Four proteins in Arabidopsis are essential for plant development and play overlapping but different roles in acetylation of cell wall polysaccharides.
Abstract
The Reduced Wall Acetylation (RWA) proteins are involved in cell wall acetylation in plants. Previously, we described a single mutant, rwa2, which has about 20% lower level of O-acetylation in leaf cell walls and no obvious growth or developmental phenotype. In this study, we generated double, triple, and quadruple loss-of-function mutants of all four members of the RWA family in Arabidopsis (Arabidopsis thaliana). In contrast to rwa2, the triple and quadruple rwa mutants display severe growth phenotypes revealing the importance of wall acetylation for plant growth and development. The quadruple rwa mutant can be completely complemented with the RWA2 protein expressed under 35S promoter, indicating the functional redundancy of the RWA proteins. Nevertheless, the degree of acetylation of xylan, (gluco)mannan, and xyloglucan as well as overall cell wall acetylation is affected differently in different combinations of triple mutants, suggesting their diversity in substrate preference. The overall degree of wall acetylation in the rwa quadruple mutant was reduced by 63% compared with the wild type, and histochemical analysis of the rwa quadruple mutant stem indicates defects in cell differentiation of cell types with secondary cell walls.
Plant cell walls are multifunctional viscoelastic networks mainly composed of polysaccharides. Many of these polysaccharides, including xylans, (gluco)mannans, xyloglucans (XyGs), and pectins, have various degrees and patterns of acetyl esterification (Gille and Pauly, 2012; Pawar et al., 2013). The biological role of cell wall acetylation is not well understood, but it is believed to be important for pathogen resistance and plant development, and the acetylation of pectin also impacts upon the mechanical properties of cell walls (Manabe et al., 2011; Orfila et al., 2012; Pogorelko et al., 2013). In vitro, acetyl groups influence susceptibility to enzymatic degradation of pectin and xylan (Selig et al., 2009; Chen et al., 2012; Gou et al., 2012; Orfila et al., 2012; Pogorelko et al., 2013), and therefore acetylation may constitute a barrier to cell wall deconstruction. Alkali treatment of wall materials, which hydrolyzes the ester bonds, is broadly used to make polysaccharides more extractable. The treatment does not only facilitate the degradation of xylan and pectins, but also improves the deconstruction of cellulose, as the depolymerization of noncellulosic polymers results in a better accessibility to cellulose by degrading enzymes (Selig et al., 2009). Low levels of acetylated polysaccharides in plant feedstocks would be desirable for downstream processing in biorefineries, firstly, because the cell wall material of plant feedstocks with low level of acetylation is expected to be more easily extracted and, secondly, because less acetate, which is highly toxic to microorganisms such as yeast (Saccharomyces cerevisiae), would be released during extraction (Manabe et al., 2011; Gille and Pauly, 2012; Pawar et al., 2013). However, although reducing the O-acetylation level of xylan by approximately 60%, as observed in the walls of the Arabidopsis (Arabidopsis thaliana) eskimo1 mutant, enhances enzymatic degradation of isolated xylan (Yuan et al., 2013), enzymatic hydrolysis yields of whole wall materials have been reported to actually be decreased (Xiong et al., 2013). This presumably results from a tighter association between these now lowly substituted xylan polymers and cellulose (Xiong et al., 2013).
Recently, we reported REDUCED WALL ACETYLATION2 (RWA2), the first protein to be involved in cell wall acetylation in planta (Manabe et al., 2011). RWA2 is a member of a small family consisting of four proteins in Arabidopsis, and its loss-of-function mutants display 20% reduction of acetylation in a range of polysaccharides that include XyG and pectins. We have hypothesized, based on phylogenetic analysis, expression pattern, moderate reduction in acetylation, and the absence of morphological phenotype, that RWA proteins have redundant functions in a biochemical reaction that occurs prior to the actual acetylation of specific polysaccharides. Independently to our research, a quadruple mutant of RWA has been reported to display reduction in xylan acetylation, secondary cell wall thickness, and mechanical strength of the stem (Lee et al., 2011). Meanwhile, Gille et al. (2011) have discovered a new family of proteins involved in the acetylation of specific polysaccharides: the plant-specific DOMAIN OF UNKNOWN FUNCTION (DUF) 231 family (also known as TRICHOME BIREFRINGENCE-LIKE [TBL] family). The loss-of-function mutants altered xyloglucan4 (axy4)/tbl27 and axy4L/tbl22 lack O-acetylation specifically of XyG in certain tissues, while eskimo1/tbl29 mutants contain reduced O-acetylation of xylan (Xiong et al., 2013; Yuan et al., 2013). The TBL/DUF231 family proteins and the RWA proteins have sequence similarity to the N-terminal and C-terminal regions of the fungal protein Cas1p, respectively (Anantharaman and Aravind, 2010). This could suggest that the TBL and RWA proteins function in protein complexes where the determinants of substrate specificity reside in the TBL partner (Manabe et al., 2011). However, because there are many more TBL proteins than RWA proteins (e.g. 46 TBL proteins versus four RWA proteins in the genome of Arabidopsis), it is likely that they do not form discrete and invariable complexes. Crossing of rwa2-3 and a leaky allele of axy4, axy4-1, resulted in a double mutant with partially additive phenotype (Gille et al., 2011). Its XyG acetylation is lower compared with either single mutant. From this analysis, RWA2 and AXY4 have been hypothesized to work in synergy, although the function of RWA2 might be substituted by other RWAs (Gille et al., 2011). Here, we have generated all the combinations of double, triple, and quadruple mutants of all four members of RWA family to further investigate the functional diversity and redundancy and to explore the function of cell wall acetylation and the role of RWAs in the network of acetylation-related enzymes. The triple and quadruple mutants we have obtained displayed severe and distinct phenotypes such as extreme dwarfism. This contrasts with the very mild phenotypes reported by Lee et al. (2011). Taken together, RWAs have partially redundant functions in the process of cell wall acetylation and show distinct impacts upon different cell wall polysaccharides.
