Abstract
PDCD2 is an evolutionarily conserved eukaryotic protein with unknown function. The Drosophila PDCD2 ortholog Zfrp8 has an essential function in fly hematopoiesis. Zfrp8 mutants exhibit marked lymph gland hyperplasia that results from increased proliferation of partially differentiated hemocytes, suggesting Zfrp8 may participate in cell growth. Based on the above observations we have focused on the role of PDCD2 in human cancer cell proliferation and hypothesized that aberrant PDCD2 expression may be characteristic of human malignancies. We report that PDCD2 is highly expressed in human acute leukemia cells as well as in normal hematopoietic progenitors. PDCD2 knockdown in cancer cells impairs their proliferation, but not viability relative to parental cells, supporting the notion that PDCD2 overexpression facilitates cancer cell growth. Prospective analysis of PDCD2 in acute leukemia patients indicates PDCD2 RNA expression correlates with disease status and is a significant predictor of clinical relapse. PDCD2’s role in cell proliferation and its high expression in human malignancies make it an attractive, novel potential molecular target for new anti-cancer therapies.
Keywords: PDCD2, acute myelogenous leukemia, acute lymphoblastic leukemia, cellular proliferation, hematopoietic stem cells
Introduction
Programmed Cell Death 2 (PDCD2) is an evolutionarily conserved eukaryotic protein with unknown function. The original PDCD2 clone (RP-8) was isolated as a death-associated cDNA from a subtractive library enriched for sequences expressed in dexamethasone-treated primary rat thymocytes.1 However, PDCD2 involvement in apoptosis remains unclear, as other studies including genetic analysis in Drosophila failed to uncover additional evidence for PDCD2 apoptotic function.2-6 Mammalian PDCD2 proteins share 85% identity and also show 53% homology to their Drosophila ortholog Zfrp8 (Zinc finger protein RP-8) along the entire protein. Zfrp8 is an essential gene and mutants exhibit a marked hyperplasia of the lymph gland, the site of hematopoiesis in Drosophila.2 The primary function of Zfrp8 is in the maintenance of hematopoietic stem cells (HSCs), but it is dispensable in pluripotent hemocyte precursors.7 PDCD2 has also been suggested to play a role in zebrafish hematopoiesis.8 PDCD2 knock down in normal human bone marrow progenitors resulted in a decrease in the total number of colony-forming units and inhibited erythropoiesis, implying a role for PDCD2 in normal human hematopoietic cell growth and differentiation.9 In mice, PDCD2 also plays an essential role in cell growth and development. Pdcd2−/− embryos harboring a disrupted PDCD2 allele exhibit early embryonic, peri-implantation lethality in vitro with failure of outgrowth of Pdcd2−/− blastocysts ex vivo.10 These observations implicate PDCD2 as playing an indispensible role in development, how this is manifested mechanistically has yet to be explained.
Human PDCD2 maps to chromosomal band 6q27, a region associated with cytogenetic changes found in a number of B-cell malignancies,11-13 and has been identified as a target of the transcriptional repressor BCL6.14,15 Although PDCD2 was identified as a putative tumor suppressor,16 no systematic investigation of its expression in tumors has been published. Biochemically, PDCD2 has been shown to physically interact in a yeast two-hybrid assay with host cell factor 1 (HCF-1), a protein whose function is required for cell cycle progression and contributes to the activation of E2F- responsive promoters via its association with mixed-lineage leukemia (MLL) and Set-1 histone H3 lysine 4 methyltransferases.17-20 Interestingly, both proteins are targets of ubiquitination, a common strategy to finely regulate the levels of proteins that govern cell cycle progression.21,22
The biological function of PDCD2 remains controversial. Although PDCD2 was initially cloned as a cell death associated cDNA, our observations in several human cancer cell lines indicated high PDCD2 expression in most cell lines studied. Based on these observations we have focused on the role of PDCD2 in human malignancies. PDCD2 is highly expressed in human hematopoietic progenitor cells relative to differentiated mononuclear cells from peripheral blood and unfractionated bone marrow. Similar high PDCD2 expression is present in clinical isolates obtained from patients with acute leukemia. PDCD2 knockdown in two different cancer cell lines (Jurkat leukemia cells and A549 lung carcinoma cells) impairs their proliferation, but not viability relative to parental cells, supporting the notion that PDCD2 overexpression facilitates cancer cell growth. PDCD2 protein expression is associated with cell proliferation with PDCD2 levels being highest in actively growing cultures, then declining with increasing cell density and increasing contact inhibition. Prospective analysis of PDCD2 expression in acute leukemia patients indicates near universal high PDCD2 expression at diagnosis that falls dramatically as patients respond to therapy. The fold reduction in PDCD2 RNA levels after treatment is a statistically significant predictor of clinical relapse in acute leukemia patients.
Our results indicate two important aspects of PDCD2 and cancer. First, we show that rather than controlling cell death PDCD2 facilitates cancer cell growth, suggesting that this evolutionarily conserved protein may represent a new molecular target for cancer therapies. Second, we provide evidence that PDCD2 expression may serve as a correlate for disease status to monitor in acute leukemia patients.
