Abstract
Background
Glioblastoma multiforme (GBM) is the most aggressive brain tumor in adults, and despite state-of-the-art treatment, survival remains poor and novel therapeutics are sorely needed. The aim of the present study was to identify new synergistic drug pairs for GBM. In addition, we aimed to explore differences in drug-drug interactions across multiple GBM-derived cell cultures and predict such differences by use of transcriptional biomarkers.
Methods
We performed a screen in which we quantified drug-drug interactions for 465 drug pairs in each of the 5 GBM cell lines U87MG, U343MG, U373MG, A172, and T98G. Selected interactions were further tested using isobole-based analysis and validated in 5 glioma-initiating cell cultures. Furthermore, drug interactions were predicted using microarray-based transcriptional profiling in combination with statistical modeling.
Results
Of the 5 × 465 drug pairs, we could define a subset of drug pairs with strong interaction in both standard cell lines and glioma-initiating cell cultures. In particular, a subset of pairs involving the pharmaceutical compounds rimcazole, sertraline, pterostilbene, and gefitinib showed a strong interaction in a majority of the cell cultures tested. Statistical modeling of microarray and interaction data using sparse canonical correlation analysis revealed several predictive biomarkers, which we propose could be of importance in regulating drug pair responses.
Conclusion
We identify novel candidate drug pairs for GBM and suggest possibilities to prospectively use transcriptional biomarkers to predict drug interactions in individual cases.
Keywords: drug combination responses, glioblastoma therapy, glioblastoma stem cell cultures, predictive medicine
Glioblastoma multiforme (GBM) is the most common primary brain tumor in adults and the highest grade of astrocytoma (World Health Organization grade IV).1,2 Clinically, GBM is characterized by rapid progression and poor response to therapy. The median life expectancy is 14 months after diagnosis, and state-of-the-art therapy for GBM involves a combination of surgery, radiation, and chemotherapy using the orally administered DNA alkylating agent temozolomide (TMZ).2 The survival benefit of TMZ treatment, however, is relatively modest (estimated 2.5 mo3). In addition, TMZ treatment is fraught with side effects, such as hematological toxicity, vomiting, fatigue, and nausea.4 Further, many GBM patients develop resistance to the drug.5,6 There is thus a need to identify novel strategies for pharmacological treatment with improved efficacy, lower variability across patients, reduced rate of resistance development, and fewer side effects.
One frequently discussed approach to enhance efficacy and delay the onset of drug resistance is to apply drug combinations.7–9 For GBM, a current strategy seen in clinical trials is to combine TMZ with an additional agent such as an inhibitor of epidermal growth factor receptor (EGFR) and of vascular endothelial growth factor receptor (erlotinib and bevacizumab, respectively) or the protease inhibitor nelfinavir.10,11 In many instances, a drug combination is preferably synergistic, meaning that the combined effect is more effective than single drugs alone.12 However, there is limited knowledge about which cellular pathways can be targeted in combination to produce synergistic anticancer effects in GBM. In addition, there is little empirical data on how the effect of different drug combinations might be modulated by the transcriptional profile of an individual tumor.
Here, we performed a drug pair interaction screen across 5 different GBM cell lines. In our screening experiment, we tested all pairs from a set of 31 drugs forming a matrix of 465 unique pairs and calculated an interaction score, as previously described,13 for each pair. In contrast to the traditional design of a screen (matrix of pairs in a single system), the experiment conducted here analyzed multiple GBM-derived cancer cell lines in parallel. Selected hits were subsequently retested using additional criteria, including the combination index and the α interaction metric.14–16 Apart from finding novel anticancer drug pairs, this extended experimental design had 2 key benefits. First, it enabled the estimation of interaction variability across GBM cell lines, which made it possible to assess the degree to which an interaction effect was context specific or likely to work across a broader range of cases. Second, the variability of interaction effects among the GBM cell lines made it possible to associate interactions with cell line molecular profiles, whereby we could define novel biomarkers predicting anticancer drug-drug interactions in GBM.
Key results of our study include 4 drug pairs displaying a strong synergy and robust effect (exhibiting a synergistic interaction in several cell lines) in the majority of the cell lines and glioblastoma-initiating cell (GIC) cultures tested. Of particular interest is an interaction between the sigma receptor ligand rimcazole and the antidepressant sertraline. These are approved compounds that have been implicated as single agents against breast cancer,17 colon cancer,18 and GBM,19 but to our knowledge a synergistic interaction in GBM has not previously been reported. In addition, we defined subsets of robust and more “variable” interactions (the latter meaning that these agents had effects in a small number of the cell lines tested), which we, alongside microarray experiments, utilized to propose a statistical method to predict drug pair interactions in individual cases.
Taken together, we identified a set of novel drug pairs as possible treatments for GBM. We also propose a method to predict drug-drug interactions, which provide a roadmap for extended efforts to find both robust and more tailored anti-GBM therapies.
Material and Methods
Cells
T98G was obtained from American Type Culture Collection, while A172, U343MG, U373MG, and U87MG were obtained from Cell Lines Services. These cell lines are commonly used as models of GBM and present a spectrum of different genetic lesions20 (Supplementary Table S1). All cell lines were grown in monolayer and maintained in high-glucose (4.5 g/L) Dulbecco's modified Eagles's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 2 mM l-glutamine (Fisher Scientific). Cells were incubated at 37°C with 5% CO2.
