Abstract
Background
Altered expression of micro(mi)RNAs has been shown to be associated with tumorigenesis and tumor progression. The expression of phosphatase and tensin homolog (PTEN) plays an important role in glioma and is regarded as a prognostic marker of glioma patients. The goal of this study was to investigate the function of lethal (let)-7a miRNA in glioma cell lines with different PTEN phenotypes.
Methods
One hundred ninety-eight glioma tissues were used to profile miRNA expression.
Results
Let-7a was shown to have lower expression in high-grade glioma than in low-grade glioma. Low expression of let-7a was correlated with poor prognosis of primary glioblastoma patients. We demonstrated that K-ras was a functional target for let-7a to induce cell cycle arrest, apoptosis, and inhibition of cell migration and invasion in vitro. Our further results showed no difference in malignancy inhibition induced by let-7a in 4 glioma cells, including U87 (PTEN null), U251 (PTEN mutant), LN229 (PTEN wild type), and LN229 (PTEN small interfering RNA). The phosphatidylinositol-3 kinase/Akt and mitogen-activated protein kinase/extracellular signal-regulated kinase pathways were inhibited by let-7a, and the inhibition effects had no difference in 4 glioma cells. We demonstrated that let-7a could induce suppression of glioma in vivo by generating a glioma xenograft model.
Conclusion
Our results indicated that let-7a suppresses its target transcript K-ras and inhibits glioma malignancy independent of PTEN expression.
Keywords: glioma, let-7a, K-ras, miRNA, PTEN
MicroRNAs (miRNAs) are short single-stranded RNA molecules, 20–25 nucleotides in length, which posttranscriptionally regulate gene expression by binding to complementary sequences in the 3′ untranslated region (UTR) of target mRNAs.1 Approximately 30% of human genes are predicted to be regulated by miRNAs.2 Recently, the important role of miRNAs in cancer has attracted attention. Many miRNAs have been shown to participate in human tumorigenesis and/or invasion by targeting oncogenes or tumor suppressor genes.3 For example, miRNA-181a and miRNA-181b were reported to inhibit cell growth, induce cell apoptosis, and suppress cell invasion in the U87, TJ905, and U251 human glioma cell lines.4 Otherwise, miR-221/222 induced cell survival by targeting PUMA (p53 upregulated modulator of apoptosis) and considerably decreasing tumor growth in a xenograft model.5
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (K-ras) is one of the 3 members of the Ras oncogene family (comprising K-ras, H-ras, and N-ras) that encodes small GTPases that are involved in cellular signal transduction.6 Ras, activated by a complicated signaling cascade, in turn triggers downstream signaling pathways, including the mitogen-activated protein kinase (MAPK) pathway and the phosphatidylinositol 3-kinase (PI3K)/Akt pathway to modulate various cellular processes, including cell growth, survival, migration, differentiation, and apoptosis. In gliomas, tumorigenesis is correlated with activation of K-ras.7 Activation of the MAPK pathway and the PI3K/Akt pathway is found in almost all high-grade gliomas, and MAPK signaling is necessary for continued glioma maintenance.8 Recently, several miRNAs—miR-18a*, miR-96, miR-143, miR-181a, and miR-217—have been reported to suppress K-ras expression and function as tumor suppressors, suggesting that miRNAs targeting K-ras have important roles in carcinogenesis and are potential therapeutic agents for human cancer.9–13
As a tumor suppressor gene, phosphatase and tensin homolog (PTEN) plays an important role in glioma development and aggressive behavior. Different glioma cell lines present mutations or deletions of the PTEN gene.14 The different PTEN phenotypes show distinct prognoses in glioma patients.15 PTEN could inhibit the PI3K/Akt and MAPK/extracellular signal-regulated kinase (ERK) pathways, both of which are downstream of the Ras signaling pathway, to suppress glioma malignancy.16
In this study, we focused on the function of lethal (let)-7a, a member of the let-7 family, and its target K-ras, in glioma. We analyzed the expression of let-7a in different grades of glioma and verified that low expression of let-7a was correlated with poor prognosis of glioma patients. Overexpression of let-7a reduced cell proliferation, induced G1/G0 phase arrest, increased apoptosis, and decreased the cell invasion and migration capacities of human glioblastoma cells. Furthermore, let-7a directly targeted K-ras to suppress glioma cells, independently of PTEN status. Our data identify let-7a as possibly a critical therapeutic target for glioblastoma intervention.
Materials and Methods
Human Tissue Samples and Cell Lines
For quantitative real-time PCR and immunohistochemistry assay, the Department of Neurosurgery of the First Affiliated Hospital of Nanjing Medical University provided us with human glioma tissue samples after informed consent was obtained from 30 adult patients diagnosed with World Health Organization (WHO) grades II–IV gliomas (10 grade II, 10 grade III, and 10 grade IV). After collection, every glioma tissue was separated into 2 pieces—one was immediately frozen in liquid nitrogen for subsequent total RNA extraction and the other was embedded in paraffin for immunohistochemistry testing. Normal brain tissues were obtained during surgery for severe traumatic brain injury after informed consent was obtained. This study was approved by the institutional review boards of the hospitals, and written informed consent was obtained from all patients.