Results
High PDCD2 expression is characteristic of human hematopoietic progenitors and human malignancies
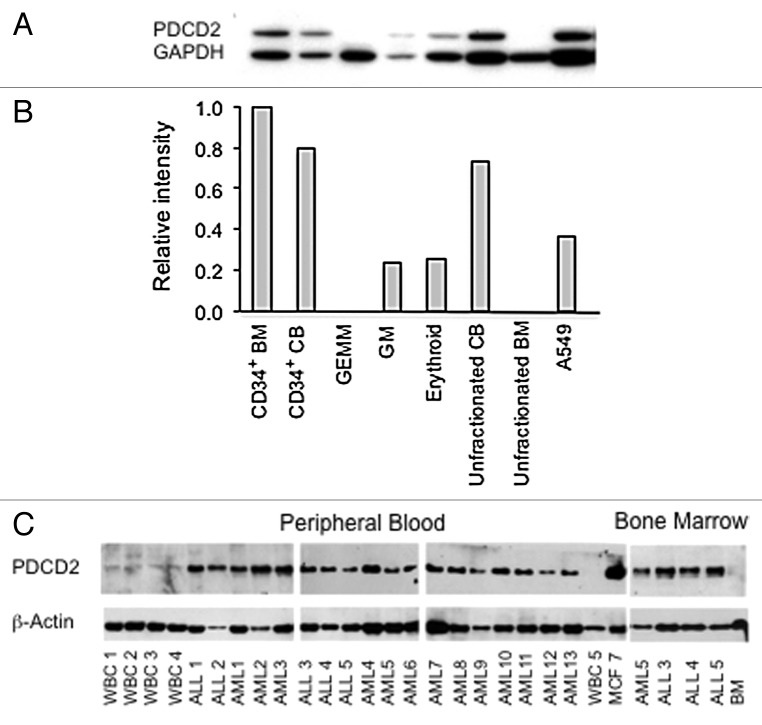
Based on experiments in Drosophila that show Zfrp8/PDCD2 is essential for the maintenance of hematopoietic stem cells (HSCs),7 we sought evidence that PDCD2 is also expressed by human hematopoietic progenitor cells. Using colony formation assays, PDCD2 protein expression was evaluated in hematopoietic progenitors of varying lineage commitment (Fig. 1A and B). PDCD2 expression was highest in the cell fraction enriched for totipotent hematopoietic stem cells, namely the bone marrow CD34+ population, and not detectable in unfractionated bone marrow, where the frequency of CD34+ positive cells is approximately 1 in 102 bone marrow cells.23 PDCD2 expression was also elevated in human cord blood progenitors, an alternate source of hematopoietic stem cells (recently reviewed in ref. 24). PDCD2 protein levels varied inversely with hematopoietic differentiation with lower PDCD2 expression present in more lineage committed progenitors (GM, erythroid and GEMM colonies).
Figure 1. PDCD2 protein expression in human hematopoietic progenitor cells. (A) Human CD34+ bone marrow cells were cultured in methylcellulose. Granulocyte, erythrocyte, macrophage, megakaryocyte (GEMM), granulocyte, macrophage (GM) and erythroid colonies were isolated and analyzed by Western analysis. Human bone marrow and cord blood, both unfractionated and CD34+ positive cells, were also analyzed for comparision. The human lung carcinoma cell line A549 was included as a positive control for PDCD2 expression. (B) Densitometry was utilized to normalize PDCD2 protein expression relative to the GAPDH loading control. (C) Western analysis of PDCD2 expression in normal peripheral blood (WBC 1–5) and bone marrow mononuclear cells (BM), in comparison to peripheral blood and bone marrow samples obtained from patients with acute lymphoblastic leukemia (ALL1–5) and acute myelogenous leukemia (AML1–13). The MCF7 breast carcinoma cell line (MCF7) was utilized as a positive control.
Given the association of Zfrp8 mutations with altered Drosophila hematopoiesis, we analyzed PDCD2 expression in peripheral blood samples obtained from patients with acute leukemia, which contained ≥ 70% leukemic blasts. Our results indicate that PDCD2 is highly expressed in all human acute leukemia samples examined (Fig. 1C). Similar to our analysis of hematopoietic progenitor cells, PDCD2 protein expression was barely detectable in differentiated peripheral blood and unfractionated bone marrow mononuclear cells obtained from normal controls. Together our results indicate that high PDCD2 protein expression is characteristic of both primitive hematopoietic progenitors and acute leukemia cells.
PDCD2 knockdown impairs cancer cell proliferation
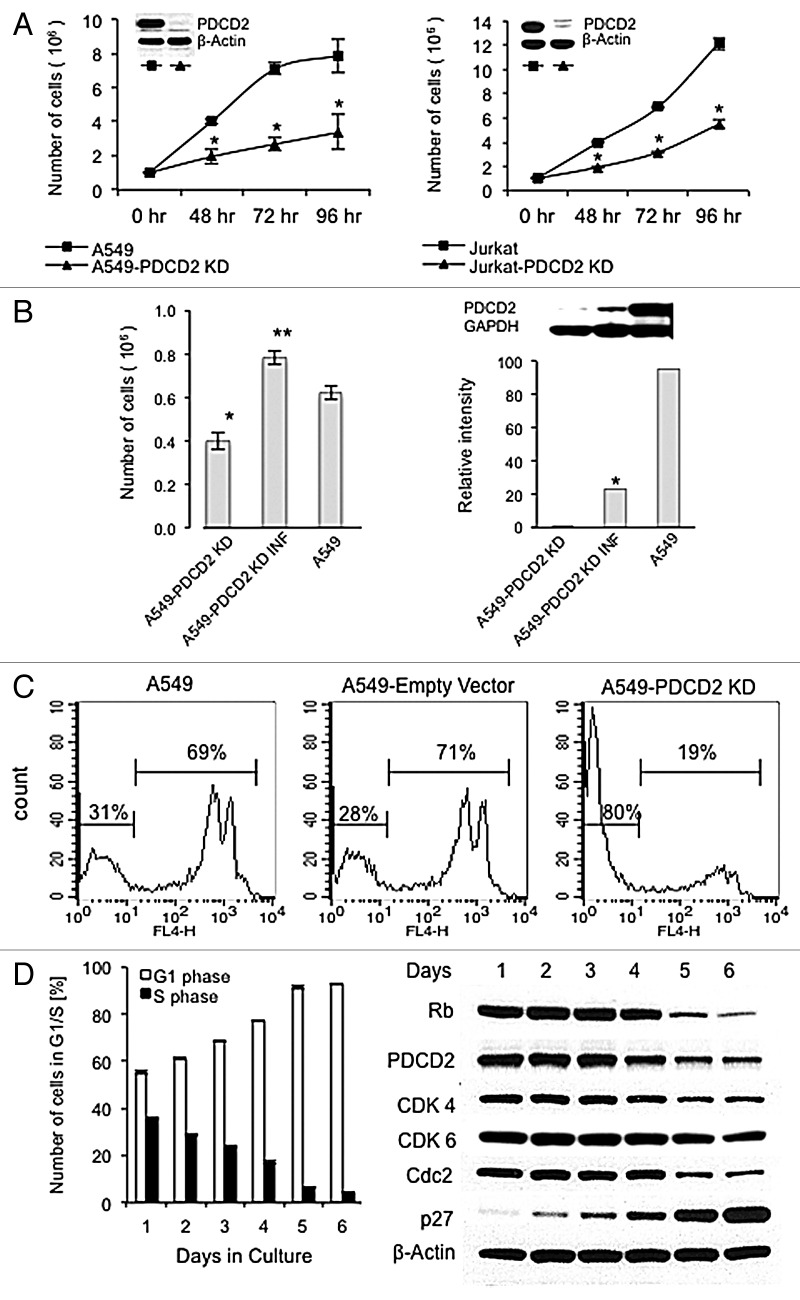
The high PDCD2 expression seen in primary acute leukemia cells suggests this protein may play a role in their malignant properties, such as high rates of cell proliferation. We investigated this possibility by transiently transfecting shRNAs targeting human PDCD2 into the readily transfectable lung carcinoma cell line A549, which has a high basal expression of PDCD2 (Fig. S1). Our results indicate PDCD2 knock down by two separate shRNAs targeting different PDCD2 sequences, impaired A549 cell proliferation in a statistically significant fashion (Fig. S2). Interestingly, PDCD2 knock down did not significantly affect cell viability, implying the decreased cell proliferation that resulted from PDCD2 repression is due to the slowing of cell growth rather than to a direct cytotoxic effect. To further analyze PDCD2 function, we stably knocked down its protein expression using a retrovirus-based shRNA expression vector in two cancer cell lines, A549 lung carcinoma cells and Jurkat leukemia cells (Fig. 2A). PDCD2 knockdown (KD) yielded similar phenotypes in both cell lines. PDCD2 KD cells exhibited significantly slower cell growth relative to parental A549 and Jurkat cells as measured by viable cell number. Again PDCD2 expression was not essential for cancer cell viability as PDCD2 KD cell cultures had the same viability over time as their parental counterparts (data not shown). In order to confirm the specificity of the PDCD2 knock down effect on lung cancer cell proliferation, the shRNA interacting site targeted by siRNA2 in PDCD2 was extensively mutated at the nucleotide level while maintaining the correct PDCD2 amino acid sequence using Wobble codons. The shRNAR PDCD2 cDNA was cloned into a murine stem cell virus vector and used to infect the A549 PDCD2 KD cell line that overexpresses siRNA2. Our results shown in Figure 2B indicate the shRNAR PDCD2 cDNA partially restored PDCD2 protein expression and rescued the effects of PDCD2 knock down on A549 lung cancer cell proliferation in a statistically significant fashion. In sum these results provide evidence that PDCD2 repression significantly impairs cancer cell growth.