Human GICs, from the Human Glioma Cell Culture project at Uppsala University (S.N., K.F.N., B.W., and L.U., manuscript in preparation) were originally derived from serum-free tumor samples and grown in defined stem cell media as previously described.19 To keep the GICs as adherent cultures, we used the laminin-based protocol developed by Pollard et al.19
Normal human fibroblasts were obtained from Coriell Cell Repository (AG07095B) and American Type Culture Collection (AG1523 and AG1518). Cells were grown in monolayer and maintained in high-glucose (4.5 g/L) DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 2 mM l-glutamine (Fisher Scientific). Cells were incubated at 37°C with 5% CO2.
Drugs
Active pharmacological agents (here referred to as “drugs”) were obtained from BioMol (Enzo Life Sciences), National Institutes of Health (NIH) Clinical Collection, National Cancer Institute/NIH Developmental Therapeutics Program Approved Oncology Drugs Set, Sigma Aldrich, and Tocris Biosciences (Table 1). The resulting list represents drugs targeting different signaling pathways—noncancer drugs chosen based on prior involvement as possible cancer therapeutics or chemotherapeutic drugs, such as (i) the chemotherapeutic mitotic inhibitor paclitaxel, (ii) the EGFR inhibitor gefitinib, and (iii) the antidepressant sertraline with a noncancer application. In addition to these manually chosen compounds, the list was supplemented with 15 randomly picked compounds from the libraries. All drugs were dissolved in dimethyl sulfoxide (DMSO), except for cefaclor, which was dissolved in water. Drug concentrations were standardized to 10 and 20 μM (see Table 1 for doses used for each drug). For most of the individual compounds, these doses produced no or a mild (>80% viability compared with control) phenotype, which is a screening strategy that enriches for synthetic lethal (synergistic) pairs.13 For all experiments, equal amounts of DMSO were used as untreated (vehicle) controls; the final concentration of DMSO in the medium following addition of the compound was 0.1%–0.4%.
Table 1.
List of drugs used in the study, dose used, and target(s)/mechanism of action
| Compound | Dose (uM) | Target(s)/Mechanism | Supplier |
|---|---|---|---|
| Erlotinib | 10 | EGFR | DTP, Enzo |
| Gefitinib | 10 | EGFR | NCC,DTP, Selleck |
| Quercetin | 10 | PI3K | BIOMOL |
| Wortmannin | 10 | PI3K | BIOMOL |
| LY294002 | 10 | PI3K | BIOMOL, Enzo |
| ZM 336 372 | 10 | cRAF | BIOMOL |
| GW 5074 | 10 | cRAF | BIOMOL |
| Retinoic acid | 10 | RAR | BIOMOL |
| Doxorubicin | 10 | Cytostatic/topoisomerase | II BIOMOL |
| Imatinib | 10 | Bcr/Abl, c-KIT, PDGFR | NCC |
| Imipramine | 10 | Tricyclic antidepressant | NCC, Sigma |
| Sertraline | 10 | SSRI | NCC |
| Temozolomide | 20 | Alkylating cytostatic | DTP, NCC |
| Olomoucine | 10 | CDK | BIOMOL |
| Roscovitine | 10 | CDK | BIOMOL, Enzo |
| Metformin | 10 | GLUT4 | Enzo, Sigma |
| Fluperlapine | 20 | Tricyclic atypical antipsychotic | NCC, Enzo |
| Ipriflavone | 20 | Isoflavone osteoclast inhibitor | NCC, Sigma |
| Physostigmine | 20 | Cholinesterase inhibitor | NCC, Enzo |
| Rapamycin | 10 | mTOR | BIOMOL, Enzo |
| Nelarabine | 10 | Purine nucleoside analogue | DTP, Tocris |
| Rimcazole | 10 | Sigma receptors | NCC, Tocris |
| Cefaclor | 20 | Cephalosporin antibiotic | NCC, Enzo |
| Pirenperone | 20 | 5-HT2 | NCC, Sigma |
| AG-490 | 10 | JAK2 | BIOMOL, Enzo |
| U-0126 | 10 | MEK1 and MEK2 | BIOMOL,Enzo |
| Paclitaxel | 10 | Microtubuli inhibitor | DTP, Enzo |
| Zolmitriptan | 20 | 5HT1B/1D | NCC, Enzo |
| Piroxicam | 20 | NSAID | NCC, Enzo |
| Pterostilbene | 20 | Phytoalexin/antioxidant | NCC, Enzo |
| PP2 | 10 | Src-family kinases | BIOMOL, Enzo |
Abbreviations: PI3K, phosphatidylinositol-3 kinase; RAR, retinoic acid receptor; CDK, cyclin-dependent kinase; GLUT4, glucose transporter type 4; mTOR, mammalian target of rapamycin; NSAID, nonsteroidal anti-inflammatory drug; NCC, National Clinical Collection from NIH; DTP, Developmental Therapeutic Program, Oncology drug plate supplied by NIH; Enzo, Enzo Biosciences; Sigma, Sigma Aldrich; BIOMOL, BIOMOL plate from Enzo; Selleck, SelleckChem; Tocris, Tocris Biosciences.
Preparations of Combinations and Cell Viability Assay
Tumor cells were plated at 1.5 × 104 cells/well in 96-well plates (TPP, Techtum Lab) 24 h prior to treatment. Cells were treated with drugs diluted in media, single or in combination, and treated cells were incubated for 48 h. Three replicates were performed with a 3-replicate negative control (DMSO). (In some plates, 4–6 replicates were used when plate space was available.) Viability studies were performed using the resazurin assay (Alamar Blue, Invitrogen) according to manufacturer's protocol. For the isobole experiments, 2000 cells/well were seeded in a 384-well plate (BD Falcon #353221) 24 h prior to treatment. Apart from this, the viability readout procedure was similar to the described setup, with the exception of the dose-titration plate plan (see Supplementary information sheet S3).
