Abstract
Objectives
To summarize all clinical studies evaluating the prognostic role of gemcitabine metabolic genes in pancreatico-biliary (PB) cancer patients receiving gemcitabine (GEM) therapy in the neoadjuvant, adjuvant or palliative settings.
Methods
Meta-analyses were performed to calculate the pooled hazard rations (HRs) for each gene by each clinical outcome (overall, disease free, and progression free survivals) using a random-effects approach.
Results
The search strategy identified 16 eligible studies, comprised of 632 PB patients total, with moderate quality. Compared to low expression, pooled hazards ratios for OS of hENT1, dCK, RRM1, RRM2, and DPD were 0.37 (95%CI, 0.28–0.47), 0.40 (95%CI, 0.20-0.80), 2.21 (95%CI, 1.12-4.36), 2.13 (95%CI, 1.00-4.52), and 1.91 (95%CI, 1.16-3.17), respectively. A similar trend was observed for each of these biomarkers in DFS and PFS prognostication. Subgroup analyses for hENT1 showed a comparable survival correlation in the adjuvant and palliative settings.
Conclusions
High expression of hENT1 in PB cancer patients receiving GEM-based adjuvant therapy is associated with improved OS and DFS and may be the best examined prognostic marker to date. Evidence for other biomarkers is limited by a small number of publications investigating these markers.
Keywords: Meta-analysis, pancreatic cancer, biliary cancer, gemcitabine, biomarkers
INTRODUCTION
Pancreatic and biliary (PB) cancers are highly lethal neoplasms with an overall 5-year survival rate between 5-15% 1, 2. Surgical resection remains the only potential therapeutic cure for patients with early stage disease, but disease relapse is frequent. In the adjuvant setting, systemic chemotherapy for pancreatic cancer (PDAC) has been shown to improve disease-specific and -free survival in phase III trials3-5. In most cases, patients are diagnosed with metastatic or locally advanced disease, and are not candidates for surgical resection. Select patients from latter group may benefit from surgical resection after a course of downstaging chemothrapy6. In the former group, systemic chemotherapy improves survival and quality of life7. In each disease-stage group, gemcitabine (GEM) is the most frequently used agent.
The present approach for selecting chemotherapeutics for the treatment of PB cancer patients depends on institutional preference. However, inter-individual variations in chemotherapeutic response is a known phenomenon. The failure to demonstrate a survival difference between different classes of chemotherapeutics in multiple phase III PDAC trials suggests that improved drug and patient matching are needed3, 4. Chemotherapeutic response heterogenity can be attributable to genetic variations in the expression level of drug-metabolizing enzymes, targets, or transporters8. GEM resistance has been linked to several key genes involved in its metabolism9, 10. These GEM-related biomarkers include nucleotide transporters such as human nucleoside transporter subunit (hENT) 1 and 2, human concentrative nucleoside transporter (hCNT) 1 and 3, metabolizing enzymes such as deoxycytidine kinase (dCK), target enzymes such as ribonucleoside-diphosphate reductase (RRM) subunits 1 and 2, deactivating enzymes such as cytidine deaminase (CDA), deoxycytidylate deaminase (DCD) and 5′ nucleotidase (5′NT), and nucleoside metabolic enzymes such as thymidylate synthase (TS), thymidine phosphorylase (TP), and orotate phosphoribosyltransferase (OPRT).
Given the widespread use of GEM in the treatment of PB cancer and the potential benefits of using biomarkers to personalize therapy, we sought to summarize all clinical studies and determine the prognostic relevance of GEM-related biomarkers in stratifying survival outcomes of PB cancer patients receiving GEM-based chemotherapy.
MATERIALS AND METHODS
Inclusion and Exclusion Criteria
We identified all publications that studied the association between survival outcomes and the expression level of GEM metabolic pathway related biomarkers in PB cancer patients treated with GEM-based regimen in either adjuvant, neoadjuvant, or palliative settings. Only studies that used patient samples/tissues to determine the expression level of GEM biomarkers were included. The IHC-based marker studies were required to satisfy the reporting recommendations by NCI-EORTC on Tumor Marker Prognostic Studies (REMARK) 11 as adapted and modified by Ansari et al with the exception of multivariable survival analysis 12. It was expanded to include all levels of survival data to comprehensively capture negative results reporting. Exclusion criteria included in vitro studies, non-GEM based therapy, or a lack of data sufficient for hazards ratio determination. In situations of insufficient data, attempts to contact primary authors were made.
Search Strategy for Identification of Studies
All studies were searched in December 2011 and abstracted from PUBMED, related-articles function in PUBMED, and citation from reference lists. The following search terms were used combined with Boolean operator with no filter applied: “pancreatic cancer,” “biliary cancer,” “cholangiocarcinoma,” “gemcitabine,” “chemoresistance,” “chemosensitivity,” “sensitivity,” “resistance,” “thymidylate synthase,” “thymidine kinase,” “TK2,” “CTP synthase,” “equilibrative nucleoside,” “hENT*,” “SLC29A1,” “hCNT*,” “CNT1,” “CNT3,” “concentrative nucleoside,” “SLC28A1,” “SLC28A3,” “CDA,” “cytidine deaminase,” “DCTD,” “deoxycytidylate deaminase,” “5′-nucleotidase,” “RRM1,” “RRM2,” “ribonucleotide reductase,” “deoxycytidine kinase,” and “dCK.”
