Abstract
Anti-epidermal growth factor receptor (EGFR) therapy has been tried in triple negative breast cancer (TNBC) patients without evaluation of molecular and clinical predictors in several randomized clinical studies. Only fewer than 20% of metastatic TNBCs showed response to anti-EGFR therapy. In order to increase the overall response rate, first step would be to classify TNBC into good or poor responders according to oncogenic mutation profiles. This study provides the molecular characteristics of TNBCs including EGFR gene copy number changes and mutation status of EGFR and KRAS gene in Korean TNBC patients. Mutation analysis for EGFR, KRAS, BRAF and TP53 from a total of 105 TNBC tissue samples was performed by direct sequencing, peptide nucleic acid-mediated PCR clamping method and real-time PCR. Copy number changes of EGFR gene were evaluated using multiplex ligation-dependent probe amplification. Out of all 105 TNBCs, 15.2% (16/105) showed EGFR copy number changes. Among them, increased or decreased EGFR copy number was detected in 13 (5 single copy gain, 2 amplification and 4 high-copy number amplification) and 3 cases (3 hemizygous deletion), respectively. The mutation frequencies of KRAS, EGFR and TP53 gene were 1.9% (G12V and G12D), 1.0% (exon 19 del) and 31.4%, respectively. There was no BRAF V600E mutation found. Future studies are needed to evaluate the clinical outcomes of TNBC patients who undergo anti-EGFR therapy according to the genetic status of EGFR.
Introduction
Triple-negative breast cancers (TNBCs) are tumors, nominally classified as a diagnosis of exclusion, that do not express clinically significant levels of the estrogen receptor (ER), progesterone receptor (PgR) and epidermal growth factor receptor 2 (Her-2) over-expression or gene amplification [1]. TNBCs account for 10–24% of all invasive breast cancers with a strong predilection for young women, but the frequency varies by race and in some racial groups reaches a frequency of up to 55% [2]–[6]. Hormonal therapies and HER2-targeted agents are not effective in these breast tumors, which tend to exhibit aggressive, metastatic behavior and have a worse prognosis than hormone receptor-positive, luminal subtypes [1], [7], [8]. In primary non–small cell lung cancer that harbors EGFR mutations, multiple randomized clinical trials comparing first-line chemotherapy to EGFR tyrosine kinase inhibitors (TKIs) have been performed and uniformly demonstrated the superiority of EGFR-TKIs [9]–[12]. In addition, patients suffering from recurrent glioblastoma with EGFR amplification and those lacking EGFRvIII expression have been treated with the EGFR-targeted monoclonal antibody cetuximab with a significantly superior progression-free and overall survival [13]. Approximately 20% of metastatic TNBCs showed response to anti-EGFR therapy in randomized clinical trials [2], [14]. Recent studies have shown no mutations in several target genes associated with the receptor tyrosine kinase/RAS/MAPK pathway, including EGFR, KRAS and BRAF, in the absence of HER2 gene amplification [15]–[17]. However, in Asians, EGFR mutations and copy number changes of the EGFR gene were detected in up to 11.4% [17], [18] and 21% of TNBCs [19], respectively. Anti-EGFR therapies are still an attractive treatment modality according to the genetic profiles of TNBCs [18]–[20]. Thus, it would be beneficial to evaluate mutations and copy number changes of EGFR in TNBC patients before treating with anti-EGFR drugs, which in turn would improve the response rates compared to previous data. Furthermore, a deliberate and clinically applicable method is also needed to evaluate EGFR mutations and copy number changes as a molecular predictor for the patients. Here, we report the mutation status of EGFR, KRAS and BRAF, and the frequency of EGFR copy number changes in Korean patients with TNBCs.
Materials and Methods
Subject selection
We obtained a total of 105 tissue samples from TNBC patients at the time of surgery. Triple negative status (negative estrogen receptor (ER), progesterone receptor (PgR) and c-erbB2) of the tumors was confirmed by immunohistochemical (IHC) staining. Briefly, all IHC staining was performed using formalin-fixed, paraffin-embedded tissue sections. After deparaffinisation/rehydration and antigen retrieval, paraffin sections were incubated with primary antibodies against ER (1∶50 dilution; Dinona, Seoul, Korea), PR (1∶100 dilution; Dinona) and Her2/neu (1∶250 dilution; Dako, Glostrup, Denmark). ER and PR IHC signal was evaluated using the Allred score [21]. A score of 0 to 2 was considered negative and a score of 3 to 8 was regarded as positive. HER2 status was determined by IHC using the HercepTest, and score of 0–1+ was regarded as negative (18). A borderline/equivocal expression of HER-2 was indicated for cerb2 when at least 10% of tumor cells demonstrated 2+ cytoplasmic membrane staining, and these samples were confirmed using fluorescence in situ hybridization with the PathVysion HER2 DNA Probe kit (Abbott, IL, USA) according to the manufacturer instructions. A HER2 gene-to-chromosome 17 ratio greater than 2 was considered positive. The study was approved by the Institutional Review Board of the Gangnam Severance Hospital and written informed consent was obtained from the patients.
