Abstract
Phylogeographic studies frequently reveal multiple morphologically cryptic lineages within species. What is not yet clear is whether such lineages represent nascent species or evolutionary ephemera. To address this question, we compare five contact zones, each of which occurs between ecomorphologically cryptic lineages of skinks from the rainforests of the Australian Wet Tropics. Although the contacts probably formed concurrently in response to Holocene expansion from glacial refugia, we estimate that the divergence times (τ) of the lineage pairs range from 3.1 to 11.5 Ma. Multi-locus analyses of the contact zones yielded estimates of reproductive isolation that are tightly correlated with divergence time and, for lineages with older divergence times (τ > 5 Myr), substantial. These results show that phylogeographic splits of increasing depth represent stages along the speciation continuum, even in the absence of overt change in ecologically relevant morphology.
Keywords: suture zone, phylogeography, hybridization, reproductive isolation, demographic reconstruction, cryptic species
1. Introduction
There is now abundant evidence for deep phylogeographic divisions within traditionally described taxa, suggesting that morphologically cryptic species are common [1]. Indeed, deep phylogeographic structure, based on mitochondrial DNA (mtDNA) and confirmed by multi-locus nuclear DNA (nDNA), is increasingly used as an initial step in species delimitation via integrative taxonomy [2]. As we grow better able to identify evolutionarily independent genetic lineages within morphologically defined species, what is often missing is both an understanding of the forces leading to this diversity and evaluation of whether these lineages are more than ephemera [3]. One way to start addressing these issues is to test for reproductive isolation (RI) among morphologically cryptic lineages. Is there substantial RI between such cryptic lineages, and if so, how does this scale with divergence time? Answers to these questions will both inform methods in systematics and contribute to our understanding of speciation processes.
It has long been supposed that phylogeographic lineages represent a step in the continuum from population divergence to speciation [4]. More generally, speciation theory posits that RI, especially post-zygotic RI, increases with divergence time, with the tempo and form of the relationship depending on the genetic architecture of Dobzhansky–Muller incompatibilities (DMIs) and the interaction of selection, drift and gene flow [5–7]. But, given this, the heterogeneity of divergent selection, whether stemming from ecological, sexual or social selective pressures, should blur the relationship between RI and time [5]. A growing body of evidence supports a general increase in RI with time; however, with few exceptions [8], these results derive from analyses of phenotypically distinct species [9,10]. As phylogeographic lineages within morphologically defined species are often found in secondary contact, comparative analyses of RI indices in zones of sympatry could provide a unique window into the dynamics of ecomorphologically cryptic speciation [11]. Such studies have the added advantage of addressing the evolution of RI in nature, in the organisms' ecological context, rather than in the laboratory crosses, as is more common in the literature.
To investigate the evolution of RI in nature, we exploit a system characterized by climate-driven fluctuations in habitat extent and connectivity during the Neogene—the rainforests of the Australian Wet Tropics (figure 1a). Extended periods of retraction of rainforests to mesic mountain tops have resulted in pronounced phylogeographic structure within endemic faunal species, but with variable levels of sequence divergence and divergence time among intraspecific lineages [12,13]. Where tested, these mtDNA lineages are generally corroborated by multi-locus nuclear gene analysis [12] (but see [14]); however, ecomorphological divergence is subtle or absent [13,15]. Following Holocene rainforest expansions, over 20 contact zones involving ecomorphologically cryptic lineage pairs formed between the historic refugia [12]. These contact zones provide a natural experiment with which to test the hypothesis that RI increases with divergence time among cryptic lineages. Previous studies of contact zones have revealed outcomes ranging from negligible to strong RI [12], including one case of speciation by reinforcement [16]. However, the cases studied to date are taxonomically and ecologically heterogeneous.
Figure 1.
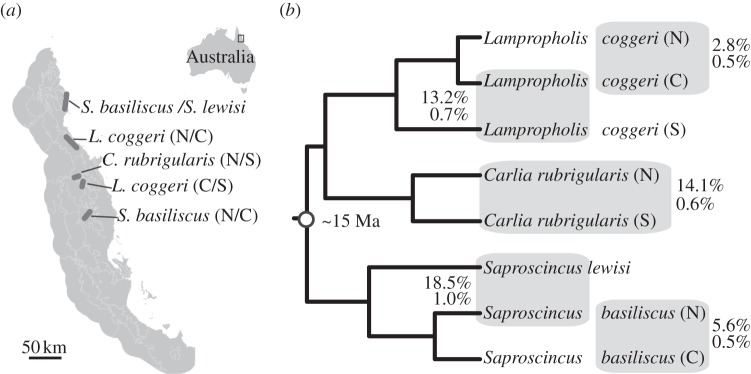
(a) A map of the Australian Wet Tropics, labelled with contact zones. A more detailed map of each contact zone is available in the electronic supplementary material, figure S1. (b) Phylogeny showing relationships among focal lineages (see the electronic supplementary material); boxes outline contact zones. Boxes are labelled with pairwise mitochondrial (top) and nuclear (bottom) divergence.