The human glioma cell lines U251, U87, and LN229 were purchased from the Chinese Academy of Sciences Cell Bank. All cell lines were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated from human glioma specimens, normal brain tissues, or cultured cells using TRIzol reagent (Invitrogen) according to a standard protocol. To detect the relative levels of let-7a in glioma samples and glioma cell lines, an ABI 7300 HT sequence detection system (Applied Biosystems) was used for TaqMan-based real-time reverse transcription (RT)–PCR assays. The primers and probes for the hsa-let-7a TaqMan miRNA assays were purchased from Applied Biosystems. Each reaction was performed in triplicate, and analysis was performed by the 2–ΔΔCt method.
Oligonucleotides and Transfection
The let-7a mimics, scrambled miRNA mimics, and PTEN–small interfering (si)RNA oligonucleotides were chemically synthesized by GenePharma. The sequences were as follows: hsa-let-7a mimics and 2′-O-methyl (2′-OMe-) hsa-let-7a mimics, 5′-UGAGGUAGUAGGUUGUAUAGUU-3′; scrambled miRNA (negative control), sense: 5′-UUCUCCGAACGUGUCACGUTT-3′; and PTEN-siRNA, 5′-GGCGUAUACAGGAACAAUATT-3′. Oligonucleotides (20 µM) were transfected into glioma cells at 60%–70% confluence using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A K-ras cDNA construct without its 3′ UTR in plasmid pReceiver-M02 (K-ras–UTR) was obtained from Fulengen.
Western Blot Assay
Total proteins were extracted from human glioma cells with radioimmunoprecipitation assay lysis buffer (Applygen) 48 h after the scrambled or let-7a oligonucleotides were transfected into cells and were quantified using a bicinchoninic acid protein assay kit (KenGEN). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis was performed on 30 μg of protein from each sample. The electrophoresed proteins were transferred to polyvinylidene difluoride membranes (Millipore) and incubated with diluted primary antibodies against K-ras (1 : 500; Santa Cruz Biotechnology); PTEN, phosphorylated (p)ERK, ERK, pAkt, or Akt (1 : 800, Cell Signaling); Bcl2-associated agonist of cell death (BAD), pBAD, mammalian target of rapamycin (mTOR), pmTOR, or matrix metalloproteinase (MMP)9 (1 : 1000; Santa Cruz), followed by incubation with a horseradish peroxidase–conjugated secondary antibody (1 : 2500; Santa Cruz). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control (KangCheng).
Luciferase Reporter Assay
For the luciferase reporter assay, the 3′ UTR of K-ras, including the predicted binding sites for let-7a, was cloned into the pGL3 luciferase reporter vector (Promega). The 3′ UTR sequences were amplified from human cDNA by PCR using the following primers: forward, 5′-GCGCGAATTCACACGATGCGTATTTTAGTT-3′, reverse, 5′-GCGCAAGCTTAAGAATCACAGTTATGCCAA-3′. A mutant K-ras 3′ UTR was constructed, carrying a mutated sequence in the let-7a binding region. Cells were cultured in 24-well plates and transfected with a complex of let-7a and the luciferase reporter vectors; the pRLSV40 Renilla luciferase construct was used for normalization. After 48 h incubation, luciferase activity was measured using a dual-luciferase reporter system according to the manufacturer's protocol (Promega).
MTT Assay
Cells in the logarithmic phase of growth were seeded into 96-well plates at 3 × 103 cells per well in 100 µL of supplemented DMEM. After 24, 48, 72, and 96 h, 50 μL of a dilution of MTT (5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; KeyGEN) was added into each well and the cells were incubated at 37°C for an additional 4 h. After discarding the supernatant, 200 μL of dimethyl sulfoxide was added to each well to dissolve the precipitate as described in specification. The optical density was measured at 490 nm wavelength, and the data were expressed as a percentage of control optical density.
Cell Cycle Assay
Cells in the logarithmic phase of growth were harvested by trypsinization after transfection for 48 h, washed with phosphate buffered saline (PBS), and fixed with 70% ethanol at –20°C for at least 20 min. After extensive washing, cells were resuspended in PBS containing 50 μg/mL propidium iodide (PI; Sigma-Aldrich), 100 μg/mL RNase A (Sigma-Aldrich), and 10 μL/mL 1% Triton X-100 for 1 h at room temperature, then analyzed by flow cytometry (FCM) using a FACScan instrument (Becton Dickinson). The data were presented as the percentage of cells in a particular phase. The experiments were performed in triplicate.
Cell and Tumor Apoptosis Assay
For cell apoptosis assay, cells were plated into 6-well plates at 1 × 105 cells per well. Forty-eight hours after transfection, the cells were harvested by trypsinization and washed with PBS. Annexin V–fluorescein isothiocyanate and PI double-staining (BD Biosciences) was used to detect and quantify cellular apoptosis by FCM. Annexin V– and PI– cells were used as controls. Annexin V+ and PI– cells were designated as apoptotic; annexin V+ and PI+ cells were necrotic. The apoptotic cell death in the xenograft tumors was examined by terminal deoxynucleotidyl transferase biotin-dUTP nick end-labeling (TUNEL) assay using an in situ cell death kit (Roche). Experimental method was in accordance with the instructions. The TUNEL-positive cells were visualized and counted using a fluorescence microscope. All the tests were performed in triplicate.