Methods of Review
Data abstraction was completed independently by C.W. Results were reviewed by C.W. and T.D. to reach consensus for queries that had arisen during the review process.
The following parameters were collected from included studies: year of publication, author, sample size, cancer type, treatment setting, biomarker detection method, type of clinical samples used, preservation methods, biomarker(s) analyzed in the study, median overall (OS), disease free (DFS), and progression free (PFS) survivals, hazards ratios (HR) and their confidence bounds (CI), response rates, and distribution of high and low biomarker expression in the cohort. Several studies analyzed multiple biomarkers but may report a lack of statistical significance for some of the biomarkers examined. Those negative results were included in the analysis.
Methodological Quality Assessment
Newcastle-Ottawa Quality Assessment Scale for cohort studies was used to assess methodological quality as recommended by the Cochrane Non-Randomized Studies Methods Working Group and has been used previously in other biomarker meta-analyses 13, 14.
Assessment of Reporting Bias Risk
Publication bias was assessed by using funnel plots on adequately sized subgroups (>=5). Trim and fill method was employed to statistically correct for publication bias 15.
Statistical Analyses
Reported HRs (comparing low vs. high marker expression on the relevant survival outcome) and their CI were recorded whenever possible. Several studies report only Kaplan Meier survival analysis. In those cases, HRs were extracted from the survival curves or rates using methods recommended by the Cochrane Handbook 16. Meta-analyses were performed to calculate the pooled HRs for each gene by each clinical outcome using a random-effects approach, which accounts for inter-study heterogeneity. Heterogeneity was evaluated by the Cochran Q statistic (significance p < .10). Z-test was performed to test the overall significance of summarized HRs (significance p < 0.05). Statistical analyses were performed using Stata 12 (College Station, Texas).
Occasionally, a study reported median survival times instead of HRs. For these studies, a hazard rate was estimated by using an exponential survival curve model. The HR was then formed by taking a ratio of these rates. The CI was estimated by simulating event times based on an the same model. In the simulation group sample sizes equaled the observed sample size in the respective publication. A HR was computed for each iteration (of 10,000) and the lower 2.5% and upper 97.5% percentiles were taken to represent the upper and lower bounds of a 95% CI.
RESULTS
Literature Search and Publication trend of GEM metabolic proteins as prognostic biomarkers in patients with PB cancers receiving GEM treatment
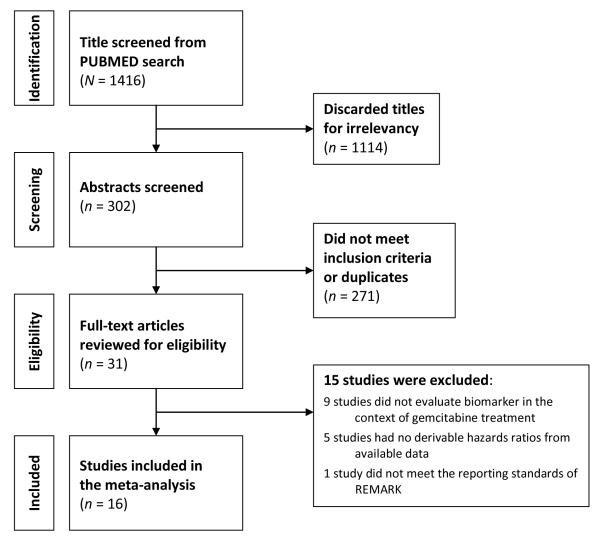
Fig. 1 illustrates the study selection flow. We identified 1416 potentially relevant titles, of which 302 were selected for abstract review. After excluding 271 studies, 31 full-texted articles were evaluated for eligibility for meta-analysis. Ten studies were subsequently excluded, because those studies did not evaluate biomarkers in the context of GEM treatment or survival outcome. This resulted in a total of 21 studies 17-38. Figure 2 summarizes the frequency each biomarker was examined and reported on survival in these 21 studies. There were 9, 8, 2, and 2 studies that examined the markers in the adjuvant 18, 23, 24, 27-31, 36, palliative 19, 20, 22, 26, 33-35, 37, both 17, 25, and neoadjuvant settings 21, 32, respectively. Ten studies examined multiple biomarkers in the same paper 18-20, 24, 25, 27-29, 32, 37. In those cases, each study result is recorded separately.
Figure 1.
Study Flow.
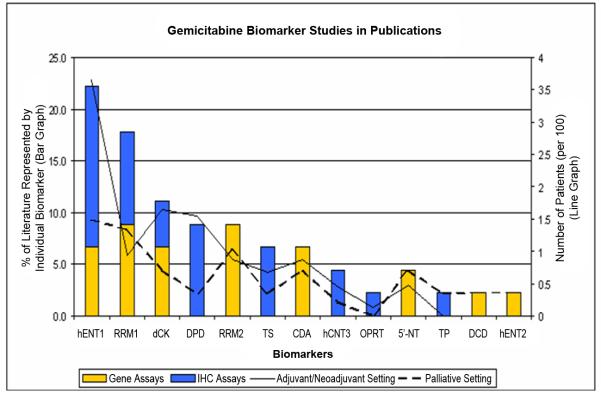
Figure 2. Publication trend for biomarker studies that examined the expression of molecular markers involved in gemcitabine metabolism.