RESULTS
Generation of Double, Triple, and Quadruple Mutants
Our previous study indicated redundant functions among four RWA genes based on their overlapping expression patterns and the fact that rwa single mutants displayed no or only modest cell wall phenotypes (Manabe et al., 2011). Therefore, we generated all combinations of double, triple, and quadruple rwa mutants to further dissect the role of RWA proteins in planta.
We found that all four triple mutants and the quadruple mutant were severely dwarfed (Figs. 1 and 2), whereas only one double mutant, rwa2 rwa4, displayed a moderate dwarf phenotype (Fig. 3; Supplemental Fig. S1). In terms of dwarfism, rwa2 rwa3 rwa4 displayed the most severe phenotype among triple mutants, followed by rwa1 rwa3 rwa4 and rwa1 rwa2 rwa4, respectively, under long-day growth condition. rwa1 rwa2 rwa3 mutant exhibited the least severe growth phenotype and was comparable with the rwa2 rwa4 double mutant. The triple mutants also displayed distinctive morphological characteristics. rwa2 rwa3 rwa4 was bushy with smaller and yellowish leaves. rwa1 rwa3 rwa4 had greener leaves compared with the wild type and looked somewhat similar in appearance to typical irregular xylem mutants.
Figure 1.
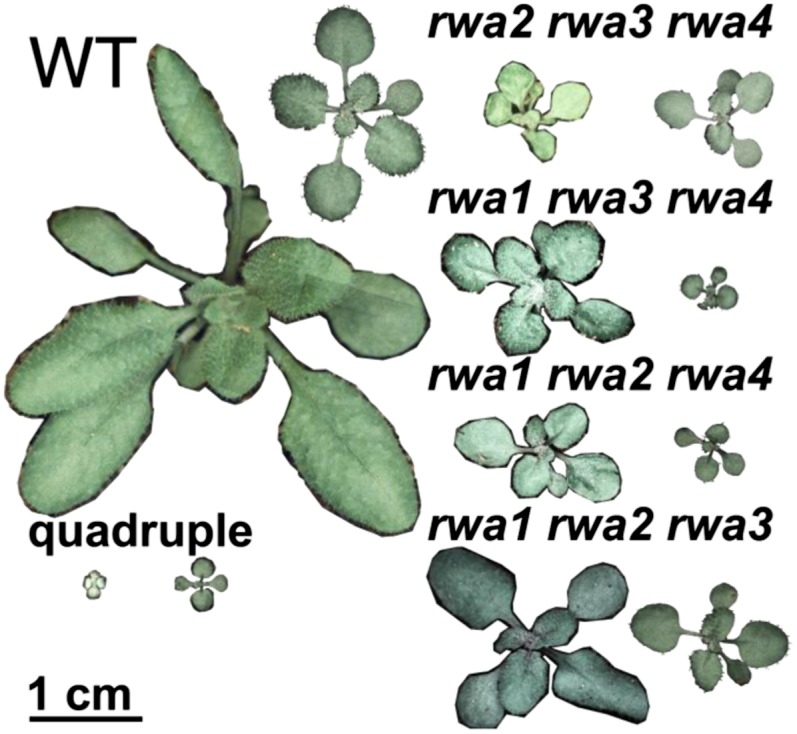
Growth phenotype of 18-d-old rwa triple and quadruple mutants grown under a regime of 12-h (right) and 16-h (left) photoperiod. The wild type and three of the triple mutants grew to larger size in long day. However, the rwa2 rwa3 rwa4 mutant and the rwa1 rwa2 rwa3 rwa4 (quadruple) mutants were light stressed and showed more extreme dwarfism under long-day than short-day conditions.
Figure 2.
The rwa quadruple mutant presents extreme dwarfism and improper flowering. A, Mature plants of the wild type (left) and the quadruple mutant (right; B). C, E, and F, The wild type. D and G, rwa1 rwa2 rwa3 rwa4. C to E, Unopened buds. F and G, Flowers. E, Dissection of the wild-type bud shown in C to show that, unlike in the quadruple mutant, pollen grains were already dehisced at this stage of flower development.
Figure 3.
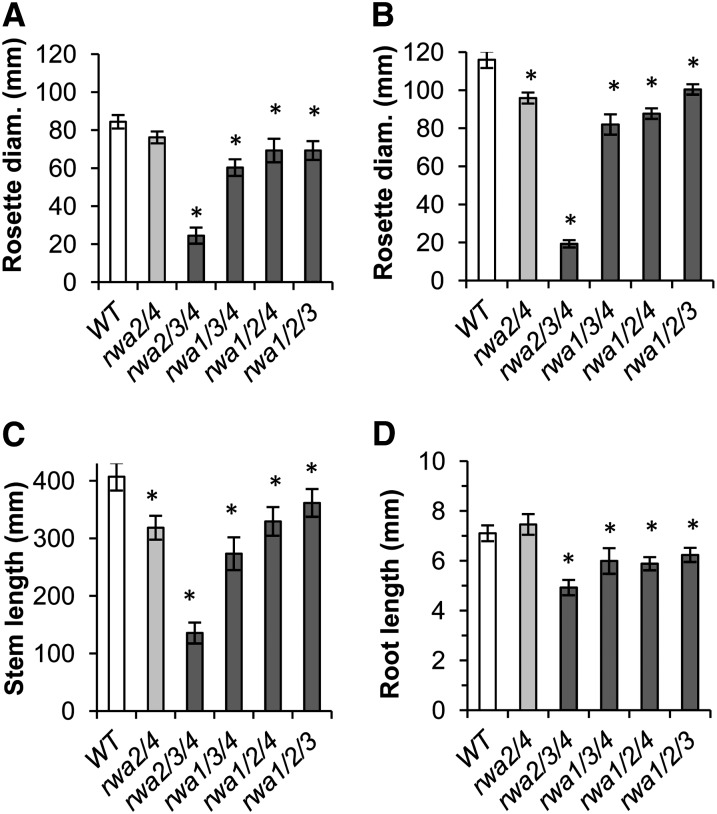
rwa mutants exhibited shoot and root growth deficiency. Shoot growth was measured for 6-week-old triple mutants along with the wild type and rwa2 rwa4 double mutant, the only double mutant with reduced growth. A and B, Diameter of leaf rosette. C, Length of inflorescence stem. A, Short day. B and C, Long day. D, The root growth of the same genotype was measured between 2 and 7 d after germination (n = 10–25). Asterisks indicate significant difference from the wild type (Student’s t test, P < 0.05). Error bars indicate se (n = 4–19).