Drug Pair Interaction Assessments
We quantified the drug response as the viability ratio W = Ytreated/Ycontrol, where Y represents the average fluorescence signal gained from all replicates. This setup was used for all types of interaction calculations. To assess drug pair interactions and possible synergies, we used ratio values and calculated interaction scores as previously described,13 derived from the Bliss independence model.21 The interaction scores presented provide a one-point quantitative measurement of the interaction, where a score less than, equal to, and more than 1 indicated synergy, additivity, and antagonism, respectively. For the follow-up experiments of the 4 interesting pairs, we used the experimentally more demanding isobologram analysis to assess possible synergies in a quantitative manner. Isobolograms are built up by isoboles, curves of constant effect. Further, the analysis of isoboles is based on the Loewe additivity model,22 which quantifies the interaction between 2 drugs based on the assumption that a drug cannot interact with itself.23 Eight different concentrations for each drug were combined in a 2-dimensional matrix (64 data points in 5 replicates/combination), where the lowest concentration was 0 and the highest concentrations were 32 μM (rimcazole and sertraline) and 64 μM (gefitinib and pterostilbene) (see Supplementary information sheet S3 for doses and Supplementary Fig. S4 for single dose response curves for the single drugs). Additional matrix entries were imputed using linear interpolation so that the resulting isobologram contained 32 × 32 entries. From each isobologram, we extracted 2 independent quantitative measurements of interaction: the combination index23,24 and the alpha (α) interaction parameter.25 The former has the value 1 as a baseline, with synergy corresponding to a combination index smaller than 1 and antagonism greater than 1, while the latter has zero as a baseline, with α smaller than zero corresponding to a synergy and α greater than zero to antagonism. We present both types of quantitative measurements (Table 2) in addition to interaction scores (Figs. 1–3). Other studies15,16,23 have compared other interaction calculations.
Table 2.
α-Values and combination indices for 4 combinations in 5 GICs
| α-Values |
Combination Index |
||||
|---|---|---|---|---|---|
| Cell Line | Average | P | Cell Line | Average | P |
| Rimcazole + sertraline | Rimcazole + sertraline | ||||
| U3005MG | 0.42 | .9040 | U3005MG | 1.06 | .9544 |
| U3013MG | −0.08 | .3558 | U3013MG | 0.98 | .1342 |
| U3024MG | −0.04 | .4549 | U3024MG | 1.00 | .5000 |
| U3034MG | −0.17 | .3304 | U3034MG | 0.97 | .1728 |
| U3068MG | −0.27 | .2865 | U3068MG | 0.95 | .0830 |
| Rimcazole + gefitinib | Rimcazole + gefitinib | ||||
| U3005MG | 0.71 | .9636 | U3005MG | 1.11 | .0041 |
| U3013MG | 0.24 | .8431 | U3013MG | 1.03 | .0584 |
| U3024MG | 0.72 | .9996 | U3024MG | 1.09 | .0009 |
| U3034MG | 0.81 | .9868 | U3034MG | 1.12 | .0021 |
| U3068MG | 0.18 | .8610 | U3068MG | 1.03 | .0130 |
| Pterostilbene + sertraline | Pterostilbene + sertraline | ||||
| U3005MG | 0.26 | .6946 | U3005MG | 1.23 | .9761 |
| U3013MG | −0.58 | .0000 | U3013MG | 0.90 | .0003 |
| U3024MG | −1.29 | .0056 | U3024MG | 0.82 | .0080 |
| U3034MG | −1.38 | .0000 | U3034MG | 0.79 | .0001 |
| U3068MG | −1.27 | .0000 | U3068MG | 0.80 | .0000 |
| Pterostilbene + gefitinib | Pterostilbene + gefitinib | ||||
| U3005MG | −0.59 | .0038 | U3005MG | 0.91 | .0010 |
| U3013MG | −0.19 | .1815 | U3013MG | 0.96 | .0486 |
| U3024MG | −0.58 | .2516 | U3024MG | 0.92 | .1048 |
| U3034MG | −0.99 | .0000 | U3034MG | 0.84 | .0001 |
| U3068MG | −0.68 | .0031 | U3068MG | 0.88 | .0008 |
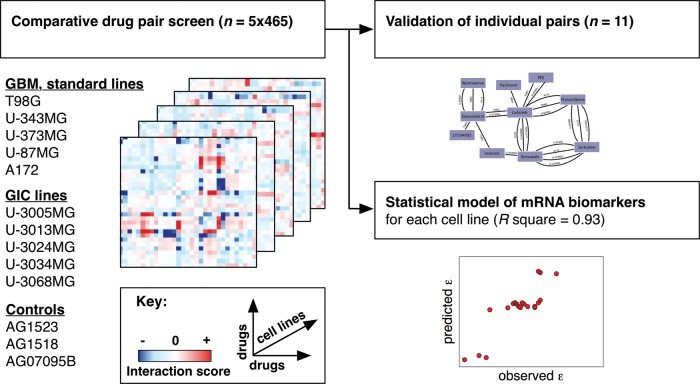
Fig. 1.
Schematic representation of comparative drug pair screening in multiple glioblastoma cell lines. We conducted a 5 × 465 drug pair screen to find novel anticancer drug pairs for GBM. Following initial screening in standard cell lines (heatmaps, left), we validated selected drug-drug interactions in GICs and develop biomarker predictors for cell line differences in drug-drug interactions (right).