Figure 2. Analysis of PDCD2 knockdown on cell proliferation. (A) Cells were harvested at the indicated time points from parental and PDCD2 KD cultures and cell number was measured by automated trypan blue exclusion. Each time point performed in triplicate; error bars represent the standard deviation from the mean; (*) denotes statistical significance, p < 0.05. The insets show PDCD2 protein expression in the A549 and Jurkat PDCD2 KD cell lines. Cells were harvested from respective cultures that were 50–70% confluent and protein lysates prepared. Western blots were probed first for PDCD2 expression, then stripped and probed for β-actin as a loading control. (B) The first panel shows PDCD2 rescue in A549 KD cells after infection with the pMSCV-huPDCD2-shRNAR retrovirus increases cell proliferation. The second panel shows partial restoration of PDCD2 protein expression by pMSCV-huPDCD2-shRNAR retroviral infection by Western analysis. PDCD2 protein expression measured by densitometry and normalized to GAPDH expression. (C) DNA synthesis, as a correlate of S phase transition, was determined by EdU incorporation detected with Alexa Flour® 647 (FL-4) measured by flow cytometry. (D) PDCD2 protein expression is associated with cell proliferation. A549 cells initially seeded at 1 × 105 per plate were cultured for the indicated time points and then cell cycle analysis was performed by propidium iodide staining followed by flow cytometry (left panel). The results presented represent the mean and standard deviations of three independent experiments. The expression of Rb, PDCD2, CDK4, CDK6, CDC2 and β-actin was analyzed in A549 at the indicated time points and increasing degrees of confluence (right panel).
The inhibitory effect of PDCD2 knockdown on cell growth was further investigated in the stable A549 PDCD2 KD lung carcinoma cells. Cell cycle analysis failed to uncover significant differences between PDCD2 KD cell lines and the parental controls (data not shown), however EdU incorporation revealed PDCD2 KD A549 cells exhibited a much lower degree of EdU uptake (19% EdU positive PDCD2 KD cells compared with A549 parental and pSuper-empty vector 69% and 71% EdU positive cells repectively; Fig. 2C) indicating a lower percentage of PDCD2 KD cells passing through S phase. The EdU uptake data, in conjunction with the lack of a specific cell cyle checkpoint block, suggests that PDCD2 knockdown results in an overall slowing of the cell cycle.
Our observations suggest that PDCD2 function is associated with cellular proliferation. As a means to investigate this possibility, PDCD2 protein expression was measured as a function of time, cell culture density and G1/S progression. A549 cells (PDCD2 intact) were initially plated at sub-confluency and allowed to reach confluence. Daily timepoints were analyzed for the percentage of cells in G1 or S phase and the expression PDCD2 along with other cell cycle regulators (Fig. 2D). Interestingly, actively dividing cultures, as indicated by the highest percentage of cells in S phase, contained the highest levels of PDCD2. As cultures reached confluency and cell growth slowed, marked by the decline in percentage of cell in S phase and increase in G1 cells, PDCD2 expression decreased precipitously. The cell cycle exit due to increasing cell density was confirmed by the induction of the cyclin dependent kinase inhibitor p27, whose expression was inversely related to PDCD2 expression. PDCD2 shared a similar expression pattern with other proteins involved with cell division, cyclin dependent kinase 1 (Cdc2), cyclin dependent kinase 4 (Cdk4), cyclin dependent kinase 6 (Cdk6) and retinoblastoma protein (Rb), whose expression also decreased with increasing cell density and subsequent cell cycle exit. Our results indicate that PDCD2 likely participates in cell growth and its high degree of expression in cancer cells may contribute to the uncontrolled growth characteristic of malignant cells.