In addition to its severe dwarf phenotype, the quadruple mutant also displayed a striking floral phenotype (Fig. 2). The wild-type stigma is enclosed in the bud before the opening of the flower, and the pollen is already dehisced at that time (Fig. 2, C and E; Alvarez-Buylla et al., 2010). In the quadruple mutant, the stigma was protruding from the bud before opening of the flower (Alvarez-Buylla et al., 2010; Fig. 2D). Moreover, the pollen did not dehisce immediately after the opening of the flower (Fig. 2G). Although the quadruple mutant can complete its life cycle and produce viable seeds, the progeny of the quadruple mutant was most often the result of outcrossing.
As only few of the quadruple mutant plants survived to maturity, we have maintained the quadruple mutant as rwa1 rwa2 rwa3 triple mutant with heterozygous rwa4 mutation. The phenotype of rwa1 rwa2 rwa3 RWA4/rwa4 plants was similar to rwa1 rwa2 rwa3 triple mutants, which are just slightly dwarfed compared with the wild type. No significant degree of outcrossing was observed in these mutants.
The Dwarf Phenotype of rwaTriple and Quadruple Mutants Is Light Period Dependent
The growth of 6-week-old triple mutants and rwa2 rwa4 double mutant was quantified as the diameter of rosettes under a regime of 12 h (short day) and 16 h (long day) of photoperiod (Fig. 3, A and B; Supplemental Fig. S1) and as the height of primary inflorescence stem under long-day condition (Fig. 3C). With the exception of rwa2 rwa3 rwa4, the 6-week-old triple mutants (Fig. 3; Supplemental Fig. S1) showed a less pronounced growth phenotype than at the earlier 18-d-old stage (Fig. 1), but all the triple mutants were still significantly smaller. As expected from the extra light exposure, the wild type and most of the triple mutants exhibited wider rosette diameters when growing under long-day condition than under short-day condition. For example, the diameter of the wild-type rosettes grown under long-day condition was, in average, 137% of that under the short-day condition. By contrast, rwa2 rwa3 rwa4 and the quadruple mutant had smaller rosettes in long-day than in short-day condition (Figs. 1 and 3). The rosette diameter of long-day-grown rwa2 rwa3 rwa4 triple mutant at 6 weeks was, for example, only 79% of its short-day-grown counterpart. Both rwa2 rwa3 rwa4 and the quadruple mutant grown under long-day condition were chlorotic, unlike the same genotypes grown under short-day condition, suggesting that they were light stressed under long-day condition (Fig. 1). As observed for the rosette diameter, rwa2 rwa3 rwa4 showed the most drastic reduction of stem height among the rwa triple mutants, whereas the other mutants presented a significant stem reduction compared with the wild type that was similar to that of the rwa2 rwa4 double mutant (Fig. 3C).