Here, we use comparative analysis of RI within five contact zones involving lineage pairs from a closely related and ecologically similar clade of terrestrial rainforest skinks, across which sequence divergence varies (figure 1b). We combine genome-scale analyses of divergence history between allopatric populations with multi-locus analysis of intensively sampled contact zones to test for increasing RI with divergence time. We assume that per-generation dispersal rates are similar across lineages, which indirect dispersal estimates from this clade support [17,18]. We further assume that the contact zones formed concurrently, because the species are ecologically similar, rainforest-edge species that probably tracked the expanding rainforest front closely [19]. Following from these assumptions, we hypothesize that RI scales closely with divergence time, especially given the limited ecomorphological divergence of these lineage pairs [13] and, for at least Carlia rubrigularis N/S, apparent absence of mate choice [20]. More specifically, we predict that, as divergence time increases, cline should become narrower and exhibit less variance in width, genetic disequilibrium should increase and the frequency of hybrids should decrease [21].
2. Methods
(a). Sampling
We sampled five contact zones in the Australian Wet Tropics from 2008 to 2011 (see the electronic supplementary material, table S1): Carlia rubrigularis N/S, Lampropholis coggeri N/C, L. coggeri C/S, Saproscincus basiliscus N/C and S. basiliscus N/Saproscincus lewisi (figure 1 and electronic supplementary material, figure S1). First, we broadly sampled throughout the species' ranges and genotyped individuals for mitochondrial lineage to identify the presumed location of the contact zones (figure 1b and electronic supplementary material, table S2). After determining the zone's location, we then collected samples from populations geographically isolated from the contact zone, which we used to infer demographic history and to develop markers. Then, for four of the five contacts, we sampled individuals non-destructively along a linear transect through the contact zone (see the electronic supplementary material, figure S1). For S. basiliscus N/S. lewisi, we sampled opportunistically because initial data suggested the lineages were not hybridizing (see the electronic supplementary material, figure S1). Data from the L. coggeri C/S hybrid zone were previously published in reference [18], and we expanded the C. rubrigularis N/S dataset collected by Phillips et al. [17], by genotyping new genes, increasing sample sizes, and adding new populations.
We sampled densely through each contact zone, averaging 20 individuals for each of 12 populations per contact (n = 55–406; electronic supplementary material, table S1), with the range of sampling determined by preliminary data on the scale of the mtDNA transition (transect length = 2–16 km).
(b). Morphological analyses
To test for ecomorphological divergence across the major phylogeographic lineages within each traditionally defined species, we measured adult lizards outside of the contact zones at four standard characters for lizards: snout–vent length, head width, head length and hind limb length. Because these species show evidence of sexual dimorphism, we analysed these data by splitting each species dataset by sex. Further, where deemed relevant by MANCOVA, we removed the effect of elevation by taking the unnormalized residuals of morphological characters against elevation. Using the first two major orthogonal axes from a scaled principal components analysis, we tested for morphological differentiation across phylogeographic lineages using MANOVAs, conducted ANOVAs on significant results, and followed significant ANOVAs with Tukey's HSD tests [22].
(c). Genetic data collection
We collected two types of genetic data: (i) transcriptomic data from populations isolated from the contact zone to infer demographic history and to develop markers, and (ii) genotypic data from populations located in the contact zone to infer the extent of RI. For (i), we collected transcriptomic data for five individuals per lineage. As described in reference [23], we filtered the data for quality, assembled and annotated a lineage's reads to create reference assemblies, mapped reads by individual to the reference assemblies and used the resulting alignments with Samtools to find variants and with ANGSD to define demographic history (see below) [24,25]. For (ii), we used PCR–RFLP markers to genotype lizards from the contact zones. From the transcriptome dataset, we selected variants that were located in untranslated regions (UTRs) of genes, fixed between the two lineages, and easily resolved by robust and inexpensive restriction enzymes. Marker details can be found in the electronic supplementary material, table S2. In total, individuals in the S. basiliscus N/S. lewisi contact zone were genotyped at six nuclear markers and mtDNA, and individuals in the other four zones at 10 nuclear markers and their mtDNA (see the electronic supplementary material, table S1). Scripts are available at https://github.com/singhal/transcriptomic and genotype data in the DataDryad package at: http://doi.org/10.5061/dryad.t838p.
(d). Genetic analysis
To analyse the data, we inferred the demographic history of the lineages and calculated several indirect measures of RI.
(i). Fitting divergence histories
Many comparative studies use genetic distance as a proxy for time since divergence [10,26]; however, genetic distance might be decoupled from divergence time, especially if migration rates are high or ancestral population sizes are large [27]. Accordingly, we inferred divergence time and other demographic parameters for each lineage pair by fitting an isolation-with-migration model to genetic data. In this model, an ancestral population splits into two daughter populations, which exchange migrants at a continuous rate until the present. Although these methods make assumptions that might not be biologically reasonable [28], such as continuous migration through time and no population structure through lineages, their ability to account for coalescent processes provides a more nuanced perspective on lineage divergence than does genetic distance itself.
