Abstract
The terminal monosaccharide of glycoconjugates on a eukaryotic cell surface is typically a sialic acid (Neu5Ac). Increased sialylation usually indicates progression and poor prognosis of most carcinomas. Here, we utilize two human mammary epithelial cell lines, HB4A (breast normal cells) and T47D (breast cancer cells) as a model system to demonstrate differential surface glycans when treated with sialic acid under nutrient deprivation. Under a starved condition, sialic acid treatment of both cells resulted in increased activities of α2→3/6 sialyltransferases as demonstrated by solid phase assay using lectin binding. However, a very strong MAL-I (Maackia amurensis agglutinin I) staining on the membrane of sialic acid treated T47D cells was observed, indicating an increase of Neu5Acα2→3Gal on the cell surface. To our knowledge, this is a first report showing the utility of lectins, particularly MAL-I, as a means to discriminate between normal and cancer cells after sialic acid treatment under nutrient deprivation. This method is sensitive and allows selectively detection of glycan sialylation on a cancer cell surface.
Keywords: sialic acid, metabolism, glycosylation, lectin, diagnosis, cancer cell
Introduction
Protein glycosylation is an important post-translational modification, which aids in protein folding and cellular secretion (1). Glycoproteins, decorate the cell surface, and often sprout sialic acid (Neu5Ac) at the termini of their glycans in a process called sialylation. Glycosylation disorders, including increased branching of N-linked glycans and sialylation are major hallmarks of cancer progression (2). Particularly, over-sialylation on the cell surface induces tumor proliferation by promoting cell detachment from primary tumors through charge repulsion. Despite the potential of variable sialylation as a diagnostic biomarker, there is presently not an early sensing platform to detect glycan sialylation constellations in diverse biological samples with high affinity and sensitivity (3). However, metabolic engineering of glycan sialylation may have the potential to advance the field of diagnosis and prognosis of cancers.
In order to assess this approach, we note that the selectively of glycan sialylation is regulated by two factors: (i) metabolic flux through the sialic acid biosynthetic pathway and (ii) differential expression level of sialyltransferases (4). However, neither of these factors has been successfully manipulated with sufficient precision to achieve specific endpoints (5). Over the past 20 years, glycoengineering approaches have provided new routes for the modulation of glycan structures on cell surfaces (6). Sugars such as N-acetyl glucosamine, N-acetyl galactosamine and sialic acid can be metabolically incorporated into glycans through salvage pathways and various constituents of the monitored glycans are studied by “lectins” (a class of carbohydrate-binding proteins) (7–9).
In humans, synthesis of Neu5Ac from its precursor, UDP-GlcNAc takes place primarily in the cytosol through a series of enzymatic reactions (10). Once Neu5Ac is synthesized, it is incorporated as a nucleotide donor substrate for subsequent transfer to an oligosaccharide structure. This substrate creation takes place in the nucleus, where Neu5Ac is attached to cytidine 5′-triphosphate (CTP) to produce the donor substrate CMP-Neu5Ac by the enzyme CMP-Neu5Ac synthetase. The CMP-Neu5Ac substrate is then transported to the cytoplasm and then to the Golgi for use by the sialyltransferases (ST), which attach Neu5Ac residues to oligosaccharides through various linkages such α2→3, α2→6, or α2→8 (11).
Although Neu5Ac and CMP-sialic acid are the most abundant immediate precursors for glycan sialylation, both offer poor cell membrane permeability. Thus, feeding of either of these sialylation precursors does not increase the intracellular pool of CMP-sialic acid (12).
Recent data indicate that the cell surface glyco-engineering is often compromised by the cell membrane, and thus, can be significantly enhanced by treating the cell membrane (13, 14). We therefore strongly suggest that stimulation of the cell membrane by the intermediate precursors for glycan sialylation through specific metabolic conditioning may allow engineering of sialoglycan metabolism and produce a distinct set of changes in glycan sialylation. In this paper, we examine the over-sialylation of cell surface glycans in a sialic acid fed-batch culture system without adequate nutrients and employing sialic acid binding lectins to probe global differences in cell-surface sialylation. We chose breast normal cells (HB4A) and breast cancer cells (T47D) because they proven to be ideal model systems for studying protein glycosylation in health and disease state (15, 16). Our method is sensitive and allows selectively detection of glycan sialylation on a cancer cell surface affording new potential applications for the study of cancer diagnosis and prognosis.
