Abstract
This unit describes ChIP-exo methodology, which involves chromatin immunoprecipitation (ChIP) combined with lambda exonuclease digestion followed by high-throughput sequencing. ChIP-exo allows us to identify a nearly complete set of binding locations of DNA-bound proteins at near single nucleotide resolution with almost no background. The process is initiated by cross-linking DNA and associated proteins. Chromatin is then isolated from nuclei and subjected to sonication. Subsequently, an antibody against a specific protein is used to immunoprecipitate specific DNA-protein complexes. ChIP DNA is purified, sequencing adaptors are ligated, and digested by lambda exonuclease. High-throughput sequencing generates 25 to 50 nucleotides sequences. The sequence of the DNA fragments is mapped back to the reference genome for determination of the binding locations. 5′ ends of DNA fragments on the forward strand indicate the left border of DNA-protein boundaries and 5′ ends of DNA fragments on the reverse strand indicate the right border of DNA-protein boundaries.
Keywords: ChIP, ChIP-exo, binding location, genome-wide, lambda exonuclease
INTRODUCTION
This unit describes a method to determine genomic locations of a known protein associated with genomic DNA sequences at near nucleotide resolution using a combination of chromatin immunoprecipitation (ChIP) and lambda exonuclease digestion (exo) followed by high-throughput sequencing. Basic protocol 1 and 2 are for yeast (S. cerevisiae) and mammalian cells, respectively. The protocol for mammalian cells is the same as for yeast, except steps 1 to 19. Intact cells are treated with formaldehyde to cross-link in vivo protein-DNA and protein-protein interactions (steps 1 to 3). After cell lysis (steps 5 to 9), chromatin is subjected to sonication to shear the associated DNA to the appropriate size (steps 10 to 13). The soluble chromatin is selectively immunoprecipitated with an antibody against a specific protein (steps 14 to 19). Since size heterogeneity of sonicated DNA fragments is too variable to precisely demarcate genomic locations of DNA-bound proteins, a 5′-3′ exonuclease is employed to trim the DNA sequences on one strand to within a few bp of the crosslinking point. DNA sequences 3′ to the crosslinking point remain intact and are sufficiently long to uniquely identify in a genome when sequenced.
While the immunoprecipitated protein-DNA adducts are still on the beads, several enzymatic steps are performed. The first step involves blunt-ended polishing of sonicated DNA fragment ends (steps 20 to 28). The second step ligates DNA adaptors to both ends of the polished DNA (steps 29 to 34). The third enzymatic stage uses lambda and RecJf exonucleases to trim the DNA (steps 35 to 42). The DNA is then eluted from antibody beads (steps 43 to 44). Protein-DNA crosslinks are reversed and the DNA is extracted (steps 45 to 49). To complete library construction for high-throughput sequencing, the DNA is primer-extended and ligated with a second sequencing adaptor (steps 50 to 54). After gel-purification and PCR (steps 55 to 60), the amplified library is ready for high-throughput sequencing. This protocol was developed for use with Applied Biosystems SOLiD sequencing instruments. Since the molecular biology of library construction in the SOLiD™ Systems 5500 Series is essentially the same as the Illumina methodology, we infer, but have not tested, that Illumina adaptors should provide equivalent results.
BASIC PROTOCOL 1
Identification of the genomic locations of DNA-binding proteins at near nucleotide accuracy in Saccharomyces cerevisiae
This protocol was designed to detect genomic locations of a DNA-binding protein in Saccharomyces cerevisiae at near single base pair resolution. First, chromatin immunoprecipitation (ChIP) is performed to purify protein-DNA complexes, then several enzymatic reactions including lambda exonuclease digestion are performed while protein-DNA complexes are still on the beads. These enzymatic reactions allow us to detect the exact boundaries of crosslinked protein-DNA complex at near nucleotide resolution. This protocol was also designed for use with next-generation sequencing instruments.
NOTE:Use only molecular biology-grade water (e.g. DNase, RNase, and Protease free, deionized, distilled) in all steps and solutions. Substitutes may be used for the indicated suppliers.