For S. basiliscus N and its sister species S. lewisi, we did not have genomic data for S. lewisi, and thus, we used previously published genetic data for eight loci [14] to infer model parameters with IMa2 [29]. For the remaining contacts, we used the transcriptomic data to fit an isolation-with-migration model using Dadi [30]. Dadi uses a diffusion approximation to fit a likelihood model for demographic history to the two-dimensional-site frequency spectrum (2D-SFS). The 2D-SFS summarizes the distribution of allele frequencies for shared and private alleles between two populations. To infer the 2D-SFS, we used the genotype caller ANGSD; ANGSD can characterize a population's SFS without calling individual genotypes, which reduces uncertainty around the SFS [25]. Here, we only used UTR sequence, because UTRs are more likely to evolve neutrally than coding sequence [31], and we restricted this analysis to high-coverage regions (greater than or equal to 20×) where we had greater confidence in genotype calling [25]. To construct the unfolded SFS, we polarized variants with sequence data from other lineages in the clade. Inferred demographic parameters were converted from coalescent units to real-time units by using estimates of the nuclear mutation rate, assuming a molecular clock and accounting for differences in total sequenced length across contacts [30]. Our estimate of the nuclear mutation rate (9 × 10−10 substitutions/bp × generation) is derived from fossil-calibrated estimates of the mitochondrial mutation rate in this broader clade of lizards [32] and estimates of the nuclear–mitochondrial substitution rate scalar as inferred from IMa2 results [14,18]. All Dadi and IMa2 input files and related scripts are available in the DataDryad package at: http://doi.org/10.5061/dryad.t838p.
(ii). Measuring reproductive isolation
To infer the strength of RI and to correlate its evolution with divergence, we calculated six indices based on the genotypic data from the contact zones: average nuclear cline width, mitochondrial cline width, coefficient of variance of cline width, Hardy–Weinberg disequilibrium (FIS), linkage disequilibrium (Rij) and per cent hybrids. Importantly, we note that these indices are independent measures of isolation, though certain biological realities, such as hybrid zones at migration–selection equilibrium [21], would lead to correlated responses [21]. We first collapsed adjoining sampling localities into geographical populations based on their Euclidean distance to their nearest neighbour. Then, we fitted clines to our data using the program Analyse [33]. Second, using Analyse, we calculated multi-locus measures of Hardy–Weinberg and linkage disequilibrium for all geographical localities across all contacts. Third, we used Structure to estimate each individual's hybrid index [34] and NewHybrids to calculate the number of hybrids in the contact zone [35].
We used our data to contrast different models for how RI evolves through time, including linear and quadratic models for the accumulation of total RI. These commonly tested models characterize the accumulation of RI as either remaining constant or accelerating through time. We restricted our analyses to three indices of RI − FIS, Rij and per cent hybrids—as we could predict starting values for these three indices. We modelled each index separately using the least-squares approach implemented in R [22], and we chose the best-fitting models by calculating the relative weight of each model based on Akaike information criterion (AIC) score.
3. Results
(a). Ecomorphological divergence
We used morphological data to test for phenotypic divergence across the major phylogeographic lineages within each traditionally defined species (mean sample size per species, 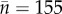 ). We summarized the data as two principal component axes that explained over 97% of the variation and, using these axes, we found little significant ecomorphological divergence across phylogeographic lineage pairs (see the electronic supplementary material, figure S2). Where there is statistically significant variation (see the electronic supplementary material, figure S2), the differences are sex-specific and of small magnitude—e.g. mean body size for female L. coggeri varies from 35.7 ± 2.7 to 38.5 ± 3.3 mm by lineage.
). We summarized the data as two principal component axes that explained over 97% of the variation and, using these axes, we found little significant ecomorphological divergence across phylogeographic lineage pairs (see the electronic supplementary material, figure S2). Where there is statistically significant variation (see the electronic supplementary material, figure S2), the differences are sex-specific and of small magnitude—e.g. mean body size for female L. coggeri varies from 35.7 ± 2.7 to 38.5 ± 3.3 mm by lineage.
(b). Fitting divergence histories
Inferring the demographic history of these lineage pairs gave two primary results. First, estimated divergence times vary nearly fourfold, from 3.1 to 11.5 Ma. Second, estimates of nuclear divergence and divergence times are tightly correlated (r2 = 0.98; p-value = 0.01), as expected given the low estimates for historical migration during divergence (M = 4 × 10−3 to 3.5 × 10−2 migrants per generation; figure 2b). Full model parameters are available in the electronic supplementary material, table S4.
Figure 2.
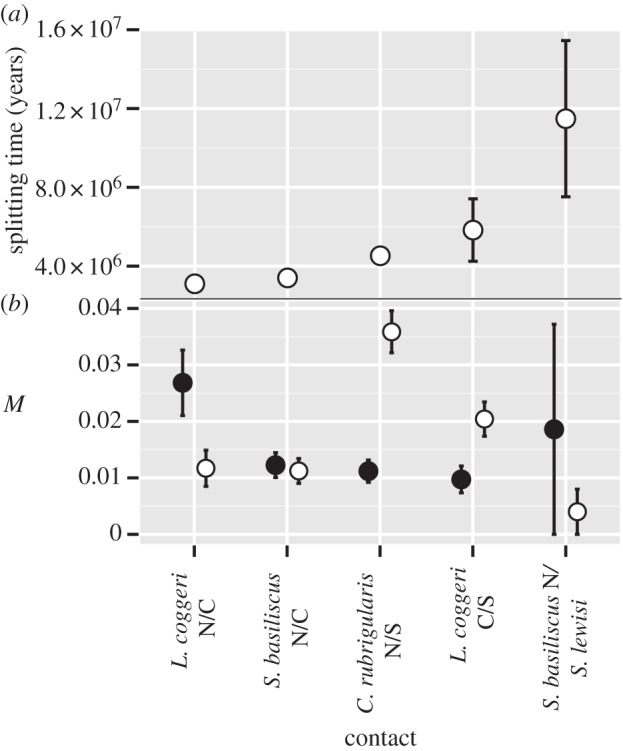
Divergence histories as inferred from the two-dimensional-site frequency spectrum by Dadi and from IMa2, (a) divergence times and (b) historical gene flow rates (as measured by number of migrants per generation, M) for lineage pairs. Black points are migration from the northern to southern lineage in each lineage pair; white points, southern to northern. Error bars reflect (as relevant) s.d. or 95% limits of posterior distribution.
