids, were spotted on media containing CuSO4 to induce expression of ClpP. (D) Hsp104 (top) and Hsp104-R830S (bottom) incubated with and without ATP (red/black) separated on a glycerol gradient by ultracentrifugation. This is one representation of three individual experiments. The top of the gradient is fraction number 1. Fractions were analyzed by SDS-PAGE and Western blot. All experiments were repeated at least three times.
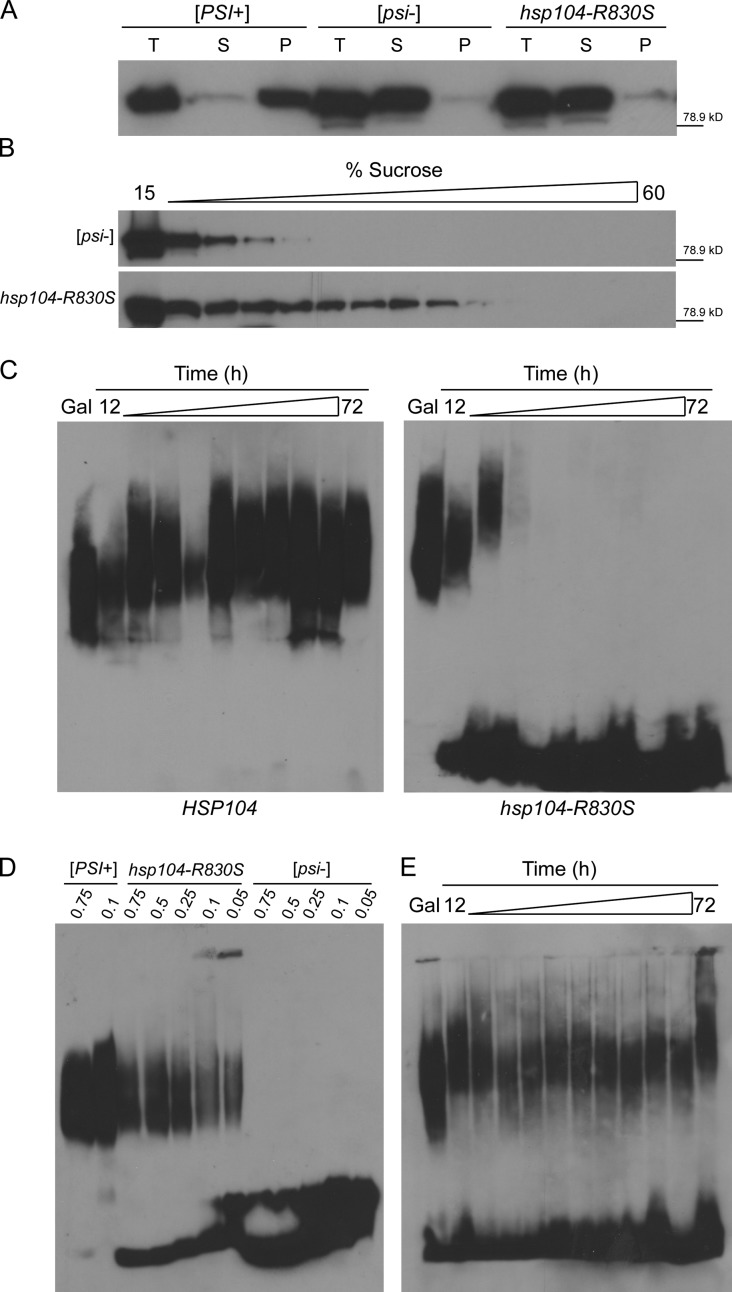
Next, we assessed the state of Sup35 in hsp104-R830S cryptic [PSI+] cells. We first performed sedimentation analysis with hsp104-R830S cryptic [PSI+] lysates and found that Sup35 was soluble (Fig. 3 A). Next, we subjected lysates of hsp104-R830S cryptic [PSI+] and HSP104 [psi−] cells to sucrose gradient fractionation. Monomeric Sup35 in [psi−] cells remained in the top fractions, while Sup35 from hsp104-R830S lysates was detected further down the gradient, demonstrating the existence of some oligomeric species (Fig. 3 B). To understand how these oligomers relate to the large SDS-resistant aggregates associated with the [PSI+] phenotype, we created a system to examine the effect of hsp104-R830S on pre-existing Sup35 aggregates. We covered hsp104-R830S cryptic [PSI+] with wild-type HSP104 expressed from a glucose-repressible promoter such that cells grown in nonrepressing galactose allowed the propagation of [PSI+] and maintained SDS-resistant aggregates (Fig. 3 C, Gal). We then shifted the cells to glucose to repress wild-type HSP104 (Fig. S3 A) and performed SDD-AGE to monitor the effect of hsp104-R830S on SDS-resistant Sup35 aggregates. Within 12 h of wild-type HSP104 repression, monomeric Sup35 was apparent in hsp104-R830S cells. Within 24 h, SDS-resistant aggregates had disappeared (Fig. 3 C). As expected, when hsp104-R830S pGAL-HSP104 cells were switched from glucose back to galactose to de-repress wild-type HSP104, they became phenotypically [PSI+] (Fig. S3 B). Therefore, despite the loss of SDS-resistant aggregates, the propagons remained. Thus, the large SDS-resistant Sup35 aggregates are not strictly required for transmission of the prion.