Materials and methods
Materials
Sialic acid (N-acetyl-5-neuraminic acid, Neu5Ac), asialofetuin, CMP-Neu5Ac, ribonuclease A, insulin, hydrocortisone, RPMI1640 medium and phosphate-buffered saline (PBS, 10 mM phosphate, 140 mM NaCl, pH 7.4) were purchased from Sigma-Aldrich, USA. Maackia amurensis agglutinin I (MAL-I) specific for Neu5Acα2,3Gal, Sambucus nigra agglutinin (SNA) specific for Neu5Acα2,6Gal, Triticum vulgaris agglutinin (WGA) specific for Neu5Ac and GlcNAc, Streptavidin-horseradish peroxidase, and biotinylated MAL-I and SNA lectins were obtained from Vector Laboratories (USA). Fetal bovine serum (FBS) was from Biosera (USA). TO-PRO-3 was obtained from Molecular Probes (USA).
Cell culture
Human normal mammary epithelial cell line HB4A and breast cancer cell line T47D (American Type Culture Collection, USA) were cultured in RPMI1640 medium supplemented with 1% FBS at 37 °C under 5% CO2. For HB4A cells, the medium also contained 10 μg/ml insulin and 5 μg/ml hydrocortisone. For all experiments, HB4A and T47D cells were used within the first three passages and cells were harvested by treatment with 5 ml of buffer containing 0.54 mM EDTA, 154 mM NaCl and 10 mM HEPES, pH 7.4 for ≤ 5 min at 37°C.
Sialic acid treatment
Cells were grown for 48 hours and harvested in the indicated conditions. Cells were then resuspended in serum-free RPMI1640 medium. After washing 3 times with warm (37 °C) PBS, 20-mL of cell suspension were treated with 10 mM sialic acid in PBS for 2 hours in shake flasks in a 37 °C incubator under 5% CO2. Control cells included only PBS. The cells were plated at a density of 3 × 105 cells mL−1. After washing, the cells were fixed in 75% ice-cold ethanol for 15 minutes and maintained at 4°C for experiments. For negative controls, cells were also treated with sialic acid in serum containing medium and harvested as described above.
Cell viability
Cells were harvested as described above without fixation. Cell pellets were resuspended in PBS supplemented with 1 mg/ml propidium iodide (PI) and incubated for 5 minutes at ambient temperature. Cells were analyzed by flow cytometry on a CyAn™ ADP flow cytometer (Beckman Coulter Inc., USA). The dead cell population was determined as the percentage of PI stained cells. MTT assay was used to determine viability of cells for 5 days after returning the SA treated and untreated cells to the complete medium. The formazan dye produced after DMSO solubilization was evaluated at 560 nm by a multiwell scanning spectrophotometer (Bio-Rad, USA).
Quantification of membrane-bound sialic acid
PBS-washed cells (1×106) were lysed by hypotonic shock in 1 ml water (15 min, 4°C). The crude membrane fraction was pelleted by centrifugation at 10000 × g for 15 min. The pellet was washed twice with water and lyophilized. The sialic acid content of the membrane fraction was determined by acid hydrolysis followed by DMB (1,2-diamino-4,5-methylenedioxybenzene) derivatization and high performance liquid chromatography (HPLC) separation as previously described (17). Briefly, sialic acids were released from lyophilized fractions by acid hydrolysis with 2 M acetic acid for 3 h at 80 °C. Samples were passed through a Microcon-10 filter and the filtrate was derivatized with DMB for analysis of sialic acids by HPLC (Perkin–Elmer, US). Separation of DMB-labeled sialic acids was performed on a Shim-pack CLC-ODS (6.0 × 150 mm) column using a mobile phase of acetonitrile-methanol-water (9:7:84, v/v) with a flow rate of 1ml/min at 22 °C and detected using an emission wavelength of 448 nm and an excitation wavelength of 373 nm.
Detection of sialyltransferase activity
The activity of ST3Gal-I and ST6Gal-I was determined by solid phase assay using asialofetuin precoated plates as previously described (18). Briefly, various cell lysates containing equal amount of protein were taken into the wells and CMP-Neu5Ac was added to initiate the reaction. After washing and blocking, the sialylated fetuin was allowed to interact with either biotinylated lectin (MAL-I or SNA) followed by binding with streptavidin-horseradish peroxidase. The negative control included only lectin binding to asialofetuin. After binding and washing, the reaction was developed with 100 μL solution of substrate (0.03% H2O2, 2 mg/mL o-phenylenediamine in 0.1 mM citrate buffer, pH 5.5) for ~10 min and terminated with 1 M H2SO4. The absorbance at 492 nm was measured using an automatic multi-well spectrophotometer (Bio-Rad, USA).