In vitro Cell Migration and Invasion Assays
A wound-healing assay was used to assess cell migration. Cells were plated 24 h before transfection at a density of 1 × 105 cells per 35 mm cell culture dish. An artificial wound was created 24 h after transfection using a 200-μL pipette tip on the confluent cell monolayer. To test cell migration and wound healing, images were taken at 0, 12, 24, and 48 h.
Transwell assays were used to test cell invasion. Cells were plated into 24-well Boyden chambers (Corning Costar) with an 8-μm-pore polycarbonate membrane, which was coated with 20 μg of Matrigel (BD Biosciences). Cells in the upper chamber were cultured in 200 μL of serum-free medium, and medium containing 20% FBS was added to the lower chamber to serve as a chemoattractant. After incubation for 24 h, noninvasive cells were removed from the top well using cotton swabs, and the invasive cells were then fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and photographed (×100) in 6 independent fields for each well. Three independent experiments were performed to calculate the fold invasion relative to a blank control.
Glioma Tumor Xenograft Model
Twelve immunodeficient female nude mice, 5–6 weeks old, were used to test whether let-7a could suppress glioma formation in vivo. All mice were bred in aseptic conditions and kept at a constant humidity and temperature according to standard guidelines under a protocol approved by Nanjing Medical University.
At first, 3 mice were injected subcutaneously with 1 × 107 U87 glioma cells in 100 μL of serum-free DMEM. When the tumors reached ∼5 mm in length, they were surgically removed, cut into pieces of 1–2 mm3, and reseeded into the inguinal region of 2 groups of 12 mice. The size of the tumors was measured every 3 days with calipers, and the simplified formula of a rotational ellipsoid (length × width2 × 0.5) was used to calculate the tumor volume. When the subcutaneous tumors reached ∼50 mm3, one group was designated the let-7a group and the other was designated the scramble group. For the let-7a group, a mixture of 200 pmol let-7a mimic oligonucleotides and 10 μL Lipofectamine 2000 was locally injected into every xenograft tumor at multiple sites. In the same way, every mouse in the scramble group was injected with 200 pmol of scrambled oligonucleotides and 10 μL Lipofectamine 2000. These injections and tumor volume measures were performed every 3 days for 15 days. The tumors were harvested 1 week after the end of the treatment for formalin fixation and preparation of paraffin-embedded sections.
Immunohistochemistry
Paraffin-embedded tissue sections (human origin and mouse origin) were used to examine K-ras, PTEN, pAkt, and pERK expression. The tissue sections were incubated with 1 : 200-diluted primary antibodies overnight at 4°C, followed by incubation with biotin-labeled immunoglobulin G (1 : 100 dilution, Gene Tech) for 1 h at room temperature. For the negative control, the primary antibody was replaced with PBS. The sections were then incubated with avidin-biotin-peroxidase complex, revealed with diaminobenzidine, counterstained with hematoxylin (Gene Tech), and visualized under a light microscope. The percentage of positive tumor cells and the staining intensity were assessed. The extent of the staining was scored based on the percentage of positive tumor cells: 0 (<5% positive cells), 1 (6%–25% positive cells), 2 (26%–50% positive cells), 3 (51%–75% positive cells), and 4 (>76% positive cells). The intensity of staining was scored in a semiquantitative manner as follows: 0 (negative), 1 (weakly positive), 2 (moderately positive), and 3 (strongly positive). Multiplication of the intensity and the percentage scores gave rise to the final staining score: – (0), + (1–4), ++ (5–8), and +++ (9–12).17 We regard the expression of specimens whose staining score was – or + as having a low expression level and those whose staining score was ++ or +++ as having a high expression level.
Let-7 Family Expression Analysis and Statistical Analysis
Let-7 family expression values of 198 glioma tissues were from the Chinese Glioma Genome Atlas (CGGA), which is a database that focuses on glioma. Then, to analyze the relation of let-7a expression and prognosis of glioma patients, whole-genome microRNA profiles of 82 primary glioblastomas were chosen from the 198 glioma tissues of the CGGA. After Z-score transformation, the top 25% of let-7a expression values were considered as having high expression. The remaining 75% were considered as having low expression.
All experiments were performed 3 times, and the data were expressed as means ± SE. Statistical evaluation of the data was performed by t-test or chi-square test using SPSS 11.0 for Windows. Survival analysis was performed using the log-rank test in GraphPad Prism 5. P < .05 was considered statistically significant.
Results
Low Expression of Let-7a Was Correlated With Poor Prognosis of Glioma Patients
We first profiled miRNA expression in 198 glioma tissues with 63 low-grade glioma specimens (WHO grade II) and 135 high-grade glioma specimens (WHO grades III and IV). Our analysis showed that expression of let-7a in high-grade glioma was more downregulated than in low-grade glioma (Fig. 1A). Then we analyzed the relation of let-7a expression and prognosis of primary glioblastoma patients. As shown in Fig. 1B, the mean overall survival time of primary glioblastoma patients with a low level of let-7a (median survival, 385 days) was significantly shorter than that of patients with a high level of let-7a (median survival, 591 days; P = .02, log-rank test).