Materials
Sample materials
Yeast culture
Reagents and solutions
37% formaldehyde
2.5 M glycine
ST buffer (see recipe)
Protease inhibitor cocktail tablets (Roche)
FA-lysis buffer (see recipe)
20% SDS (volume/volume)
Antibody against the protein of interest
Protein A- or G- Sepharose beads or equivalent (e.g. magnetic beads)
FA-high salt wash buffer (see recipe)
FA-wash 2 buffer (see recipe)
FA-wash 3 buffer (see recipe)
TE buffer (see recipe)
10 mM Tris-HCl (pH 8.0)
NEBuffer 2 (New England BioLabs)
10× BSA (1mg/ml)
3 mM dNTPs
T4 DNA polymerase (3 U/ul, New England BioLabs)
10 mM Tris-HCl (pH 7.5)
10× T4 DNA ligase buffer (New England BioLabs)
T4 Polynucleotide Kinase (10 U/ul, New England BioLabs)
3 mM dATP
Klenow Fragment (3′→5′ exo-) (5 U/ul, New England BioLabs)
T4 DNA ligase (500 U/ul, New England BioLabs)
10× phi29 DNA polymerase buffer (New England BioLabs)
phi29 DNA polymerase (10 U/ul, New England BioLabs)
10 mM Tris-HCl (pH 9.2)
10× lambda exonuclease buffer (New England BioLabs)
Lambda exonulease (5 U/ul, New England BioLabs)
RecJf exonuclease (30 U/ul, New England BioLabs)
ChIP elution buffer (see recipe)
Protease K (20 ug/ul, Roche)
25:24:1 phenol/chloroform/isoamyl alcohol (Sigma)
100% ethanol (Any supplier)
75% (volume/volume) ethanol
Glycogen (20 mg/ml, Roche)
AMPure magnetic beads (Agencourt)
25 mM dNTPs
10× standard taq reaction buffer (New England BioLabs)
Taq DNA polymerase (5 U/ul, New England BioLabs)
Library construction oligonucleotides for Applied Biosystems SOLiD™ Systems 5500 Series fragment library preparation
P1-T Adaptor, 15 uM (Life Technologies)
5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′ (41 bp)
5′-TCACCGACTGCCCATAGAGAGGAAAGCGGAGGCGTAGTGGCC-3′ (42 bp)
P2-T Adaptor, 15 uM (Life Technologies)
5′-CGCCTTGGCCGTACAGCAGCCTCTTACACAGAGAATGAGGAACCCGGGGCAGTT-3′ (55 bp)
5′-CTGCCCCGGGTTCCTCATTCTCTGTGTAAGAGGCTGCTGTACGGCCAAGGCGT-3′ (53 bp)
Library PCR Primer 1, 20 uM (Life Technologies)
5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′ (28 bp)
Library PCR Pimer 2, 20 uM (Life Technologies)
5′-CTGCCCCGGGTTCCTCATTCT-3′ (21 bp)
Oligonucleotide sequences © Copyright 2008. Applied Biosystems. All rights reserved.
Library construction oligonucleotides for Applied Biosystems SOLiD™ Systems 2.0 fragment library preparation
P1 Adaptor, 15 uM (Life Technologies)
5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′ (41 bp)
5′-ATCACCGACTGCCCATAGAGAGGAAAGCGGAGGCGTAGTGGCC-3′ (43 bp)
P2 Adaptor, 15 uM (Life Technologies)
5′-AGAGAATGAGGAACCCGGGGCAGTT-3′ (25 bp)
5′-CTGCCCCGGGTTCCTCATTCTCT-3′ (23 bp)
Library PCR Primer 1, 20 uM (Life Technologies)
5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′ (28 bp)
Library PCR Pimer 2, 20 uM (Life Technologies)
5′-CTGCCCCGGGTTCCTCATTCT-3′ (21 bp)
Oligonucleotide sequences © Copyright 2011. Life Technologies Corporation. All rights reserved.
Library construction oligonucleotides for Illumina instruments (as provided by Illumina, but not tested)
TruSeq Adaptor, Index 1, first ligation (also requires reverse complement oligo, having a 3′ T overhang)
5′-GATCGGAAGAGCACACGTCTGAACTCCAGTCAMCATCACGATCTCGTATGCCGTCTTCTGCTTG-3′
Index 1 underlined
TruSeq Universal Adaptor, second ligation
(also requires reverse complement oligo, but lacking 5′ A)
5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′
Oligonucleotide sequences © 2007-2011 Illumina, Inc. All rights reserved.
Disposables
2.0-ml Natural Conical Tube (USA Scientific)
Zirconium Silicate beads - 0.5 mm (Next Advance)
Kimwipe (Any supplier)
22-G 11/2 needles (Becton Dickinson)
Birosilicate glass culture tubes - 13×100mm (Any supplier)
1.5-ml microcentrifuge tube (Any supplier)
15-ml Conical polypropylene tube (Any supplier)
0.5-ml PCR tubes (Eppendorf)
Filtered pipettor tips (Any supplier)
Equipment
Incubator Shakers I-26R (New Brunswick)
Centrifuge Bottle with Sealing Cap (Nalgene)
Centrifuge RC6+ (Sorvall)
F10S-6X500Y rotor (Sorvall)
Centrifuge 5424R (Eppendorf)
Mini-Beadbeater-96 (BioSpec)
Centrifuge 5810R (Eppendorf)
A-4-81 swing-bucket rotor (Eppendorf)
Bioruptor® Standard (Diagenode)
Centrifuge 5424 (Eppendorf)
Rotating Wheel (Any supplier)
Thermomixer Comfort 5355R 1.5 ml (Eppendorf)
65°C Heat block (Any supplier)
Vortex Mixer (Any supplier)
Vacufuge Plus Vacuum Concentrator (Eppendof)
Centrifuge 5415R (Eppendorf)
DNA Engine® Thermal Cycler PTC-200 (Bio-Rad)
DynaMag™ (Invitrogen)
Pipettor 10 ul (Any supplier)
Pipettor 20 ul (Any supplier)
Pipettor 200 ul (Any supplier)
Pipettor 1 ml (Any supplier)
Harvesting, cross-linking, and quenching
1. For each sample, grow 250 ml yeast cells to OD600 ~0.8 to 1.0. Add 6.75 ml of 37% formaldehyde solution (1% final concentration) and grow yeast cells for 15 min while shaking in an incubator shaker at 25°C.