Publication summary of gemcitabine metabolism biomarker studies. Bar graph represents the percent distribution of each biomarker examined & reported on survival in these 21 studies. Stacked color bars represent the distribution of the index assays employed (BLUE bar: immunohistochemistry; YELLOW bar: gene-expression quantitative assay). Line graph represents the total number of patients samples (per 100) evaluated for each biomarker (solid line: adjuvant and neoadjuvant settings combined; dashed line: palliative setting).
Immunohistochemistry was the most utilized assay (n=16). To ensure an adequate number of evidence available for synthesizing a meaningful meta-conclusion, the top 5 most published biomarkers were selected as the focus of our meta-analysis. They include hENT1 (n=10), dCK (n = 4), RRM1 (n=4), RRM2 (n=3), and DPD (n=3). This restriction led to the final inclusion of the 16 studies (Table 1).
Table 1.
Summary of all eligible studies examining the association between biomarker expression and survival in gemctiabine-treated pancreatico-biliary cancer patients.
| Marker | Reference | Year | n | Assay/ Specimen |
Assay Cutpoints (n) | Cancer Type |
|---|---|---|---|---|---|---|
| hENT1 | Kondo (28) |
2011 | 86 | IHC FFPE |
Low (23) or high (63) on a 0-3 score system; high defined as score >=2 in > 50% cells. |
Pancreas |
| Morinaga (31) |
2011 | 27 | IHC FFPE |
Low (11) and high (16) on a 6-point score system; cutoff point at mid-point of the score system. |
Pancreas | |
| Murata (32) |
2011 | 55 | IHC FFPE |
Negative (16) or positive (39) on a 0-3 score system; negative defined as score =0 or =1 in >50% cells. |
Pancreas | |
| Fujita (24) |
2010 | 40 | qPCR FFPE |
Low (26) or high (14); cutoff determined by recursive descent partition analysis. |
Pancreas | |
| Farrell (23) |
2009 | 91 | IHC FFPE |
Negative (18) or positive (73); negative defined as no staining in > 50% cells. |
Pancreas | |
| Marechal (29) |
2009 | 45 | IHC FFPE |
Low (26) or high (19) on a 0-300 staining score system; cutoff point at the median. |
Pancreas | |
| Giovannetti (25) |
2006 | 81 | qPCR Frozen |
Low (27), medium (28), or high (26) gene expression level; tertile cutoff. |
Pancreas | |
| Spratlin (37) |
2004 | 21 | IHC FFPE |
Negative (12) or positive (9) on a 0-2 score system; negative defined as 0. |
Pancreas | |
| Santini (35) |
2011 | 31 | IHC FFPE |
Negative (10) or positive staining (21) on a 0-2 score system; positive defined as > 50% cell stained. |
Bile duct | |
| Borbath (22) |
2011 | 43 | IHC FFPE |
Low (9) or high (17); low defined as no staining in > 50% cells. |
Bile duct | |
|
| ||||||
| dCK | Fujita (24) |
2010 | 40 | qPCR FFPE |
Low (27) or high (13); cutoff determined by recursive descent partition analysis. |
Pancreas |
| Marechal (30) |
2010 | 45 | IHC FFPE |
Low (26) or high (19) on a 0-200 staining score system; cutoff at median. |
Pancreas | |
| Giovannetti (25) |
2006 | 81 | qPCR Frozen |
Low (25), medium (31), or high (25); tertile of gene expression level. |
Pancreas | |
| Sebastiani (26) |
2006 | 40 | IHC PE |
Low (9) or high (23) on a 0-3 score system; low defined as score <2. |
Pancreas | |
|
| ||||||
| RRM1 | Fujita (24) |
2010 | 40 | qPCR FFPE |
Low (12) or high (28); cutoff determined by recursive descent partition analysis. |
Pancreas |
| Nakahira (33) |
2007 | 18 | QPCR Frozen |
Low (9) or high (9) gene expression level; cutoff at median. |
Pancreas | |
| Giovannetti (25) |
2006 | 81 | qPCR Frozen |
Low (29), medium (25), or high (27) gene expression level; tertile cutoff. |
Pancreas | |
| Nakamura (34) |
2010 | 10 | qDFIHC PE |
Low (6) or high (4) quantitative fluroscence level; cutoff at mean. |
Bile Duct | |
|
| ||||||
| RRM2 | Fujita (24) |
2010 | 40 | qPCR FFPE |
Low (13) or high (27); cutoff determined by recursive descent partition analysis. |
Pancreas |
| Itoi (26) |
2007 | 31 | qPCR EUS- FNAB |
Low (18) or high (13) gene expression level; cutoff at median. |
Pancreas | |
| Giovannetti (25) |
2006 | 81 | qPCR Frozen |
Low (18), medium (33), or high (30) gene expression level; tertile cutoff. |
Pancreas | |
|
| ||||||
| DPD | Kondo (28) |
2011 | 86 | IHC FFPE |
Low (51) or high (35) on a 0-3 score system; high defined as score >=2 in > 30% cells. |
Pancreas |
| Murata (32) |
2011 | 55 | IHC FFPE |
Low (15) or high (40); low defined as positive staining in < 30% cells. |
Pancreas | |
| Komori (27) |
2010 | 13 | IHC FFPE |
Low (6) or high (7) expression index measuring % cell staining; cutoff at median. |
Pancreas | |
IHC = immunohistochemistry; qPCR = real time polymerase chain reaction; FFPE = formalin fixed paraffin embedded tissue; PE = paraffin embedded tissue
Study quality
Table 1 summarizes the methodologic quality of the 16 included studies. Overall, all the studies exhibited moderate to high level methodological quality. Ten directly reported HRs. HRs and their CIs were back-calculable for the remaining 6 studies. Fifteen out of 16 studies segregated comparison groups according to high/low expression groups. Only one study used a tertile cutoff point. To facilitate analysis, we used values derived from the highest and lowest tertile groups. Due to the limited number of publications examining biomarkars in the neoadjuvant setting, data derived from the neoadjuvant setting were combined with the adjuvant setting group.