DNA preparation
DNA was extracted from breast cancer tissues (ER-, PR-, and HER2-) obtained at the time of surgical resection. Genomic DNA was extracted using QIAamp DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer protocol. The concentration and quality of genomic DNA was evaluated by Nanodrop (ND-1000; Thermo Scientific, DE, USA).
Direct sequencing of EGFR, KRAS and TP53 genes
Mutation analysis for EGFR and KRAS genes was performed in duplicate using direct sequencing and the peptide nucleic acid (PNA)-mediated PCR clamping method. PCR amplification and direct sequencing of EGFR gene (exons 18–21), KRAS (exon2) and TP53 gene (exon 5–9) were performed in 105 TNBCs [22]–[27]. The primers designed to amplify exons and flanking introns of those genes are summarized in Table 1. PCR was performed using an Accu-Power™ Premix (Bioneer, Daejeon, Korea) under the following amplification conditions: 94°C for 4 min followed by 50 cycles of 94°C for 1 min, 60°C for 30 s and 72°C for 30 s, and final extension at 72°C for 15 min. Purified PCR products obtained using a QIAquick Gel Extraction kit (Qiagen, Düsseldorf, Germany) were used for sequencing with a Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA, USA). The thermal cycler conditions were as follows: 96°C for 5 min followed by 24 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 4 min, and final extension at 72°C for 5 min. The sequences were analysed using ABI 3500Dx system (Applied Biosystems). Sequences were compared with the database sequence in GenBank (http://www.ncbi.nlm.nih.gov assessed June, 2012). The GenBank accession numbers are NM_005228.3, NM_004985.3 and NC_000017.9 for the EGFR, KRAS and TP53 genes, respectively.
Table 1. PCR primers of TP53 and EGFR gene.
| Target | Exon | Forward Primer (5'→3') | Reverse Primer (5'→3') | PCR product size (bps) |
| EGFR | 18 | ATGTCTGGCACTGCTTTCCA | ACAGCTTGCAAGGACTCTGG | 277 |
| 19 | AGATCACTGGGCAGCATGT | AGCAGCTGCCAGACATGAG | 246 | |
| 20 | CATTCATGCGTCTTCACCTG | CATATCCCCATGGCAAACTC | 377 | |
| 21 | AGCCATAAGTCCTCGACGTG | ATCCTCCCCTGCATGTGTTA | 372 | |
| KRAS | 2 | TGTATTAACCTTATGTGTGACA | CTTGTAATAAGTACTCATGAAA | 279 |
| TP53 | 5&6 | CACTTGTGCCCTGACTTTCA | TTGCACATCTCATGGGGTTA | 619 |
| 7 | CCTGCTTGCCACAGGTCT | TGATGAGAGGTGGATGGGTAG | 279 | |
| 8&9 | CAAGGGTGGTTGGGAGTAGA | CCCCAATTGCAGGTAAAACA | 496 |
Abbreviations: bps, base pairs; PCR, polymerase chain reaction.
Mutation analysis of EGFR, KRAS and BRAF genes
We evaluated EGFR and KRAS mutations using the PNAClamp™ EGFR and PNAClamp™ KRAS Mutation Detection kits (Panagene, Inc., Daejeon, Korea). PNA-mediated PCR clamping is a mutant enrichment method and can detect minor mutant alleles in whole tumor cells [14], [28], [29]. All reactions were carried out in a volume of 20μl with template DNA, a primer and PNA probe set, and SYBR Green PCR master mix according to the manufacturer instructions. Real-time PCR reaction was performed using a CFX 96 (Bio-Rad, USA). PCR cycling conditions were as follows: 5 minutes at 94°C followed by 40 cycles of 94°C for 30 seconds, 70°C for 20 seconds, 63°C for 30 seconds, and then 72°C for 30 seconds. Each kit is designed to detect 29 mutations in the EGFR gene (exon 18 – 21) and 7 mutations in the KRAS gene (exon 2). Delta Ct values were calculated for defining whether there were mutations in EGFR and KRAS [28], [29]. BRAFV600E was evaluated using Real-QTM™ BRAF V600E detection kit (Biossewoom, Inc., Seoul, Korea). All optical reaction tubes which contained 10 µL genomic DNA and 15 µL RQ-PCR reaction mixture were placed in a CFX96 (Bio-Rad, USA). The thermal cycler protocol used was 50°C for 120 minutes, 95°C for 10 minutes and then 40 cycles of 95°C for 15 seconds and 58°C for 45 seconds. Data analysis was done according to the manufacturer instructions.