Growth deficiency was also observed in the root system of rwa triple mutants. To prevent inaccuracy due to variability in seed germination, root growth was measured as the expansion of root length between the second and the seventh day post germination. Root growth of the double mutant did not significantly differ from that of the wild type (Fig. 3D). On the contrary, all rwa triple mutants showed a significant reduction in root length, with rwa2 rwa3 rwa4 being once again the most affected mutant. Together, these results demonstrate that RWAs are important determinants of plant development at the whole plant level.
Cell Differentiation Is Disrupted in the rwa Quadruple Mutant
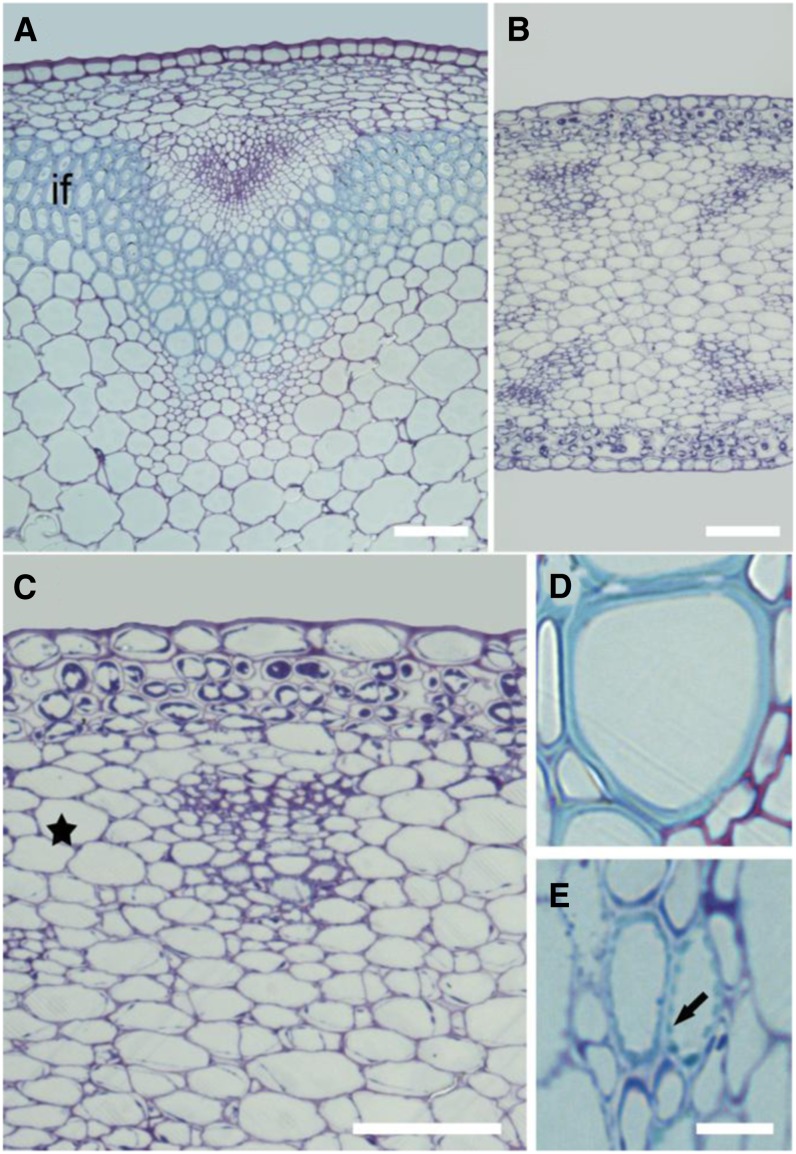
Transverse sections of mature stems stained with toluidine blue revealed improper cell differentiation in the quadruple mutant. By comparison with the wild type, interfascicular fibers and xylem cells, both cell types with secondary cell walls, were respectively absent and aberrant in the quadruple mutant (Fig. 4). In the wild type, the xylem cells were entirely surrounded by a secondary cell wall (Fig. 4D). Instead of fully developed xylem cells, the quadruple mutant contained cells with dots of secondary cell wall thickening in its xylem region (Fig. 4E), indicating that the secondary cell wall deposition in these cells cannot be initiated normally. Similar patterns of cell wall thickening can be observed in longitudinal sections of wild-type protoxylem cells. We hypothesize that the secondary cell wall machinery is misoriented in the quadruple mutant. Unfortunately, we have not been able to confirm this, as the viability of the mutant and the presence of xylem cells were extremely limited, making it difficult to visualize these cells in longitudinal sections. In addition to the absence of interfascicular fibers, the reduced stem diameter of the quadruple mutant (about 40% of the wild-type stem diameter) also resulted from a reduced radial expansion of the cells. For example, the average diameter of parenchyma cells in the quadruple mutant was 43 μm compared with 102 μm in the wild type. It is also to note that the cortical parenchyma cells in rwa1 rwa2 rwa3 rwa4 contained an abundant amount of unknown cytoplasmic corpuscles that might reflect growth stress.
Figure 4.
The quadruple rwa mutant shows an absence of interfascicular fibers and has abnormal xylem cells. Representative transverse stem sections of the mature wild type and quadruple rwa mutant stained with toluidine blue. A and D, Wild type. B, C, and E, rwa1 rwa2 rwa3 rwa4. C, A magnified micrograph of B that highlights a vascular bundle. The interfascicular fibers (marked by “if” in the wild type; A) is absent in the quadruple mutant (equivalent region to “if” is indicated by a star in C). D and E, Xylem cells located in the most inner region of xylem. The arrow in E highlights a secondary cell wall thickening. Bars = 50 µm (A–C) and 10 µm (D and E).
Degree of Acetylation in the Quadruple Mutant Is Reduced to One-Third Compared with the Wild Type