(c). Measuring reproductive isolation
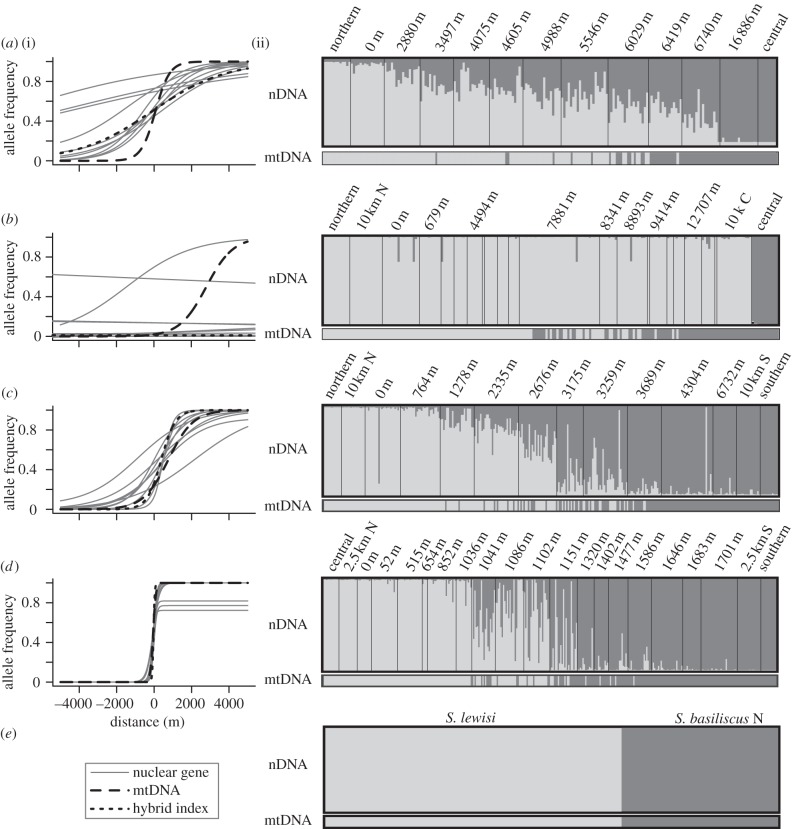
We used fine-scale spatial sampling and multi-locus estimates of hybridization, disequilibrium and introgression to infer the strength of RI. We first describe general patterns at each contact zone before summarizing across all the zones, stepping from the least to most divergent contact (figure 3). In the L. coggeri N/C contact, we see widespread introgression that extends throughout the sampled transect and evidence for two general patterns of introgression in nuclear loci relative to mtDNA: clines whose centre and width is similar to the mtDNA cline, and clines which show broad introgression of the central (C) alleles into the northern (N) lineage (figure 3a). In the S. basiliscus N/C contact, we were only able to infer clines at one of the nuclear loci; it appears that the northern nuclear genome has almost completely introgressed into the central lineage (figure 3b). The asymmetric introgression in both the L. coggeri N/C and S. basiliscus N/C contacts could stem from stochastic, demographic or selective processes; disentangling the causes of asymmetry is not possible here so we focus on consensus patterns. Both the C. rubrigularis N/S and L. coggeri C/S show similar clines across all loci, and both show limited introgression beyond the contact zone (figure 3c,d). Finally, there is no evidence for hybridization between S. basiliscus N and its ecomorphologically similar sister species S. lewisi, even when sampled in sympatry (figure 3e).
Figure 3.
Cline fitting (i) and genetic clustering results (ii) for contacts in the Australian Wet Tropics suture zone: (a) Lampropholis coggeri N/C, (b) Saproscincus basiliscus N/C, (c) Carlia rubrigularis N/S, (d) L. coggeri C/S, and (e) S. basiliscus N/S. lewisi. For showing cline fitting results, distances along transects were recalculated, so that each hybrid zone centre was centred at 0 m. Scale for genetic clustering results differs among contacts.
Examining the correlation of divergence time with the six indices of RI, we see significant and strong correlations for all indices but mitochondrial cline width (figure 4). As predicted, with increasing divergence time, we see decreased cline width and less variance in cline width, fewer hybrids and increased disequilibrium between and within loci. Given our limited cline data for S. basiliscus N/C, we also calculated correlations for cline measures without this contact; correlations remained significant (see the electronic supplementary material, figure S4). Further, these results are robust to our estimates of splitting times; using pairwise nuclear divergence gives quantitatively similar results (see the electronic supplementary material, figure S5). Interestingly, the correlations between mitochondrial divergence and RI are weak but non-significant (see the electronic supplementary material, figure S6). Note that not all indices of RI could be estimated for all contacts. We did not infer cline indices for the S. basiliscus N/S. lewisi contact zone because of insufficient sampling, and we did not estimate either disequilibrium or hybridization measures for S. basiliscus N/C as most nuclear loci were nearly monoallelic throughout the sampled contact zone.
Figure 4.