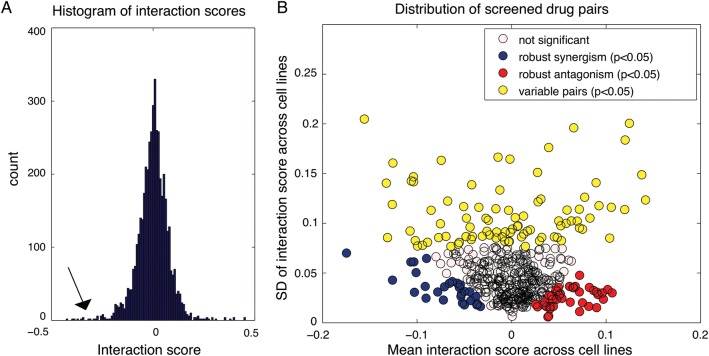
Fig. 2.
Distribution of interaction scores for screened drug pairs. (A) Distribution of drug-drug interaction scores, pooled for all 5 cell lines. Histogram of the interaction score distribution where tails on both sides are visible, indicating negative (arrow) and positive interactions. (B) Characterization of interaction score mean vs interaction score variance of all 465 screened drug pairs. The mean interaction scores across the 5 cell lines (x-axis) vs the SD of interaction scores across the 5 cell lines (y-axis). The graph reveals the significant (P < .05) most variable pairs (yellow) (both synergistic and antagonistic), most robust synergistic pairs (blue), and most robust antagonistic pairs (red).
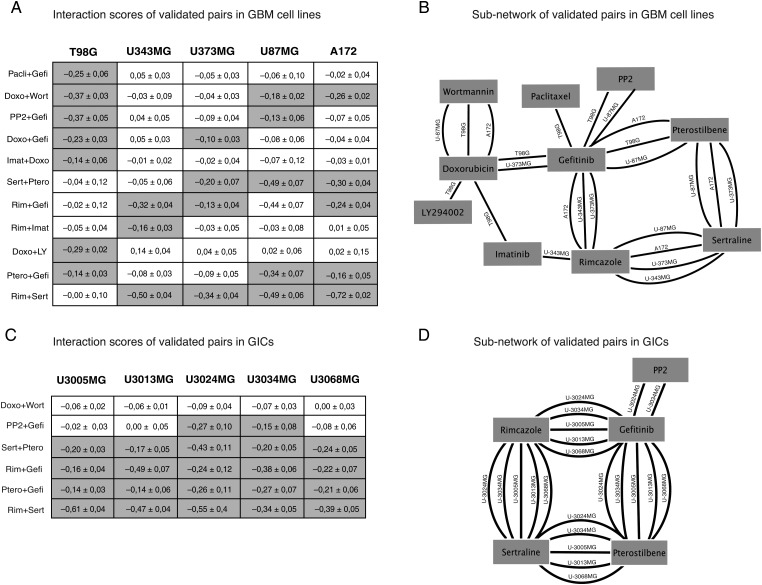
Fig. 3.
Core set of retested drug-drug interactions in standard GBM cell lines and GIC cultures. (A) Chart of 11 combination responses in 5 GBM cell lines. Interaction scores ± 95% estimated confidence intervals. Values highlighted in gray indicate an interaction score < −0.1 (P < .05). (B) Subnetwork of validated pairs; edges indicate synergistic interactions in specific cell lines. Note that rimcazole + sertraline stands out as the most robust acting combination with 4 connections, followed by pterostilbene + sertraline, pterostilbene + gefitinib, and doxorubicin + wortmannin with 3 connections. EGFR inhibitor gefitinib stands out as the most connected drug (C and D). Six selected combinations were further validated in a panel of GICs. This demonstrated that the synergistic interaction scores, thus indicating a synergy, seen in the cell lines could be reproduced, resulting in a similar network around gefitinib, rimcazole, sertraline, and pterostilbene. (Pacli = paclitaxel, Gefi = gefitinib, Doxo = doxorubicin, Wort = wortmannin, Imat = imatinib, Sert = sertraline, Ptero = pterostilbene, Rim = rimcazole, LY = LY294002).
Transcript Profiling
All cells were seeded at 5 × 105 cells/well in 6-well TPP plates 24 h prior to harvest. RNA for all microarray experiments was extracted and purified, according to protocol, using the RNeasy Plus Mini-kit (Qiagen). RNA was prepared from untreated cells. Affymetrix 1.1 ST was used and experiments were performed by the Bioinformatics and Expression Analysis core facility at Karolinska Institutet, Stockholm, Sweden.