Gene expression analyses by RT-PCR and quantitative RT-PCR
Total RNA was extracted from cells using Trizol reagent and 1 μg of total RNA was reverse-transcribed to cDNA using GoScript™ Reverse Transcription System (Promega, USA). The resulting cDNA was amplified in triplicate using GoTaq qPCR Master Mix (Promega, USA) on a Real-Time PCR System (Applied Biosystems, USA). β-Actin was used as the internal control. The relative expression levels were analyzed in Microsoft Excel using the comparative 2−ΔΔCT method as per the instructions of the manufacturer (Applied Biosystems). The primer sequences for ST3Gal-I and ST6Gal-I were previously reported (19).
Flow cytometric analysis
Cells were fixed with 75% ice-cold ethanol for 15 minutes, washed twice with cold PBS and placed in 96-well plates (1×104 cells per well). The cells were then stained with the fluorescein isothiocyanate-labeled lectins (MAL-I, SNA, and WGA). For the comparison of mean fluorescence intensities, the instrument settings for fluorescence and compensation were the same for all experiments. Data were collected from at least 10000 cells for each sample.
Lectin staining and confocal microscopy
Cells were grown in 6 well plates and the adherent cells were then fixed with 75% ice-cold ethanol for 15 minutes. After washing, the fixed cells were stained with various lectin-conjugates (10 μg/ml) in 0.05 M Tris-buffered saline (TBS), 1 mM CaCl2, 1 mM MgCl2, pH 7.6 for 1 hour. After staining, cells were further treated with ribonuclease A (50 μg/ml) and the nuclei were counter-stained with TO-PRO-3. Images were acquired by sequential scanning using a Leica TCS SP2 confocal system (Leica Microsystems, Milton Keynes, UK) and a X63 ceramic dipping objective at 1024×1024 format and scanning speed of 400 Hz with a line average of 2. A 488 nm (Intensity 25%) laser was used for the excitation of FITC and a 633 nm laser (Intensity 35%) was used for TO-PRO-3. Emission was recorded over the bandwidth of 500–550 nm (FITC) and 650–720 nm (TO-PRO-3).
Statistical analysis
Data are expressed as the mean S.E. for at least three independent experiments. Statistical significance of differences between means was determined by analysis of variance. The differences were considered significant when p<0.05.
Results
Effects of sialic acid treatment on the cell viability
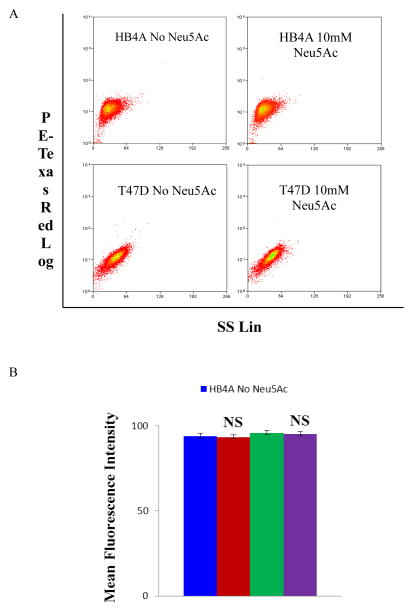
To investigate if sialic acid treatment affects cell viability under nutrient deprived condition, cells were stained with PI and analyzed on a flow cytometry (Fig. 1A). Using a fixed number of cells (3 × 105 cells mL−1) the percentage of viable cells were 93.45% and 95.27% for untreated HB4A and T47D cells (PBS control), respectively (Fig. 1B). For sialic acid treated cells, the cell viability was 94.16% and 96.05% for HB4A and T47D, respectively (see Fig. 1B). Moreover, the cell proliferation ability of each treated and untreated cell line (when the cells were put back into the complete medium after 2 h nutrient-deprived condition) was also unchanged as demonstrated by the MTT assay (Suppl. Fig. 1), suggesting that the exposure to sialic acid at the indicated conditions did not affect cell viability.