Figure 3.
hsp104-R830S propagates SDS-sensitive soluble oligomers of Sup35. (A) Ultracentrifugation separated lysates into total (T), soluble (S), and pellet (P) fractions. (B) Fractionation of [psi−] and hsp104-R830S cryptic [PSI+] lysates separated by a 3-h centrifugation through a linear sucrose gradient. (C) Hsp104 and hsp104-R830S cells carrying p416GAL-HSP104 grown in galactose were switched to glucose to repress p416GAL-HSP104, and p416GAL-HSP104 was efficiently repressed by 12 h in glucose as shown by Western blot in an hsp104Δ control (Fig. S3). Aliquots were taken from galactose and between 12 and 72 h in glucose and subjected to SDD-AGE. The shift in aggregate distribution occurs when changing carbon sources. (D) Lysates of the indicated strains were analyzed by SDD-AGE. The HSP104 [PSI+] and [psi−] and hsp104-R830S samples were incubated in sample buffer containing the indicated percentages of SDS. (E) Same as D, but samples were subjected to SDD-AGE containing 0.01% SDS. All experiments were repeated at least three times and protein molecular weight markers (kD) are indicated.
We therefore reasoned that a less stable subpopulation of Sup35 might be capable of prion transmission. Indeed, that the addition of SDS to Sup35 prion aggregates results in smaller SDS-resistant polymers suggests that some less SDS-resistant species exists within the large prion aggregates (Kryndushkin et al., 2003). We investigated whether the soluble oligomers in hsp104-R830S cryptic [PSI+] cells were less resistant to SDS, as has been reported for another prion (Taneja et al., 2007). We again performed SDD-AGE on hsp104-R830S cryptic [PSI+] lysates but decreased the SDS concentration, and only then did we observe the presence of oligomeric species (Fig. 3 D). To determine how these species related to prion propagons and the cryptic [PSI+] phenotype, we repeated the wild-type HSP104 repression time course with hsp104-R830S cells using a low-SDS SDD-AGE. We observed the maintenance of less SDS-resistant, oligomeric species throughout the time course (Fig. 3 E), suggesting that the soluble, less SDS-resistant oligomers are the transmissible propagons in hsp104-R830S cryptic [PSI+] cells.