Prospective analysis of PDCD2 in acute leukemia patients
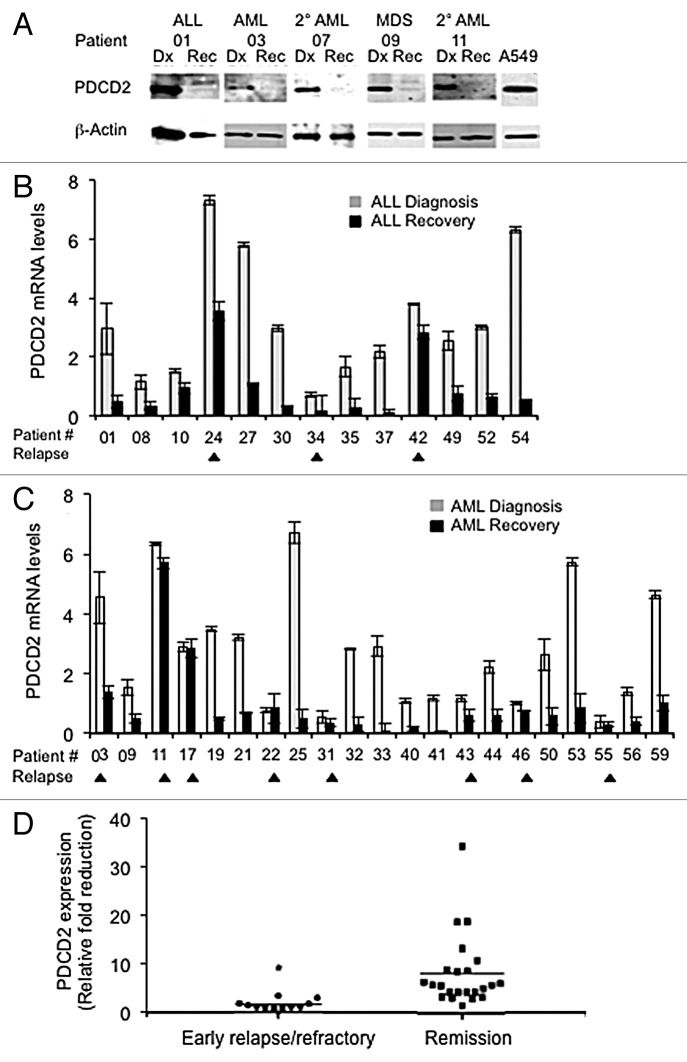
We next sought to further implicate PDCD2 in human malignancies by prospectively analyzing its expression in patients with acute leukemia in whom its expression could be followed temporally as patients present and undergo treatment. Peripheral blood and/or bone marrow samples were collected at the time of diagnosis and at complete remission or, in patients with persistent disease, once best hematologic recovery was attained usually approximately Day 30 after induction chemotherapy. Patient bone marrow (BM) samples were initially analyzed by Western analysis with a representative panel of acute leukemia/advanced myelodysplasia patient samples presented in Figure 3A. At diagnosis PDCD2 protein levels were high, similar to what we found in our initial survey of clinical isolates (Fig. 1C). After treatment PDCD2 protein levels were markedly lower, barely detectable by Western analysis, suggesting PDCD2 protein expression may correlate with disease status.
Figure 3. PDCD2 expression in acute leukemia patients ongoing therapy. (A) PDCD2 expression in patient samples pre and post-treatment. Western analysis was performed on bone marrow aspirates obtained from acute leukemia and advanced myelodysplasia patients pre- (Dx) and post-treatment (Rec). Blots were probed first for PDCD2 expression, then β-actin as a loading control. (B and C) Quantitative Real Time RT-PCR was used to measure PDCD2 transcripts in blood and/or bone marrow of acute leukemia patients at diagnosis and once hematologic recovery was attained. PDCD2 transcript levels are presented relative to basal PDCD2 RNA levels present in normal unfractionated bone marrow. (B) Acute lymphoblastic leukemia (ALL) patients; (C) Acute myelogenous leukemia (AML) patients. Each RT-PCR was performed in triplicate. (Arrowheads below x axis denote relapsed/refractory patients; error bars represent the standard deviation from the mean). (D) Overall reduction in PDCD2 RNA expression in relapsed/refractory leukemia patients vs. those in complete remission after induction chemotherapy (*p = 0.006).
To more precisely measure PDCD2 expression, a quantitative Real Time RT-PCR (qRT-PCR) assay was developed and utilized to monitor PDCD2 RNA expression in subsequent patients. PDCD2 RNA is expressed at low levels in normal, unfractionated BM, which provided a means to quantitate the elevated PDCD2 RNA expression present in leukemia patient samples. PDCD2 RNA levels were normalized to β-actin message as a loading control and presented relative to the level of PDCD2 RNA in normal total bone marrow (e.g., a value of 1 would indicate equivalent PDCD2 RNA levels to normal bone marrow). PDCD2 transcripts were amplified at diagnosis and at hematopoietic recovery post-induction chemotherapy in 34 acute leukemia patients. Our qRT-PCR results indicate that PDCD2 RNA is expressed at much higher levels at diagnosis than at hematopoietic recovery post-treatment in the vast majority of patients analyzed (Fig. 3 and Table 1). The qRT-PCR proved to be more sensitive than Western analysis as illustrated by patient 11, where Day 30 PDCD2 protein levels were not detectable (Fig. 3A) due to the paucity of bone marrow cells remaining as a result of treatment, but PDCD2 RNA was still detectable and relatively unchanged by qRT-PCR (Fig. 3C).