Copy number analysis of EGFR
EGFR copy number changes in TNBCs were evaluated using SALSA® MLPA® Probemix P315-B1 EGFR kit (MRC-Holland, Amsterdam, the Netherlands) that contains 12 reference probes and 30 probes in the exons of the EGFR gene, including specific probes for the L858R and T790M mutations. Probe sequences of SALSA® MLPA® P315-B1 EGFR probemix are available at http://www.mlpa.com. Denatured DNA samples were hybridized with probemix for 16 hours at 60°C. Ligation of probes was performed with the Ligase-65 enzyme at 54°C for 15 min, followed by 5 minutes at 98°C for heat inactivation of the Ligase-65 enzyme. PCR was performed with the specific SALSA PCR primers for 35 cycles (95°C for 30 s, 60°C for 30 s and 72°C for 1 min) using the GeneAmp® PCR System 9700 (Applied Biosystems, USA). All MLPA analysis of 105 samples was performed in duplicate. Three cancer cell lines (HCC827, H2279, and H1975) were used as the positive controls for MLPA analysis. According to results from fluorescence in situ hybridization and/or the nanofluidic digital PCR array, HCC827, H2279 and H1975 were considered to have high-copy number amplifications, amplifications and single copy gain with L858R and T790M mutations, respectively [30], [31]. To determineanalytical sensitivity of MLPA for EGFR copy number changes in tumor cells, DNA from HCC827 was serially diluted with wild type DNA. Each spiked sample was assayed up to five times using SALSA®MLPA® Probemix P315-B1 EGFR kit (MRC-Holland).
MLPA fragment analysis data were generated on Applied Biosystems 3500Dx system (Applied Biosystems). The data were analyzed using the GeneMarker® software (SoftGenetics, PA, USA). The copy number of the EGFR gene was determined on the basis of the relative peak height ratio of the probes. Seventy-eight good quality samples (the SD of all 44 probes were within 0.1) without gain and/or loss were used as a reference group to construct the mean peak of each probe. The SD of all control probes in 105 samples, except by random errors due to capillary electrophoresis conditions (ex. SPG11 gene, 439 nt, 16538-L21570) and disease-associated control probe (ex.COL11A1 gene, 196 nt, 13232-L14565 etc.) [32], was within 0.1. Based on our previous report with MLPA copy number detection, we used thresholds of 1.2 and 0.8 for the detection of single copy gain (GAIN) and hemizygous deletion (HETD), respectively. Furthermore, the ratios above 2.0 were considered as amplifications (AMP) and those exceeding 10 as high-copy number amplifications (HLAMP) [33]. The probe fractions are then divided by the average probe fraction of the reference group, resulting in a probe ratio [34].
Statistical Analysis
Patient follow-up periods were calculated as time between surgery date and the date of last follow-up (months). Relapse free survival (RFS) included loco-regional recurrence, distant metastasis, and death from any cause. Breast cancer-specific survival (BCSS) included only patients who died from any breast cancer-related cause. TP53 mutations were classified as missense, nonsense, and frameshift mutations. In addition, missense mutations were further subdivided as missense mutations in DNA binding motifs (DBM) and outside the DBM according to mutation position, nature, and suspected effect on protein structure and activity [23], [35]. Kaplan-Meier survival curves with log-rank tests were performed to compare RFS and BCSS according to the type of TP53 mutation. Cox proportional hazard models were performed to assess the influence of prognostic factors on RFS. Univariate analyses for age at surgery, tumor size, node status, Ki67 labeling index and TP53 mutations were performed. Any factors from the univariate analysis with a p value less than 0.10 was included in the multivariate analysis. All statistical analyses were performed using SPSS 20.0 (SPSS Inc, Chicago, IL, USA). For all statistical analyses except the univariate analysis, a p value less than 0.05 was regarded significant.
Results
Patient characteristics
The clinicopathological characteristics of TNBCs are listed in Table 2. The median age of patients was 49 years old (range, 28–76) and the median tumor size was 22 mm (range, 5–100 mm). The majority of TNBCs (84/105, 78.5%) were invasive ductal carcinomas with nuclear grade 2–3 and histologic grade II-III. Of 105 TNBCs, 33 (31.4%) carried mutations in exon 5–9 of the TP53. Sixteen (48.5%) carried a missense mutation, and 17 (51.5%) harbored a frameshift, nonsense, or splicing mutation.