Defining Predictive Transcripts by Sparse Canonical Correlation Analysis
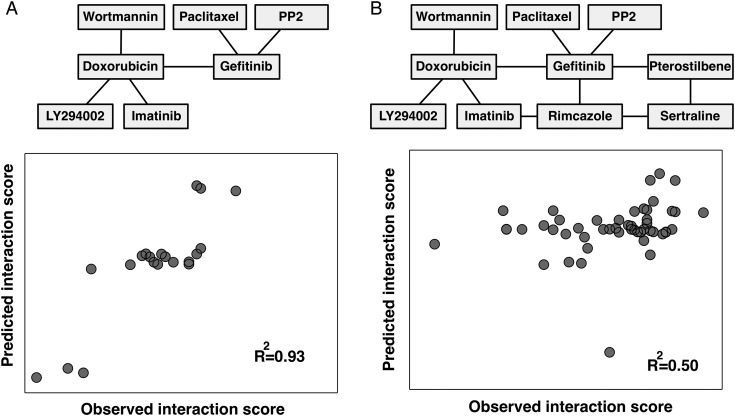
The goals of the analysis performed in this section were to (i) assess the possibility of predicting drug-drug interaction scores from mRNA biomarkers and (ii) define predictive mRNAs for drug-drug interaction effects in GBM. To do this, we used a multivariate method, sparse canonical correlation analysis (sCCA), as follows. We selected as our data the top 200 most variable transcripts, determined as the variance across the 5 cell lines, and data for the 11 validated drug pairs (Fig. 3A). The mRNA data were arranged into a 200-row (mRNA) × 5-column (cell lines) matrix X and the drug pair data were arranged into an 11-row (interaction scores) × 5-column (cell lines) matrix Z (Supplementary Table S2). In detail, we subsequently applied the sCCA function in the “PMA” R package,26 using default settings and other parameters (“penaltyx” and “penaltyz”) determined by 10-fold cross-validation simulations. The sCCA function returned sparse vectors u and v (of lengths 200 and 11, respectively), such that Corr(uTX, vTZ) was maximal. From the results, we obtained 3 pieces of information. First, the nonzero elements of u indicated predictive mRNAs (Supplementary Table S2). Second, the nonzero elements of v indicated a subset of interaction scores that were well predicted by the elements in u (Fig. 4A and Supplementary Table S2). Third, given an expression pattern x, the u and v vectors could be used to predict the interaction scores by Zpredicted = vuTX. The sCCA procedure can be used to extract either a single pair (u,v) or a sequence of vector pairs (u1, v1), (u2, v2) to predict increasing proportion of the interaction network (Fig. 4).
Fig. 4.
Transcriptional biomarkers can predict drug-drug interaction in GBM. We used sCCA to select subsets of interaction links (interaction scores) that are well predicted by subsets of mRNAs. (A) Results for a single component model, where a subset of 6 interactions are predicted by 82 transcripts (see main text), obtaining excellent model fit (R2 = 0.93). (B) Results for a 3-component model where the full core network of 11 interactions is predicted by 200 transcripts (R2 = 0.50).
Significance Testing
To statistically test for effects on viability, we used Student's 2-sample t test, assuming equal variance, to quantify differences between combination (ratio) and control (ratio) effects. We also tested the experimental replicate interaction score distribution and thus performed nonparametric bootstrapping (resampling data points, 500 iterations from all replicates in all experiments) to obtain empirical 95% confidence intervals for the interaction scores (Fig. 3A and C). In Fig. 2B, potential robust (blue, red) interactions are operationally defined as interactions with a t test nominal P value <.05, and variable (yellow) interactions with a variance exceeding the 95th percentile of the simulated sampling distribution under the null hypothesis (constant interaction). Combination index and α-values were tested for significance using a single sample 2-tailed t test, with 5 independent measurements of combination index and α-value done in each of the cell lines tested.
Results
Drug-Drug Interaction Scores Across 5 GBM Cell Lines
We performed a screen of 465 drug combinations in the GBM cell lines T98G, A172, U87MG, U343MG, and U373MG (Supplementary Figure S1). The drug pairs analyzed represent all unique pairs formed from a set of 31 selected drugs (Table 1). The key concept behind selecting these drugs was to strike a reasonable balance between (i) cytotoxic drugs and tyrosine kinase inhibitors with glioblastoma relevance and (ii) several noncancer drugs that may be of interest for drug repurposing (Methods). The screening protocol included measurement of viability (% of control) after 48 h of compound exposure, using both single and combination treatments, followed by calculation of interaction scores. Following estimation of interaction scores from triplicate measurements (Methods), the interaction score data were organized as 5 matrices of dimension 31 × 31, each containing 465 drug pair interaction scores (=2325 interaction measurements) used for downstream statistical analysis (Fig. 1).
Distribution and Within-Pathway Correlations of Interaction Scores
The empirical distribution of interaction scores for the 465 drug pairs in the 5 GBM cell lines is bell shaped, with extended tails (Fig. 2A, arrow). A similar distribution has been both predicted and observed in other biological systems and appears to be a generic property of pairwise functional interactions.25,27 In addition to showing these characteristic statistical trends, our data were consistent with the biological expectation that drugs targeting the same pathway produce correlating interaction scores in relation to other drugs (Supplementary Fig. S2), a trend that was observed in reported genetic and pharmacological interaction data,25,28 which we here demonstrate for the first time in GBM.
Interaction Scores Reveal Robust and Variable Synergies
Unlike interaction screens in single cell lines or experimental systems,13,25,28 the comparative experimental design used here made it possible to characterize the interaction scores between the 465 drug pairs across 5 cell lines. For this, we plotted the mean vs the standard deviation of the cell-specific interaction score. The resulting graph (Fig. 2B) thus separated the drug pairs along 2 dimensions: pairs that on average had a synergistic interaction score vs a positive one (x-axis) and pairs whose interaction was robust (consistent) across cell lines vs variable (y-axis). The graph thus reveals drug pairs with robust synergy (blue, t test P< .05), robust antagonism (red, t test P < .05), and more variable interaction behavior (yellow, bootstrapping simulation test P < .05). We conclude that the tested drug pairs vary in interaction scores across the cell lines, but a subset of drug pairs have consistently synergistic interaction scores, potentially indicating a synergy that is robust to cellular context. In the following experiments, we therefore sought to (i) validate the initial screening data in the same and additional cell lines, (ii) characterize interactions with a complementary interaction analysis model, and (iii) apply molecular profiling to define biomarkers predicting interaction score variability in GBM.