Table 1. Prospective analysis of PDCD2 RNA expression: patient characteristics.
| Patient | Disease | Cytogenetics/mutations | Diagnosis† | Recovery† | Fold‡ reduction |
|---|---|---|---|---|---|
| 01 |
pre-B-ALL |
t(9;22) |
2.96 |
0.48 |
6.1 |
| 08* |
pre-B-ALL |
trisomy 21; T(5;11) |
1.15 |
0.34 |
3.3 |
| 10 |
pre-B-ALL |
normal |
1.52 |
0.95 |
1.6 |
| 24* |
pre-B-ALL |
t(9;22) |
7.34 |
3.54 |
2 |
| 27 |
pre-B-ALL |
t(9;22) |
5.8 |
1.08 |
5.3 |
| 30* |
pre-B-ALL |
t(1;19); T(9,12) |
2.99 |
0.33 |
9.0 |
| 34* |
pre-B-ALL |
11q23; T(4;11) |
0.71 |
0.19 |
3.7 |
| 35 |
pre-B-ALL |
t(9;22) |
1.67 |
0.28 |
5.9 |
| 37 |
pre-B-ALL |
trisomy 9 |
2.17 |
0.11 |
19.7 |
| 42 |
pre-B-ALL |
normal |
3.8 |
2.8 |
1.3 |
| 49 |
pre-B-ALL |
t(9;22) |
2.55 |
0.75 |
3.4 |
| 52 |
pre-B-ALL |
normal; del(12p) by FISH |
3.02 |
0.67 |
4.5 |
| 54 |
pre-B-ALL |
t(9;22); +der(22)T(9;22) |
6.32 |
0.55 |
11.4 |
| 03 |
AML |
normal |
4.55 |
1.38 |
3.2 |
| 09 |
RAEB Type 2 |
complex |
1.54 |
0.49 |
3.1 |
| 11 |
2° AML |
complex |
6.35 |
5.7 |
1.1 |
| 17 |
AML |
complex |
2.89 |
2.86 |
1.0 |
| 19 |
AML |
trisomy 8 |
3.48 |
0.51 |
6.0 |
| 21 |
2° AML |
trisomy 4; trisomy 5; del within 5 |
3.23 |
0.69 |
4.6 |
| 22 |
AML |
minus Y |
0.77 |
0.84 |
0.91 |
| 25* |
AML |
normal; NPM1+ |
6.71 |
0.48 |
13.9 |
| 31 |
AML |
normal; FLT3+ |
0.56 |
0.36 |
1.5 |
| 32 |
AML |
normal |
2.83 |
0.31 |
9.1 |
| 33 |
AML |
normal; NPM1+ |
2.92 |
0.08 |
36 |
| 40 |
AML |
normal |
1.09 |
0.17 |
6.4 |
| 41 |
AML |
normal; NPM1+ |
1.18 |
0.06 |
19.6 |
| 43 |
AML |
11q23; T(11;19), inv(3) |
1.17 |
0.6 |
1.9 |
| 44 |
AML |
normal; NPM1+ |
2.23 |
0.62 |
3.5 |
| 46 |
AML |
trisomy 8, NPM1+ |
1 |
0.76 |
1.3 |
| 50 |
AML |
normal |
2.63 |
0.58 |
4.5 |
| 53 |
AML |
trisomy 21; NPM1+ |
5.72 |
0.85 |
6.7 |
| 55 |
AML |
Complex |
0.39 |
0.31 |
1.26 |
| 56 |
AML |
Normal; NPM1+ |
1.40 |
0.40 |
3.5 |
| 59 | AML | Normal; NPM1+ | 4.64 | 1.03 | 4.5 |
†PDCD2 mRNA fold change in comparison to normal bone marrow; ‡Fold reduction of PDCD2 pre/post treatment; *Peripheral blood analyzed at diagnosis.
In 13 acute lymphoblastic leukemia (ALL) patients, PDCD2 RNA levels are consistently high at diagnosis and markedly decreased after treatment (Fig. 3B). There have been two relapses (patients 24 and 34) and one patient with refractory disease (42) in this group thus far and in 2/3 of these relapsed/refractory cases, PDCD2 RNA levels remained elevated post-treatment (24 and 42). In AML patients, more patients have relapsed, and a trend of PDCD2 RNA expression emerges where the fold reduction in PDCD2 RNA levels appears to be important (Fig. 3C). Thus far 7/20 AML patients have relapsed (03, 17, 22, 31, 43, 46 and 55) and one patient (11) had refractory disease. AML patients that have remained in clinical remission to date exhibit an average 9.34 fold reduction in PDCD2 RNA levels at hematologic recovery after receiving chemotherapy. However, AML patients that relapsed or had refractory disease exhibited only an average reduction in PDCD2 RNA levels of 1.52 fold after treatment. This fold reduction in PDCD2 RNA level does not appear to correlate with a specific cytogenetic abnormality in this small sample of AML patients, with relapses roughly distributed between intermediate and poor prognostic cytogenetic categories. Combining the ALL and AML data, the difference in PDCD2 RNA reduction after treatment in relapsed/refractory patients vs. those who maintained a complete remission reached statistical significance. Patients with refractory disease or who relapsed had an average fold reduction of 1.74 after treatment, whereas patients who achieved a complete remission had a much greater average fold reduction in PDCD2 RNA levels of 8.33 (Fig. 3D; p = 0.006). These data suggest that PDCD2 RNA expression correlates with disease status and the fold reduction in PDCD2 RNA after initial induction chemotherapy appears to be a significant predictor of relapse in acute leukemia patients.
Discussion
Multiple lines of evidence suggest that PDCD2 may play an important role in human oncogenesis. Zfrp8 mutations in Drosophila cause an enormous overgrowth of the lymph gland, the primary site of fly hematopoiesis. This overgrowth phenotype, although suggestive of Zfrp8 acting as a tumor suppressor, was found to be the result of changed cellular homeostasis and a delay in prohemocyte differentiation. Genetic analyses have revealed the loss of Zfrp8 affected the stem and precursor cells in the lymph gland without changing the fate of pluripotent progenitors and cells with limited mitotic potential.7 These results indicate that Zfrp8 has a stem cell function and its loss affects HSC self-renewal and not the differentiation of more committed hematopoietic progenitors. The results presented here suggest that PDCD2 may have a similar HSC-specific function in human hematopoiesis. PDCD2 expression is high in HSC-enriched populations (cord blood and CD34+ bone marrow cells) and diminishes with further differentiation. Likewise PDCD2 overexpression in acute leukemia cells is likely a reflection of the primitive nature of this malignant population.