Fig. 1.
Cell viability as measured by propidium iodide exclusion. Cells were stained with 1 mg/ml propidium iodide (PI) on ice for 20 min and analyzed by flow cytometry in PE-Texas Red setting. The dead cells population was determined as the percentage of PI stained cells. The cytograms show the live and dead cells for the HB4A and T47D cells in the presence or absence of Neu5Ac (A) and the live cells are shown in a bar diagram (B). The data are representative of three independent experiments with S.D. indicated by error bars. P values are determined by two-tailed student t-test and <0.05, <0.01 and <0.001 are indicated by one, two and three asterisks, respectively; NS, not significant.
Effects of sialic acid treatment on cell membrane composition
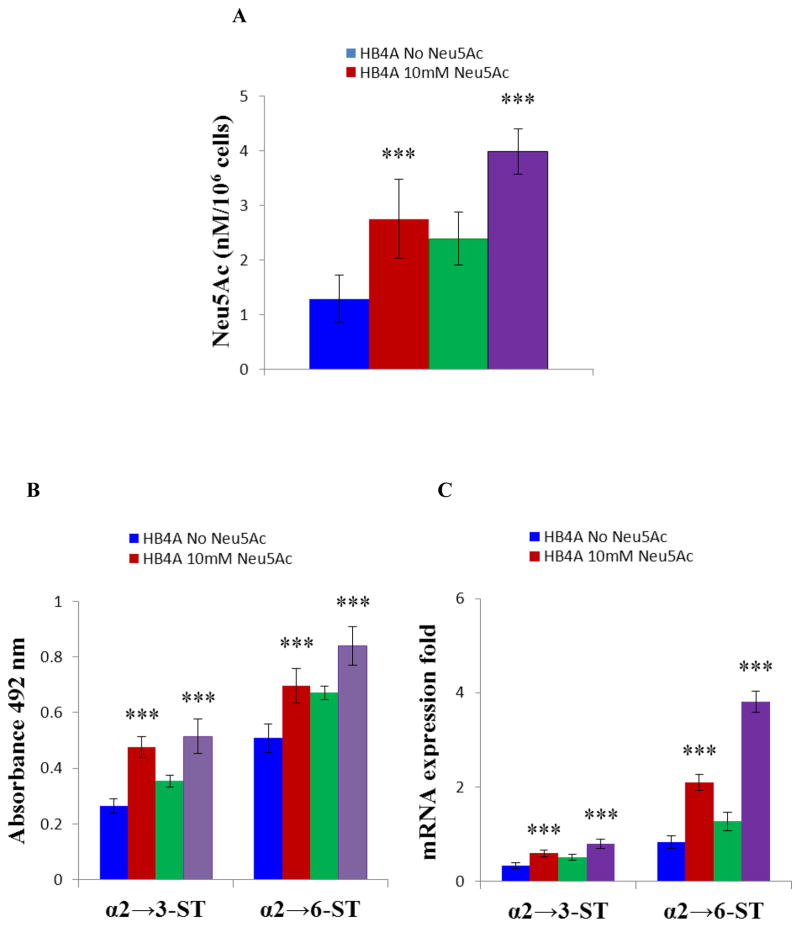
The sialic acid content of the membrane fraction was determined by acid hydrolysis on HPLC. The untreated T47D cancer cells (2.39 nM/106 cells) contain more membrane-bound sialic acid as compared to HB4A normal cells (1.29 nM/106 cells) (Fig. 2A, Suppl. Fig. 2A). The membrane-bound sialic acid is further increased in both cell types when treated with sialic acid under nutrient deficient condition (2.75 nM/106 HB4A sialic acid treated cells, p<0.001 and 3.98 nM/106 T47D sialic acid treated cells, p<0.001) (Fig. 2A, Suppl. Fig. 2A). The results clearly demonstrated the increase of cell membrane bound sialic acid upon sialic acid treatment at nutrient deprived condition.
Fig. 2.