Given the dramatic growth reduction in the triple and quadruple mutants, it was of interest to determine how much the acetylation level was affected. The overall degree of wall O-acetylation was measured as acetic acid released upon saponification of destarched alcohol insoluble residue (AIR). We selected green inflorescence stems from 6-week-old plants to prepare AIR from double and triple mutants. However, leaves from 6-week-old plants were used to analyze the quadruple mutant, as it was the only material that could be collected in sufficient amounts for cell wall analysis.
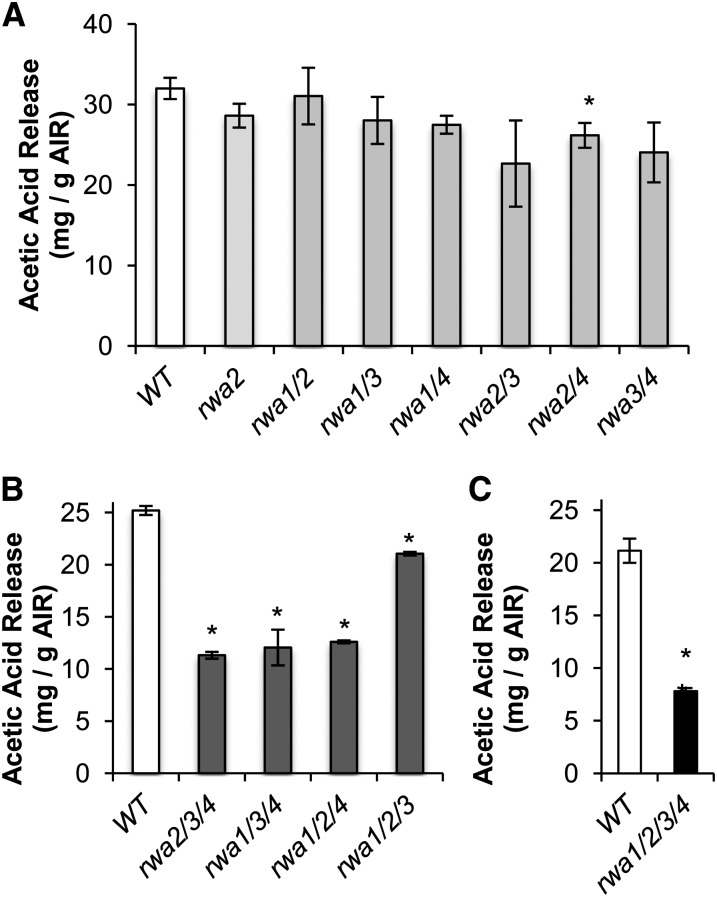
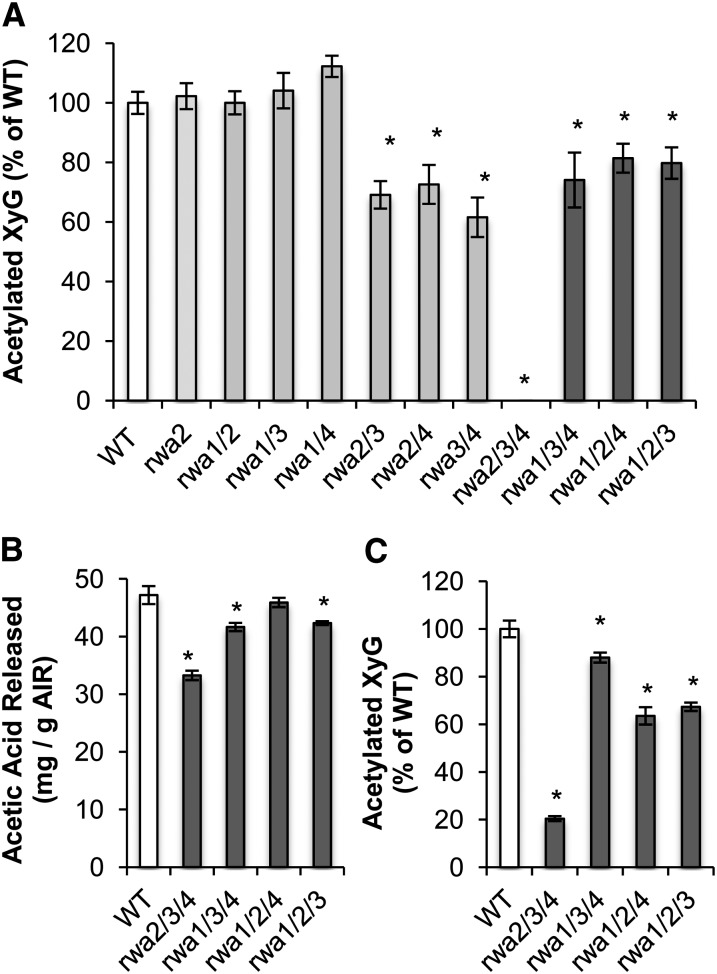
Among the double mutants, only rwa2 rwa4 had a significantly lower amount of acetylation compared with the wild type (Fig. 5A). The triple and quadruple mutants showed a highly significant reduction of cell wall acetylation (Fig. 5, B and C). Among the triple mutants, rwa1 rwa2 rwa3 had the smallest decrease in acetylation, consistent with the mild phenotype of this mutant. The largest reduction in acetylation level occurred in the quadruple mutant, where the acetic acid released upon saponification was only 37% of the wild-type level (Fig. 5C). These results indicate that the severe morphological changes observed for triple and quadruple mutants are associated with reduction in wall O-acetylation. However, rwa2 rwa3 rwa4, rwa1 rwa3 rwa4, and rwa1 rwa2 rwa4 showed almost the same reduction in total acetylation at this growth stage (Fig. 5C) but very different growth phenotypes, with rwa2 rwa3 rwa4 much more dwarfed and stressed than the other two genotypes (Fig. 3; Supplemental Fig. S1). Hence, the growth phenotype does not show a simple correlation with overall acetylation.
Figure 5.
Cell wall acetylation is depleted in double, triple, and quadruple rwa mutants. The amount of released acetic acid upon saponification of AIR was measured. A, AIR prepared from green stems of 6-week-old double mutants (n = 3). B, AIR prepared from green stems of 6-week-old triple mutants (n = 2; statistic analysis performed assuming same sd as for the wild type in A). C, AIR prepared from 6-week-old rosette leaves of the wild type and quadruple mutant (n = 3). Asterisks above each bar indicate statistically significant difference compared with the wild type (Student’s t test, P < 0.05). Error bars indicate se.
The rwa1 rwa2 rwa3 rwa4 Quadruple Mutant Can Be Complemented with RWA2
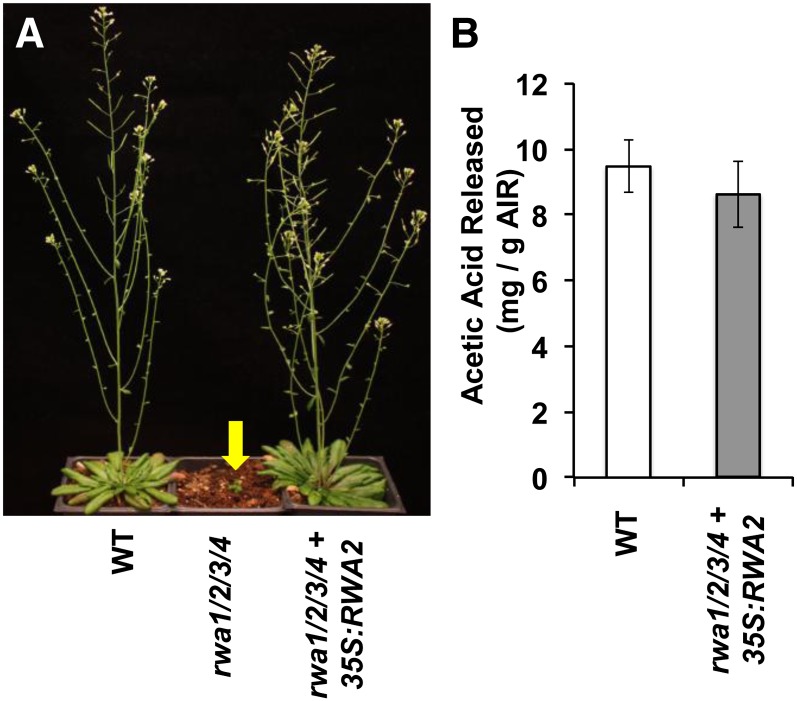
To bring additional evidence regarding the relationship between rwa genotype, reduced acetylation phenotype, and the morphological alteration observed in rwa1 rwa2 rwa3 rwa4, the quadruple mutant was complemented with a p35S:RWA2 construct (Fig. 6). Because the quadruple mutant is extremely dwarfed and produced only a few flowers, we transformed the rwa1 rwa2 rwa3 RWA4/rwa4 mutant. The recovered transformants were genotyped using PCR to find a transformant with homozygous insertions in all four genes. The morphological phenotype of the rwa quadruple mutant was completely reverted to the wild type, and the degree of acetylation was not significantly different from that of the wild type (Fig. 6).