Our clinical study albeit in a small panel of acute leukemia patients, showed that the fold reduction PDCD2 RNA levels after initial treatment may be a predictor of relapse in these patients. Specifically, patients with persistent levels of PDCD2 expression were statistically more likely to relapse than those exhibiting marked decreases in expression. Moreover, the high level of PDCD2 expression in leukemia cells is reminiscent of PDCD2 expression in primitive hematopoietic progenitors and may be reflective of the proliferative potential of these cellular populations relative to normal, differentiated hematopoietic cells. These observations suggest PDCD2 or its overexpression may play an active role in leukemogenesis and represent a new therapeutic target for acute leukemia. Patients who relapsed or had refractory disease had a much lower fold reduction in PDCD2 RNA levels (mean ratio = 1.74); while patients in complete remission had a much higher fold reduction PDCD2 RNA levels after induction chemotherapy (mean ratio = 8.33). This relationship between relapse and the ratio of before- and after-treatment PDCD2 levels will require validation in prospective analysis of a larger panel of acute leukemia patients. PDCD2 expression levels may provide a means to not only identify patients early in the course of treatment who are at higher risk for relapse, but also may be a biomarker that can be followed in surveillance for early relapse once a complete remission is obtained for both ALL and AML patients.
Consistent with the high level of PDCD2 expression characteristic of acute leukemia clinical isolates, we have identified a new biological PDCD2 function as a facilitator of cancer cell growth. PDCD2 knockdown significantly impairs cancer cell proliferation and the proportion of cells progressing through S phase. The PDCD2 protein expression pattern is similar to other cell cycle regulators that function in G1/S transition, suggesting that PDCD2 may also function in the cellular decision to enter the cell cycle or affect the efficiency the G1/S transition. Additional work is required to ascertain if PDCD2 functions to facilitate cell cycle progression at the transcriptional level, perhaps as a binding factor for other transcriptional regulators such as HCF-1, or via other as yet undescribed protein-protein interactions.
In sum, we have shown that PDCD2 is a marker for normal early hematopoietic progenitor cells and PDCD2 also participates in cancer cell proliferation, consistent with its high level of expression in clinical samples obtained from patients with cancer. Moreover, we provide preliminary evidence that PDCD2 expression, based on its ability to predict poor outcomes, may be exploited as a biomarker to aid surveillance for recurrent disease in acute leukemia patients. PDCD2’s role in cell proliferation and its high expression in human malignancies make it an attractive, novel molecular target for new anti-cancer therapies.
Materials and Methods
Human hematopoietic progenitor cell culture
Human CD34+ bone marrow cells were cultured in methylcellulose according to manufacturer’s recommendations (STEMCELL Technologies Inc.; #ABM017F). To insure an appropriate number of hematopoietic colonies per plate, varying cell numbers ranging from 0.25−1 × 104 cells were added to 3 ml of complete Methocult® medium (STEMCELL Technologies Inc.; #H4434) per 35 mm culture dish. After a 10 d period, the Colony-Forming Unit-Erythroid (CFU-E), Granulocyte, Macrophage (CFU-GM) and Granulocyte, Erythrocyte, Macrophage, Megakaryocyte (CFU-GEMM) colonies were identified based on morphology in a dissecting microscope and harvested. Multiple colonies of the same lineage were pooled in IMDM with 2% FBS and processed for protein isolation.
Cell lines
The A549 lung carcinoma cell line was obtained from the American Type Culture Collection and maintained in Dulbecco’s modified Eagle’s (DMEM)/F-12 medium supplemented with 10% heat inactivated fetal bovine serum (FBS) plus 2 mM glutamine, penicillin 100 U/mL and streptomycin 100 U/mL (Invitrogen Corporation). Jurkat T cells were maintained in RPMI medium with the same supplements. Cells were incubated at 37°C with 5% CO2.
PDCD2 knockdown constructs and transfection
A panel of five Dicer Substrate RNAi duplexes were identified using the IDT SciTools RNAi Design algorithm (Integrated DNA Technologies), two of which corresponding to human PDCD2 nucleotides 738−762 (siRNA2) and nucleotides 768−792 (siRNA1) (GeneBank accession number NM_002598.2) yielded the greatest knockdown of PDCD2 protein expression in transient transfection assays and were used for subsequent experiments. Stable PDCD2 knockdown was achieved using a retrovirus-based mammalian RNAi expression system (pSuper RNAi System, OligoEngine; #VEC-pRT-006). Complementary sense and antisense oligonucleotide sequence corresponding to nucleotides 738−762 (siRNA2) of human PDCD2 was synthesized, annealed and then ligated into pSuper (pSUPER-shPDCD2). A PDCD2 cDNA resistant to siRNA2 was constructed by changing the nucleotide sequence of the siRNA2 interacting site in PDCD2 while maintaining the PDCD2 amino acid sequence. The human PDCD2 coding sequence was mutated to add a unique BamH1 site at nucleotide 760 at the 3′ end of the siRNA2 interacting site. This was accomplished by site-directed mutagenesis (QuikChange II-E-Site Directed Mutagenesis Kit; Agilent Technologies; #200555) using the following oligonucleotides (5′ to 3′): PDCD2 Bam sense-GAA GAT TAC TCA GAG ATT ATA GGA TCC ATG GGT GAA GCA CTT GAG G; PDCD2 Bam reverse-CCT CAA GTG CTT CAC CCA TGG ATC CTA TAA TCT CTG AGT AAT CTT C. The remaining siRNA2 PDCD2 interacting site was mutated using the following oligonucleotides (5′ to 3′): siRNA-resistant sense oligo: TGA GGT TGT GGA AAA AGA GGA CTA TAG TGA AAT CAT CG; shRNA-resistant anti-sense oligo: GAT CCG ATG ATT TCA CTA TAG TCC TCT TTT TCC ACA ACC. These oligos were annealed and then ligated into the PDCD2-BamH1 construct restricted with Bsu36I and BamH1 generating the siRNA2-resistant PDCD2 construct where 12/25 nucleotides of the siRNA2 interacting site are mutated while the PDCD2 amino acid sequence is maintained. The resultant siRNA-resistant PDCD2 cDNA was amplified by PCR adding a 5′ BglII site and a 3′ XhoI site to facilitate cloning into the Murine Stem Cell Virus vector pMSCVpuro (Clontech; #6311461) yielding the pMSCV-huPDCD2-shRNAR construct.