T47D cells contains more membrane bound sialic acids as compared to HB4A, would be good model to compare as there are striking differences between the lectin binding characteristics of the different cell types. Cells were treated in the presence or absence of 10 mM Neu5Ac for 2 hours under nutrient deprivation, lysed by hypotonic shock in 1 ml water (15 min, 4 °C) and surface Neu5Ac content was determined by HPLC analysis. (A) Bar diagram showing quantitation of surface Neu5Ac. (B) Detection of ST3Gal-I and ST6Gal-I activities. Cells were lysed and the same amounts of proteins were used on solid phase assays. The α-2,3 and α-2,6 sialylated glycans of resultant fetuin was detected with biotinylated MAL-I and biotinylated SNA, respectively. (C) Quantitative RT-PCR of ST3Gal-I and ST6Gal-I. Cells were treated in the presence or absence of 10 mM Neu5Ac for 2 hours under nutrient deprivation. The relative mRNA expression of ST3Gal-I and ST6Gal-I was assessed by quantitative RT-PCR and the results were normalized to β-actin expression. The data are representative of four independent experiments with S.D. indicated by error bars. P values are determined by two-tailed student t-test and <0.05, <0.01 and <0.001 are indicated by one, two and three asterisks, respectively.
Influence of sialic acid treatment on the sialyltransferase activity
To investigate if sialic acid treatment under nutrient deprived condition influences sialyltransferase activity, cell extract was incubated with asialofetuin (acts as acceptor) and the resultant sialo-fetuin was quantitated with linkage specific lectins as described in the Experimental Procedures. Results showed that both α2→3 sialyltransferase (ST3Gal-I) and α2→6 sialyltransferase (ST6Gal-I) activities were present in HB4A and T47D cells as demonstrated by positive binding with MAL-I and SNA binding (Fig. 2B). As expected, negative control (lectin binding to asialofetuin only) resulted no binding or in negligible binding (not shown). Interestingly, sialic acid treatment of HB4A and T47D cells increased ST3Gal-I activity as measured by MAL-I binding by 60% (p<0.001) and 45% (p<0.001), respectively. Similarly, ST6Gal-1 activity of the sialic acid treated HB4A and T47D cells as measured by SNA binding was increased by 37% (p<0.001) and 25% (p<0.001), respectively. To determine whether the increase in the sialic acid is related to gene expression profiles, we examined the mRNA expression levels of sialyltransferases by quantitative RT-PCR. The mRNA expression level of ST3Gal-I and ST6Gal-I in sialic acid-treated cells was also increased at least 1.5 to 4-fold when compared to the corresponding untreated cells (Fig. 2C and Suppl. Fig. 2B). Together, these results show that sialic acid treatment under nutrient deficient condition increased the expression of α2→3 sialyltransferase (ST3Gal-I) and α2→6 sialyltransferase (ST6Gal-I).
Cell surface sialoglycoconjugates changes
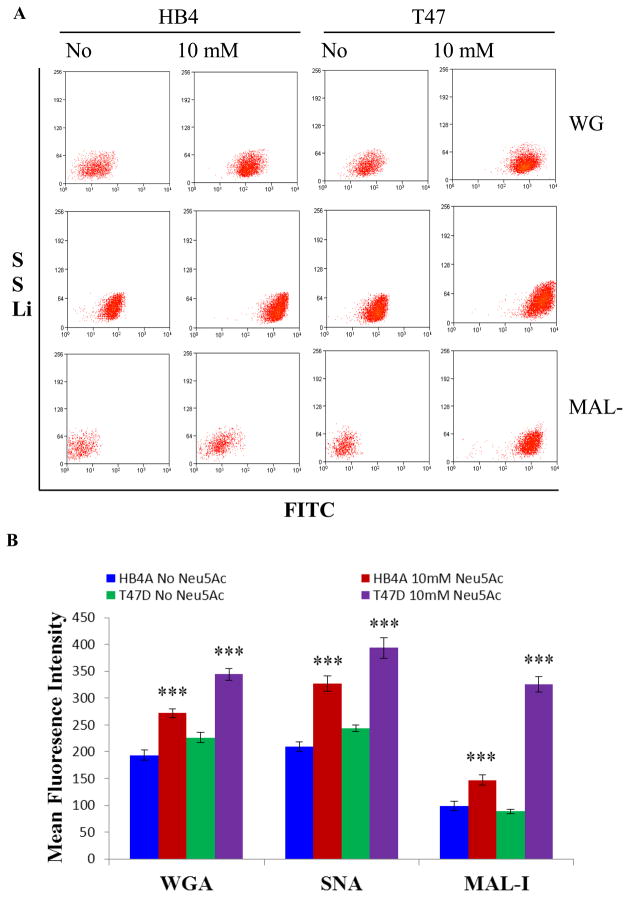