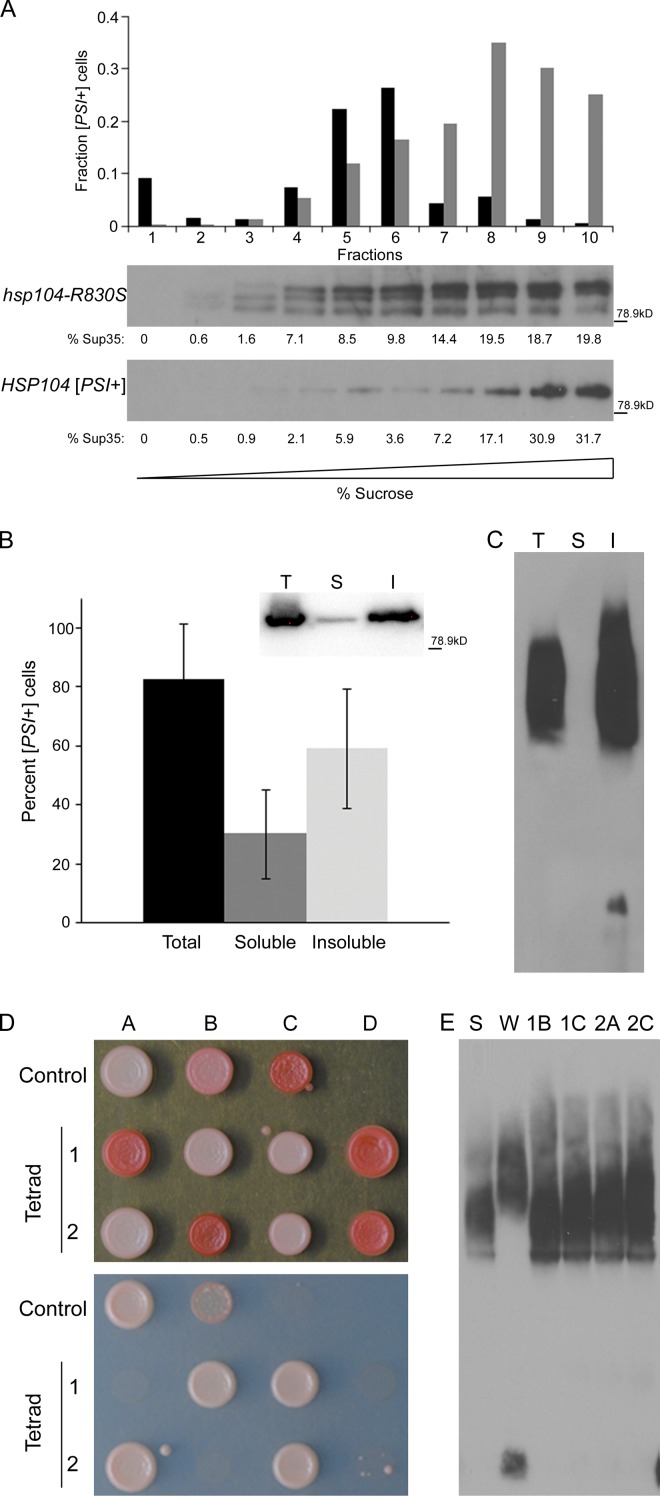
Although we observed the unstable, soluble oligomers in the absence of insoluble aggregates, it is likely that both species exist in wild-type cells, and the more SDS-sensitive, soluble oligomers are more readily detectable in cells lacking insoluble aggregates. Therefore, we separated the Sup35 species in both hsp104-R830S cryptic [PSI+] and wild-type [PSI+] cells by sucrose gradient and performed protein transformation to determine whether the soluble fractions contained infectious propagons. Both hsp104-R830S and HSP104 cells contained soluble Sup35 oligomers that were infectious to [psi−] cells (Fig. 4 A). Although much of the Sup35 species from wild-type lysates migrated to the bottom of the gradient (35% of the total Sup35 which correspond to 54% of the infectivity), there were highly infectious oligomers in wild-type [PSI+] cells that did not migrate with the heavy-sedimenting aggregates. This soluble pool of Sup35 contains 46% of the infectivity in wild-type [PSI+] lysates. Curiously, the high-molecular weight aggregates observed in hsp104-R830S cells were not very infectious, suggesting that these aggregates are not the same high-molecular weight aggregates present in wild-type [PSI+] cells and are not efficient [PSI+] propagons. We then performed a simple solubility assay to separate wild-type [PSI+] lysates into soluble and insoluble fractions (see Fig. 3 A), and then performed infectivity assays and SDD-AGE analysis. Strikingly, we found much of the infectivity in the soluble fraction (32.8% of the total Sup35, which corresponds to 30% of the infectivity) from sedimentation analysis (Fig. 4 B). Additionally, by SDD-AGE, we found that there were no Sup35 oligomers resistant to 2% SDS in the soluble fraction (Fig. 4 C). As such, these data support our hypothesis that insoluble aggregates are not strictly required for prion transmission.
Figure 4.