2. To quench cross-linking, add 15.4 ml of 2.5 M glycine (0.15 M final concentration) for 5 min while shaking in the incubator shaker at 25°C. Transfer yeast culture to a Centrifuge Bottle with Sealing Cap. Centrifuge cells for 3 min in Centrifuge RC6+ with F10S-6X500Y rotor at 2,820 × g, 4°C, and discard supernatant.
NOTE: Specific centrifuges and rotors we have used are listed for convenience; any centrifuge that uses similar sized bottles and reaches the same g force can be used.
3. Place cells on ice and resuspend in 1 ml ice-cold ST buffer containing protease inhibitors. Transfer cells to a 2.0-ml Natural Conical tube on ice. Centrifuge the cells for 2 min in Centrifuge 5424R at 2,300 × g, 4°C, and discard supernatant. Repeat.
Freeze cells in liquid nitrogen. Store cells at -80°C until use.
Cell lysis
5. Thaw cells on ice. Add 1 ml of ice-cold FA-lysis buffer containing protease inhibitors to the cell pellet. Add 1 ml of 0.5-mm Zirconium Silicate beads.
6. Lyse cells in a Mini-Beadbeater-96 for 3 min twice (total 6 min). Be sure to keep samples chilled.
7. Place samples on ice. Quickly wipe off ice or water from the outside of the 2.0-ml Natural Conical tube with a Kimwipe, invert, and puncture the bottom with a red hot 22-G 11/2 needle. Immediately place the 2.0-ml Natural Conical tube inside a Borosilicate glass culture tube (13×100mm) on ice so that the 2.0-ml Natural Conical tube is hanging at the opening of the larger tube.
8. Centrifuge for 1 min at 200 × g, 4°C in Centrifuge 5810R with A-4-81 swing-bucket rotor. Then place samples on ice. After confirming that the entire sample except for the 0.5-mm Zirconium Silicate beads has been transferred to the 1.5-ml microcentrifuge tube (step 9), discard the 2.0-ml Natural Conical tube containing the 0.5-mm Zirconium Silicate beads.
9. Resuspend the disrupted cells into the liquid layer by pipetting up and down with a 1-ml micropipettor and transfer suspension to a 1.5-ml microcentrifuge tube on ice.
Sonication
10. Centrifuge the cells for 3 min in a Centrifuge 5424R at 2,300 × g, 4°C, and discarded supernatant. Add 1 ml of ice-cold FA-lysis buffer containing protease inhibitors. Resuspend the pellet by pipetting up and down with a 1-ml micropipettor. Centrifuge the cells for 2 min in Centrifuge 5424R at 2,300 × g, 4°C, and discard supernatant. Repeat.
11. Add 1 ml of ice-cold FA-lysis buffer containing 0.2% SDS and protease inhibitors to the washed chromatin pellet. Resuspend the pellet by pipetting up and down with a 1-ml micropipettor. Transfer samples to a 15-ml conical polystyrene tube on ice.
NOTE: Be sure that samples are transferred to a polystyrene tube (not to a polypropylene tube).
12. Shear the chromatin to a median size of 250-300 bp by sonication: using a Diagenode Bioruptor® Standard (power setting: high, 30 seconds on / 30 seconds off, 30 cycles). Incubate on ice-cold water bath during sonication. Transfer to a fresh 1.5-ml microcentrifuge tube on ice.
13. Centrifuge the cells for 10 min in Centrifuge 5424R at 2,300 × g, 4°C, to pellet unbroken cells and debris, and transfer supernatant to a fresh 1.5-ml microcentrifuge tube on ice. Repeat.
NOTE: Supernatant (soluble chromatin) should be transferred after centrifuging and unbroken cells (pellets) should be discarded.
Chromatin immunoprecipitation (ChIP)
14. Transfer to a 15-ml conical polypropylene tube and add ice-cold FA-lysis buffer to dilute SDS concentration to 0.05%. Add antibody against the protein of interest to the chromatin extract and mix. Incubate overnight at 4°C.
15. Using a wide-bore pipette tip (e.g. a 200-ul pipette tip with the narrow end cut off), add 20 ul of sepharose beads (40 ul of 50% beads slurry) to each sample and mix. Incubate on a rotating wheel 1.5 hr at 4°C.
16. Centrifuge for 1 min in Centrifuge 5424 at 94 × g, room temperature, and remove the supernatant by aspiration. Add 0.5 ml ice-cold FA-lysis buffer containing protease inhibitors. Using a wide-bore pipette tip, resuspend the beads by pipetting up and down, and transfer to a fresh 1.5-ml microcentrifuge tube. Centrifuge for 1 min in Centrifuge 5424 at 94 × g, room temperature. Remove supernatant by aspiration.
17. Wash the beads by adding ice-cold 1 ml FA-lysis buffer containing protease inhibitors and incubating 5 min at room temperature on a rotating wheel. Pellet beads by centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature. Remove the supernatant by aspiration. Repeat (total two washes).
18. Repeat step 17 with the following buffers containing protease inhibitors in this order: ice-cold 1 ml FA-high salt wash buffer, FA-wash 2 buffer, FA-wash 3 buffer, and TE buffer.
19. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 8.0). After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
Enzymatic reactions on beads
20. Add the buffer listed on Table 1, and incubate with mixing for 20 min at 12°C.
Table 1.