Fig. 1.
Let-7a is correlated with glioma grade and patient outcome and targets K-ras. (A) Identification of let-7 miRNA family expression profile in glioma. The heatmap shows let-7 miRNA family expression profile in 63 low-grade specimens and 135 high-grade specimens. (B) Kaplan–Meier survival analysis of overall survival duration in 82 primary glioblastoma patients according to let-7a expression. The log-rank test was used to calculate P-values. (C) The relative levels of let-7a in glioma samples and glioma cells were measured using real-time quantitative RT-PCR. Normal brain tissues were used as controls. *Significant difference, P < .05. (D) Potential let-7a binding sites in the K-ras mRNA 3′ UTR, predicted by miRanda. Luciferase activities were analyzed in 2 glioma cell lines 48 h after cotransfection of let-7a mimics with either mutant (Mut) or wild-type (WT) K-ras luciferase reporter vectors. *Significant difference, P < .05. (E) Western blots showed that let-7a inhibited K-ras expression in the U87 and U251 glioma cell lines. (F) Immunohistochemistry showed the expression of K-ras in glioma. Chi-square test analyzed let-7a and K-ras expression. The inverse correlation is significant, P < .05. Magnification, ×200.
K-ras Is a Target of Let-7a in Glioma Cell Lines and Tissues
Quantitative real-time PCR was used to analyze let-7a levels in 30 human glioma samples, U87 cells, U251 cells, LN229 cells, and 10 normal human brain tissues. Let-7a was downregulated in glioma tissues and 3 glioma cell lines versus normal brain tissues (Fig. 1C), and furthermore, its expression was significantly lower in high-grade (WHO III–IV) glioma tissues than in low-grade (WHO II) glioma tissues (Fig. 1C).
To explore whether K-ras was a target of let-7a in glioma cells, we used the miRanda miRNA tool (http://www.microrna.org/microrna/home.do) to predict the target sites in the K-ras 3′ UTR (Fig. 1D). Then, we cloned the wild-type or mutant K-ras 3′ UTR downstream of the luciferase open reading frame to construct a luciferase reporter vector. We cotransfected this reporter vector with scrambled or let-7a mimics into glioma cells. The relative luciferase activity was decreased in glioma cells transfected with let-7a and a reporter vector containing the wild-type K-ras 3′ UTR compared with that in cells transfected with scramble or a mutant 3′ UTR (Fig. 1D). Western blotting showed that K-ras was downregulated upon let-7a overexpression in U87 and U251 cells (Fig. 1E). We concluded that let-7a can bind to the K-ras 3′ UTR to inhibit its expression.
Then we separated the glioma specimen into 2 groups by the expression level of let-7a, which we had tested with quantitative real-time PCR. The high let-7a expression group (17 specimens) and low let-7a expression group (13 specimens) were distinguished by mean value of all 30 glioma specimens (Fig. 1F). Immunohistochemical analysis indicated that 13 specimens had high K-ras protein expression and 17 specimens had low K-ras protein expression in these 30 glioma specimens (Fig. 1F). We analyzed the correlation of let-7a level and K-ras expression and found the inverse correlation of expression of let-7a and K-ras in glioma (Fig. 1F).
K-Ras Is a Functional Target of Let-7a That Affects Proliferation, Apoptosis, Migration, and Invasion in Glioma Cells
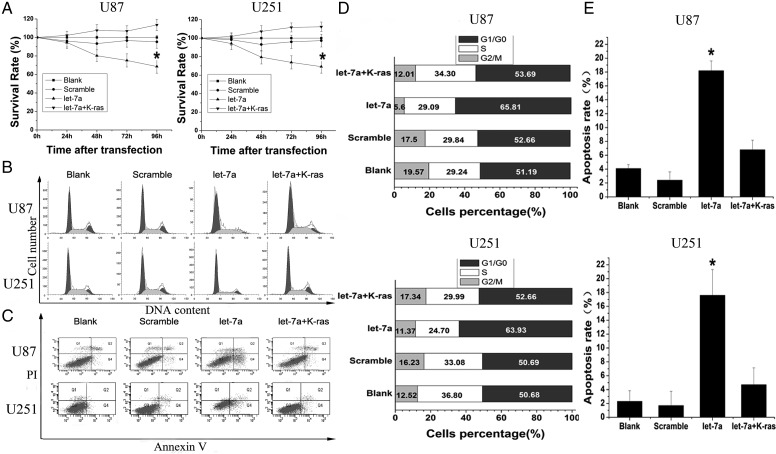
Having demonstrated K-ras as a target of let-7a, we further investigated whether K-ras was a functional target of let-7a by cotransfection with let-7a mimics and K-ras–UTR in glioma cells. We first tested whether let-7a affected glioma cell proliferation via its target K-ras. By MTT assay, the growth of cells transfected with let-7a mimics was inhibited by 31.40 ± 7.21% and 30.79 ± 6.99% 96 h after transfection in U87 and U251 cells, respectively. Cells cotransfected with let-7a mimics and the K-ras–UTR did not show significant growth inhibition (113.6 ± 6.05%) in U87 cells and in U251 cells (112.27 ± 5.2%) compared with the blank group or the scramble group (Fig. 2A).