Table 2. Clinicopathological Characteristics of Triple Negative Breast Cancers (n = 105).
| Numbers | Percentage (%) | ||
| Age (years) | |||
| Median/Range | 49/28–76 | ||
| Tumor size | |||
| Median/Range | 22 mm/5–100 mm | ||
| <20 mm/≥20 mm | 41/64 | 39.0/61.0 | |
| Histolopathological features | |||
| Invasive ductal carcinoma | 88 | 83.8 | |
| Medullary carcinoma | 5 | 4.8 | |
| Invasive lobular carcinoma | 2 | 1.9 | |
| Metaplastic carcinoma | 2 | 1.9 | |
| Specialized typesa | 8 | 7.6 | |
| T stage | |||
| 1/2/3/4/ | 43/57/2/3/ | 41/54.3/1.9/2.9 | |
| N stage | |||
| 01/2/3 | 65/28/7/5 | 61.9/26.7/6.7/4.7 | |
| Nuclear gradeb | |||
| 1/2/3 | 2/28/70 | 1.9/26.7/66.7 | |
| Histologic gradec | |||
| I/II/III | 7/25/68 | 6.7/23.8/64.8 | |
| Ki67 labeling index | |||
| <5% | 17 | 16.2 | |
| 5% ∼49% | 67 | 63.8 | |
| ≥50% | 21 | 20 | |
| TP53 gene mutations | |||
| Missense DBM | 11 | 10.5 | |
| Missense non-DBM | 5 | 4.8 | |
| Frameshift | 8 | 7.6 | |
| Nonsense | 7 | 6.7 | |
| Splicing | 2 | 1.9 | |
Adenoid cystic carcinoma, ductal carcinoma in situ, invasive cribriform carcinoma, invasive micropapillary carcinoma, invasive tubular carcinoma, neuroendocrine carcinoma and mucinous carcinoma. b,c Five samples did not have a defined histologic grade and nuclear grade.
Mutation analysis of EGFR, KRAS, BRAF and TP53 genes
The mutation frequency of KRAS and EGFR gene were 1.9% (2/105) and 1.0% (1/105), respectively. One activating mutation (exon 19 del) was detected using PNA-mediated clamping PCR in a commonly deleted region (codons 746–753) of the EGFR gene. Two KRAS mutations (G12V and G12D) were identified using PNA-mediated clamping PCR and sequencing. There was no BRAFV600E mutation found (Table 3 & Table S1).
Table 3. Molecular Characteristics of Triple Negative Breast Cancers (n = 105).
| Number of cases (%) | EGFR copy number change | EGFR mutation | KRAS mutation | TP53 mutation |
| 61 (58.1%) | N | − | − | − |
| 25 (23.8%) | N | − | − | + |
| 2 (1.9%) | N | − | + a | + |
| 1 (1.0%) | N | + b | − | − |
| 6 (5.7%) | I | − | − | − |
| 5 (4.8%) | I | − | − | + |
| 2 (1.9%) | I† | − | − | − |
| 2 (1.9%) | D | − | − | − |
| 1 (1.0%) | D | − | − | + |
| 105 | 1 (1.0%) | 2 (1.9%) | 33 (31.4%) |
'−', wild type; '+', mutant type; 'D', decreased copy number (hemizygous deletion); ‘N’, normal copy number; 'I', increased EGFR copy number (single copy gain, amplification and high-copy number amplification); † single exon amplification; a p.Gly12Val and p.Gly12Asp; b Deletion on Exon 19. BRAF gene mutation was not detected in all cases
Frequencies and spectrum of EGFR gene copy number changes
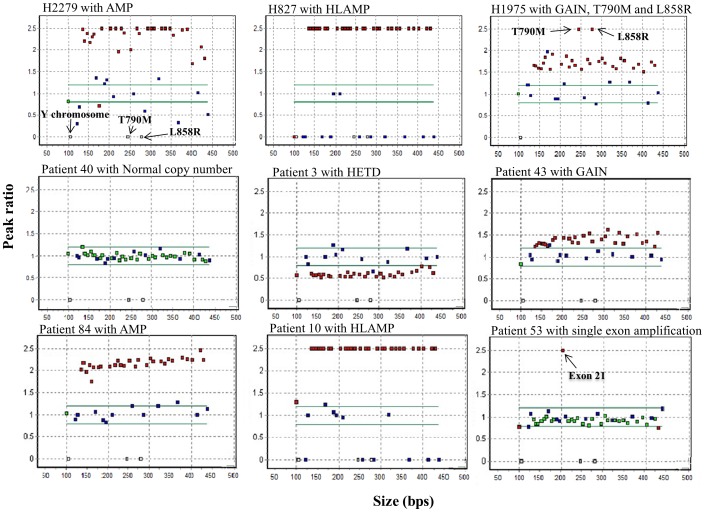
The frequency of EGFR copy number changes was noted in 15.2% of total 105 TNBCs. Among them, increased or decreased EGFR copy number was detected in 13 (12.4%) and 3 cases (2.9%), respectively. Of 13 cases, two cases (1.9%) showed an increased copy number of a single exon (one in exon 1 and the other in exon 21) and 11 cases (10.5%) showed increased copy number changes consists of five single copy gain (GAIN) (4.8%), two amplification (AMP) (1.9%) and four high-copy number amplification (HLAMP) cases (3.8%) (Figure 1 & Table 3).