Buffer for Polishing Reaction on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| Chromatin on beads | - | 20 ul |
| 10 mM Tris-HCl (pH 8.0) | 10 mM | 27 ul |
| 10x NEBuffer 2 | 1x | 6 ul |
| 10x BSA (1mg/ml) | 1x | 3 ul |
| 3 mM dNTPs | 150 uM | 3 ul |
| T4 DNA polymerase (3 U/ul) | 3 U | 1 ul |
Total volume = 60 ul
NOTE: We use a Thermomixer Comfort 5355R for incubation. If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
21. Repeat step 17 and 18.
22. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 7.5) containing protease inhibitors. After centrifuging for 1 min in a Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
NOTE: The Kinase reaction on steps 23 to 25 is optional. This kinase reaction may improve the yield of ligation, but this reaction (steps 23 to 25) can be skipped.
23. Add the buffer listed on Table 2, and incubate on a Thermomixer Comfort 5355R for 30 min at 37°C.
Table 2.
Buffer for Kinase Reaction on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| Chromatin on beads | - | 20 ul |
| 10 mM Tris-HCl (pH 7.5) | 10 mM | 33 ul |
| 10x T4 DNA ligase buffer | 1X | 6 ul |
| T4 Polynucleotide Kinase (10 U/ul) | 10 U | 1 ul |
Total volume = 60 ul
NOTE: If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
24. Repeat steps 17 and 18 to wash samples.
25. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 8.0) containing protease inhibitors. After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
NOTE: Use 1 ml of 10 mM Tris-HCl buffer (pH 7.5) in step 25 for SOLiDTM Systems 2.0 fragment library preparation instead of 1 ml of 10 mM Tris-HCl buffer (pH 8.0).
NOTE: Perform steps 26-28 for SOLiDTM Systems 5500 Series fragment library preparation. If preparing SOLiDTM Systems 2.0 fragment library, skip steps 26-28.
26. Add the buffer listed on Table 3, and incubated on a Thermomixer Comfort 5355R for 30 min at 37°C.
Table 3.
Buffer for A-Tailing Reaction on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| Chromatin on beads | - | 20 ul |
| 10 mM Tris-HCl (pH 8.0) | 10 mM | 31 ul |
| 10x NEBuffer 2 | 1X | 6 ul |
| 3 mM dATP | 100 uM | 2 ul |
| Klenow Fragment (3′→5′ exo-) (5 U/ul) | 5 U | 1 ul |
Total volume = 60 ul
NOTE: If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
27. Repeat steps 17 and 18 to wash samples.
28. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 7.5) containing protease inhibitors. After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
NOTE: Use P2-T Adaptor for SOLiDTM Systems 5500 Series fragment library preparation. If preparing SOLiDTM Systems 2.0 fragment library, use P2 Adaptor.
29. Add the buffer listed on Table 4, and incubated on a Thermomixer Comfort 5355R for 1.5 hr at 25°C.
Table 4.
Buffer for P2-T Adaptor Ligation on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| Chromatin on beads | - | 20 ul |
| 10 mM Tris-HCl (pH 7.5) | 10 mM | 28 ul |
| 10x T4 DNA ligase buffer | 1x | 6 ul |
| P2-T Adaptor or P2 Adaptor (15 uM) | 1.25 pM | 5 ul |
| T4 DNA ligase (500 U/ul) | 500 U | 1 ul |
Total volume = 60 ul
NOTE: If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
30. Repeat steps 17 and 18.
31. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 7.5) containing protease inhibitors. After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
32. Add the buffer listed on Table 5, and incubated on a Thermomixer Comfort 5355R for 20 min at 30°C.
Table 5.
Buffer for Filling In Reaction on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| DNA on Resin | - | 20 ul |
| 10 mM Tris-HCl (pH 7.5) | 10 mM | 18 ul |
| 10x BSA (1mg/ml) | 2x | 12 ul |
| 10x phi29 DNA polymerase buffer | 1x | 6 ul |
| 3mM dNTPs | 150 uM | 3 ul |
| phi29 DNA polymerase (10 U/ul) | 10 U | 1 ul |
Total volume = 60 ul
NOTE: If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
33. Repeat steps 17 and 18.
34. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 7.5) containing protease inhibitors. After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
NOTE: The kinase reaction on steps 35 to 37 can be skipped if 5′-phosphorylated adaptors are used in the first adaptor ligation (steps 29 to 31).
35. Add the buffer listed on Table 6, and incubated on a Thermomixer Comfort 5355R for 30 min at 37°C.
Table 6.
Buffer for Kinase Reaction on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| Chromatin on beads | - | 20 ul |
| 10 mM Tris-HCl (pH 7.5) | 10 mM | 33 ul |
| 10x T4 DNA ligase buffer | 1X | 6 ul |
| T4 Polynucleotide Kinase (10 U/ul) | 10 U | 1 ul |
Total volume = 60 ul
NOTE: If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
36. Repeat steps 17 and 18.
37. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 9.2) containing protease inhibitors. After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
38. Add the buffer listed on Table 7, and incubated on a Thermomixer Comfort 5355R for 30 min at 37°C.
Table 7.