Soluble oligomeric [PSI+] propagons are infectious and maintain the prion variant structure. (A) Fractions of [PSI+] and hsp104-R830S cryptic [PSI+] lysates separated on a linear sucrose gradient by a 22-h ultracentrifugation step were analyzed by Western blot and the amount of Sup35 (presented as the percent of soluble Sup35) in each fraction was quantified by ImageJ (National Institutes of Health) and is indicated below each fraction in the Western blot. Equal volumes of each fraction were transformed into [psi−] cells. The fraction of infected [PSI+] cells obtained from each gradient fraction is indicated for hsp104-R830S (black) and HSP104 (gray) cells. The fraction of infected [PSI+] cells was generated by compiling the infectivity data from four (HSP104) or five (hsp104-R830S) separate sucrose gradients and transformations. Sup35 in hsp104-R830S lysates appears to be more susceptible to proteolysis and frequently shows degradation products. Protein molecular weight markers (kD) are indicated. (B) Total, soluble, and insoluble fractions of HSP104 [PSI+] lysates from sedimentation analysis were transformed into [psi−] cells. The relative infectivity of the total, soluble, and insoluble fractions was calculated as above and is indicated. (Inset) A Western blot for Sup35 in total (T), soluble (S), and insoluble (I) fractions from one sedimentation assay in this experiment with protein molecular weight marker (kD). Of the thousands of red colonies resulting from transformation of the soluble and insoluble fractions from wild-type [psi−] cells as a negative control, further analysis verified that 48 from each fraction were [psi−] in each of three independent experiments. (C) Total (T), soluble (S), and insoluble (I) fractions of HSP104 [PSI+] lysates from sedimentation analysis were analyzed by SDD-AGE. The soluble fraction is from the top of the supernatant to prevent contamination from the pellet. This experiment was repeated three times. (D) Two representative tetrads from the mating of hsp104-R830S cryptic [PSI+] to [psi−] were spotted on YPD (top) and SD-Ade plates (bottom) to assess the level of nonsense suppression. Strong [PSI+] (Control A), weak [PSI+] (Control B), and [psi−] (Control C) strains were spotted for reference. (E) The lysates of the four white haploids from the tetrads in C were analyzed by SDD-AGE and compared with strong (St) and weak (Wk) [PSI+] controls.
Propagons contain the variant-specific properties required for inheritance and propagation of distinct prion phenotypes (Tanaka et al., 2004; Satpute-Krishnan and Serio, 2005). Therefore, the soluble oligomers in hsp104-R830S cryptic [PSI+] cells should retain the properties necessary to propagate the parental strong [PSI+] phenotype. If hsp104-R830S altered the prion structure, instead of cryptic [PSI+], weakened nonsense suppression and enhanced mitotic loss could be expected. After crossing hsp104-R830S cryptic [PSI+] cells to [psi−] cells, we compared the [PSI+] meiotic progeny to the parental strong [PSI+] variant used initially in the screen. We found that the [PSI+] progeny were indeed strong [PSI+] phenotypically (Fig. 4 D). We then performed SDD-AGE analysis of the resulting [PSI+] haploids as compared with the strong [PSI+] and a weak [PSI+] variant (Derkatch et al., 1996; Kryndushkin et al., 2003; Bagriantsev and Liebman, 2004). The SDS-resistant Sup35 aggregates in the [PSI+] progeny appear the same as strong [PSI+] (Fig. 4 E). We also noted that all the [PSI+] transformants from infection of hsp104-R830S lysates into [psi−] cells were strong [PSI+]. Thus, Hsp104-R830S did not alter the properties of the original variant phenotypically or biochemically, suggesting that there was no change in prion structure.
We have shown, for the first time, the presence of soluble, prion-competent, less SDS-resistant oligomers of Sup35 in vivo. The less SDS-resistant, soluble oligomers are sufficient for transmission of the prion conformation, but are not sufficient to produce the nonsense suppression phenotype associated with [PSI+]. We have also shown that the insoluble aggregates that characterize [PSI+] cells are not required for prion transmission or infectivity. Furthermore, we have uncovered two novel features of yeast prion propagons. First, they can be more SDS sensitive, where previously only SDS-resistant aggregates of Sup35 were shown to be infectious (Bagriantsev et al., 2008). This sensitivity parallels the previously described PK-sensitive PrP species, which can also transmit the prion (Sajnani et al., 2012). Second, soluble oligomers were able to act as propagons in vivo. Oligomers of several amyloidogenic proteins have been described and can cause toxicity in the absence of large aggregates (Haass and Selkoe, 2007). Indeed, our data correlate well with previous reports that visible Sup35 aggregates are lost in the mother cell and yet the prion is faithfully propagated (Taguchi and Kawai-Noma, 2010). Perhaps the SDS-resistant, insoluble Sup35 aggregates function as a reservoir for the continued renewal of transmissible propagons. This role for SDS-resistant yeast prion aggregates mirrors the proposed role for amyloid aggregates as traps for oligomers (Kirkitadze et al., 2002). As such, we clearly show separate functions of two distinct aggregate species that are structurally related components of the same prion propagation pathway. A mutant in Hsp104 enabled us to initially tease out these species. We favor a model whereby the decreased activity of hsp104-R830S results in reduced fragmentation of aggregates, but our data do not exclude the possibility that Hsp104 normally plays a role in generating the large insoluble aggregates.