To examine if sialic acid treatment could influence the display of specific sialoglycans, we performed cell binding of SA treated and untreated cells (with and without nutrient) with FITC conjugated lectins (MAL-I, SNA, and WGA) on a flow cytometer (Fig. 3A). We found that sialic acid treatment under nutrient deprivation promotes differential expression of cell surface sialoglycoconjugates and reveals the utility of MAL-I lectin binding as a means to discriminate between normal and cancer cells. In the case of SA treatment under nutrient deprivation, both normal (HB4A) and cancer (T47D) cells display sialoglycoconjugates as measured by the above lectin-FITC binding (Fig. 3A, B). As expected, no binding was observed with FITC alone with either cell (normal or cancer, untreated or treated) (Suppl. Fig. 3). However, sialic acid treatment of each cell under nutrient deprivation increased lectin binding, suggesting increased surface expression of Neu5Acα2→3(6)Gal. Particularly, WGA-FITC binding to sialic acid treated normal and cancer cells was increased by 41% (p<0.001) and 35% (p<0.001), respectively. Similarly, SNA-FITC binding to sialic acid treated normal and cancer cells was enhanced by 56% (p<0.001) and 62% (p<0.001), respectively. The sialic acid treated normal cells also showed enhanced binding (by 43% (p<0.001) to MAL-I-FITC. However, MAL-I-FITC binding to sialic acid treated cancer cells provided interesting results, as the lectin binding to the treated cells was increased by almost 200% (p<0.001) (Mean fluorescence intensity 326 for the treated cells vs. 117 for the untreated cells) (Fig. 3B). Conversely, decreased lectin-binding (as maximum as mean fluorescence intensity 218) was observed on the surface of both normal and cancer cells when treated with sialic acid for 48 h under nutrient full medium (Suppl. Fig. 4).
Fig. 3.
Flow cytometric analysis of lectin binding of cells. Cells were treated in the presence or absence of 10 mM Neu5Ac under nutrient deprivation, and fluorescence associated with fluorescein isothiocyanate (FITC)-lectin binding to α-2,6 and α-2,3 sialylated surface moieties was ascertained by flow cytometry using bivariate sideward scattering (SS Lin) vs. FITC log settings. The data are representative of three independent experiments with S.D. indicated by error bars. P values are determined by two-tailed student t-test and <0.05, <0.01 and <0.001 are indicated by one, two and three asterisks, respectively.
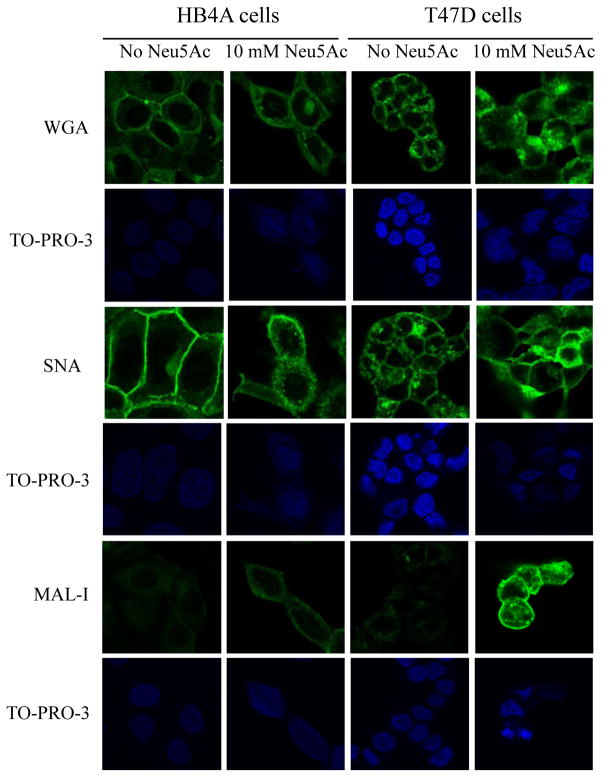
We next performed lectin staining of the cells followed by confocal assessment. In both normal and cancer cells, FITC-conjugated WGA and SNA lectins showed a strong fluorescence signal, which was further enhanced in sialic acid treated cells (Fig. 4). However, a very weak staining was observed with MAL-I lectin in the untreated HB4A and T47D cells. Consistent with flow analyses, sialic acid treated normal cells showed a slight increase in MAL-I staining, but a very strong staining was observed in sialic acid treated cancer cells (see Fig. 4). MAL-I-binding of sialic acid treated cancer cells, as revealed through either flow analysis or confocal imaging, suggests that cancer cells, upon sialic acid treatment, differentially display Neu5Acα2→3Gal, which makes the cancer cells distinguishable from the normal cells.