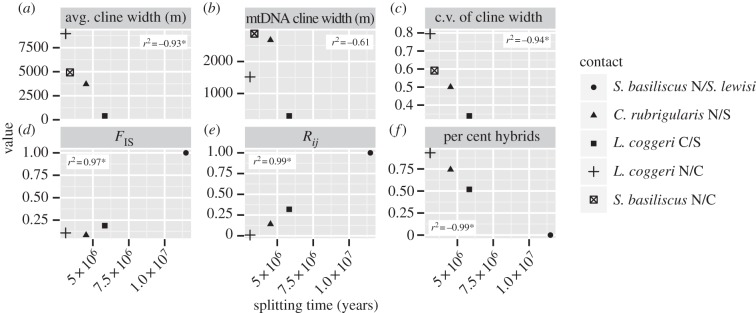
Comparative results showing the correlation between divergence time (as measured in years) and indices of reproductive isolation: (a) average nuclear cline width, (b) mitochondrial cline width, (c) coefficient of variance in cline width, (d) Hardy–Weinberg disequilibrium (FIS), (e) linkage disequilibrium (Rij) and (f) per cent of hybrids in the contact zone. Graphs are labelled with correlation coefficients; asterisks indicate significant correlations.
To make our data comparable with other published datasets [9], we fit linear and quadratic models to the increase of RI (as measured by FIS, Rij and per cent hybrids) through time. Using relative weights from AIC scores, we determined that total RI, as measured by each of these three indices, best fits a model of quadratic growth with time (see the electronic supplementary material, figure S7).
4. Discussion
By looking across five contacts in a clade of closely related and ecologically similar skinks in the Australian Wet Tropics, we find strong support for the prediction of increasing RI with divergence time. To the best of our knowledge, this is the first comparative study of the strength of lineage boundaries across ecomorphologically similar lineages. These data support the view that phylogeographic splits of increasing depth can represent stages along the speciation continuum, including genetically cohesive lineages with long-term potential for persistence.
Interestingly, most datasets comparing RI with divergence time show significantly more noise than ours [9,10], even though these datasets were collected in controlled laboratory settings. In comparison, the strength of our correlations is unexpected, especially given that biases with sampling design (i.e. using a sole linear transect, selecting only diagnostic alleles for genotyping) and stochastic processes (i.e. extinction and recolonization of local populations, variance in dispersal length) can greatly influence inference of hybrid zone structure and dynamics [8,36]. We speculate that the close fit between RI and divergence time in our study stems from the ecological similarity of the species studied, the shared biogeographic history of the contact zones, and most importantly, the lack of overt divergent selection on ecomorphology, as varying strengths and forms of selection would be expected to introduce heterogeneity in the rate at which RI accumulates [5]. After all, selection can lead to the evolution of RI even without concomitant widespread genetic divergence [37]—as seen in the Laupala crickets of Hawaii [38] or the Gambusia fishes of the Bahamas [39]. For the taxa examined here, the relationships between different indices of RI and divergence time are so strong that we can use them as a general metric for predicting the progress of speciation in this group. In their influential review, Kirkpatrick & Ravigne [40] considered that populations achieve ‘species status’ when linkage disequilibrium (measured here as Rij) is 0.5. Using this threshold, we find that our lineage pairs are predicted to show Rij = 0.5 at 7 or 8.1 Myr after divergence (and at 0.79% or 0.81% nDNA divergence), under a linear versus quadratic model for accumulation of RI, respectively. Although this specific calibration is contingent on our divergence time calibration and unlikely to be generalizable to other taxa, it does provide a yardstick for the tempo of cryptic speciation in this group.
Given this rate of speciation—and noting that C. rubrigularis N/S and L. coggeri C/S show substantial isolation with even less time—we suggest that the accumulation of RI here is faster relative to a purely drift-driven model of the evolution of intrinsic RI via DMIs. A simplistic model of drift-driven accumulation of DMIs in allopatry suggests that the waiting time to speciation is approximately the number of substitutions needed for RI divided by the substitution rate [5], which, given the skinks' estimated mutation rate, could be in the order of hundreds of millions of years. Alternative processes for accumulating substantial RI in the absence of ecomorphological divergence on the time scale inferred here include: (i) drift-driven rapid divergence along ‘holey adaptive landscapes’ [5], (ii) parallel selection driving mutation-order speciation [41], and/or (iii) natural or sexual selection acting on more cryptic phenotypes, such as chemiosensory production and perception. Indeed, sexual isolation appears to have evolved between S. basiliscus N/S. lewisi, as there is no evidence that this lineage pair mates successfully. As such, future research should investigate the evolution of pre-zygotic isolation in this group. As yet, we lack the fine-scale ecological data necessary to characterize the barriers to gene flow acting in this group and to determine which of these hypothetical drivers of divergence are relevant [42]. However, these data do confirm cryptic speciation among phylogeographic lineages and suggest that this could be common, in contrast to the present focus on speciation between ecomorphologically diverged lineages [43].
Looking beyond the velocity of RI accumulation to its acceleration, we find that the total strength of RI increases exponentially through time in this clade (see the electronic supplementary material, figure S7). The pattern of exponentially increasing RI emerges when individual barriers to gene flow combine multiplicatively rather than additively [6], as seen in other studies [44]. This result suggests that as barriers to gene flow start to evolve, the cumulative effect of these barriers can grow quickly. Thus, species formation can be thought of as an accelerating process—particularly as RI decreases gene flow, which typically further promotes divergence [45].