The soluble oligomers that exist in many protein conformational disorders are challenging to characterize due to their dynamic and metastable nature. In addition, conflicting reports on the properties and structure of oligomers complicate the elucidation of the important oligomeric species (Haass and Selkoe, 2007). The properties and mechanisms associated with yeast prion propagons may be similar to those associated with amyloidogenic proteins involved in self-propagating protein conformational disorders. Therefore, investigating these features may lead to a better understanding of the function and structure of soluble, self-templating oligomers, as well as the role of amyloid in disease.
Materials and methods
Strains and yeast cultivation
The yeast strains used in this study are derivatives of Saccharomyces cerevisiae 74-D694 and were grown using standard culture techniques. YPD is rich yeast medium, whereas SD is synthetic medium lacking amino acids as needed to select for maintenance of plasmids or screen for a nonsense suppression phenotype (SD-Ade). The strain used in the screen, 74-D694 [PSI+] can1Δ::MFA1pr-HIS3-MFa1p r-LEU2 ade1-14 ura3-52 lys2Δ::KanMX4 pRS315CUP1-SUP35, was transformed with 10 independent plasmid pools of the mini-transposon (3XHA/lacZ(mTn3)) mutagenized library (provided by M. Snyder, Stanford University, Stanford, CA) and selected on media containing 350 µM copper sulfate to induce expression of SUP35. Candidates that rescued toxicity associated with increased expression of SUP35 were passaged a second time on copper sulfate media and were examined for the effect on [PSI+] phenotypically by color and genetically by mating to unmutagenized 74-D694 [PSI+] and [psi−] isogenic strains. Several mutants, not linked to the transposon, were analyzed and determined to be in Hsp104 through genetic analysis. The mutant characterized here, hsp104-R830S, was recreated and these strains were used for further characterization. Phenotypic assays are based on the amount of functional Sup35 to terminate the premature stop codon in ade1-14, which results in the accumulation of a red-pigmented intermediate in the adenine biosynthesis pathway (Liebman and Derkatch, 1999).
SDD-AGE protein analysis
Cells were lysed by bead-beating in buffer A (25 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol plus mini EDTA-free protease inhibitors [Roche], Aprotinen [Sigma-Aldrich], and PMSF [Sigma-Aldrich]). Samples were treated at room temperature for 7 min in sample buffer containing 2% SDS, then electrophoresed through a 0.1% SDS, 1.5% agarose gel. The gels were subjected to Western blot with anti-Sup35 antibody. The experiments done with lower SDS concentrations contained 0.01% SDS in the gel and between 0.05 and 0.1% SDS in the sample buffer, as indicated. Molecular weight markers were not used because nondenatured protein aggregates were being analyzed.
Fluorescence microscopy
Cells were transformed with pRS426CUP1-SUP35NM-GFP and grown in media lacking uracil. 50 µM CuSO4 was added to logarithmically growing cells and grown for 4 h to induce expression of SUP35NM-GFP. Images of cells expressing Sup35NM-GFP, in water and at room temperature, were captured on a microscope (Bmax-60F; Olympus) containing a 1.35 NA/100× UPlanApo objective lens, spinning disc Confocal Scanner Unit (CSU10), and an ICCD camera (XR-Mega10; Stanford Photonics). Images were acquired using QED software and analyzed by ImageJ (National Institutes of Health). In ImageJ, images were colored green for presentation.
Protein transformation
Recipient [psi−] cells from logarithmically growing cultures were spheroplasted with lyticase. Samples from cell lysis, sedimentation analysis, and sucrose gradient fractionation were added to the spheroplasts in 1 M sorbitol along with a vector containing the URA3 gene for selection on media lacking uracil. The cells were incubated at room temperature then recovered at 30°C. Media containing glucose, sorbitol, and agar was added to the cells before plating on sorbitol plates lacking uracil. Transformants were replica plated to YPD plates, and individual colonies were picked from those YPD plates and spotted onto YPD and both media lacking adenine and containing GdHCl for quantification of infectivity.