Cells were transiently transfected with Dicer Substrate RNAi duplexes (Integrated DNA Technologies) using Lipofectamine 2000 (Invitrogen Corporation; #11668−019) following manufacturer’s recommendations. A549 cells were seeded at a density of 0.2 × 106 cells per well in a 6 well plate, then 24 h later transfected with Dicer Substrate RNAi duplexes (final concentration 10 nM), The transfection was repeated 24 h later with a lower concentration of Dicer Substrate RNAi duplexes (5 nM). Transfected cells were split one day after the final transfection and cultured for five additional days. At the end of the 5 d period, cells were harvested for automated trypan blue exclusion using a Vi-Cell™ Cell Viability Analyzer (Beckman-Coulter) and Western analysis. Each transfection was performed in triplicate and the overall transfection experiment was repeated at least twice. The siRNA duplexes for human PDCD2 are as follows: siRNA1, 5′-CAG TTC TTC CTC AAG TGC TTC ACC CAT-3′, siRNA2, 5′-CCC TAT AAT CTC TGA GTA ATC TTC CTT-3′.
A549 cells were stably transfected with the pSuper-shPDCD2 construct using Lipofectamine 2000 as per manufacturer’s recommendations. Stable G418R colonies were selected and screened for PDCD2 knockdown by western blotting. For Jurkat cells, cells were transfected with pSuper-shPDCD2 using the Amaxa Nucleofector II program X-001, then subjected to G418 selection and cloned by limiting dilution. The clonal Jurkat cell lines were screened for PDCD2 knockdown (KD) by Western analysis. Jurkat PDCD2 KD cells were maintained at a density of 1 × 105 cells/ml and the A549 PDCD2 KD at a maximum confluence of 25%.
Retroviral particles were produced by transient transfection of pMSCV-huPDCD2-shRNAR into the Phoenix packaging cell line as described previously.25 Media conditioned by the transfected Phoenix packaging cells was used to infect A549 and A549-PDCD2 KD cells in the presence of 4 μg/ml polybrene. Infected cultures were maintained in 1.5 μg/ml puromycin. After selection, 0.25 × 106 puromycinR cells were plated per well of 6-well plates and allowed to grow. Cells were harvested 72 h later for cell counts and western analysis. Triplicate wells were counted using a Vi-Cell™ Cell Viability Analyzer (Beckman-Coulter).
Quantitative real time RT-PCR analysis
Acute leukemia patients were enrolled in a clinical trial to prospectively measure PDCD2 expression. All patients provided informed assent/consent approved by the Robert Wood Johnson Medical School Institutional Review Board. Peripheral blood and/or bone marrow samples were obtained at diagnosis of acute leukemia and at complete remission, indicated by recovery of peripheral blood counts [absolute neutrophil count (ANC) ≥ 1000; platelet count ≥ 100,000] and no morphological evidence of persistent leukemia, or once best hematopoietic recovery was achieved in cases of refractory disease. The clinical characteristics of the 34 patients analyzed by Quantitative real time RT-PCR (qRT-PCR) is presented in Table 1. All patient peripheral blood and/or bone marrow samples were subjected to red blood cell lysis and if adequate cell numbers were present based on the complete blood cell count; samples were further purified by histoplaque centrifugation (Sigma-Aldrich; #10771) using standard methodologies. Total RNA was initially isolated from patient samples using Trizol Reagent (Invitrogen Corporation; #15596–026), according to the manufacturer’s instructions. To remove potential genomic DNA contamination, RNA was treated with RNase-free DNase Turbo DNA-free kit (Ambion/Applied Biosystems; #AM1907) prior to amplification. DNase-treated total RNA was isolated from later samples using the RNeasy® Mini Kit (Qiagen, Inc.; #74104). Quantitative Real-Time RT-PCR was performed using the AgPath-ID One-Step RT-PCR Kit (Ambion/Applied Biosystems; #AM1005) as per manufacturer’s recommendations. The β-actin levels were measured using the following primers: Left primer 5′-GTC TTC CCC TCC ATC GTG-3′; Right primer 5′-CAT GTC GTC CCA GTT GGT G-3′; probe (5′6-FAM) CAT CCT CAC CCT GAA GTA CC-(3′ Iowa Black®FQ) (Integrated DNA Technologies). The PDCD2 primers used are as follows (5′ to 3′): Left GCT GCA TCT TCC TCT TCT GC; Right GGG GAG GGA TTC TCA GAA GT; and probe (5′-6-FAM) CCT GCG AGT TTT TAG GAA TCA (3′Iowa Black® FQ) (Integrated DNA Technologies). Real-time PCR data was acquired on a Stratagene Mx4000 or a Mx3005P instrument (Agilent Technologies). Samples were analyzed in triplicate. The level of PDCD2 RNA, normalized to the housekeeping gene β-actin, was compared with PDCD2 transcripts levels in normal bone marrow (STEMCELL Technologies Inc.; #ABM008F) using the comparative C(T) method.26
Cell cycle analysis
EdU (5-ethynyl-2’- deoxyuridine; Invitrogen Corporation; #C10424) incorporation was used to measure G1/S progression. Relevant cell lines were seeded in triplicate at 0.5 × 106 cells per 10 cm plate then 72 h later were pulsed for 2 h with EdU (10 μM). Cells were harvested, fixed and processed for EdU incorporation by Alexa Fluor® 647 detection with flow cytometry according to the manufacturer’s protocols. For cell proliferation, the percentage of newly synthesized DNA was determined as percentage of Alexa Fluor® 647 stained cells using the CellQuest software. A minimum of 10,000 cells per sample was analyzed. The results presented are representative of at least three experiments. Cell cycle analysis was performed using standard methodologies [Propidium Iodide (PI) RNase staining buffer, BD PharMingen; #550825]. Fluorescent intensities were acquired on a BD FACSCalibur™ and the percentage of cells in each phase of the cell cycle was calculated using the ModFit v3.0 software (BD Biosciences).
Protein expression
Western analysis was done using standard methods. Polyclonal antibody against PDCD2 was obtained from the Sharp Laboratory,17 mouse monoclonal Rb (556538) and p27 (554069) were purchased from BD Biosciences; Cdk4 (sc-601), Cdk6 (sc-177) and Cdc2 (sc-54) antibodies purchased from Santa Cruz Biotechnology, Inc. Monoclonal antibody against β-actin (A1978) was purchased from Sigma-Aldrich and anti-glyceraldehyde-3-phosphate dehydrogenase-horseradish peroxidase (HRP) conjugate (GAPDH; 3683) was purchased from Cell Signaling, Inc.