Fig. 2.
K-ras is a functional target of let-7a to inhibit cell growth and induce apoptosis in U87 and U251 glioma cells. (A) Cell proliferation was measured by MTT assay after transfection with let-7a or cotransfection with let-7a and K-ras–UTR. (B and D) Cells were transfected with let-7a or cotransfected with let-7a and K-ras–UTR and analyzed by FCM cell cycle distribution test. (C and E) After transfection with let-7a or cotransfection with let-7a and K-ras–UTR, apoptosis was measured with annexin V–fluorescein isothiocyanate and PI double-staining by FCM.
To examine the mechanism of cell growth inhibition, we used FCM to analyze the cell cycle distribution of transfected cells after 48 h. In U87 cells, the percentage of let-7a transfected cells in G1/G0 phase, 65.81 ± 3.02%, was higher than that in the blank and scramble groups (51.19 ± 2.23% and 52.66 ± 2.50%, respectively). This was also the case in U251 cells (63.93 ± 3.11%, 50.68 ± 3.14%, and 50.69 ± 2.35% for the let-7a transfected, blank, and scramble groups, respectively; Fig. 2B and D). Then, cells were cotransfected with let-7a and K-ras–UTR to test whether K-ras could prevent the increase of cells in G1/G0 phase. In these circumstances, the percentage of let-7a transfected cells in G1/G0 phase was reduced to 53.69 ± 2.76% in U87 cells and 52.66 ± 3.57% in U251 cells (Fig. 2B and D). This demonstrates that the let-7a–induced cell cycle block is mediated by downregulation of K-ras.
We also analyzed cell apoptosis induced by let-7a via its regulation of K-ras using annexin V and PI double-staining. For U87 cells, 18.2 ± 1.41% of let-7a transfected cells were apoptotic compared with 4.1 ± 0.56% and 2.4 ± 1.21% in the blank and scramble groups, respectively (Fig. 2C and E). Let-7a transfection also caused increased apoptosis in U251 cells: 17.60 ± 3.71% were apoptotic in the let-7a group compared with 2.3 ± 1.53% in the blank group and 1.7 ± 2.06% in the scramble group. In contrast, there was no increased apoptosis of cells cotransfected with let-7a mimics plus K-ras–UTR in U87 cells (6.8 ± 1.37%) and U251 cells (4.7 ± 2.44%) (Fig. 2C and E).
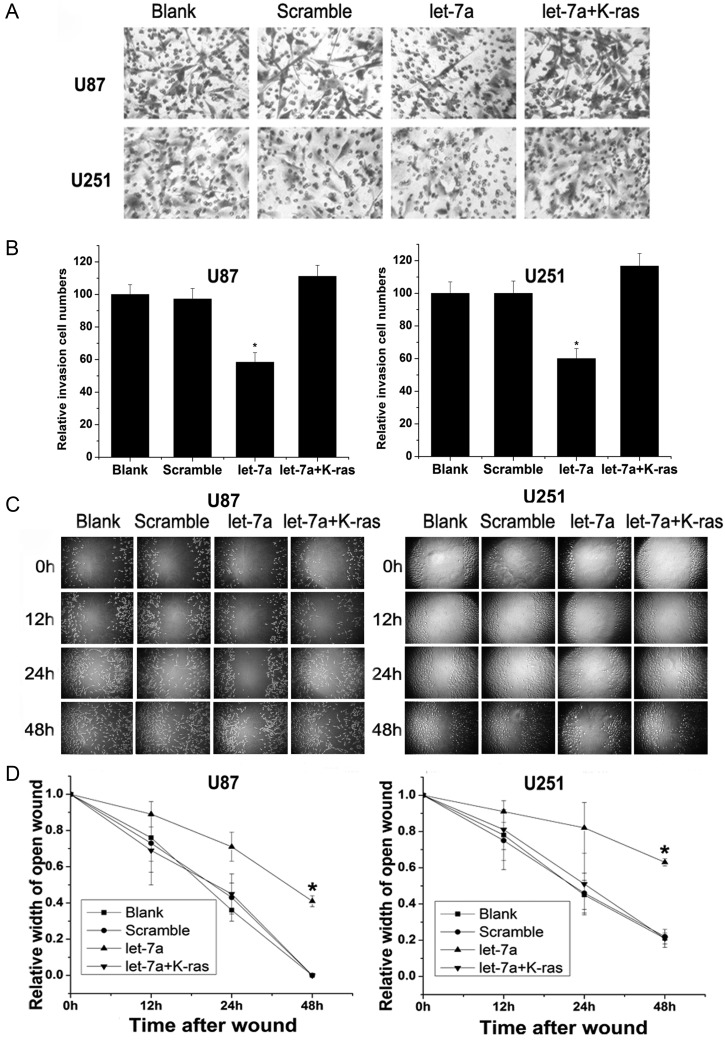
Subsequently, we tested the effect of let-7a on cell invasion and migration via its target K-ras. Overexpressed let-7a inhibited the invasion and migration of glioma cells, while cotransfection with K-ras–UTR moderated the effect of let-7a. Transfection with let-7a led to a reduction of invasion of 41.67 ± 5.92% and 40.00 ± 6.18% in U87 cells and U251 cells, respectively, compared with control cells, while K-ras–UTR cotransfection restored the cells' invasive capacity (Fig. 3A and B). The same effect was observed in a wound-healing assay: The relative widths of the open wounds of cells were 0.41 ± 0.03 in U87 cells and 0.43 ± 0.02 in U251 cells 48 h after transfection with let-7a compared with 0 in U87 scramble cells and 0.27 ± 0.04 in U251 scramble cells, while the relative widths of open wounds were 0 in U87 cells and 0.32 ± 0.05 in U251 cells cotransfected with let-7a and K-ras–UTR. The migration capacity of U87 cells and U251 cells was significantly decreased after transfection with let-7a, while there was no significant change in migration capacity after cotransfection with let-7a and K-ras–UTR (Fig. 3C and D).