Meta-analysis
Overall Survival
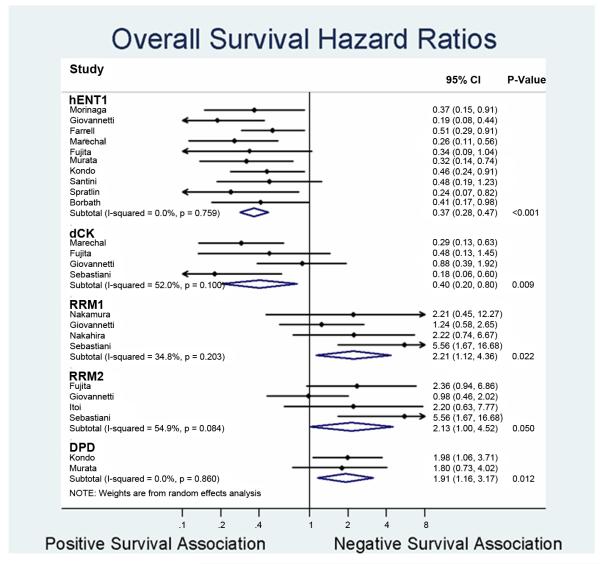
The prognostic value of hENT1, dCK, RRM1, RRM2, and DPD for overall survival in PB cancer were evaluated in 10, 4, 4, 3, and 2 studies, respectively (Figure 3). High hENT1 (HR=0.37; 95%CI 0.28 – 0.47) and dCK (HR=0.40; 95%CI 0.20 – 0.80) expression level were associated with improved OS. In contrast, high expression level of RRM1 (HR=2.21; 95%CI 1.12 – 4.36), RRM2 (HR=2.13; 95%CI 1.00 – 4.52), and DPD (HR=1.91; 95%CI 1.16 – 3.17) were negatively associated with OS. As hENT1 had the most number of publications, further study was carried out to increase the stringency of the analysis. Subgroup analysis that examined the prognosticative role of hENT1 in the adjuvant and palliative settings demonstrated that hENT1 is equally prognosticative in each disease stage (HR=0.39; 95%CI 0.29 – 0.54 vs HR=0.39; 95%CI 0.22-0.68), which remained consistent after controlling for publication bias using trim-and-fill statistical methodology (OS: HR=0.44; 95%CI 0.34-0.57 and DFS: HR=0.45; 95%CI 0.34-0.60). There were low (hENT1, RRM1, RRM2, and DPD) to moderate (dCK) level of heterogeneity among the pooled studies (Figure 3).
Figure 3. Forest plot summarizing hazards ratios comparing high versus low expression levels of the individual biomarkers.
Forest plot for overall survival. Data from each study are summarized. Hazards ratios and their 95% confidence bounds are reported. Study heterogeneity are represented by p-val derived from the Cochrane Q test (p >0.1 denotes significance), with corresponding magnitude represented by I2 value. P-value column denotes statistical significance of the summarized HRs.
Disease free survival
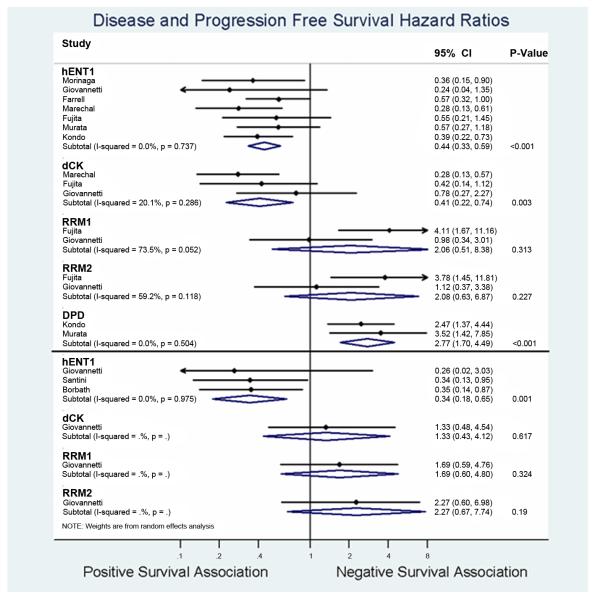
The prognostic role of hENT1, dCK, RRM1, RRM2, and DPD for DFS after PB cancer resection for early stage patients were evaluated in 7, 3, 2, 2, 2 studies, respectively (Figure 4a). High expression of hENT1 (HR=0.44; 95%CI 0.33-0.59) and dCK (HR=0.41; 95%CI 0.22-0.74) were associated with improved DFS. In contrast, high expression of DPD was associated with decreased DFS (HR 2.77; 95%CI 1.70-4.49). Minimal heterogenity was found among all the pooled studies on DFS for hENT1, dCK, and DPD. RRM1 and RRM2 were not significant prognosticators of DFS.