Figure 1. Examples of representative scatter plot patterns of multiplex ligation-dependent probe amplification (MLPA) analysis for the EGFR gene.
Gain or loss (peak ratio cut off, <0.8 or >1.2) of exons in EGFR gene were represented by red squares. Green square indicated within neutral peak ratio of EGFR gene. Three white squares indicate specific probe for Y chromosome (105 bps), the L858R (281 bps) and the T790M mutations (246 bps). Blue squares represent control probes. ‘HETD’, hemizygous deletion; ‘GAIN’, copy gain; ‘AMP’, amplifications; ‘HLAMP’, high-copy amplifications.
Association between TP53 mutations and prognosis in 105 TNBC patients
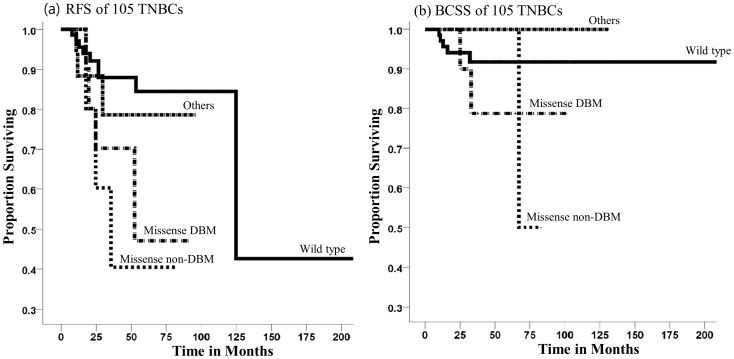
Of 105 TNBC patients with a median follow up period of 35 months, there were 20 (19.0%) cases of breast cancer relapse, and 8 cases (7.6%) were due to breast cancer-related death. Kaplan-Meier survival analysis of TNBC patients grouped according to the type of TP53 mutation showed that missense mutations in DBM (P = 0.036) and non-DBM (P = 0.011) were associated with a higher relapse rate compared with patients without the mutations, whereas other types of mutations including frameshift, nonsense, and splicing mutations (P = 0.462) was not significant. The relapse incidence per 100 persons at 25 months for non-mutations, DBM mutations and non-DBM missense mutation groups were 8, 30, 40, respectively (Figure 2A). The DBM mutations (P = 0.311), non-DBM missense mutation (P = 0.449) and other mutations (P = 0.274) were not significantly associated with BCSS compared with wild type (figure 2B). Prognostic factors associated with relapse free survival in the univariate analysis had higher T and N stage and TP53 mutation status. However, in the multivariate analysis, only N stage with grade 2 or 3 was associated with relapse rate (Table 4).
Figure 2. Kaplan-Meier relapse free survival (RFS) and breast cancer specific survival (BCSS) curves of patients with triple negative breast cancer.
(a) RFS and (b) BCSS curves stratified by non-mutations, missense mutations in the DBM, missense mutations outside the DBMs and others including nonsense, frameshift and splicing mutations in the TP53 gene.
Table 4. Univariate and Multivariate analysis of Relapse Free Survival in 105 TNBCs.
| Univariate | Multivariate | |||||
| HR | 95% CI of HR | P | HR | 95% CI of HR | P | |
| T status | ||||||
| T≤1 | 1 | 1 | ||||
| T≥3 | 6.118 | 1.381–27.109 | 0.017 | 3.648 | 0.789–16.868 | 0.098 |
| N status | ||||||
| N≤1 | 1 | 1 | ||||
| N≥2 | 4.16 | 1.481–11.687 | 0.007 | 2.945 | 1.010–8.588 | 0.048 |
| TP53 mutations | ||||||
| Wild type | 1 | 1 | ||||
| Mutations | 2.742 | 1.080–6.960 | 0.034 | 2.193 | 0.852–5.643 | 0.104 |
HR, hazard ratio.
Sensitivity analysis of EGFR MLPA
Detection of the EGFR amplifications in HCC827 cell line was possible in spiked samples containing 6.25% of HCC827 DNA (Table 5). All five replicates of spiked samples with 6.25% of HCC827 DNA showed EGFR amplifications.
Table 5. Analytical Sensitivity of MLPA for EGFR copy number changes in the HCC827 cell line.
| Concentrations of HCC827 (%)a | Number of replicates | MLPA |
| 100 | 3 | + b |
| 50 | 3 | + |
| 25 | 5 | + |
| 12.5 | 5 | + |
| 6.25 | 5 | + |
| 3.13 | 3 | – |
| Wild type DNA | 3 | – |
MLPA, multiplex ligation-dependent probe amplification. aDNA of the HCC827 was serially diluted with wild type DNA. b All spiked samples analyzed using MLPA three or five times, and all replicates with from 100% to 6.25% of HCC827 DNA showed EGFR amplifications.