Fig. 3.
K-ras is a functional target of let-7a to suppress invasion and reduce migration in U87 and U251 glioma cells. (A and B) Transwell assays were used to evaluate glioma cell invasion ability after transfection with let-7a or cotransfection with let-7a and K-ras–UTR. (C and D) Glioma cell migration ability was detected by wound-healing assays after transfection with let-7a or cotransfection with let-7a and K-ras–UTR. *Significant difference, P < .05.
Let-7a–Mediated Suppression of Glioma Cell Malignancy Is Independent of PTEN
Loss or mutation of PTEN, an important tumor suppressor gene, is commonly found in gliomas. PTEN exhibits a tumor-suppressor function via inhibition of the PI3K/Akt and MAPK signaling pathways, which are both downstream of Ras. To explore whether PTEN participates in the suppression of glioma cells induced by let-7a via its targeting of K-ras, we transfected PTEN-siRNA into LN229 cells to downregulate PTEN expression. Western blot analysis indicated that PTEN expression was significantly downregulated (Fig. 4A). The gene phenotypes were PTEN null in U87 cells, mutant in U251 cells, and wild-type in LN229 cells. To study whether different PTEN gene phenotypes affect the suppression induced by let-7a via its target K-ras, let-7a mimics were transfected into U87 cells, U251 cells, LN229 cells, and LN229 (PTEN-siRNA) cells that had different PTEN phenotypes or status. Then we used MTT assays, FCM, annexin V and PI double-staining, wound-healing, and transwell assays to analyze the suppression effects of let-7a in glioma cells of different PTEN status (Fig. 4B–F). The cell survival rates 96 h after transfection, as judged by MTT assay, were 68.9 ± 6.23% in U87 cells, 67.8 ± 6.15% in U251 cells, 67.90 ± 6.72% in LN229 cells, and 65.60 ± 6.69% in LN229 (PTEN-siRNA) cells. Using FCM, the percentage of cells in G1/G0 phase 48 h after transfection was 61.82 ± 2.12% in U87 cells, 73.19 ± 3.06% in U251 cells, 62.11 ± 3.77% in LN229 cells, and 63.78 ± 3.15% in LN229 (PTEN-siRNA) cells. The apoptosis rates 48 h after transfection were 17.8 ± 2.89% in U87 cells, 17.9 ± 2.73% in U251 cells, 14.90 ± 4.35% in LN229 cells, and 16.9 ± 3.22% in LN229 (PTEN-siRNA) cells. The relative invasion cells after transfection, tested by transwell assay, were 60.4 ± 7.18% in U87 cells, 65.2 ± 7.02% in U251 cells, 54.69 ± 6.17% in LN229 cells, and 54.28 ± 5.28% in LN229 (PTEN-siRNA) cells. Measured with wound-healing assay, the relative widths of open wounds were 0.61 ± 0.07 in U87 cells, 0.67 ± 0.09 in U251 cells, 0.46 ± 0.05 in LN229 cells, and 0.48 ± 0.04 in LN229 (PTEN-siRNA) cells. These findings suggest that let-7a could inhibit glioma cell malignancy independently of PTEN.
Fig. 4.
Let-7a inhibits glioma cells by targeting K-ras independently of PTEN. (A) PTEN expression was decreased on western blots of LN229 cells transfected with a PTEN-siRNA. (B) MTT assays showing the suppression of U87, U251, LN229, and LN229 (PTEN-siRNA) cell proliferation induced by transfection with let-7a. (C) The G1/G0 inhibition in cell cycle distribution show no statistical difference in U87, U251, LN229, and LN229 (PTEN-siRNA) cells after transfection with let-7a. (D) The apoptosis rate in U87, U251, LN229, and LN229 (PTEN-siRNA) cells induced by transfection with let-7a. (E) The cell invasion ability was evaluated by transwell assay in U87, U251, LN229, and LN229 (PTEN-siRNA) cells induced by transfection with let-7a. (F) The cell migration was measured by wound-healing assay in U87, U251, LN229, and LN229 (PTEN-siRNA) cells induced by transfection with let-7a. The data are shown as the mean ± SE of 3 independent experiments. *Significant difference, P < .05.