Buffer for Lambda Exonuclease Digestion on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| Chromatin on beads | - | 20 ul |
| 10 mM Tris-HCl (pH 9.2) | 10 mM | 32 ul |
| 10x lambda exonuclease buffer | 1x | 6 ul |
| Lambda exonulease (5 U/ul) | 10 U | 2 ul |
Total volume = 60 ul
NOTE: If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
39. Repeat step 17 and 18.
40. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 8.0) containing protease inhibitors. After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
41. Add the buffer listed on Table 8, and incubated on a Thermomixer Comfort 5355R for 30 min at 37°C.
Table 8.
Buffer for RecJf Exonuclease Digestion on Beads
| Components | Final concentration | Per reaction |
|---|---|---|
| Chromatin on beads | - | 20 ul |
| 10 mM Tris-HCl (pH 8.0) | 10 mM | 33 ul |
| 10x NEBuffer 2 | 1x | 6 ul |
| RecJf Exonuclease (30 U/ul) | 30 U | 1 ul |
Total volume = 60 ul
NOTE: If using a heat block instead of the Thermomixer Comfort 5355R, tubes should be periodically vortexed to resuspend beads.
42. Repeat steps 17 and 18.
Chromatin elution from antibody beads
43. Add 450 ul of ChIP elution buffer, mix, and incubate 15 min in a 65°C heat block to elute precipitate from antibody beads.
NOTE: Other ChIP elution buffers can be used instead of the ChIP elution buffer described in this protocol.
44. Centrifuge for 1 min in Centrifuge 5424 at 94 × g, room temperature, and transfer eluate (450 ul) to a fresh 1.5-ml microcentrifuge tube.
Reverse cross-linking and PCIA extraction
45. Add 1 ul of Protease K (20 ug/ul) and incubate at a 65°C heat block overnight to reverse cross-link.
46. Add 450 ul of ice-cold phenol/chloroform/isoamyl alcohol. Vortex vigorously for 1 min and separate phases by centrifuging for 6 min in Centrifuge 5424 at 18,400 × g, room temperature.
47. Transfer aqueous (upper) phase to a fresh 1.5-ml microcentrifuge tube and add 1 ml of ice-cold 100% ethanol. Add 1 ul of 20 mg/ml glycogen to help precipitation. Mix thoroughly, and incubate at -80°C for 1 hr.
48. Centrifuge for 15 min in a Centrifuge 5415R at 16.2 × g, 4°C. Discard supernatant.
Note: The pellet in the precipitates will be very small and nearly invisible.
49. Wash the pellets with 500 ul of ice-cold 75% ethanol, centrifuge for 5 min in a Centrifuge 5415R at 16.2 × g, 4°C, and discard supernatant. Allow the sample to dry for 20 min in a Vacufuge Plus Vacuum Concentrator. Resuspend the precipitate in 11 ul of TE and transfer 0.5-ml PCR tube. Store samples at -20°C until use.
Primer extension and 2nd adaptor ligation
50. Thaw the precipitate and add buffer listed on Table 9 to the DNA in TE from step 49 for a total of 19 ul total volume in a tube, and incubated on a thermal cycler for 5 min at 95°C to denature to single-stranded DNA, then for 5 min at 62°C to anneal primer, then leave cool at room temperature.
Table 9.
Buffer for Denaturing and Primer Extension
| Components | Final concentration | Per reaction |
|---|---|---|
| 10x phi29 DNA polymerase buffer | 1x | 2 ul |
| 10x BSA (1 mg/ml) | 1x | 4 ul |
| 3mM dNTPs | 75 uM | 1 ul |
| Library PCR Primer 2 (20 uM) | 1 uM | 1 ul |
Note: We use a DNA Engine® Thermal Cycler PTC-200, any similar cycler should also work.
51. Add 1 ul of phi29 DNA polymerase (10 U/ul) and incubate on a thermal cycler for 20 min at 30°C to make double-strand DNA by primer extension, then for 10 min at 65°C for heat inactivation.
NOTE: Perform step 52 for SOLiDTM Systems 5500 Series fragment library preparation. If preparing SOLiDTM Systems 2.0 fragment library, skip step 52.
52. Add the buffer listed on Table 10 to the reaction mixture from step 51 for a total of 30 ul total volume in a tube, and incubated on a thermal cycler for 30 min at 37°C.
Table 10.
Buffer for A-Tailing Reaction
| Components | Final concentration | Per reaction |
|---|---|---|
| TE | - | 5 ul |
| 10x NEBuffer 2 | 1X | 3 ul |
| 3 mM dATP | 100 uM | 1 ul |
| Klenow Fragment (3′→5′ exo-) (5 U/ul) | 5 U | 1 ul |
NOTE: Perform step 53 for SOLiDTM Systems 5500 Series fragment library preparation. If preparing SOLiDTM Systems 2.0 fragment library, skip step 53.
53. Leave cool at room temperature and add the buffer listed on Table 11 to the reaction mixture from step 52 for a total of 40 ul total volume in a tube, and incubated on a thermal cycler for 1 hr at 25°C.
Table 11.
Buffer for P1-T Adaptor Ligation
| Components | Final concentration | Per reaction |
|---|---|---|
| TE | - | 4 ul |
| 10x T4 DNA ligase buffer | 1x | 4 ul |
| P1-T Adaptor (15 uM) | 0.4 uM | 1 ul |
| T4 DNA ligase (500 U/ul) | 500 U | 1 ul |
NOTE: Skip step 54 for SOLiDTM Systems 5500 Series fragment library preparation. If preparing SOLiDTM Systems 2.0 fragment library, perform step 54.