Fig. 4.
Lectin-mediated staining followed by confocal imaging. Cells were treated for 2 h in the presence or absence of 10 mM Neu5Ac under nutrient deprivation and stained for 1 hour with FITC-labeled lectins (WGA, SNA, and MAL-I) at concentration of 5 μg/ml (green fluorescence). Cells were further treated with 50 μg/ml ribonuclease A and the nuclei were counter-stained with TO-PRO-3 (blue fluorescence). Magnification of images 10X.
Discussion
Cancer cell has properties different than normal cell in grow, divide and death. Cancer cell must reprogram cellular metabolism to satisfy the demands of growth and persistence. The rapid cancer cell growth is also fundamentally dependent upon generating a metabolic condition that satisfies the sum of these requirements. Ultimately, efforts to unravel metabolic alterations may enable the identification of novel therapeutic and diagnostic targets (20, 28). Our approach is useful when seeking to differentiate sialylation epitopes that are quantitative or linkage specific through the glycan sialylation expression on a tumor cell surface arising from altered metabolism. Therefore, comparative studies of the expression levels of glycan sialylation in normal and cancer cells in a specific metabolic conditioning might aid in the understanding of their roles in cancer development and help to improve cancer diagnosis, prognosis, and treatment.
Here we showed the treatment of nutrient deprived cells with sialic acid enhanced sialyltransferase activities. The enhanced sensitivity of these enzymes is reflected through dysregulation of sialoglycan metabolism during breast cancer transformation and progression. Overexpression of sialyltransferases and increased sialylation in breast cancer cells, relative to non-malignant cells, has been documented (21). Increased sialylation on the cell surface promotes cell detachment from primary tumors through charge repulsion, thereby inducing tumor proliferation and migration (22). Recently, the overexpression of ST3Gal-I has been shown to promote mammary tumorigenesis in transgenic mice (23). Moreover, expression of α2→6 sialyltransferase I (ST6GalNAc-I) in MDA MB231 breast cancer cells enhances the tumorigenicity of breast cancer cells (24).
Treatment of nutrient deprived cells, particularly cancer cells with sialic acid, resulted in enhanced binding with the sialic acid specific lectins as demonstrated in two settings: flow analysis and confocal imaging. The examined lectins are distributed mostly in the cell surface and partly in the cytoplasm. Confocal imaging of the lectins, particularly MAL-I and SNA, confirmed a membrane-associated fluorescence. Therefore, the increase of SNA and MAL-I staining can be attributed to an increase of sialic acids on the outer leaflet of the cellular membranes (25). Previous studies have reported that MAL-I, SNA, and WGA recognize sialic acid on the terminal branches (26, 27). In particular, MAL-I detects glycans containing Neu5Ac-Gal-GlcNAc with sialic acid at the 3 position of galactose, while SNA binds preferentially to sialic acid attached to terminal galactose in α2→6 and, to a lesser degree, in α2→3 linkage. WGA binds to almost all the isomers of sialylated glycans where its signal translates into a general level of sialylation. This is consistent with the observation that both untreated cells normally show abundant α2→6 linked sialic acid, whereas terminal α2→3 linked sialic acid residues are highly restricted. More importantly, comparison of the treated cells profiles revealed that T47D cells enforced a much broader range of α2→3 and α2→6 sialylated glycans compared with those in HB4A cells which expressed a high level of α2→6 sialylated glycans, and retained restrictions of α2→3 sialylated glycans. Thus, the results of the current study suggest that Neu5Ac treatment promotes the display of α2→3 sialylated glycans structures on the surface of cancer cells and not on those of normal cells. To our knowledge, this is the first report showing that lectins, in particular MAL-I, can be used in a metabolic-specific manner to discriminate between normal and cancer glycans positioning this approach for rapid translation to clinical settings. The result is significant as this approach can be applied to identify and quantitate circulating tumor cells (CTC) for prognosis or therapeutic management of cancer. However, a larger number of cancer cell lines must be investigated to determine the breadth of this conclusion. Overall, our findings will enable the development of additional sialic acid engineering strategies essential for cancer diagnostic product development and optimization.