Robust Synergistic Effects Against GBM Growth in a Subnetwork of Approved Drugs
In the next set of experiments, and as a secondary screen of hits found in the primary screen, we retested all drug pairs that met combined criteria of (i) a synergistic interaction score (less than −0.1975, corresponding to 2 standard deviations from the interaction score distribution for all cell lines), (ii) significant inhibition of viability (P < .01), and (iii) a sufficient magnitude of the inhibition (viability ratio <0.8). In this secondary screen, we retested a total of 45 pairs that met these criteria to obtain a stringent network of 11 interacting drug pairs for which a synergistic interaction score, here defined as an interaction score less than −0.1975, was reproduced across multiple experimental replicates in GBM cell lines (Fig. 3A–B).
The 11 reproduced combinations involved several known cancer drugs, including kinase inhibitors gefitinib and imatinib, mitotic inhibitor paclitaxel, and doxorubicin (Fig. 3A). We also noted a number of synergistic interactions between/among noncancer drugs—for example, among PP2 (an Src inhibitor), rimcazole (a sigma receptor ligand), sertraline (a selective serotonin reuptake inhibitor [SSRI]), and pterostilbene (an antioxidant) (Fig. 3A). The most interaction-prone drugs were gefitinib, sertraline, rimcazole, and pterostilbene (6–9 synergistic interactions; Fig. 3B). Of these, rimcazole and sertraline were recently shown to exhibit single drug activity against GBM-derived stem cells at 10 μM,19 and a function of sertraline as an inhibitor of translation initiation in cancer cells has been demonstrated.29 Taken together, the results demonstrate numerous pairwise interactions between noncancer drugs in the 5 tested GBM cell lines.
As a point of comparison, we tested 4 combinations on a normal fibroblast panel, consisting of 3 human fibroblast cell lines. In contrast to the GBM cells, the fibroblasts showed variable responses to the top 4 combinations (Supplementary Table S3).
Drug Pair Interactions Were Consistently Reproduced in a Set of Glioblastoma-Initiating Cell Cultures
To address the generality of our findings, we chose to further characterize drug pairs (Fig. 3C) from the 11 hits in the GIC cultures U3005MG, U3013MG, U3024MG, U3034MG, and U3068MG. The GICs were originally derived from serum-free tumor samples and maintained as adherent cultures using the protocol developed by Pollard et al.19 Previous analyses have shown that GICs more closely reflect the genotype and phenotype of the primary tumor cells,30 which makes them highly relevant and valid in vitro models of GBM. In our experiments, we could reproduce the synergistic interaction scores seen in the original 5 cell lines; for example, gefitinib + rimcazole and rimcazole + sertraline were found to interact with synergistic interaction scores also in the GICs (Fig. 3C and D). Of note, the more “variable” combination PP2 + gefitinib (strong synergy in T98G only) was found to have synergistic interaction scores in only 2 of the GICs (U3024MG and U3034MG), which supports that this particular drug pair would produce a synergistic interaction only in a particular subset of GBM. The drug pair doxorubicin + wortmannin, which produced a synergistic interaction score in the cell lines, was not replicated in any of the GICs (Fig. 3C and D), indicating that this pair is an unlikely candidate for synergistic combination targeting of GBM. Thus, the synergistic interaction scores seen in the original 5 cell lines broadly generalize to GICs.
Validation of Interaction Scores Using Isobole Analysis
While the initial, high-throughput tests were performed using a well-established metric of effect-based interaction,13 we went on to characterize a selected set of combinations using dose-based interaction criteria (combination index14 and α-values25). For this, we selected 4 of the top robust combinations with negative interaction scores from previous experiments (Fig. 3C and D) for isobole-based experimental follow-up (rimcazole + sertraline, rimcazole + gefitinib, pterostilbene + sertraline, and pterostilbene + gefitinib). For each of these pairs, we measured the response to each of 7 × 7 dose combinations of the 2 drugs. Based on the data, we obtained isoboles (Methods) from which we estimated the combination index and α-values (Table 2). We generally observed several negative α-values and low combination indices (suggesting a synergy) for pterostilbene + sertraline and pterostilbene + gefitinib in most GICs (Table 2). However, some additive or even antagonistic trends were seen in a few GICs for rimcazole + sertraline and rimcazole + gefitinib. These results suggest that this type of analysis can capture more subtle interaction effects not seen in the initial screen.
GBM Drug-Drug Interactions Predicted From mRNA Biomarkers
In a final analysis, we asked whether the drug-drug interactions of each GBM cell line could be predicted from its transcriptional profile (Fig. 4 and Supplementary Table S2). For this, we used the sCCA. This is a multivariate method that can be used to simultaneously select and associate sets of variables in 2 data sets,26 here applied to select mRNA features that correlated with drug-drug interactions across the GBM cell lines. Applied to our data, we obtained a good model fit for 6 selected interactions (R2 = 0.93) using a subset of 82 mRNAs selected by the sCCA algorithm (Fig. 4A, all mRNAs in Supplementary Table S2) and a reasonable fit (R2 = 0.50) for the full set of 11 interactions (Fig. 4B). The 82 selected mRNAs comprised the differentiation markers of glial fibrillary acidic protein and sex determining region Y-box (Sox)21 (Supplementary Table S2). Further, we identified cell-cell interaction and cell adhesion markers such as myelin protein zero-like 3, integrin beta 4 (ITGB4), protocadherin beta 5, cadherin (CDH)1, CDH6, and markers involved in the Wnt signaling pathway, including prickle homolog 1 and secreted frizzled-related protein (SFRP)1. We could also identify glycoprotein markers such as peripheral myelin protein 22, fibulin-1, microfibrillar associated protein 5, fibronectin 1, and markers previously reported to be associated with oncogenesis, including ITGB4,31 LIM domain only protein 3,32 Sox2, Sox21,33 and SFRP1.34 Of note, our method selected carboxyl esterase 1, for which mutations are associated with altered metabolism of this enzyme's substrates, such as certain drugs.35 The data taken together, sCCA is a promising tool to define mRNA biomarkers for GBM drug-drug interactions in extended studies; however, a larger data set would most likely increase the power of this method, something that is reserved for future work.