Discussion
Recently, oncogenic mutations linked to the AKT and MEK/MAPK pathways have been evaluated as possible predictive markers associated with a good response to EGFR-targeted agents in TNBCs [15], [17], [18], [36]. Grob et. al., have reported that no mutation was found in EGFR, KRAS, and BRAF among 63 TNBCs, and only one EGFR amplification case was detected [15], consistent with other results in European patients [16], [17]. However, mutations and copy number changes of the EGFR gene were more frequently observed in Asian patients with a detection rate up to 11.4% [18] and 21% [19], respectively. Out of all 105 TNBCs, 10.5% showed increased EGFR copy number changes including five GAIN, four HLAMP, two AMP, and one case harbored activating mutation (exon 19 del). These genetic aberrations were thought to be susceptible to either EGFR-TKIs or EGFR-blocking monoclonal antibody therapy in primary non–small cell lung cancer [9]–[12] or in recurrent glioblastoma [13]. The frequency of activating EGFR mutations in our study was lower than that of a recent Asian study among patients in Singapore (8/70). However, the frequencies of activating EGFR mutations and copy number changes in our study were comparable to that of a study including Caucasian patients [20], which reported four HLAMP cases (6.2% 4/65) and one EGFR gene mutation case (1.5%, 1/65).
In a total of 105 TNBCs, two KRAS point mutations and three HETD cases were detected (4.8%, 5/105). Mutations of the KRAS gene are known to be negative predictive genetic markers for EGFR targeted therapy and were significantly associated with resistance to EGFR-blocking monoclonal antibody (cetuximab) in colon cancer [27], [37]. In our study, three HETD cases were detected, but a partial deletion variant of EGFR gene, EGFRvIII (in-frame deletion of exon 2–7) reported frequently in glioblastoma [13], [38] and breast cancer [39], was not observed. Two cases showed only single exon copy number amplifications. Tang et al. showed that EGFRvIII enhanced the tumorigenic potential of breast cancer cells in mice [40] and Sok et al. also showed that xenografts expressing EGFRvIII grew more rapidly than tumors derived from vector-transfected control cells [38]. However, the functions of the whole gene deletion of EGFR or the single exon copy number amplifications in tumor cells were not evaluated. Therefore, further investigation is needed to characterize the functional and clinical significance of the whole gene deletion of EGFR and the single exon copy number changes.
TP53 mutations were more prevalently detected in tumors with high grade, large size, and node-positivity. The relative risk of deaths with TP53 mutations was 2 to 3 times higher than those without TP53 mutations, especially for the group with missense mutation in DBM was associated with shortest survival [23]. In our study, patients with missense mutations showed reduction in relapse free survival, but the effect of the missense mutation in the DBM was not stronger than the missense mutation outside the DBM on breast cancer relapse.
Among 105 TNBCs, 5 of 12 tumors with increased EGFR copy number or EGFR mutation also had TP53 mutations (Table 3 & Table S1). Several studies have reported that amplification of EGFR and inactivation of TP53 are associated with sensitivity to anti-EGFR monoclonal antibodies in metastatic colorectal cancer [41], [42]. However, in TNBCs, the TP53 mutation has been limited as one prognostic marker that can identify tumors with more aggressive behavior [43], [44]. Therefore, it is necessary to further evaluate the correlation between TP53 mutation and sensitivity of anti-EGFR therapy in TNBCs with EGFR amplification or EGFR mutation.
The frequency of TNBCs varies by race. In African Americans, up to 55% of breast cancers were TNBCs, which was 2–3 times more frequent than in other racial groups [2], [5]. Among Asian populations, TNBCs account for 8.0 to 24.1% of all breast cancers (24.1% in Korean patients; 18.6% in Chinese patients; 8% in Japanese patients) [3], [4], [6]. Copy number changes of the EGFR gene were detected in 6.2% (in Western patients) to 21% (in Asian patients) of TNBCs [19], [20].
Several randomized clinical studies of anti-EGFR therapy reported that fewer than 20% of metastatic TNBCs showed response to anti-EGFR therapy [45], [46] and the use of these therapies for targeted breast cancer treatment has been controversial. However, these studies had not clearly validated the genetic predictive markers that can modulate sensitivity or resistance to anti-EGFR therapy as described in other cancers [9]–[13], [47].
According to previous studies, TKIs may be more promising in specific subsets of patients with activating EGFR mutations, whereas EGFR-blocking monoclonal antibodies may be more effective in TNBC cases where the whole EGFR gene copy number is increased [47], [48]. Therefore, analysis of EGFR mutations and copy number changes could offer new therapeutic options for TNBC patients. Moreover, analysis of EGFR mutations and copy number changes may be more beneficial in the racial groups with high frequencies of TNBCs and with EGFR gene aberrations.
This study provides the spectrum of copy number changes and mutation statuses of the EGFR gene in Korean TNBC patients. Future studies should include evaluation of clinical outcomes of TNBC patients who undergo anti-EGFR therapy according to the genetic status of EGFR.