Figure 4a and 4b. Forest plot summarizing hazards ratios comparing high versus low expression levels of the individual biomarkers.
Forest plot for disease free survival (a) and progression free survival (b). Data from each study are summarized. Hazards ratios and their 95% confidence bounds are reported. Study heterogeneity are represented by p-val derived from the Cochrane Q test (p >0.1 denotes significance), with corresponding magnitude represented by I2 value. P-value column denotes statistical significance of the summarized HRs.
Progression free survival
Figure 4b summarizes the meta-analysis results. In summary, there is a limited number of studies reporting the association of biomarkers with PFS in patients with advanced stage disease who were treated palliatively. The pooled hazard ratios for for hENT1 was 0.34 (95%CI 0.18 –0.65), with low inconsistency among the 3 studies pooled (I2=0.0%). In contrast, dCK, RRM1, and RRM2 were not significantly associated with PFS.
Other biomarkers
There were limited number studies that examined the prognostic significance of TS, CDA, hCNT3, 5-NT, OPRT, TP, DCD, hENT2 biomarkers in the context of GEM-based therapy. No OS correlation was found for biomarkers CDA, OPRT, TP, and 5-NT 18, 19. One study reported a statistically significant association between low TS expression and longer DFS on univariate analysis (median DFS 15.9 vs 7 months; logrank p=0.03) in the adjuvant setting of pancreatic cancer 27. Two other studies found no association between survival and TS expression levels in the neoadjuvant and palliative settings 20, 32. For hCNT3, one study reported an association between high hCNT3 expression level and OS on multivariable analysis (HR: 2.65, 95% CI: 1.19-5.87; p=0.017) and DFS (HR: 2.09, 95%CI: 0.99-4.42; p=0.052) 29. However, another study reported no association between hCNT3 expression level and overall survival in one publication on advanced stage PDAC patients receiving GEM-based therapy 37. Ashida et al reported a lack of prognostic value of TP and hENT2 for response to GEM-based therapy by the RECIST criteria in advanced stage PDAC 19.
Publication Bias Assessment
The plots for hENT1 OS and DFS were symmetric and the effect size did not appear to depend on the standard error of the reported HRs (Fig. 1). In conjuction with the results of Trim and Fill analysis performed on the same subgroups, these methods showed that publication bias is probably not present.
DISCUSSION
Our meta-analysis provides a summary of existing evidence on the prognostic biomarkers involved in the GEM metabolic pathways for GEM-based therapy in PB cancers. The majority of the publications on molecular biomarkers of GEM therapy evaluated hENT1 expression, which provided the strongest evidence to date for its prognostic value in the adjuvant and palliative settings. These results hold true after statistical correction for publication bias. Other biomarkers, such as dCK, RRM1, RRM2, and DPD, are also prognosticative in selected treatment settings and survival endpoints; although the evidence is limited by a small number of publications investigating these markers. We believe that disease-stage subgroup analysis for hENT1 is necessary since tumor genetic landscapes are highly dynamic during cancer progression, and may result in nonlinear protein expression pattern changes.
Our systematic literature search on GEM metabolic biomarkers revealed that the prognostic role of hENT1 is most consistently shown in pancreatic cancer patients treated with GEM. The association between hENT1 expression level and survival in patients receiving non-GEM chemotheradpy is controversial. In one study, Farrell et al showed that hNET1 expression levels is not associated with survival in patients receiving 5-flurouracil 23. In contrast, Kim et al showed that hENT1 is prognosticative in patients receiving non-GEM adjuvant chemotherapy38. Therefore, the predictive role of hNET1 for identifying GEM-responsive patient subgroups is unclear. The prognostic role of hENT1 appears to be restricted to patients undergoing chemotherapy treatment, since hENT1 expression level did not correlate with survival in patients who did not receive adjuvant therapy 24.
By restricting our scope of analysis to gene and protein expression, we have excluded other classes of molecular prognosticators such as single nucleotide polymorphisms (SNPs). Interestingly, intracoding region SNPs have not been associated with functional changes in several GEM-metaboic gene biomarkers 9. Three independent studies that examined SNPs of gemcitabine metabolic genes reported a lack of association between these genomic markers with survival 39-41. The lack of association was confirmed in one genome-wide association study 42. However, genetic polymorphisms in CDA are linked to gemcitabine clearance and possibility toxicities 40, 43. Our current limited understanding of the signficance of these genetic variants restrict their clinical utility.
There are other molecular biomarkers of GEM resistance that are not involved in its metabolic pathway. They include the activation of PI3K/AKT/NFkB and stem cell maintenance 8. However, these molecular pathways are also implicated in chemoresistance to other chemotherapeutic drugs such as 5-flurouracil 8. Thus, they are more likely to represent markers for chemoresistance in general with less specificity as prognostic biomarkers for patients receiving GEM therapy. Another emerging biomarker class is the microRNAs. For example, miR-21 has been associated with GEM-resistance 44. A combinatorial appraoch of using different classes of biomarkers may synergistically increase their overall prognosticative values and thus merit further investigation into this area.