Discussion
We performed a first integrated investigation of drug-drug interactions across GBM cell lines and identified a core set of reproducible drug pair interactions. The most striking finding is that a subset of drug-drug interactions—such as rimcazole + sertraline and pterostilbene + gefitinib—produce synergistic interactions over many cell lines that are also reproduced in the highly more clinically relevant GICs. This class of robust interactions may be of interest to the development of GBM therapies with consistent response across broad subtypes of the disease, whereas the more “variable” interactions, such as PP2 + gefitinib (Fig. 3), showing synergistic interactions in only a few cell lines, is an example of a combination that could act more efficiently on subtypes of GBM. All single drugs included in the validated combinations have previously been shown to exhibit an anticancerous effect, including the noncancer drugs pterostilbene,36 rimcazole,17,19 and sertraline.19 These results indicate that drug repurposing could pose an interesting alternative to GBM therapy. Previous studies have identified the phosphatidylinositol-3 kinase inhibitors wortmannin and LY294002 as enhancers of doxorubicin-induced apoptosis.37,38 Apart from this, no previous studies have to our knowledge shown the combination effects we present in this study.
Strikingly, some of the strongest synergies were between noncancer drugs (eg, the SSRI sertraline, the sigma receptor antagonist rimcazole, the antioxidative phytoalexin pterostilbene), showing the possibility of drug repurposing for treating GBM.
Similar to previous work in bacteria,13 yeast,25 and cancer cell lines,8 our interaction data contain correlations between functionally related targets (Supplementary Fig. S2). These observations, together with the retesting using the isobologram method, serve to validate our approach of a single dose first-pass screen.
The analysis of isobolograms resulting in α-values and combination indices revealed dose dependencies not captured by the interaction scores from the screen. For example, rimcazole + sertraline and rimcazole + gefitinib seemed to exhibit less synergy using α-values and combination indices, while pterostilbene + sertraline and pterostilbene + gefitinib were confirmed to be synergistic using the same measurements. Nevertheless, both rimcazole and sertraline represent safe and tested psychiatric drugs, which will penetrate the blood–brain barrier, and the combination will therefore be interesting as a therapeutic candidate for further study. Direct use of isoboles and calculations from these in a primary screen are valuable interaction measurements. However, this needs to be weighed against the experimental load accompanying this approach in multi–cell line experiments. Although it is beyond the scope of this study, to gain full insight into a particular drug-drug interaction, one must analyze multidose interactions by isobologram analysis, curve-shift analysis, combination index, or universal surface response analysis, all of which are derivatives from the Loewe additivity model.15,22
The presented sCCA model shows promise as a new approach to predict drug pair interactions based on the transcriptional profile of a cell line. In the extension, this type of approach may have applications in GBM research (defining functional biomarker genes associated with changes in drug-drug interactions) and GBM therapy (personalizing therapy strategies based on biomarker profiles). One limitation of the current sCCA analysis was that the number of transcripts analyzed (the top 200 most variable transcripts) and the number of unique drug pairs (11) were relatively high in comparison with the number of cell lines (5). To deal with this “underdetermined” statistical setting, the version of sCCA that we applied used stringent variable selection, which rendered the method robust against small samples (Methods). In addition, we restricted the selection of mRNAs to the first sCCA component only, to avoid overinterpretation. In the future, a larger set of cell lines, derived from different molecular subtypes of GBM,39,40 could reveal mRNA biomarkers predicting the interactions of individual pairs with even better accuracy.
Taken together, the results provide 3 main implications for the future development of anti-GBM combination therapies. First, approved drugs not developed for cancer therapy produce synergistic anti-GBM responses. Second, consistent synergistic drug pairs may elicit responses across a broader spectrum of patients. Third, transcriptional biomarkers of combination effects can be applicable to stratification of GBM patients. Further studies will help reveal additional robust targets for GBM and uncover principles of robustness and variability.
Supplementary Material
Funding
This work was supported by the Swedish Cancer Society; the Swedish Research Council, and Science for Life Laboratory, the Assar Gabrielssons Fund; and the Swedish Childhood Cancer Foundation NBCNS.
Conflict of interest statement. None declared.
Supplementary Material
Acknowledgments
Special thanks to Marianne Kastemar (Rudbeck laboratory, Uppsala) and Therese Carlsson (Sahlgrenska Cancer Center, Gothenburg) for technical assistance on GIC cultures. Thanks to Wilhelm and Martina Lundgrens Vetenskapsfond for supporting this work. A part of this work was presented as a poster at the Hallmarks of Cancer Cell Symposia, San Francisco, in 2012.