Purification of Hsp104
Hsp104 was purified as described previously (Lum et al., 2004). Recombinant Hsp104 tagged with a 6x-Histidine tag was expressed in Escherichia coli cells and affinity purified using a Ni2+-Sepharose column. Next, the 6x-Histidine tag was removed by cleavage with the TEV protease and the untagged Hsp104 was separated from the uncleaved, tagged Hsp104 by affinity purification. The untagged Hsp104 was further purified using anion exchange via a Q-Sepharose column. The pure Hsp104 collected from the Q-Sepharose column was then separated on an S-300 gel filtration column to isolate monomeric Hsp104. Purified, monomeric Hsp104 was pooled, concentrated, and frozen at −80°C. Multiple purification preparations were used for all assays in repeat experiments.
ATP hydrolysis assays
ATP hydrolysis was monitored by the Malachite Green assay. 2 µg purified Hsp104 was equilibrated in buffer (40 mM Tris-HCl, pH 7.5, 175 mM NaCl, 5 mM MgCl2, and 0.02% Triton X-100) at 37°C before adding 5 mM ATP (Sigma-Aldrich). During incubation of Hsp104 with ATP at 37°C, samples were taken and free phosphate was measured by addition of Malachite Green dye (Sigma-Aldrich). Color development occurred over 1 min and was terminated by the addition of 34% citric acid (Sigma-Aldrich). Absorbance was measured at 650 nm.
Glycerol gradients
Purified protein at 75 µg was incubated in buffer (20 mM Tris, pH 8, 100 mM NaCl, 10 mM MgCl2, 2 mM EDTA, and 10% glycerol) ± ATP for 5 min on ice before loading onto linear glycerol gradients (15–35%). The gradients were spun at 34,000 rpm for 18 h in a rotor (SW55; Beckman Coulter). Gradients were fractionated and the fractions analyzed by SDS-PAGE and Western blot with anti-Hsp104 antibody.
Size-exclusion chromatography
Purified protein at 1 mg/ml was incubated in buffer (50 mM Tris, pH 7.5, 200 mM KCl, 10 mM MgCl2, 2 mM DTT, and 2 mM EDTA) ± ATP for 5 min. The sample was applied to an S-300HR column (GE Healthcare) and fractions were collected. The concentration of Hsp104 was measured by Bradford analysis (Bio-Rad Laboratories).
Thermotolerance assays
Cells were grown to mid-log phase at 30°C. An equal number of cells from each sample were aliquoted and pretreated at 37°C for 30 min. Samples were heat shocked at 50°C for the indicated amounts of time and then diluted fivefold before plating onto solid rich medium.
In vivo luciferase refolding assay
In vivo luciferase refolding assays were performed as described previously (Tipton et al., 2008). In brief, cells carrying pRS316GPD-Lux (provided by B. Bukau, ZMBH, Ruprecht-Karls-Universitat Heidelberg, Germany) were pretreated at 37°C for 1 h to induce expression of HSP104, then heat shocked at 44°C for 1 h. After 50 min of heat shock, cycloheximide was added to prevent new protein synthesis. After heat shock, the cells recovering at 30°C were aliquoted at 15-min intervals and D-luciferin (Sigma-Aldrich) was added. Luminescence was measured and plotted as a percentage of total luciferase activity before heat shock.
Hsp104 shutoff experiments
The diploid strains, HSP104/hsp104Δ::leu2 and HSP104/hsp104-R830S were transformed with pRS416GAL-HSP104, sporulated, and dissected on CSM-Ura + 0.25% galactose + 2% raffinose. Low levels of galactose were used to avoid curing [PSI+] by overexpression of HSP104. Progeny were selected that contained the plasmid and were pink in color as evidence that [PSI+] was maintained. Cells were grown in CSM-Ura + 0.25% galactose + 2% raffinose for 48 h before switching the cells to glucose media lacking uracil to repress expression of the HSP104 plasmid. At 6-h intervals, aliquots of the sample were taken and both pelleted for SDD-AGE and plated on CSM-Ura + 0.25% galactose + 2% raffinose plates. SDD-AGE was performed as described above. Gels were transferred to PVDF membrane and probed with anti-Sup35 antibody.
Sup35 solubility assays
Cells were lysed by bead-beating in buffer (10 mM NaPO4, pH 7.5, 250 mM NaCl, 2% SDS, 1% Triton X-100 plus mini EDTA-free protease inhibitors [Roche], and PMSF [Sigma-Aldrich]). Lysates were subjected to ultracentrifugation at 100,000 rpm in a rotor (TLA-100; Beckman Coulter) for 1 h. The supernatant was collected and the pellet resuspended in lysis buffer. Total, supernatant, and pellet fractions were subjected to SDS-PAGE and Western blot with anti-Sup35 antibody as well as protein transformation.