Supplementary Material
Acknowledgments
This study was supported by the grant 0068GF for Scientific Research on Priority Areas Cancer from the Ministry of Science and Education (Science and Technology Program Kazakhstan) to L.B.D. and grants R15ES021079-01 and R41CA141970-01A2 from the National Institute of Health (U.S.A.) to C.-Z. Li. and H.A., respectively.
References
- 1.Varki A. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 2.Ohtsubo K, Marth JD. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Service RF. Science. 2012;338:321–323. doi: 10.1126/science.338.6105.321. [DOI] [PubMed] [Google Scholar]
- 4.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essential of Glycobiology. 2. Cold Spring Harbor Laboratory Press; New York: 2009. [PubMed] [Google Scholar]
- 5.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VDP, Yarema KJ. Glycobiology. 2009;12:1382–1401. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agard NJ, Bertozzi CR. Acc Chem Res. 2009;42:788–797. doi: 10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariño K, Bones J, Kattla JJ, Rudd PM. Nat Chem Biol. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 8.Prescher JA, Bertozzi CR. Cell. 2006;126:851–854. doi: 10.1016/j.cell.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Sharon N, Lis H. Science. 1989;246:227–234. doi: 10.1126/science.2552581. (1989) [DOI] [PubMed] [Google Scholar]
- 10.Paulson JC, Rademacher C. Nat Struct Mol Biol. 2009;16:1121–1122. doi: 10.1038/nsmb1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dube DH, Bertozzi CR. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 12.Ghaderi D, Zhang M, Hurtado-Ziola N, Varki A. Biotechnol Genet Eng Rev. 2012;28:147–75. doi: 10.5661/bger-28-147. [DOI] [PubMed] [Google Scholar]
- 13.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 14.DeBerardinis RJ, Thompson CB. Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrasekaran EV, Xue J, Xia J, Locke RD, Patil SA, Neelamegham S, Matta KL. J Proteome Res. 2012;11:2609–2618. doi: 10.1021/pr201108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramessur KT, Greenwell P, Nash R, Dwek MV. Br J Biomed Sci. 2010;67:189–96. doi: 10.1080/09674845.2010.11730318. [DOI] [PubMed] [Google Scholar]
- 17.Ghaderi D, Taylor RE, Karavani VP, Diaz S, Varki A. Nat Biotechnol. 2010;28:863–867. doi: 10.1038/nbt.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiani J, Zhu C, Tang S, Shen A, Ai J, Li J, Geng M, Ding J. Acta Pharm Sinic. 2009;30:1039–1045. doi: 10.1038/aps.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recchi MA, Lepers AH, Marer YB, Verbert A, Delannoy P. Glycoconjugate J. 1998;15:19–27. doi: 10.1023/a:1006983214918. [DOI] [PubMed] [Google Scholar]
- 20.Vander Heiden MG. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 21.Meany DL, Chan DW. Clin Proteomics. 2011;8:1–14. doi: 10.1186/1559-0275-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulson JC, Blixt O, Collins BE. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 23.Picco G, Julien S, Brockhausen I, Beatson R, Antonopoulos A, Haslam S, Mandel U, Dell A, Pinder S, Papadimitriou J, Burchell J. Glycobiology. 2010;20:1241–1250. doi: 10.1093/glycob/cwq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julien S, Adriaenssens E, Ottenberg K, Furlan A, Courtand G, Vercoutter-Edouart AS, Hanisch FG, Delannoy P, Le Bourhis X. Glycobiology. 2006;16:54–64. doi: 10.1093/glycob/cwj033. [DOI] [PubMed] [Google Scholar]
- 25.Varki NM, Varki A. Lab Invest. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geisler C, Jarvis DL. Glycobiology. 2011;21:988–993. doi: 10.1093/glycob/cwr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwefel D, Maierhofer C, Beck JG, Seeberger S, Diederichs K, Moller HM, Welte W, Wittmann V. J Am Chem Soc. 2010;25:8704–8719. doi: 10.1021/ja101646k. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Lei T, Kosuke I, Matsue T, Tao N, Li CZ. Chem Comm. 2012;48:10389–10391. doi: 10.1039/c2cc34853e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.