Supporting Information
Forty-four TNBCs with Abnormal Status of EGFR, KRAS or/and TP53.
(DOCX)
Acknowledgments
We gratefully acknowledge Dr. Mitsuo Sato (Nagoya University Graduate School of Medicine, Japan) and Dr. Sun-Young Kong (National Cancer Center, Korea) for the supply of adenocarcinoma line HCC827, H1975 and H2279.
Funding Statement
This study was supported by a faculty research grant from Yonsei University College of Medicine for 2012. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Reis-Filho JS, Tutt AN (2008) Triple negative tumours: a critical review. Histopathology 52: 108–118. [DOI] [PubMed] [Google Scholar]
- 2. Huo D, Ikpatt F, Khramtsov A, Dangou JM, Nanda R, et al. (2009) Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer. J Clin Oncol 27: 4515–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim MJ, Ro JY, Ahn SH, Kim HH, Kim SB, et al. (2006) Clinicopathologic significance of the basal-like subtype of breast cancer: a comparison with hormone receptor and Her2/neu-overexpressing phenotypes. Hum Pathol 37: 1217–1226. [DOI] [PubMed] [Google Scholar]
- 4. Kurebayashi J, Moriya T, Ishida T, Hirakawa H, Kurosumi M, et al. (2007) The prevalence of intrinsic subtypes and prognosis in breast cancer patients of different races. Breast 16 Suppl 2S72–77. [DOI] [PubMed] [Google Scholar]
- 5. Stead LA, Lash TL, Sobieraj JE, Chi DD, Westrup JL, et al. (2009) Triple-negative breast cancers are increased in black women regardless of age or body mass index. Breast Cancer Res 11: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yin WJ, Lu JS, Di GH, Lin YP, Zhou LH, et al. (2009) Clinicopathological features of the triple-negative tumors in Chinese breast cancer patients. Breast Cancer Res Treat 115: 325–333. [DOI] [PubMed] [Google Scholar]
- 7. Carey L, Winer E, Viale G, Cameron D, Gianni L (2010) Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 7: 683–692. [DOI] [PubMed] [Google Scholar]
- 8. Hudis CA, Gianni L (2011) Triple-negative breast cancer: an unmet medical need. Oncologist 16 Suppl 11–11. [DOI] [PubMed] [Google Scholar]
- 9. Fukui T, Mitsudomi T (2008) Mutations in the epidermal growth factor receptor gene and effects of EGFR-tyrosine kinase inhibitors on lung cancers. Gen Thorac Cardiovasc Surg 56: 97–103. [DOI] [PubMed] [Google Scholar]
- 10. Sequist LV, Bell DW, Lynch TJ, Haber DA (2007) Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol 25: 587–595. [DOI] [PubMed] [Google Scholar]
- 11. Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, et al. (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 12. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 13.Lv S, Teugels E, Sadones J, De Brakeleer S, Duerinck J, et al.. (2012) Correlation of EGFR, IDH1 and PTEN status with the outcome of patients with recurrent glioblastoma treated in a phase II clinical trial with the EGFR-blocking monoclonal antibody cetuximab. Int J Oncol. [DOI] [PubMed]
- 14. Araki T, Shimizu K, Nakamura T, Baba M, Kawai Y, et al. (2011) Clinical screening assay for EGFR exon 19 mutations using PNA-clamp smart amplification process version 2 in lung adenocarcinoma. Oncol Rep 26: 1213–1219. [DOI] [PubMed] [Google Scholar]
- 15. Grob TJ, Heilenkotter U, Geist S, Paluchowski P, Wilke C, et al. (2012) Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancer. Breast Cancer Res Treat 134: 561–567. [DOI] [PubMed] [Google Scholar]
- 16. Sanchez-Munoz A, Gallego E, de Luque V, Perez-Rivas LG, Vicioso L, et al. (2010) Lack of evidence for KRAS oncogenic mutations in triple-negative breast cancer. BMC Cancer 10: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacot W, Lopez-Crapez E, Thezenas S, Senal R, Fina F, et al. (2011) Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res 13: R133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teng YH, Tan WJ, Thike AA, Cheok PY, Tse GM, et al. (2011) Mutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapy. Breast Cancer Res 13: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toyama T, Yamashita H, Kondo N, Okuda K, Takahashi S, et al. (2008) Frequently increased epidermal growth factor receptor (EGFR) copy numbers and decreased BRCA1 mRNA expression in Japanese triple-negative breast cancers. BMC Cancer 8: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shah SP, Roth A, Goya R, Oloumi A, Ha G, et al. (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harvey JM, Clark GM, Osborne CK, Allred DC (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 17: 1474–1481. [DOI] [PubMed] [Google Scholar]
- 22. Mitsudomi T, Yatabe Y (2010) Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 277: 301–308. [DOI] [PubMed] [Google Scholar]
- 23. Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, et al. (2006) The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12: 1157–1167. [DOI] [PubMed] [Google Scholar]