(a). Implications for cryptic speciation
Because we can quickly and cheaply query geographical variation within species using mtDNA, deep mtDNA divergence is often used as an initial hypothesis for species delimitation. However, deep mtDNA divergence is neither necessary nor sufficient to delimit species [46], and it is often discordant with other genetic or phenotypic measures of divergence [47]. In this study, we provide additional evidence that mtDNA can present an idiosyncratic perspective on historical dynamics, finding that mitochondrial cline width is the sole index of RI that had a non-significant correlation with divergence time (figure 4), and that mitochondrial divergence and RI are non-significantly correlated (see the electronic supplementary material, figure S6). Given our and others' findings of mtDNA's idiosyncracy [48], we concur that the observation of deeply divergent mtDNA phylogroups is a useful start of taxonomic and phylogeographic studies, but it certainly should not be the end.
Starting since researchers began to catalogue diversity within species, most species have been found to show geographically restricted variation [49]. Why do some of these intraspecific lineages continuously diverge and exhibit RI from sister lineages, whereas others collapse [3,50]? As suggested by many, geography is a powerful determinant [49]. But, as we see here, RI can take millions of years to accumulate, particularly in the absence of strong ecologically driven divergence, suggesting that isolation must be sustained across millions of years to lead to genetically independent lineages. In the absence of such long-term stability, geography can be insufficient, leading to the extensive introgression and discordance often reported in other systems [51,52]. Thus, as a working hypothesis, we suggest that climatically and geomorphologically stable regions, such as the major refugia of the Australian Wet Tropics, are more likely to accumulate cryptic diversity than more spatio-environmentally dynamic regions. Indeed, broad-range introgression and discordance are exceedingly rare in this system, except among lineages endemic to the relatively unstable southern rainforest isolates [14,53].
Finally, these data add to a growing body of literature that support a Darwinian perspective on species formation [54] and extend this perspective to cryptic diversification. Whether from lineages that probably diverged with gene flow [55] or lineages that diverged in allopatry [56], these datasets show that the accumulation of RI is often a gradual process and that species boundaries are dynamic. Indeed, divergence is a continuous, reversible process [50]. For these lineage pairs, we find that the evolution of RI followed a predictable timeline during their divergence in allopatry (figure 4 and electronic supplementary material, figure S3). Now that lineages have expanded following the Last Glacial Maximum, and the original barrier to gene flow (i.e. geography) has disappeared, these lineage pairs will move again along the continuum. For L. coggeri N/C and S. basiliscus N/C, this initial divergence will probably be reversed, because the lineage pairs seem to be hybridizing freely in the apparent absence of RI. We can estimate the strength of selection against hybrids by assuming a tension zone model [21]; given the dispersal lengths of these lizards [17,18] and the cline widths for these lineage pairs, selection against hybrids is estimated to be weak, of the order of 0.08–0.3%. However, L. coggeri C/S and C. rubrigularis N/S appear to be more strongly isolated, because the extent of introgression between these lineage pairs is very limited. Given the limited ecomorphological differentiation of these lineages, we hypothesis this RI is intrinsic and not environmentally dependent and, thus, likely to maintain lineage boundaries even in changing environments. As such, these lineage pairs will probably continue to diverge. That said, RI between these lineages is not complete, and such ‘leaky’ species boundaries can serve as a source of novelty, whether through the evolution of reinforcement or through the selective introgression of adaptive alleles [16,57].
Acknowledgements
For advice and discussions, we gratefully acknowledge C. Ellison, T. Linderoth, R. Pereira and members of the Moritz Laboratory; and for comments on previous versions of this manuscript, we thank R. Damasceno, C. DiVittorio, R. Harrison, J. Patton, R. Von May, D. Wake and two anonymous reviewers. Additionally, T. Linderoth provided scripts used to infer the 2D-SFS from ANGSD output. For assistance in the field, we thank A. Blackwell, D. Glas, E. Hoffmann, C. Hoskin, B. Phillips, M. Tonione, S. Williams and Y. Williams. Supercomputing resources used in this study were provided by the grid resources at the Texas Advanced Computing Center and Pittsburgh Supercomputing Center.
All animal use was approved by the Animal Ethics Committee at James Cook University in Australia and sampling permits were obtained from the Queensland Parks and Wildlife Services.
Funding statement
Support for this work was provided by the Museum of Vertebrate Zoology, National Geographic Society, NSF-GRFP and NSF-DDIG, and the SSE Rosemary Grant Award.