Reporting bias in tumor biomarker studies due to preferential reporting of “positive results” is well recognized. This is compounded by cutpoint manipulation to inflate effect sizes 11. Such reporting imbalance invariably limits our meta-analysis, potentially resulting in quantitative summation of optimistic reportings. This issue was addressed in our study using two strategies. First, we endeavored to comprehensively capture all reported results. We observed that studies simultaneously examining multiple biomarkers were more likely to report negative results in a subset of biomarkers in conjunction with one or more positive results. In those cases, the negative results were recorded to increase their recovery. Second, we used trim and fill to mathematically correct for publication bias. This approach resulted in a more conservative estimate of the summated results. A national biomarker study registry that allows users to deposit biomarker study data may be one solution to facilitae unbiased biomarker discovery. There is great diversity in tumor biomarker studies in terms of detection method, qantitation and scoring methods, and cutpoint levels, which contribute to heterogenity in prognostic effect sizes 11. Interestingly, we found low to moderate level of heterogeneity among our pooled studies. This may be attributable to our inclusion of only those IHC-based studies that met the REMARK criteria and established a baseline equivalency in methodological standards.
Previous reviews on tissue biomarkers have consistently reported the promise of GEM metabolism proteins as prognostic biomarkers in patients with PDAC 12, 45-47. However, none of the published reviews performed a quantitative meta-analysis on the prognosticative role of GEM-metabolism related biomarkers specifically in the context of patients receiving GEM treatment. To our knowledge, only Jamieson et la, Ansari et al and our group restricted the analyses to those IHC-based studies meeting REMARK material and methods reporting standards 12, 47.
Multiple phase III randomized adjuvant treatment trials comparing GEM- to 5-fluorouracil based regimens have largely failed to show statistically significant differences in survival outcomes in resected PDAC patients 3, 4. The lack of efficacy difference may be explained by a baseline variation in the expression levels of transporters and enzymes involved in the GEM-metabolic pathway. Currently there are no prognostic biomarkers available to stratify survival outcomes for PB cancer patients receiving gemcitabine. The results of our study indicate that these biomarkers may be useful for guiding selection of the most optimal chemotherapy regimen on an individual basis.
Table 2.
Newcastle-Ottawa quality assessment of primary studies.
| Reference | Year | Selection (4 stars max) |
Comparability (2 stars max) |
Outcome (3 stars max) |
Quality Points |
|---|---|---|---|---|---|
| Borbath (22) | 2011 | 3 | 2 | 2 | 7 of 9 |
| Kondo (28) | 2011 | 3 | 2 | 3 | 8 of 9 |
| Morinaga (31) | 2011 | 3 | 2 | 2 | 7 of 9 |
| Murata (32) | 2011 | 3 | 2 | 2 | 7 of 9 |
| Santini (35) | 2011 | 3 | 1 | 2 | 6 of 9 |
| Fujita (24) | 2010 | 3 | 2 | 3 | 8 of 9 |
| Komori (27) | 2010 | 3 | 1 | 2 | 6 of 9 |
| Marechal (30) | 2010 | 3 | 2 | 3 | 8 of 9 |
| Nakamura (34) | 2010 | 3 | 1 | 2 | 6 of 9 |
| Farrell (23) | 2009 | 3 | 2 | 3 | 8 of 9 |
| Marechal (29) | 2009 | 3 | 2 | 3 | 8 of 9 |
| Itoi (26) | 2007 | 3 | 1 | 3 | 7 of 9 |
| Nakahira (33) | 2007 | 3 | 1 | 2 | 6 of 9 |
| Giovannetti (25) | 2006 | 3 | 2 | 3 | 8 of 9 |
| Sebastiani (36) | 2006 | 3 | 1 | 2 | 6 of 9 |
| Spratlin (37) | 2004 | 3 | 1 | 2 | 6 of 9 |
Acknowledgments
Source of Financial Support: None.