Sucrose gradients
Cells were lysed by bead-beating in buffer A. Total protein was normalized to 1 mg/ml and loaded onto a 4-mL linear sucrose gradient (15–60%) and centrifuged at 32,000 rpm for either 3 or 22 h. To compare infectivity of soluble and insoluble Sup35 from HSP104 [PSI+] by sucrose gradient, we increased the time of centrifugation to a time that reproducibly showed a large fraction of pelletable Sup35. Thus, after a 22-h spin, we are able to distinguish the soluble Sup35 and the insoluble Sup35 that migrates to the bottom of the gradient. Gradients were fractionated and the fractions were analyzed by SDS-PAGE, Western blot, and protein transformation.
Online supplemental material
Fig. S1 shows that the mating of hsp104-R830S cells to HSP104 [psi−] cells results in cells that are phenotypically [PSI+]. Fig. S2 shows that Hsp104-R830S forms unstable hexamers by size-exclusion chromatography. Fig. S3 A shows that growing the cells containing the pRS416-GAL-HSP104 construct in glucose efficiently represses wild-type HSP104 on the plasmid. Fig. S3 B shows that hsp104-R830S cells maintain [PSI+] propagons throughout the shutoff time course as de-repression of wild-type HSP104 results in pink [PSI+] colonies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307040/DC1.
Supplementary Material
Acknowledgments
We thank J.R. Glover, B. Bukau, and J. Weissman for generously providing plasmids and protocols; and J. Cooper for use of his confocal microscope. We also thank members of the True laboratory and C. Weihl, A. Cashikar, O. Mooren, and D. Sanders for comments on the manuscript.
This work was supported by funding from the National Institutes of Health (grant GM072778) and the National Science Foundation.
The authors declare no competing financial interests.
Author contributions: The screen was performed by R.E. Bouttenot. J.E. Dulle, R.E. Bouttenot, and H.L. True designed the experiments. J.E. Dulle and R.E. Bouttenot carried out all the experiments except for the HAP-R830S experiments, which were carried out by L.A. Underwood. J.E. Dulle, R.E. Bouttenot, and H.L. True analyzed the data. J.E. Dulle and H.L. True wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- SD
- synthetic defined media
- SDD-AGE
- semi-denaturing detergent-agarose gel electrophoresis
- YPD
- yeast extract peptone dextrose
References
- Bagriantsev S., Liebman S.W. 2004. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J. Biol. Chem. 279:51042–51048 10.1074/jbc.M410611200 [DOI] [PubMed] [Google Scholar]
- Bagriantsev S.N., Gracheva E.O., Richmond J.E., Liebman S.W. 2008. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol. Biol. Cell. 19:2433–2443 10.1091/mbc.E08-01-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Lansbury P.T. 2003. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26:267–298 10.1146/annurev.neuro.26.010302.081142 [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist S.L., Ono B., Inge-Vechtomov S.G., Liebman S.W. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 268:880–884 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75:333–366 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- Conway K.A., Harper J.D., Lansbury P.T. 1998. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat. Med. 4:1318–1320 10.1038/3311 [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Chernoff Y.O., Kushnirov V.V., Inge-Vechtomov S.G., Liebman S.W. 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 144:1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 94:73–82 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- Glover J.R., Lum R. 2009. Remodeling of protein aggregates by Hsp104. Protein Pept. Lett. 16:587–597 10.2174/092986609788490087 [DOI] [PubMed] [Google Scholar]
- Haass C., Selkoe D.J. 2007. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 8:101–112 10.1038/nrm2101 [DOI] [PubMed] [Google Scholar]
- Kirkitadze M.D., Bitan G., Teplow D.B. 2002. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J. Neurosci. Res. 69:567–577 10.1002/jnr.10328 [DOI] [PubMed] [Google Scholar]
- Klyubin I., Walsh D.M., Lemere C.A., Cullen W.K., Shankar G.M., Betts V., Spooner E.T., Jiang L., Anwyl R., Selkoe D.J., Rowan M.J. 2005. Amyloid beta protein immunotherapy neutralizes Abeta oligomers that disrupt synaptic plasticity in vivo. Nat. Med. 11:556–561 10.1038/nm1234 [DOI] [PubMed] [Google Scholar]
- Kryndushkin D.S., Alexandrov I.M., Ter-Avanesyan M.D., Kushnirov V.V. 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278:49636–49643 10.1074/jbc.M307996200 [DOI] [PubMed] [Google Scholar]