Figure 6.
35S:RWA2 complements phenotype of quadruple mutant. A, Mature wild-type (WT) plants (left), quadruple mutant rwa1 rwa2 rwa3 rwa4 (center), and quadruple mutant transformed with 35S:RWA2 (right). B, The level of acetylation in 6-week-old rosette leaves from the wild type and quadruple mutant transformed with 35S:RWA2. Values are mean ± se (n = 4).
The Specificity of RWAs
An important question of RWA function is the functional diversity among the family members. We have hypothesized in our previous study that RWA proteins do not have a substrate preference because the decrease in degree of acetylation was similar for AIR, extracted pectin, residues after pectin extraction, and XyG in the rwa2 mutant (Manabe et al., 2011). However, because only rwa2, which displays moderate reduction of wall acetylation in leaves and hypocotyls, was available at that time, a definitive conclusion could not be reached. Therefore, we have addressed this question again with the combinations of multiple rwa mutants.
We first assessed the degree of acetylation of XyG, a major hemicellulose in the primary wall, but only contributing to 2.8% of the total wall acetylation level in l5-week-old leaf tissue (Gille et al., 2011). It is expected that in tissues with a higher proportion of secondary walls, such as Arabidopsis stems, XyG content will be lower, thus contributing even less to overall wall polymer acetylation. However, the method employed, oligosaccharide mass profiling (OLIMP; Lerouxel et al., 2002; Obel et al., 2009), is very sensitive, and even minute amounts of XyG can be assessed. Seedlings of double and triple mutants (Fig. 6) were treated with a XyG-specific endo-1,4-β-glucanase, and the solubilized XyG oligosaccharides (XyGOs) were analyzed by mass spectrometry. The degree of acetylation was measured as the ratio of O-acetylated XyGOs to total XyGOs. To our surprise, the rwa2 rwa3 rwa4 mutant etiolated seedlings had no detectable levels of O-acetylated XyGOs, whereas the other triple mutants only exhibited moderate reduction in O-acetylated XyGOs (Fig. 7A). We then did an OLIMP analysis on senesced dry stems from triple mutants and found that the rwa2 rwa3 rwa4 mutant had a much lower level of XyG acetylation than the other triple mutants (Fig. 7C). To allow direct comparison with the total level of acetylation, this was determined in the exact same senesced stem samples from the triple mutant collection (Fig. 7B). The decrease in acetylation in senesced stems was less pronounced than in green stems (Fig. 5B), especially for rwa1 rwa3 rwa4 and rwa1 rwa2 rwa4. The pattern of XyG acetylation was very different from that of overall acetylation. For example, the rwa2 rwa3 rwa4 triple mutant showed a much more moderate decrease in overall acetylation than in XyG acetylation, and the rwa1 rwa2 rwa4 mutant had no significant decrease in overall acetylation, while XyG acetylation was dramatically reduced. This result indicates that acetylation of XyG did not directly correlate with overall degree of acetylation. In other words, there seems to be a substrate preference of the RWA proteins in contrast to our initial hypothesis. Whole-wall two-dimensional NMR (2D-NMR) was employed to dissect the substrate specificity, as this method allows an estimation of xylan and glucomannan O-acetylation (Supplemental Fig. S2). In all triple mutants investigated, overall xylan O-acetylation was reduced to between 57% and 47%, while mannan O-acetylation was reduced to between 54% and 50%, again providing evidence that not all polymers are affected in their O-acetylation status in the same way.
Figure 7.
XyG acetylation is disproportionally decreased in the rwa2 rwa3 rwa4 mutant. XyG-specific acetylation was measured by OLIMP in etiolated seedlings (A) and senesced dry stem (C). Total acetylation level of the same senesced stem samples was determined to compare with XyG acetylation (B). Asterisks above each bar indicate statistically significant difference when compared with the wild type (Student’s t test, P < 0.05). Error bars indicate se (A, n = 10; B and C, n = 5).