Statistical analysis
Statistical analyses were performed using the two tailed Student’s t-test. A p value less than 0.05 was considered to be statistically significant. All statistical analyses were conducted using the GraphPad Prism® software program (version 5, Graphpad Software, Inc.).
Supplementary Material
Acknowledgments
This work is dedicated to the late Sonya Greenwood whose outstanding clinical research support made this work possible. This work was funded by a research grant (09–1079-CCR-EO) provided to D.G.S by The New Jersey Commission on Cancer Research. The authors thank Dr Joseph Bertino for his critical reading of this manuscript.
Submitted
01/28/2013
Revised
02/14/2013
Accepted
03/28/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/nucleus/article/24484.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/24484
References
- 1.Owens GP, Hahn WE, Cohen JJ. Identification of mRNAs associated with programmed cell death in immature thymocytes. Mol Cell Biol. 1991;11:4177–88. doi: 10.1128/mcb.11.8.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minakhina S, Druzhinina M, Steward R. Zfrp8, the Drosophila ortholog of PDCD2, functions in lymph gland development and controls cell proliferation. Development. 2007;134:2387–96. doi: 10.1242/dev.003616. [DOI] [PubMed] [Google Scholar]
- 3.Chen Q, Qian K, Yan C. Cloning of cDNAs with PDCD2(C) domain and their expressions during apoptosis of HEK293T cells. Mol Cell Biochem. 2005;280:185–91. doi: 10.1007/s11010-005-8910-z. [DOI] [PubMed] [Google Scholar]
- 4.D’Mello SR, Galli C, Ciotti T, Calissano P. Induction of apoptosis in cerebellar granule neurons by low potassium: inhibition of death by insulin-like growth factor I and cAMP. Proc Natl Acad Sci U S A. 1993;90:10989–93. doi: 10.1073/pnas.90.23.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guénal I, Mignotte B. Studies of specific gene induction during apoptosis of cell lines conditionally immortalized by SV40. FEBS Lett. 1995;374:384–6. doi: 10.1016/0014-5793(95)01157-A. [DOI] [PubMed] [Google Scholar]
- 6.Vaux DL, Häcker G. Cloning of mouse RP-8 cDNA and its expression during apoptosis of lymphoid and myeloid cells. DNA Cell Biol. 1995;14:189–93. doi: 10.1089/dna.1995.14.189. [DOI] [PubMed] [Google Scholar]
- 7.Minakhina S, Steward R. Hematopoietic stem cells in Drosophila. Development. 2010;137:27–31. doi: 10.1242/dev.043943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramer J, Granier CJ, Davis S, Piso K, Hand J, Rabson AB, et al. PDCD2 Controls Hematopoietic Stem Cell Differentiation During Development. Stem Cells Dev. 2012; 22:58–72. doi: 10.1089/scd.2012.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kokorina NA, Granier CJ, Zakharkin SO, Davis S, Rabson AB, Sabaawy HE. PDCD2 knockdown inhibits erythroid but not megakaryocytic lineage differentiation of human hematopoietic stem/progenitor cells. Exp Hematol. 2012;40:1028–42, e3. doi: 10.1016/j.exphem.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mu W, Munroe RJ, Barker AK, Schimenti JC. PDCD2 is essential for inner cell mass development and embryonic stem cell maintenance. Dev Biol. 2010;347:279–88. doi: 10.1016/j.ydbio.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amiel A, Mulchanov I, Elis A, Gaber E, Manor Y, Fejgin M, et al. Deletion of 6q27 in chronic lymphocytic leukemia and multiple myeloma detected by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1999;112:53–6. doi: 10.1016/S0165-4608(98)00255-6. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami T, Furukawa Y, Sudo K, Saito H, Takami S, Takahashi E, et al. Isolation and mapping of a human gene (PDCD2) that is highly homologous to Rp8, a rat gene associated with programmed cell death. Cytogenet Cell Genet. 1995;71:41–3. doi: 10.1159/000134058. [DOI] [PubMed] [Google Scholar]
- 13.Stilgenbauer S, Bullinger L, Benner A, Wildenberger K, Bentz M, Döhner K, et al. Incidence and clinical significance of 6q deletions in B cell chronic lymphocytic leukemia. Leukemia. 1999;13:1331–4. doi: 10.1038/sj.leu.2401499. [DOI] [PubMed] [Google Scholar]
- 14.Baron BW, Anastasi J, Thirman MJ, Furukawa Y, Fears S, Kim DC, et al. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: implications for a role of BCL6 in the down-regulation of apoptosis. Proc Natl Acad Sci U S A. 2002;99:2860–5. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baron BW, Zeleznik-Le N, Baron MJ, Theisler C, Huo D, Krasowski MD, et al. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of human B and T cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:7449–54. doi: 10.1073/pnas.0701770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinemann D, Gesk S, Zhang Y, Harder L, Pilarsky C, Hinzmann B, et al. Identification of candidate tumor-suppressor genes in 6q27 by combined deletion mapping and electronic expression profiling in lymphoid neoplasms. Genes Chromosomes Cancer. 2003;37:421–6. doi: 10.1002/gcc.10231. [DOI] [PubMed] [Google Scholar]
- 17.Scarr RB, Sharp PA. PDCD2 is a negative regulator of HCF-1 (C1) Oncogene. 2002;21:5245–54. doi: 10.1038/sj.onc.1205647. [DOI] [PubMed] [Google Scholar]
- 18.Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, et al. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–37. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 19.Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 2003;22:2360–9. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–19. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 21.Fukae J, Sato S, Shiba K, Sato K, Mori H, Sharp PA, et al. Programmed cell death-2 isoform1 is ubiquitinated by parkin and increased in the substantia nigra of patients with autosomal recessive Parkinson’s disease. FEBS Lett. 2009;583:521–5. doi: 10.1016/j.febslet.2008.12.055. [DOI] [PubMed] [Google Scholar]
- 22.Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–92. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins RH., Jr. CD34+ selected cells in clinical transplantation. Stem Cells. 1994;12:577–85. doi: 10.1002/stem.5530120605. [DOI] [PubMed] [Google Scholar]
- 24.Mayani H. Umbilical cord blood: lessons learned and lingering challenges after more than 20 years of basic and clinical research. Arch Med Res. 2011;42:645–51. doi: 10.1016/j.arcmed.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–6. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.