Footnotes
Disclosure: Nothing to disclose.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Khan SA, Davidson BR, Goldin R, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: consensus document. Gut. 2002;51(Suppl 6):VI1–9. doi: 10.1136/gut.51.suppl_6.vi1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philip PA, Mooney M, Jaffe D, et al. Consensus report of the national cancer institute clinical trials planning meeting on pancreas cancer treatment. J Clin Oncol. 2009;27:5660–9. doi: 10.1200/JCO.2009.21.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neoptolemos JP, Stocken DD, Bassi C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304:1073–81. doi: 10.1001/jama.2010.1275. [DOI] [PubMed] [Google Scholar]
- 4.Regine WF, Winter KA, Abrams RA, et al. Fluorouracil vs gemcitabine chemotherapy before and after fluorouracil-based chemoradiation following resection of pancreatic adenocarcinoma: a randomized controlled trial. JAMA. 2008;299:1019–26. doi: 10.1001/jama.299.9.1019. [DOI] [PubMed] [Google Scholar]
- 5.Oettle H, Post S, Neuhaus P, et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–77. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- 6.Lim KH, Chung E, Khan A, et al. Neoadjuvant therapy of pancreatic cancer: the emerging paradigm? Oncologist. 2012;17:192–200. doi: 10.1634/theoncologist.2011-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glimelius B, Hoffman K, Sjoden PO, et al. Chemotherapy improves survival and quality of life in advanced pancreatic and biliary cancer. Ann Oncol. 1996;7:593–600. doi: 10.1093/oxfordjournals.annonc.a010676. [DOI] [PubMed] [Google Scholar]
- 8.Voutsadakis IA. Molecular predictors of gemcitabine response in pancreatic cancer. World J Gastrointest Oncol. 2011;3:153–64. doi: 10.4251/wjgo.v3.i11.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Candelaria M, de la Cruz-Hernandez E, Perez-Cardenas E, et al. Pharmacogenetics and pharmacoepigenetics of gemcitabine. Med Oncol. 2010;27:1133–43. doi: 10.1007/s12032-009-9349-y. [DOI] [PubMed] [Google Scholar]
- 10.Wong A, Soo RA, Yong WP, et al. Clinical pharmacology and pharmacogenetics of gemcitabine. Drug Metab Rev. 2009;41:77–88. doi: 10.1080/03602530902741828. [DOI] [PubMed] [Google Scholar]
- 11.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 12.Ansari D, Rosendahl A, Elebro J, et al. Systematic review of immunohistochemical biomarkers to identify prognostic subgroups of patients with pancreatic cancer. Br J Surg. 2011;98:1041–55. doi: 10.1002/bjs.7574. [DOI] [PubMed] [Google Scholar]
- 13.Gong W, Zhang X, Wu J, et al. RRM1 expression and clinical outcome of gemcitabine-containing chemotherapy for advanced non-small-cell lung cancer: a meta-analysis. Lung Cancer. 2011;75:374–80. doi: 10.1016/j.lungcan.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Schoenleber SJ, Kurtz DM, Talwalkar JA, et al. Prognostic role of vascular endothelial growth factor in hepatocellular carcinoma: systematic review and meta-analysis. Br J Cancer. 2009;100:1385–92. doi: 10.1038/sj.bjc.6605017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutton AJ, Song F, Gilbody SM, et al. Modelling publication bias in meta-analysis: a review. Stat Methods Med Res. 2000;9:421–45. doi: 10.1177/096228020000900503. [DOI] [PubMed] [Google Scholar]
- 16.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akita H, Zheng Z, Takeda Y, et al. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. 2009;28:2903–9. doi: 10.1038/onc.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyake K, Imura S, Yoshizumi T, et al. Role of thymidine phosphorylase and orotate phosphoribosyltransferase mRNA expression and its ratio to dihydropyrimidine dehydrogenase in the prognosis and clinicopathological features of patients with pancreatic cancer. Int J Clin Oncol. 2007;12:111–9. doi: 10.1007/s10147-006-0634-x. [DOI] [PubMed] [Google Scholar]
- 19.Ashida R, Nakata B, Shigekawa M, et al. Gemcitabine sensitivity-related mRNA expression in endoscopic ultrasound-guided fine-needle aspiration biopsy of unresectable pancreatic cancer. J Exp Clin Cancer Res. 2009;28:83. doi: 10.1186/1756-9966-28-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh DY, Lee KW, Lee KH, et al. A phase II trial of erlotinib in combination with gemcitabine and capecitabine in previously untreated metastatic/recurrent pancreatic cancer: combined analysis with translational research. Invest New Drugs. 2011;30:1164–74. doi: 10.1007/s10637-011-9651-3. [DOI] [PubMed] [Google Scholar]
- 21.Wakai T, Shirai Y, Sakata J, et al. Ribonucleotide reductase M1 expression in intrahepatic cholangiocarcinoma. Hepatogastroenterology. 2011;58:1659–63. doi: 10.5754/hge11175. [DOI] [PubMed] [Google Scholar]
- 22.Borbath I, Verbrugghe L, Lai R, et al. Human equilibrative nucleoside transporter 1 (hENT1) expression is a potential predictive tool for response to gemcitabine in patients with advanced cholangiocarcinoma. Eur J Cancer. 2011;48:990–6. doi: 10.1016/j.ejca.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Farrell JJ, Elsaleh H, Garcia M, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136:187–95. doi: 10.1053/j.gastro.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 24.Fujita H, Ohuchida K, Mizumoto K, et al. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12:807–17. doi: 10.1593/neo.10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovannetti E, Del Tacca M, Mey V, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66:3928–35. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 26.Itoi T, Sofuni A, Fukushima N, et al. Ribonucleotide reductase subunit M2 mRNA expression in pretreatment biopsies obtained from unresectable pancreatic carcinomas. J Gastroenterol. 2007;42:389–94. doi: 10.1007/s00535-007-2017-0. [DOI] [PubMed] [Google Scholar]