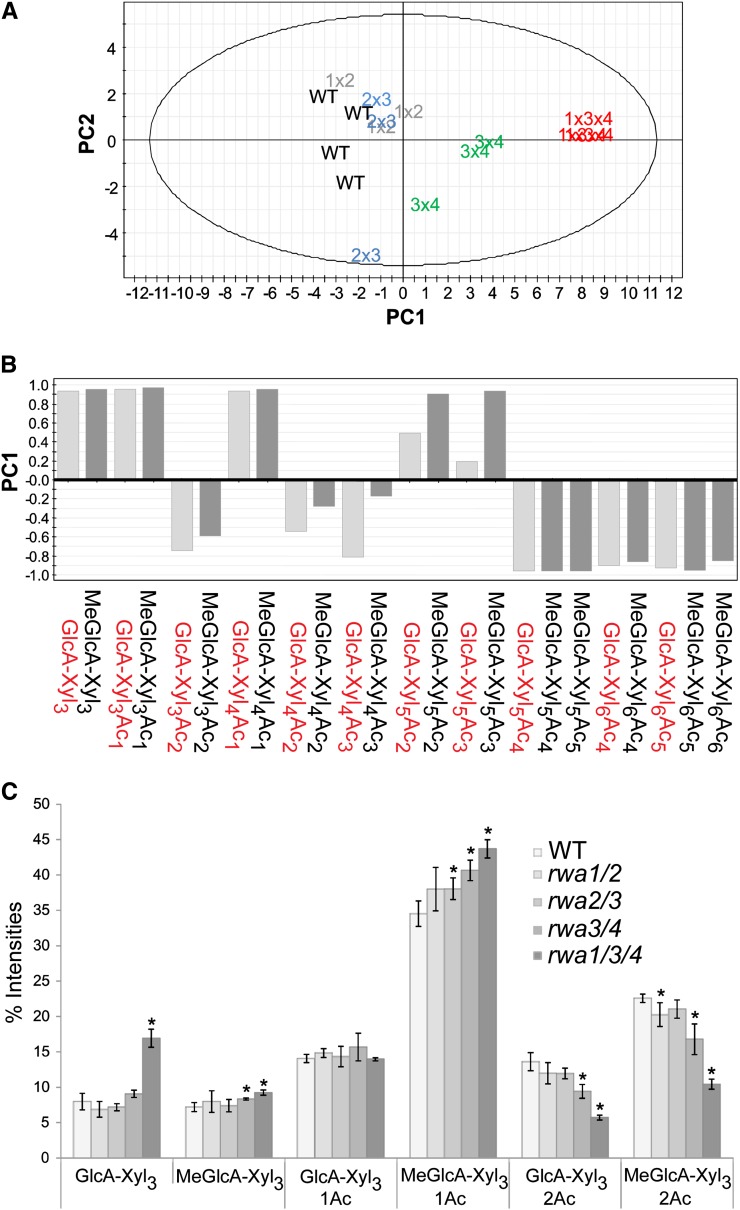
To further investigate the effect of RWA mutations on the pattern of xylan acetylation, we did an OLIMP analysis of xylan from stems from selected mutants. The rwa1 rwa2 double mutant was chosen for comparison because it had little overall change in acetylation level, while rwa2 rwa3 and rwa3 rwa4 were chosen as the most strongly affected double mutants and rwa1 rwa3 rwa4 as the mutant with the strongest effect on xylan acetylation (Fig. 4; Supplemental Fig. S2). The samples were treated with GH10 endo-1,4-β-xylanase to specifically release xylo-oligosaccharides (XOSs) for the mass spectrometry analysis (Chong et al., 2011). The percentage peak intensity was calculated by dividing intensity of a single acidic XOS peak over intensity of all acidic XOS peaks (Supplemental Fig. S3). The principal component analysis of all identified acidic XOSs in the mass spectrometry spectra separated rwa3 rwa4 and rwa1 rwa3 rwa4 from the other double mutants and the wild type in the scores scatterplot (Fig. 8A). The first component, which contributed to 67% of variance in the analysis, shows that acidic XOSs having three to five xylopyranosyl (Xylp) residues with lower acetylation level are relatively more abundant in rwa3 rwa4 and rwa1 rwa3 rwa4 (Fig. 8B), confirming that xylan acetylation in these mutants is reduced. Thus, the rwa2 rwa3 and rwa3 rwa4 double mutants differ in their pattern of xylan acetylation, even though they have the same decrease in XyG acetylation. Clearly, the RWA proteins have overlapping functions, in the sense that either one of the four proteins can support some level of acetylation of all polysaccharides in the wall. However, it is apparent that RWA2 is particularly important for XyG O-acetylation, while RWA3/RWA4 impact mainly xylan O-acetylation. RWA1 alone is relatively efficient in xylan acetylation in the rwa2 rwa3 rwa4 mutant, while the total acetylation level is more decreased in this mutant than in the other triple mutants.
Figure 8.
Acidic XOS liberated from Arabidopsis AIR samples by GH10 endoxylanase hydrolysis. A, Principle component analysis scores scatterplot showing the separation of the wild type, rwa1 rwa2, rwa2 rwa3, rwa3 rwa4, and rwa1 rwa3 rwa4. B, Loading plot for Principal Component 1 (PC1; 67% variance) showing mass peaks separating rwa3 rwa4 and rwa1 rwa3 rwa4 (positive) from the wild type and other double mutants (negative). C, Peak intensities for MeGlcA (1→2)-linked xylotriose with varying acetylation level. Error bars represent sd with three to four biological replicates. Asterisk indicates P value < 0.05 (Student’s t test). Xyl, (1→4)-Linked β-d-Xyl; Ac, acetyl residue. Numeric symbol indicates number of Xyl or acetyl residues. [See online article for color version of this figure.]
A more detailed comparison of the xylan OLIMP data on the acidic xylotrioses showed surprising changes in the pattern of XOSs produced (Fig. 8C). Due to reduced acetylation, less xylotriose with one GlcA and two acetyl substituents (GlcA-Xyl3-Ac2) and 4-O-methyl-α-d-GlcA (MeGlcA)-Xyl3Ac2 were present, especially in the rwa1 rwa3 rwa4 mutant. Respectively, the level of MeGlcA-Xyl3Ac was increased. Interestingly, the level of GlcA-Xyl3Ac was not similarly elevated. However, the level of nonacetylated GlcA-Xyl3 was substantially increased in the rwa1 rwa3 rwa4 mutant, while the level of the 4-O-methylated counterpart MeGlcA-Xyl3 was only mildly affected compared with the wild type. In the wild-type hydrolyzate, these acidic oligosaccharides without acetyl are present in low amounts, as most of the Xylp carrying (1→2)-linked GlcA or MeGlcA also carry 3-O-Ac (S. Chong and M. Tenkanen, unpublished data). Thus, it seems that reduced acetylation results in formation of more GlcA-Xyl3 than MeGlcA-Xyl3. The overall ratio of GlcA to MeGlcA is about 2:3 in Arabidopsis xylan (Peña et al., 2007; Chong et al., 2013), resulting in higher amounts of MeGlcA-substituted XOSs; however, even in the wild type, the ratio between GlcA-Xyl3 and MeGlcA-Xyl3, is about 1:1, i.e. the nonmethylated oligosaccharide lacking acetyl esters is overrepresented. In the triple mutant, the ratio was closer to 2:1. These observations suggest that lack of Ac in Xylp carrying GlcA impacts the methylation of GlcA, which is known to take place at the polymer level (Lee et al., 2012; Urbanowicz et al., 2012). The observation of reduced methylation of GlcA in the rwa1 rwa3 rwa4 mutant is consistent with results of Lee et al. (2011) reporting increased GlcA level in their rwa1 rwa2 rwa3 rwa4 mutant. The data indicate interplay between acetylation and methylation, where methylation of GlcA is more efficient if the GlcA-substituted Xyl residue is acetylated.