- 24. Olivier M, Hainaut P (2001) TP53 mutation patterns in breast cancers: searching for clues of environmental carcinogenesis. Semin Cancer Biol 11: 353–360. [DOI] [PubMed] [Google Scholar]
- 25. Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, et al. (2009) Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol 6: 519–527. [DOI] [PubMed] [Google Scholar]
- 26. Langer CJ (2011) Roles of EGFR and KRAS Mutations in the Treatment Of Patients With Non-Small-Cell Lung Cancer. P T 36: 263–279. [PMC free article] [PubMed] [Google Scholar]
- 27. Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, et al. (2007) Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer 96: 1166–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwon MJ, Lee SE, Kang SY, Choi YL (2011) Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: Comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract 207: 762–768. [DOI] [PubMed] [Google Scholar]
- 29. Han HS, Lim SN, An JY, Lee KM, Choe KH, et al. (2012) Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol 7: 355–364. [DOI] [PubMed] [Google Scholar]
- 30. Wang J, Ramakrishnan R, Tang Z, Fan W, Kluge A, et al. (2010) Quantifying EGFR alterations in the lung cancer genome with nanofluidic digital PCR arrays. Clin Chem 56: 623–632. [DOI] [PubMed] [Google Scholar]
- 31. Helfrich BA, Raben D, Varella-Garcia M, Gustafson D, Chan DC, et al. (2006) Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res 12: 7117–7125. [DOI] [PubMed] [Google Scholar]
- 32.Vargas AC, Reed AE, Waddell N, Lane A, Reid LE, et al.. (2012) Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Res Treat. [DOI] [PubMed]
- 33. Jeuken J, Cornelissen S, Boots-Sprenger S, Gijsen S, Wesseling P (2006) Multiplex ligation-dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J Mol Diagn 8: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jeuken J, Sijben A, Alenda C, Rijntjes J, Dekkers M, et al. (2009) Robust detection of EGFR copy number changes and EGFR variant III: technical aspects and relevance for glioma diagnostics. Brain Pathol 19: 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cho Y, Gorina S, Jeffrey PD, Pavletich NP (1994) Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265: 346–355. [DOI] [PubMed] [Google Scholar]
- 36.Nakajima H, Ishikawa Y, Furuya M, Sano T, Ohno Y, et al.. (2012) Protein expression, gene amplification, and mutational analysis of EGFR in triple-negative breast cancer. Breast Cancer. [DOI] [PubMed]
- 37. Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, et al. (2006) KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 66: 3992–3995. [DOI] [PubMed] [Google Scholar]
- 38. Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, et al. (2006) Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res 12: 5064–5073. [DOI] [PubMed] [Google Scholar]
- 39. Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, et al. (1995) Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res 55: 5536–5539. [PubMed] [Google Scholar]
- 40. Tang CK, Gong XQ, Moscatello DK, Wong AJ, Lippman ME (2000) Epidermal growth factor receptor vIII enhances tumorigenicity in human breast cancer. Cancer Res 60: 3081–3087. [PubMed] [Google Scholar]
- 41. Di Fiore F, Sesboue R, Michel P, Sabourin JC, Frebourg T (2010) Molecular determinants of anti-EGFR sensitivity and resistance in metastatic colorectal cancer. Br J Cancer 103: 1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oden-Gangloff A, Di Fiore F, Bibeau F, Lamy A, Bougeard G, et al. (2009) TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br J Cancer 100: 1330–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Powell B, Soong R, Iacopetta B, Seshadri R, Smith DR (2000) Prognostic significance of mutations to different structural and functional regions of the p53 gene in breast cancer. Clin Cancer Res 6: 443–451. [PubMed] [Google Scholar]
- 44. Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, et al. (2007) Prognostic markers in triple-negative breast cancer. Cancer 109: 25–32. [DOI] [PubMed] [Google Scholar]
- 45. Dickler MN, Cobleigh MA, Miller KD, Klein PM, Winer EP (2009) Efficacy and safety of erlotinib in patients with locally advanced or metastatic breast cancer. Breast Cancer Res Treat 115: 115–121. [DOI] [PubMed] [Google Scholar]
- 46.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, et al.. (2012) TBCRC 001: Randomized Phase II Study of Cetuximab in Combination With Carboplatin in Stage IV Triple-Negative Breast Cancer. J Clin Oncol. [DOI] [PMC free article] [PubMed]
- 47. Brand TM, Iida M, Wheeler DL (2011) Molecular mechanisms of resistance to the EGFR monoclonal antibody cetuximab. Cancer Biol Ther 11: 777–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moran T, Sequist LV (2012) Timing of Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Therapy in Patients With Lung Cancer With EGFR Mutations. J Clin Oncol. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forty-four TNBCs with Abnormal Status of EGFR, KRAS or/and TP53.
(DOCX)