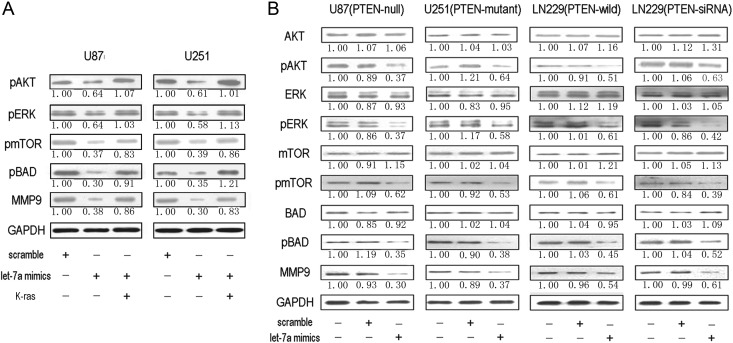
Overexpressed Let-7a MiRNA Inhibits Downstream Signaling Pathways of K-ras Independently of PTEN
It is well known that upregulation of Ras leads to the activation of numerous downstream signaling pathways to result in malignant transformation of normal cells. Here, as shown in Fig. 5A, pAkt, pERK, pmTOR, pBAD, and MMP9 were inhibited by let-7a in U87 and U251 cells. Furthermore, expression of K-ras largely abrogated let-7a–mediated cell malignancy inhibition. So let-7a inhibited K-ras to downregulate activation of downstream signaling pathways.
Fig. 5.
Overexpressed let-7a miRNA inhibits glioma cells via inhibition of signaling pathways downstream of K-ras independently of PTEN. (A) Western blots showing that let-7a inhibited downstream signaling pathway protein activation of K-ras, including pAkt, pERK, pmTOR, pBAD, and MMP9, in glioma cell lines U87 and U251. (B) Overexpression of let-7a led to decreased levels of K-ras, pAkt, pERK, pmTOR, pBAD, and MMP9 expression in U87, U251, LN229, and LN229 (PTEN-siRNA) cells by western blotting. Quantification of the bands was calculated by densitometric analysis. Data were adjusted to blank control, and fold changes were indicated.
To assess whether PTEN affected the downstream signaling pathways involved in K-ras regulation by let-7a, we employed U87, U251, LN229, and LN229 (PTEN-siRNA) cells. Phosphorylated Akt, pERK, pmTOR, pBAD, and MMP9 were downregulated when overexpressed let-7a inhibited K-ras in glioma cells, independently of PTEN (Fig. 5B), but there was no statistically significant reduction of Akt, ERK, mTOR, or BAD. These results indicate that let-7a suppresses glioma malignancy via targeting K-ras to inhibit the PI3K/Akt and MAPK/ERK pathways independently of PTEN.
Let-7a Inhibits Glioma Growth In vivo
To test the function of let-7a in vivo, we generated a glioma xenograft model by subcutaneously injecting U87 glioma cells into mice and then treated the animals with Lipofectamine-mediated let-7a when the tumor size reached ∼50 mm3. The growth of tumors in the let-7a treated group was significantly suppressed compared with that in the scramble group. As shown in Fig. 6A, the tumor volume was 2703.5 ± 376.0 mm3 in the scramble group and 1118.0 ± 186.0 mm3 in the let-7a treated group after 5 let-7a treatments and a further week of tumor growth. Then the apoptosis rate of let-7a treated xenograft tumors was much more than scramble xenograft tumors by TUNEL assay of xenograft tumors after harvesting (Fig. 6B). Finally, we examined the expression of K-ras and its downstream signaling pathway proteins, including pAkt and pERK, in the glioma xenograft model. As shown in Fig. 6C, the expression of K-ras, pAkt, and pERK was reduced in the let-7a treated group.
Fig. 6.
Overexpressed let-7a inhibits glioma growth in vivo by suppressing (A) difference in tumor volume between the scramble group and the let-7a group. *P < .05; **P < .01. (B) TUNEL assay in xenograft tumor sections indicated that let-7a induced cell apoptosis. (C) Expression of K-ras, pAkt, and pERK was downregulated in mice of the let-7a group compared with those of the scramble group, as measured by immunohistochemistry. Magnification, ×200. *P < .05; **P < .01.
Discussion
We have indicated that the expression level of let-7a is downregulated in high-grade gliomas relative to low-grade gliomas, and a low level of let-7a is correlated with poor prognosis in glioma patients. We have also demonstrated that let-7a inhibits glioma malignancy by targeting K-ras and affecting the downstream pathways of Ras, including the MAPK and Akt pathways, both in vivo and in vitro. In particular, our results suggest that the important glioma tumor suppressor PTEN has no effect on let-7a–mediated regulation in glioma cells.
In recent years, some or all of the let-7 family members have been found to be downregulated in various cancers and correlated with the survival of tumor cells.18 The expression of let-7a has been reported to be downregulated in lung, breast, and pancreatic cancers.19 And we demonstrated that expression of let-7a was downregulated in high-grade gliomas, while low expression of let-7a was correlated with poor prognosis of glioma patients. Let-7a may be one of the important prognostic biomarkers of glioma.