54. Leave cool at room temperature and add the buffer listed on Table 12 to the reaction mixture from step 51 for a total of 30 ul total volume in a tube, and incubated on DNA Engine® Thermal Cycler PTC-200 for 1 hr at 25°C.
Table 12.
Buffer for P1-T Adaptor Ligation
| Components | Final concentration | Per reaction |
|---|---|---|
| TE | - | 5 ul |
| 10x T4 DNA ligase buffer | 1x | 3 ul |
| P1 Adaptor (15 uM) | 0.4 uM | 1 ul |
| T4 DNA ligase (500 U/ul) | 500 U | 1 ul |
AMPure purification
55. Transfer DNA samples to 1.5-ml microcentrifuge tube. Gently shake AMPure bottle to resuspend magnetic particles.
56. Add 72 ul of AMPure beads for SOLiDTM Systems 5500 Series fragment library preparation and mix thoroughly by pipette mixing 10 times. Place the 1.5-ml microcentrifuge tube onto a DynaMagTM for 2 min to separate beads from the solution.
NOTE: Add 52 ul of AMPure beads for SOLiDTM Systems 2.0 fragment library preparation instead of 72 ul of AMPure beads.
57. Aspirate the cleared solution from the tube on a magnetic rack and discard. Dispense 200 ul of 70% ethanol to the sample and incubate for 30 sec at room temperature and mix thoroughly by pipette mixing 10 times. Place the tube onto a DynaMagTM for 2 min to separate beads from the solution. Repeat twice.
58. Aspirate the cleared solution from the tube on a DynaMagTM and dry beads at room temperature for 20 min (be sure to remove all of the ethanol from the bottom of the tube).
59. Off a DynaMagTM, add 30 ul of TE (pH 8.0) and pipette mix 10 times. Place the tube onto a DynaMagTM for 2 min to separate beads from the solution. Transfer the 30 ul eluate to a fresh 0.5-ml PCR tube and store samples at -20°C until use.
PCR amplification
60. Set up a PCR reaction with the buffer listed on Table 13 to the eluate from step 58 for a total of 40 ul total volume in a tube. Carry out hot-start PCR on a DNA Engine® Thermal Cycler PTC-200 using the following parameters:
| Initial step: | 5 min | 95°C |
| 12-25 cycles: | 15 sec | 95°C |
| 15 sec | 62°C | |
| 1 min | 72°C | |
| Final step: | 5 min | 72°C |
Table 13.
Buffer for PCR reaction
| Components | Final concentration | Per reaction |
|---|---|---|
| TE | - | 3 ul |
| 10x standard taq reaction buffer | 1x | 4 ul |
| 25 mM dNTPs | 0.3 mM | 0.5 ul |
| Library PCR Primer 1 (20 uM) | 0.5 uM | 1 ul |
| Library PCR Primer 2 (20 uM) | 0.5 uM | 1 ul |
| Taq DNA polymerase (5 U/ul) | 2.5 U | 0.5 ul |
NOTE: Minimal cycling is desirable for avoiding over-amplification. The number of cycles should be decided based on the amount of starting material used for ChIP.
61. Perform next-generation sequencing by following the manufacture’s instruction.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps
ST buffer
10 mM Tris-HCl (pH 7.5)
100 mM NaCl
Store up to 1 year at -4°C
FA-lysis buffer
50 mM HEPES
150 mM NaCl
2 mM EDTA (pH 8.0)
1% Triton X-100
0.1% NaDeoxycholate
Store up to 1 year at -4°C
FA-high salt wash buffer
50 mM HEPES
1 M NaCl
2 mM EDTA (pH 8.0)
1% Triton X-100
0.1% NaDeoxycholate
Store up to 1 year at -4°C
FA-wash 2 buffer
50 mM Hepes/KOH
0.5 M NaCl
2 mM EDTA (pH 8.0)
1% TritonX-100
0.1% NaDeoxycholate
Store up to 1 year at -4°C
FA-wash 3 buffer
25 mM LiCl
1% NP40-Nonidet (IPEGAL)
1% NaDeoxycholate
2 mM EDTA
10 mM Tris-Cl (pH 8.0)
Store up to 1 year at -4°C
Tris-EDTA buffer (TE)
10 mM Tris-HCl (pH 8.0)
1 mM EDTA (pH 8.0)
ChIP elution buffer
25 mM Trizma 2 mM EDTA (pH 8.0)
200 mM NaCl
0.5% SDS
Store at room temperature
BASIC PROTOCOL 2
Identification of the genomic location of DNA-binding proteins at near nucleotide accuracy in mammalian cells
This protocol was designed to detect genomic locations of a DNA-binding protein in mammalian cells at near single base pair resolution. First, chromatin immunoprecipitation (ChIP) is performed to purify protein-DNA complexes, then several enzymatic reactions including lambda exonuclease digestion are performed while protein-DNA complexes are still on the beads. These enzymatic reactions allow us to detect the exact boundaries of crosslinked protein-DNA complex at near nucleotide resolution. This protocol was also designed for use with next-generation sequencing instruments.
NOTE: Use only molecular biology-grade water (e.g. DNase, RNase, and Protease free, deionized, distilled) in all steps and solutions.