References
- 1.Bickford D, Lohman D, Sodhi N, Ng P, Meier R, Winker K, Ingram K, Das I. 2006. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155 (doi:10.1016/j.tree.2006.11.004) [DOI] [PubMed] [Google Scholar]
- 2.Padial J, Miralles A, De la Riva I, Vences M. 2010. The integrative future of taxonomy. Front. Zool. 7, 16 (doi:10.1186/1742-9994-7-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenblum E, Sarver B, Brown J, Des Roches S, Hardwick K, Hether T, Eastman J, Pennell M, Harmon L. 2012. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol. Biol. 39, 255–261 (doi:10.1007/s11692-012-9171-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avise J, Walker D, Johns G. 1998. Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc. R. Soc. Lond. B 265, 1707–1712 (doi:10.1098/rspb.1998.0492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gavrilets S. 2004. Fitness landscapes and the origin of species. Cambridge, MA: Princeton University Press [Google Scholar]
- 6.Gourbiere S, Mallet J. 2009. Are species real? The shape of the species boundary with exponential failure, reinforcement and the ‘missing snowball’. Evolution 64, 1–24 (doi:10.1111/j.1558-5646.2009.00844.x) [DOI] [PubMed] [Google Scholar]
- 7.Orr H. 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139, 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan-Richards M, Wallis G. 2003. A comparison of five hybrid zones of the weta Hemideina thoracica (Orthoptera: Anostomatidae): degree of cytogenetic differentiation fails to predict zone width. Evolution 57, 849–861 [DOI] [PubMed] [Google Scholar]
- 9.Bolnick D, Near T. 2005. Tempo of hybrid inviability in Centrarchid fishes (Teleostei: Centrarchidae). Evolution 59, 1754–1767 [PubMed] [Google Scholar]
- 10.Sasa M, Chippindale P, Johnson N. 1998. Patterns of postzygotic reproductive isolation in frogs. Evolution 52, 1811–1820 (doi:10.2307/2411351) [DOI] [PubMed] [Google Scholar]
- 11.Harrison R. 1993. Hybridization and hybrid zones: historical perspective. In Hybrid zones and the evolutionary process (ed. R Harrison), pp. 3–12 Oxford, UK: Oxford University Press [Google Scholar]
- 12.Moritz C, Hoskin C, MacKenzie J, Phillips B, Tonione M, Silva N, VanDerWal J, Williams S, Graham C. 2009. Identification and dynamics of a cryptic suture zone in tropical rainforest. Proc. R. Soc. B 276, 1235–1244 (doi:10.1098/rspb.2008.1622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell R, Parra J, Tonione M, Hoskin C, MacKenzie J, Williams S, Moritz C. 2010. Patterns of persistence and isolation indicate resilience to climate change in montane rainforest lizards. Mol. Ecol. 19, 2531–2544 [DOI] [PubMed] [Google Scholar]
- 14.Singhal S, Moritz C. 2012. Testing hypotheses of genealogical discordance in a rainforest lizard. Mol. Ecol. 21, 5059–5072 (doi:10.1111/j.1365-294X.2012.05747.x) [DOI] [PubMed] [Google Scholar]
- 15.Hoskin C, Tonione M, Higgie M, MacKenzie J, Williams S, VanDerWal J, Moritz C. 2011. Persistence in peripheral refugia promotes phenotypic divergence and speciation in a rainforest frog. Am. Nat. 178, 561–578 (doi:10.1086/662164) [DOI] [PubMed] [Google Scholar]
- 16.Hoskin C, Higgie M, McDonald K, Moritz C. 2005. Reinforcement drives rapid allopatric speciation. Nature 437, 1353–1356 (doi:10.1038/nature04004) [DOI] [PubMed] [Google Scholar]
- 17.Phillips B, Baird S, Moritz C. 2004. When vicars meet: a narrow contact zone between morphologically cryptic phylogeographic lineages of the rainforest skink, Carlia rubrigularis. Evolution 58, 1536–1548 [DOI] [PubMed] [Google Scholar]
- 18.Singhal S, Moritz C. 2012. Strong selection maintains a narrow hybrid zone between morphologically cryptic lineages in a rainforest lizard. Evolution 66, 1474–1489 (doi:10.1111/j.1558-5646.2011.01539.x) [DOI] [PubMed] [Google Scholar]
- 19.Williams S, et al. 2010. Distributions, life history characteristics, ecological specialization and phylogeny of the rainforest vertebrates in the Australian Wet Tropics bioregion. Ecology 91, 2493 (doi:10.1890/09-1069.1) [Google Scholar]
- 20.Dolman G. 2009. Evidence for differential assortative female preference in association with refugial isolation of rainbow skinks in Australia's tropical rainforests. PLoS ONE 3, e3499 (doi:10.1371/journal.pone.0003499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barton N, Gale K. 1993. Genetic analysis of hybrid zones. In Hybrid zones and the evolutionary process (ed. R Harrison), pp. 13–45 Oxford, UK: Oxford University Press [Google Scholar]
- 22.R Development Core Team 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- 23.Singhal S. 2013. De novo transcriptomic analyses for non-model organisms: an evaluation of methods across a multi-species data set. Mol. Ecol. Res. 13, 403–416 (doi:10.1111/1755-0998.12077) [DOI] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. and 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079 (doi:10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. 2012. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS ONE 7, e37558 (doi:10.1371/journal.pone.0037558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyle L, Nakazato T. 2010. Hybrid incompatibility ‘snowballs’ between Solanum species. Science 329, 1521–1523 (doi:10.1126/science.1193063) [DOI] [PubMed] [Google Scholar]
- 27.Nielsen R, Wakeley J. 2001. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strasburg J, Rieseberg L. 2010. How robust are ‘isolation with migration’ analyses to violations of the IM model? a simulation study. Mol. Biol. Evol. 27, 297–310 (doi:10.1093/molbev/msp233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hey J. 2010. Isolation with migration models for more than two populations. Mol. Biol. Evol. 27, 905–920 (doi:10.1093/molbev/msp296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutenkunst R, Hernandez R, Williamson S, Bustamante C. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (doi:10.1371/journal.pgen.1000695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williamson S, Hernandez R, Fiedel-Alon A, Zhu L, Nielsen R, Bustamante C. 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl Acad. Sci. USA 102, 7882–7887 (doi:10.1073/pnas.0502300102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandley M, Wang Y, Guo X, Nieto Montes de Oca A, Ortiz MF, Hikida T, Ota H. 2011. Accommodating locus-specific heterogeneity in molecular dating methods: an example using inter-continental dispersal of Plestiodon (Eumeces) lizards. Syst. Biol. 60, 3–15 (doi:10.1093/sysbio/syq045) [DOI] [PubMed] [Google Scholar]
- 33.Barton N, Baird S.1998. Analyse 1.10. See http://archive.bio.ed.ac.uk/software/Mac/Analyse/index.html .