- Lasmézas C.I., Deslys J.P., Robain O., Jaegly A., Beringue V., Peyrin J.M., Fournier J.G., Hauw J.J., Rossier J., Dormont D. 1997. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science. 275:402–405 10.1126/science.275.5298.402 [DOI] [PubMed] [Google Scholar]
- Lesné S., Koh M.T., Kotilinek L., Kayed R., Glabe C.G., Yang A., Gallagher M., Ashe K.H. 2006. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 440:352–357 10.1038/nature04533 [DOI] [PubMed] [Google Scholar]
- Liebman S.W., Derkatch I.L. 1999. The yeast [PSI+] prion: making sense of nonsense. J. Biol. Chem. 274:1181–1184 10.1074/jbc.274.3.1181 [DOI] [PubMed] [Google Scholar]
- Lum R., Tkach J.M., Vierling E., Glover J.R. 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279:29139–29146 10.1074/jbc.M403777200 [DOI] [PubMed] [Google Scholar]
- Ness F., Ferreira P., Cox B.S., Tuite M.F. 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22:5593–5605 10.1128/MCB.22.15.5593-5605.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhashi Y., Ito K., Toyama B.H., Weissman J.S., Tanaka M. 2010. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat. Chem. Biol. 6:225–230 10.1038/nchembio.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino M.M., Liu J.J., Glover J.R., Lindquist S. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 273:622–626 10.1126/science.273.5275.622 [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov V.V., Smirnov V.N., Ter-Avanesyan M.D. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15:3127–3134 [PMC free article] [PubMed] [Google Scholar]
- Sajnani G., Silva C.J., Ramos A., Pastrana M.A., Onisko B.C., Erickson M.L., Antaki E.M., Dynin I., Vázquez-Fernández E., Sigurdson C.J., et al. 2012. PK-sensitive PrP is infectious and shares basic structural features with PK-resistant PrP. PLoS Pathog. 8:e1002547 10.1371/journal.ppat.1002547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez I., Mahlke C., Yuan J. 2003. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 421:373–379 10.1038/nature01301 [DOI] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Serio T.R. 2005. Prion protein remodelling confers an immediate phenotypic switch. Nature. 437:262–265 10.1038/nature03981 [DOI] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Langseth S.X., Serio T.R. 2007. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 5:e24 10.1371/journal.pbio.0050024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio T.R., Cashikar A.G., Kowal A.S., Sawicki G.J., Moslehi J.J., Serpell L., Arnsdorf M.F., Lindquist S.L. 2000. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 289:1317–1321 10.1126/science.289.5483.1317 [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 304:1793–1797 10.1126/science.1098007 [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. 2006. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell. 23:425–438 10.1016/j.molcel.2006.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira J.R., Raymond G.J., Hughson A.G., Race R.E., Sim V.L., Hayes S.F., Caughey B. 2005. The most infectious prion protein particles. Nature. 437:257–261 10.1038/nature03989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi H., Kawai-Noma S. 2010. Amyloid oligomers: diffuse oligomer-based transmission of yeast prions. FEBS J. 277:1359–1368 10.1111/j.1742-4658.2010.07569.x [DOI] [PubMed] [Google Scholar]
- Tanaka M., Chien P., Naber N., Cooke R., Weissman J.S. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature. 428:323–328 10.1038/nature02392 [DOI] [PubMed] [Google Scholar]
- Taneja V., Maddelein M.L., Talarek N., Saupe S.J., Liebman S.W. 2007. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol. Cell. 27:67–77 10.1016/j.molcel.2007.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P., Mogk A., Bukau B. 2008. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 68:87–97 10.1111/j.1365-2958.2008.06135.x [DOI] [PubMed] [Google Scholar]
- Tipton K.A., Verges K.J., Weissman J.S. 2008. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell. 32:584–591 10.1016/j.molcel.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J.M., Glover J.R. 2004. Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J. Biol. Chem. 279:35692–35701 10.1074/jbc.M400782200 [DOI] [PubMed] [Google Scholar]
- Tzaban S., Friedlander G., Schonberger O., Horonchik L., Yedidia Y., Shaked G., Gabizon R., Taraboulos A. 2002. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry. 41:12868–12875 10.1021/bi025958g [DOI] [PubMed] [Google Scholar]
- Vishveshwara N., Bradley M.E., Liebman S.W. 2009. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol. Microbiol. 73:1101–1114 10.1111/j.1365-2958.2009.06836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.