Materials
NOTE: Only materials which are different from the basic protocol 1 are listed here.
Sample materials
Mammalian cell culture
Reagents and solutions
Mixed micelle buffer (see recipe)
High salt buffer (see recipe)
Detergent buffer (see recipe)
Chromatin immunoprecipitation (ChIP)
1. Harvest, cross-link, lyse and sonicate the DNA of mammalian cells following the protocols listed in Unit 21.YY.
Note: The protocol is the same as used for doing any standard chromatin immunoprecipitation up to this point, and any normal protocol that works for ChIP in mammalian cells might be used through the sonication step. Use the following from when the antibody is added and for all subsequent steps.
2. Transfer 2.5×106 cell-equivalents to 15-ml polypropylene tube, and add antibody against the protein of interest to the chromatin extract and mix. Incubate on a rotating wheel overnight at 4°C.
3. Using a wide-bore pipette tip (e.g. a 200-ul pipette tip with the narrow end cut off), add 20 ul of sepharose beads (40 ul of 50% beads slurry) to each sample and mix. Incubating on a rotating wheel 1.5 hr at 4°C.
4. Centrifuge for 1 min in Centrifuge 5424 at 94 × g, room temperature, and remove the supernatant by aspiration. Add 0.5 ml ice-cold IP buffer containing protease inhibitors. Using a wide-bore pipette tip, resuspend the beads by pipetting up and down, and transfer to a fresh 1.5-ml microcentrifuge tube. Centrifuge 1 min in Centrifuge 5424 at 94 × g, room temperature. Remove supernatant by aspiration.
5. Wash the beads by adding ice-cold 1 ml Mixed micelle buffer containing protease inhibitors and incubating 5 min at room temperature on a rotating wheel. Precipitate beads by centrifuging 1 min in Centrifuge 5424 at 94 × g, room temperature. Remove the supernatant by aspiration. Repeat (total two washes).
6. Repeat step 4 with the following buffers containing protease inhibitors in this order: ice-cold 1 ml Mixed micelle buffer, High salt buffer, Detergent buffer, and TE buffer.
7. Add ice-cold 1 ml of 10 mM Tris-HCl buffer (pH 8.0) containing protease inhibitors. After centrifuging for 1 min in Centrifuge 5424 at 94 × g, room temperature, remove as much wash solution as possible without disturbing the beads.
Enzymatic reactions on beads
8. Follow the basic protocol 1 from this step to the end.
NOTE: Perform steps 5 and 6 in the basic protocol 2 instead of step 17 and 18 in the basic protocol 1 when washing after each enzymatic reaction on beads.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps
Mixed micelle buffer
150 mM NaCl
0.2% SDS
20 mM Tris-Cl (pH 8.0)
5 mM EDTA
5.2% sucrose
1% Triton X-100
Store up to 1 year at -4°C
High salt buffer
250 mM NaCl
5 mM Tris-Cl (pH 8.0)
0.5 mM EDTA
0.05% sodium deoxycholate
25 mM HEPES (pH 8.0)
0.5% Triton X-100
Store up to 1 year at -4°C
Detergent buffer
250 mM LiCl
10 mM Tris-Cl (pH 8.0)
10 mM EDTA
0.5% NP40-Nonidet (IPEGAL)
0.5% Sodium Deoxycholate
Store up to 1 year at -4°C
COMMENTARY
Background Information
Proteins bind to DNA to regulate genes. Thus, it is important to determine where proteins are bound in the genome to understand how they regulate gene expression (Lee et al., 2002; Ptashne and Gann, 1997; Struhl, 1995). Chromatin immunoprecipitation (ChIP) is the most widely used method to identify where DNA-binding proteins are located on the genome. However, ChIP contains non-specifically bound DNA during immunoprecipitation, so ChIP creates background signal (Peng et al., 2007; Rozowsky et al., 2009; Tuteja et al., 2009). Moreover, size heterogeneity of randomly sheared ChIP DNA technically limits mapping resolution. ChIP-exo methodology described here can precisely map genomic binding locations of a protein at near base resolution and eliminate most of background signal (Rhee and Pugh, 2011).
Critical Parameters
Although lambda exonuclease digests 5′ DNA from sonicated ends of DNA to protein-DNA cross-linking points, smaller starting sizes of sonicated DNA fragments are better. In yeast cells, a median size of 250-300 bp DNA fragments is preferred after sonication.
Lambda exonuclease is a highly processive enzyme that acts in the 5′ to 3′ direction to remove 5′ mononucleotides from double-stranded DNA (Little, 1981). Therefore, it degrades only one strand of double-stranded DNA, leaving a single-stranded DNA product. Since the substrate is preferred to be 5′-phosphorylated double-stranded DNA, a kinase reaction (step 35 in protocol 1) before lambda exonuclease is optimal for lambda exonuclease activity.
RecJf is a single-stranded specific exonuclease that acts in the 5′ to 3′ direction to remove 5′ mononucleotide (Lovett and Kolodner, 1989). Therefore it acts on the product generated by lambda exonuclease. Unbound DNA can nonspecifically adsorb to sepharose beads during immunoprecipitation, producing “background” sequencing tags throughout the genome, which decrease the sensitivity of ChIP. The combination of lambda exonuclease and RecJf exonucleases (step 41 in protocol 1) removes a substantial portion of this background.