- 34.Pritchard J, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 145, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson E, Thompson E. 2002. A model-based method for identifying species hybrids by using multilocus genetic data. Genetics 160, 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dufkova P, Macholan M, Pialek J. 2011. Inference of selection and stochastic effects in the house mouse hybrid zone. Evolution 65, 993–1010 (doi:10.1111/j.1558-5646.2011.01222.x) [DOI] [PubMed] [Google Scholar]
- 37.Doebeli M, Dieckmann U. 2003. Speciation along environmental gradients. Nature 16, 259–264 (doi:10.1038/nature01274) [DOI] [PubMed] [Google Scholar]
- 38.Parsons Y, Shaw K. 2001. Species boundaries and genetic diversity among Hawaiian crickets of the genus Laupala identified using amplified fragment length polymorphism. Mol. Ecol. 10, 1765–1772 (doi:10.1046/j.1365-294X.2001.01318.x) [DOI] [PubMed] [Google Scholar]
- 39.Langerhans R, Gifford M, Joseph E. 2007. Ecological speciation in Gambusia fishes. Evolution 61, 2056–2074 (doi:10.1111/j.1558-5646.2007.00171.x) [DOI] [PubMed] [Google Scholar]
- 40.Kirkpatrick M, Ravigne V. 2002. Speciation by natural and sexual selection: models and experiments. Am. Nat. 159, S22–S35 (doi:10.1086/338370) [DOI] [PubMed] [Google Scholar]
- 41.Nosil P, Flaxman S. 2011. Conditions for mutation order speciation. Proc. R. Soc. B 278, 399–407 (doi:10.1098/rspb.2010.1215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobel J, Chen G, Watt L, Schemske D. 2009. The biology of speciation. Evolution 54, 295–315 [DOI] [PubMed] [Google Scholar]
- 43.Schluter D. 2009. Evidence for ecological speciation and its alternative. Science 323, 737–741 (doi:10.1126/science.1160006) [DOI] [PubMed] [Google Scholar]
- 44.Mendelson T, Inouye B, Rausher M. 2004. Quantifying patterns in the evolution of reproductive isolation. Evolution 58, 1424–1433 [DOI] [PubMed] [Google Scholar]
- 45.Endler J. 1977. Geographic variation, speciation and clines. Princeton, NJ: Princeton University Press; [PubMed] [Google Scholar]
- 46.Hickerson M, Meyer C, Moritz C. 2006. DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 55, 729–739 (doi:10.1080/10635150600969898) [DOI] [PubMed] [Google Scholar]
- 47.Toews D, Brelsford A. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930 (doi:10.1111/j.1365-294X.2012.05664.x) [DOI] [PubMed] [Google Scholar]
- 48.Ballard J, Whitlock M. 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744 (doi:10.1046/j.1365-294X.2003.02063.x) [DOI] [PubMed] [Google Scholar]
- 49.Avise J. 2000. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press [Google Scholar]
- 50.Seehausen O. 2006. Conservation: losing biodiversity by reverse speciation. Curr. Biol. 16, 334–347 (doi:10.1016/j.cub.2006.03.080) [DOI] [PubMed] [Google Scholar]
- 51.McGuire J, Linkem C, Koo M, Hutchinson D, Lappin A, Orange D, Lemos-Espinal J, Riddle B, Jaeger J. 2007. Mitochondrial introgression and incomplete lineage sorting through space and time: phylogenetics of Crotaphytid lizards. Evolution 61, 2878–2897 (doi:10.1111/j.1558-5646.2007.00239.x) [DOI] [PubMed] [Google Scholar]
- 52.Keck B, Near T. 2010. Geographic and temporal aspects of mitochondrial replacement in Nothonotus darters (Teleostei: Percidae: Etheostomatinae). Evolution 64, 1410–1428 [DOI] [PubMed] [Google Scholar]
- 53.Bell R, MacKenzie J, Hickerson M, Chavarria K, Cunningham M, Williams S, Moritz C. 2012. Comparative multi-locus phylogeography confirms multiple vicariance events in co-distributed rainforest frogs. Proc. R. Soc. B 279, 991–999 (doi:10.1098/rspb.2011.1229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mallet J. 1995. A species definition for the modern synthesis. Trends Ecol. Evol. 10, 294–299 (doi:10.1016/0169-5347(95)90031-4) [DOI] [PubMed] [Google Scholar]
- 55.Mallet J, Beltran M, Neukirchen W, Linares M. 2008. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol. Biol. 7, 28 (doi:10.1186/1471-2148-7-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pereira R, Monahan W, Wake D. 2011. Predictors for reproductive isolation in a ring species complex following genetic and ecological divergence. BMC Evol. Biol. 11, 194 (doi:10.1186/1471-2148-11-194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson E. 1949. Introgressive hybridization. New York, NY: Wiley [Google Scholar]