Many oncogenes, including Ras, c-Myc, high-mobility group AT-hook 2, and signal transducer and activator of transcription 3, have been found to be targets of let-7 that suppress tumor cell malignancy.19,20 K-ras is a member of the Ras family, whose mutation and overexpression are considered to result in malignant cell transformation. Moreover, K-ras may play an important role in gliomagenesis. The activation of K-ras in neural progenitors could induce glioma generation combined with Akt in mice.21 K-ras had been demonstrated to be a target of let-7 to protect from growth of lung carcinoma.22,23 Then studies reported let-7 had antitumorigenic and antimigratory effects on glioblastoma cells, and these may be because of the reduced expressions of Ras.24 Our results showed that K-ras is a direct target of let-7a in glioma cells. Furthermore, as a direct target of let-7a, K-ras displayed an important functional role affecting proliferation, apoptosis, migration, and invasion in the U87 and U251 glioma cell lines.
Mutation or loss of expression of PTEN is common in glioma and occurs in about 40% of glioblastoma multiforme.14 The genetic background of PTEN is closely correlative with prognosis of glioma patients. The median survival times for cases with PTEN mutation were 4.4 months, while those without PTEN mutation were 34.4 months.15 PTEN is an inhibitor of the Ras/PI3K/Akt pathway, which has been reported to be required for glioma maintenance in vivo.25 PTEN can also suppress the activity of Ras to inhibit the MAPK pathway.16 Furthermore, it has been reported that miR-221 and miR-222 can regulate the expression of PTEN to affect glioma cell malignancy.26 Thus, we focused on the effect of PTEN on let-7a modulation by employing 4 glioma cell lines with different PTEN phenotypes and status, including U87 (PTEN null), U251 (PTEN mutant), LN229 (PTEN wild type), and LN229 (PTEN-siRNA) cells. Our data showed that let-7a could suppress K-ras and its downstream pathways and induces glioma cell cycle arrest, cell apoptosis, and inhibition of cell migration and invasion, regardless of PTEN status.
In summary, our results suggest that let-7a is associated with glioma grade and patient outcome. And let-7a targets K-ras and its downstream pathways and then inhibits glioma cell malignant phenotypes, regardless of PTEN status. Thus, the role of let-7a in gliomagenesis needs to be further investigated.
Conflict of interest statement. None declared.
Funding
This study was supported by National High Technology Research and Development Program 863 (2012AA02A508), the China Natural Science Foundation (91229121, 81072078, 81101901 and 81172389), Jiangsu Province's Key Provincial Talents Program (RC2011051), Jiangsu Province's Key Discipline of Medicine (XK201117), the Program for Development of Innovative Research Team in the First Affiliated Hospital of NJMU, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- 1.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Slack FJ, Weidhaas JB. MicroRNA in cancer prognosis. N Engl J Med. 2008;359:2720–2722. doi: 10.1056/NEJMe0808667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 4.Shi L, Cheng Z, Zhang J, et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 5.Zhang CZ, Zhang JX, Zhang AL, et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall A. The cellular functions of small GTP-binding proteins. Science. 1990;249:635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- 7.Abel TW, Clark C, Bierie B, et al. GFAP-Cre-mediated activation of oncogenic K-ras results in expansion of the subventricular zone and infiltrating glioma. Mol Cancer Res. 2009;7:645–653. doi: 10.1158/1541-7786.MCR-08-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson JP, Van Brocklin MW, Guilbeault AR, Signorelli DL, Brandner S, Holmen SL. Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene. 2010;29:335–344. doi: 10.1038/onc.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 10.Shin KH, Bae SD, Hong HS, Kim RH, Kang MK, Park NH. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2011;404:896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- 11.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–959. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 12.Yu S, Lu Z, Liu C, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 13.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 14.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 15.Smith JS, Tachibana I, Passe SM, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 16.Gu J, Tamura M, Yamada KM. Tumor suppressor PTEN inhibits integrin- and growth factor-mediated mitogen-activated protein (MAP) kinase signaling pathways. J Cell Biol. 1998;143:1375–1383. doi: 10.1083/jcb.143.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Hao L, Wang L, et al. Elevated IGFIR expression regulating VEGF and VEGF-C predicts lymph node metastasis in human colorectal cancer. BMC Cancer. 2010;10:184. doi: 10.1186/1471-2407-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7:an emerging next-generation cancer therapeutic. Curr Oncol. 2010;17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:19–36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Lu Y, Toh ST, et al. Lethal-7 is down-regulated by the hepatitis B virus × protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 21.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 22.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 23.He XY, Chen JX, Zhang Z, Li CL, Peng QL, Peng HM. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer Res Clin Oncol. 2009;136:1023–1028. doi: 10.1007/s00432-009-0747-5. [DOI] [PubMed] [Google Scholar]
- 24.Soon-Tae L, Kon C, Hyun-Jung O, et al. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J Neurooncol. 2010;102:19–24. doi: 10.1007/s11060-010-0286-6. [DOI] [PubMed] [Google Scholar]
- 25.Robinson JP, Vanbrocklin MW, McKinney AJ, Gach HM, Holmen SL. Akt signaling is required for glioblastoma maintenance in vivo. Am J Cancer Res. 2011;1:155–167. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Kang C, Wang P, et al. MicroRNA-221 and -222 regulate radiation sensitivity by targeting the PTEN pathway. Int J Radiat Oncol Biol Phys. 2011;80:240–248. doi: 10.1016/j.ijrobp.2010.12.049. [DOI] [PubMed] [Google Scholar]