References
- 1.Louis DN, Holland EC, Cairncross JG. Glioma classification: a molecular reappraisal. Am J Pathol. 2001;159(3):779–786. doi: 10.1016/S0002-9440(10)61750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson C, Kanu OO, Mehta AI, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18(8):1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Marosi C. Chemotherapy for malignant gliomas. Wien Med Wochenschr. 2006;156(11–12):346–350. doi: 10.1007/s10354-006-0307-4. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens A-C, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Stevens MF, Bradshaw TD. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2012;5(1):102–114. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann GR, Lehar J, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12(1–2):34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Lehar J, Krueger AS, Avery W, et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27(7):659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVita VT, Jr., Hellman S, Rosenberg SA. Principles of Cancer Management: Chemotherapy. Philadelphia: Lippincott;: 1997. pp. 333–347. [Google Scholar]
- 10.University of California San Francisco, Study of Bevacizumab Plus Temodar and Tarceva in Patients With Glioblastoma or Gliosarcoma (AVF4120s) ClinicalTrials.gov [Internet], National Library of Medicin (US) 2012–05–04. Michael D. Prados. (MD)( NCT00525525)
- 11.Oncology, M.U.M.C.M.R., Study With Nelfinavir and Combined Radiochemotherapy for Glioblastoma. ClinicalTrials.gov [Internet], National Library of Medicin (US), 2012-05-04. Brigitta Baumert (MD)( NCT00694837)
- 12.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4(1):71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 13.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38(4):489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 14.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 15.Zhao L, Au JL, Wientjes MG. Comparison of methods for evaluating drug-drug interaction. Front Biosci (Elite Ed) 2010;2:241–249. doi: 10.2741/e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47(2):331–385. [PubMed] [Google Scholar]
- 17.Spruce BA, Campbell LA, McTavish N, et al. Small molecule antagonists of the sigma-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 2004;64(14):4875–4886. doi: 10.1158/0008-5472.CAN-03-3180. [DOI] [PubMed] [Google Scholar]
- 18.Gil-Ad I, Zolokov A, Lomnitski L, et al. Evaluation of the potential anti-cancer activity of the antidepressant sertraline in human colon cancer cell lines and in colorectal cancer-xenografted mice. Int J Oncol. 2008;33(2):277–286. [PubMed] [Google Scholar]
- 19.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Ishii N, Maier D, Merlo A, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9(3):469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 22.Loewe SMH. Effects of combinations: mathematical basis of problem. Arch Exp Pathol Pharmakol. 1926;114:313–326. [Google Scholar]
- 23.Berenbaum MC. What is synergy? Pharmacol Rev. 1989;41(2):93–141. [PubMed] [Google Scholar]
- 24.Berenbaum MC. The expected effect of a combination of agents: the general solution. J Theor Biol. 1985;114(3):413–431. doi: 10.1016/s0022-5193(85)80176-4. [DOI] [PubMed] [Google Scholar]
- 25.Cokol M, et al. Systematic exploration of synergistic drug pairs. Mol Syst Biol. 2011;7:544. doi: 10.1038/msb.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witten DM, Tibshirani RJ. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat Appl Genet Mol Biol. 2009;8(1) doi: 10.2202/1544-6115.1470. 1–27. Article28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segre D, Deluna A, Church GM, et al. Modular epistasis in yeast metabolism. Nat Genet. 2005;37(1):77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 28.Lehár J, Zimmermann GR, Krueger AS, et al. Chemical combination effects predict connectivity in biological systems. Mol Syst Biol. 2007;3:80. doi: 10.1038/msb4100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin CJ, Robert F, Sukarieh R, et al. The antidepressant sertraline inhibits translation initiation by curtailing mammalian target of rapamycin signaling. Cancer Res. 2010;70(8):3199–3208. doi: 10.1158/0008-5472.CAN-09-4072. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 31.Lipscomb EA, Mercurio AM. Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev. 2005;24(3):413–423. doi: 10.1007/s10555-005-5133-4. [DOI] [PubMed] [Google Scholar]
- 32.Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469(7329):216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferletta M, Caglayan D, Mokvist L, et al. Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer. 2011;129(1):45–60. doi: 10.1002/ijc.25647. [DOI] [PubMed] [Google Scholar]
- 34.Huang J, Zhang Y-L, Teng X-M, et al. Down-regulation of SFRP1 as a putative tumor suppressor gene can contribute to human hepatocellular carcinoma. BMC Cancer. 2007;7:126. doi: 10.1186/1471-2407-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu HJ, Patrick KS, Yuan HJ, et al. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am J Hum Genet. 2008;82(6):1241–1248. doi: 10.1016/j.ajhg.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Ding L, Wang X, et al. Pterostilbene simultaneously induces apoptosis, cell cycle arrest and cyto-protective autophagy in breast cancer cells. Am J Transl Res. 2012;4(1):44–51. [PMC free article] [PubMed] [Google Scholar]
- 37.Opel D, Westhoff MA, Bender A, et al. Phosphatidylinositol 3-kinase inhibition broadly sensitizes glioblastoma cells to death receptor- and drug-induced apoptosis. Cancer Res. 2008;68(15):6271–6280. doi: 10.1158/0008-5472.CAN-07-6769. [DOI] [PubMed] [Google Scholar]
- 38.Wang YA, Johnson SK, Brown BL, et al. Enhanced anti-cancer effect of a phosphatidylinositol-3 kinase inhibitor and doxorubicin on human breast epithelial cell lines with different p53 and oestrogen receptor status. Int J Cancer. 2008;123(7):1536–1544. doi: 10.1002/ijc.23671. [DOI] [PubMed] [Google Scholar]
- 39.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.