- 27.Komori S, Osada S, Mori R, et al. Contribution of thymidylate synthase to gemcitabine therapy for advanced pancreatic cancer. Pancreas. 2010;39:1284–92. doi: 10.1097/MPA.0b013e3181dec17d. [DOI] [PubMed] [Google Scholar]
- 28.Kondo N, Murakami Y, Uemura K, et al. Combined analysis of dihydropyrimidine dehydrogenase and human equilibrative nucleoside transporter 1 expression predicts survival of pancreatic carcinoma patients treated with adjuvant gemcitabine plus S-1 chemotherapy after surgical resection. Ann Surg Oncol. 2011;19(Suppl 3):S646–55. doi: 10.1245/s10434-011-2140-2. [DOI] [PubMed] [Google Scholar]
- 29.Marechal R, Mackey JR, Lai R, et al. Human equilibrative nucleoside transporter 1 and human concentrative nucleoside transporter 3 predict survival after adjuvant gemcitabine therapy in resected pancreatic adenocarcinoma. Clin Cancer Res. 2009;15:2913–9. doi: 10.1158/1078-0432.CCR-08-2080. [DOI] [PubMed] [Google Scholar]
- 30.Marechal R, Mackey JR, Lai R, et al. Deoxycitidine kinase is associated with prolonged survival after adjuvant gemcitabine for resected pancreatic adenocarcinoma. Cancer. 2010;116:5200–6. doi: 10.1002/cncr.25303. [DOI] [PubMed] [Google Scholar]
- 31.Morinaga S, Nakamura Y, Watanabe T, et al. Immunohistochemical analysis of human equilibrative nucleoside transporter-1 (hENT1) predicts survival in resected pancreatic cancer patients treated with adjuvant gemcitabine monotherapy. Ann Surg Oncol. 2011;19(Suppl 3):S558–64. doi: 10.1245/s10434-011-2054-z. [DOI] [PubMed] [Google Scholar]
- 32.Murata Y, Hamada T, Kishiwada M, et al. Human equilibrative nucleoside transporter 1 expression is a strong independent prognostic factor in UICC T3-T4 pancreatic cancer patients treated with preoperative gemcitabine-based chemoradiotherapy. J Hepatobiliary Pancreat Sci. 2012;19:413–25. doi: 10.1007/s00534-011-0440-3. [DOI] [PubMed] [Google Scholar]
- 33.Nakahira S, Nakamori S, Tsujie M, et al. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer. 2007;120:1355–63. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura J, Kohya N, Kai K, et al. Ribonucleotide reductase subunit M1 assessed by quantitative double-fluorescence immunohistochemistry predicts the efficacy of gemcitabine in biliary tract carcinoma. Int J Oncol. 2010;37:845–52. [PubMed] [Google Scholar]
- 35.Santini D, Schiavon G, Vincenzi B, et al. Human equilibrative nucleoside transporter 1 (hENT1) levels predict response to gemcitabine in patients with biliary tract cancer (BTC) Curr Cancer Drug Targets. 2011;11:123–9. doi: 10.2174/156800911793743600. [DOI] [PubMed] [Google Scholar]
- 36.Sebastiani V, Ricci F, Rubio-Viqueira B, et al. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: relationship to molecular mechanisms of gemcitabine resistance and survival. Clin Cancer Res. 2006;12:2492–7. doi: 10.1158/1078-0432.CCR-05-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spratlin J, Sangha R, Glubrecht D, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. 2004;10:6956–61. doi: 10.1158/1078-0432.CCR-04-0224. [DOI] [PubMed] [Google Scholar]
- 38.Kim R, Tan A, Lai KK, et al. Prognostic roles of human equilibrative transporter 1 (hENT-1) and ribonucleoside reductase subunit M1 (RRM1) in resected pancreatic cancer. Cancer. 2011;117:3126–34. doi: 10.1002/cncr.25883. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka M, Javle M, Dong X, et al. Gemcitabine metabolic and transporter gene polymorphisms are associated with drug toxicity and efficacy in patients with locally advanced pancreatic cancer. Cancer. 2010;116:5325–35. doi: 10.1002/cncr.25282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farrell JJ, Bae K, Wong J, et al. Cytidine deaminase single-nucleotide polymorphism is predictive of toxicity from gemcitabine in patients with pancreatic cancer: RTOG 9704. Pharmacogenomics J. 2011;12:395–403. doi: 10.1038/tpj.2011.22. [DOI] [PubMed] [Google Scholar]
- 41.Kasuya K, Tsuchida A, Nagakawa Y, et al. Prediction of a side effect and efficacy of adjuvant chemotherapy with gemcitabine for post operative patient of pancreatic cancer by a genetic polymorphism analysis. Hepatogastroenterology. 2012;59:1609–13. doi: 10.5754/hge11729. [DOI] [PubMed] [Google Scholar]
- 42.Innocenti F, Owzar K, Cox NL, et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin Cancer Res. 2012;18:577–84. doi: 10.1158/1078-0432.CCR-11-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugiyama E, Kaniwa N, Kim SR, et al. Population pharmacokinetics of gemcitabine and its metabolite in Japanese cancer patients: impact of genetic polymorphisms. Clin Pharmacokinet. 2010;49:549–58. doi: 10.2165/11532970-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 44.Giovannetti E, Funel N, Peters GJ, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–38. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 45.Garcea G, Neal CP, Pattenden CJ, et al. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer. 2005;41:2213–36. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 46.Ghaneh P, Kawesha A, Evans JD, et al. Molecular prognostic markers in pancreatic cancer. J Hepatobiliary Pancreat Surg. 2002;9:1–11. doi: 10.1007/s005340200000. [DOI] [PubMed] [Google Scholar]
- 47.Jamieson NB, Carter CR, McKay CJ, et al. Tissue biomarkers for prognosis in pancreatic ductal adenocarcinoma: a systematic review and meta-analysis. Clin Cancer Res. 2011;17:3316–31. doi: 10.1158/1078-0432.CCR-10-3284. [DOI] [PubMed] [Google Scholar]