When PCR is performed, minimal cycling is desirable for avoiding over-amplification (i.e. selective amplification of certain sequences more so than others). Although The number of cycles should be decided based on the amount of starting material used for ChIP, 12, 15, 18, 21, and 25 cycles can be tested and PCR products can be checked on an agarose gel.
Troubleshooting
If the most of material seems to be not converted into double-stranded DNA after the primer extension (steps 50 and 51 in protocol 1), the efficiency of the first adaptor ligation (step 29 in protocol 1) can be checked by ligation mediated (LM)-PCR using PCR primer for the first adaptor. In addition, the presence of ChIP material can be checked by locus specific PCR for known binding locations. If different primer sequences are used for the primer extension (steps 50-51 in protocol 1), annealing temperature (62°C in this protocol) can be changed by titration at various temperatures. Furthermore, annealing temperature (62°C in this protocol) in PCR amplification (step 60 in protocol 1) may be need to changed if different PCR primer sequences are used.
If the sequencing data show relatively low complexity and have many repeat sequencing reads, more amount of cells may be used for ChIP, although this protocol suggests to use 250 ml yeast cells (OD600 ~0.8 to 1.0) and 2.5×106 mammalian cells for ChIP. Moreover, the ratio of cells to antibody can be titrated, which depends on the protein of interest, the antibody efficiency, and the accuracy of the concentration of antibody. Fewer PCR cycles might also help reduce repeat sequencing reads.
If there is a too strong signal from adaptor dimers on the final PCR product (step 59 in protocol 1), the size selection is recommended to reduce it. The size selection on polyacrylamide gels can be performed after AMPure purification step (between step 59 and 60 in protocol 1). It allows for the exclusion of unligated adaptor DNA. Fewer PCR cycles might also help reduce selective amplification of adaptor dimer DNA sequences.
Anticipated Results
Starting with the sonicated chromatin with a median size of 250-300 bp fragments, the median size of the final PCR product (step 60 in protocol 1) should be about 225-250 bp (125-150 bp of DNA fragment and 97 bp of ligation adaptors) (Rhee and Pugh, 2012). It is possible that other minor products from adaptor dimers will also be visible.
Time Considerations
The basic protocol 1 can be completed in three and half days. Harvesting cells, cross-linking, and quenching require one day. Cell lysis and sonication require a half day. Chromatin immunoprecipitation (ChIP), enzymatic reactions on beads, and elution require one day. Reverse cross-linking, PCIA extraction, adaptor ligation, and PCR require one day.
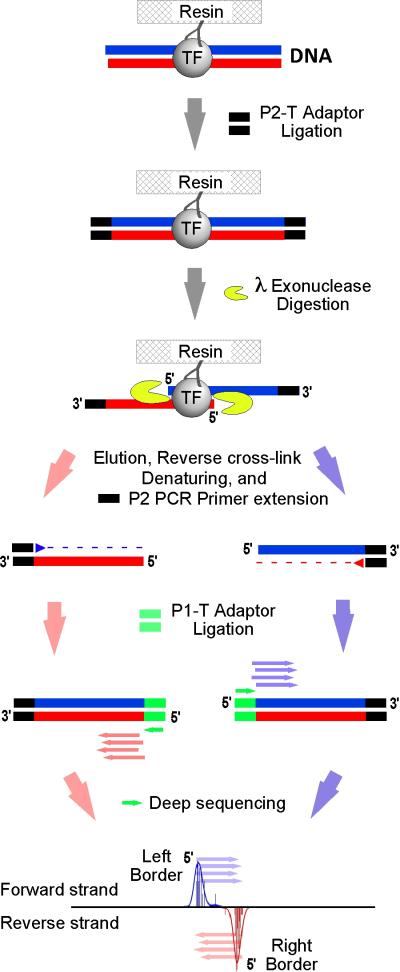
Figure 1.
Scheme for ChIP-exo. After ChIP, 3′ sonicated ends are demarcated from the eventual exonuclease-treated 5′ end, by ligating P2-T Adaptor to the ChIP DNA on the resin prior to exonuclease digestion. 5′ to 3′ exonuclease trimming up to the site of cross-linking selectively eliminates the P2-T adaptor sequence attached at the 5′ end of each strand. After crosslink reversal, eluted single-stranded DNA is made double-stranded by P2 PCR Primer extension. Then, a P1-T Adaptor was ligated to the exonuclease-treated end. The resulting library is subjected to high-throughput sequencing. Mapping the 5′ ends of the resulting sequencing tags to the reference genome demarcates the exonuclease barrier and thus the precise site of protein-DNA cross-linking (Rhee and Pugh, 2011).
Literature Cited
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Little JW. Lambda exonuclease. Gene Amplif Anal. 1981;2:135–145. [PubMed] [Google Scholar]
- Lovett ST, Kolodner RD. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Alekseyenko AA, Larschan E, Kuroda MI, Park PJ. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic preinitiation complexes. Nature. 2012 doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Key Reference
- Rhee HS, Pugh BF. This is the first paper, which described a ChIP-exo method for various transcription factors in yeast to human. Moreover, this paper shows the proof of principle of ChIP-exo, which identified a nearly complete set of genomic bound locations at near nucleotide resolution. 2011 [Google Scholar]