Mass Distribution of Xylans Is Not Affected in the Mutants
To investigate if the reduced stem height phenotypes observed in the triple mutants (Fig. 3C) were due to a decrease in molecular mass of the xylan, the mass distribution of extracted xylan was determined. Two pools of xylans could be detected, a quantitatively small, high Mr fraction (1,000 kD) and a quantitatively dominant peak around 60 kD (Supplemental Fig. S4). The observed mass distribution is comparable to that reported by Wu et al. (2010). No significant differences between rwa triple mutants and the wild type could be detected.
DISCUSSION
Previously, we have identified four homologs of the fungal Cas1p in Arabidopsis and demonstrated the function of RWA2 in plant cell wall acetylation (Manabe et al., 2011). Here, we have further investigated the role of the RWA protein family in cell wall acetylation by generating double, triple, and quadruple rwa mutants. The triple and quadruple mutants displayed severely dwarfed phenotypes with striking morphological changes (Figs. 1 and 2). The growth phenotype and the cell wall acetylation phenotype depended not only on the genotype, but also on environmental conditions and developmental stage. Several different growth stages and organs were analyzed in the course of this study, and an overview with indication of the main results is given in Supplemental Table S1. Through this study, we have demonstrated the importance of RWA proteins in cell expansion, developmental processes, and secondary cell wall deposition. The quadruple mutant was barely able to survive but, nevertheless, able to produce viable mutant seeds. Surprisingly, most of the offspring was heterozygotic, demonstrating an exceptionally high rate of outcrossing under our growth conditions.
An earlier report (Lee et al., 2011) of an rwa quadruple mutant, rwa1-2 rwa2-3 rwa3-1 rwa4-2, indicated a phenotype remarkably different from the phenotype of our quadruple mutant, rwa1-1 rwa2-1 rwa3-1 rwa4-1, reported here. Lee et al. (2011) used a mutant nomenclature that is different from the one we have introduced in our earlier paper (Manabe et al., 2011). To facilitate the comparison and discussion, we have listed the different mutant alleles in Supplemental Table S2. Although the authors do not specifically discuss morphology at the whole-plant level, the stem section does not show any signs of cell number reduction for their quadruple mutant in contrast to the drastic reduction observed here (Fig. 4). Likewise, they reported reduction in secondary wall thickness by 38%, whereas secondary walls of our quadruple mutant were virtually absent. We cannot explain the difference between the two studies. The mutations used by Lee et al. (2011) are reported to result in absence of detectable transcript. Nevertheless, a possible explanation could be that one of the alleles used by Lee et al. (2011) allowed enough transcript to be generated to allow recovery of some RWA function. Another possibility could be that either of the mutants have a second site mutation that reduces the severity of the phenotype in the Lee et al. (2011) study or exacerbates the phenotype described here. Finally, although we have observed a severe developmental phenotype under several different growth conditions, the phenotype is quite variable depending on the growth conditions, and the possibility remains that the difference in phenotype between the two mutants is a result of different growth conditions.
Through the phenotypic study of rwa mutants, we have shown that acetylation of polysaccharides plays a vital role in normal plant development. Still, plants seem to be able to tolerate a reduction of wall polysaccharide O-acetylation to a certain threshold without an apparent impact on plant development. Based on the results shown here, we are not able to determine if acetylation of particular polymers is more important. The rwa2 rwa3 rwa4 mutant, which is disproportionally affected in XyG acetylation, has a strong growth phenotype. However, the axy4-1 and axy4like-1 mutants did not show any visible symptoms, despite a complete lack of O-acetyl substituents on XyG in most tissues (Gille and Pauly, 2012). Xylan acetylation is known to be important for plant development, and tbl29 mutants with a 60% decrease specifically in xylan acetylation display a reduced-growth phenotype with collapsed xylem cells (Xiong et al., 2013). None of the rwa triple mutants had such a low level of xylan acetylation, but the rwa1 rwa3 rwa4 mutant, which had the largest decrease in xylan acetylation, did exhibit characteristics resembling known irregular xylem mutants with smaller rosette size and darker green leaves with a dome shape rather than flat shape (Shao et al., 2004; Wu et al., 2009), suggesting that vascular function was impaired. However, we did not detect irregular xylem vessels in this mutant or reduction in molecular mass of extracted xylan.
The role of both RWA and TBL proteins in acetylation of cell wall polysaccharides is now clearly established (Gille et al., 2011; Lee et al., 2011; Manabe et al., 2011; Xiong et al., 2013; Yuan et al., 2013). However, the mechanism of acetylation and how the RWA and TBL proteins function together is far from clear. Apparently, the RWA proteins operate at a biosynthetic step preceding that of the TBL proteins, because the RWA proteins have a much wider and overlapping specificity. One possibility is that RWA proteins acetylate one or more intermediate products, which then function as substrate for the TBL proteins. Another possibility is that RWA and TBL proteins form complexes where the substrate specificity is governed by the TBL protein. Fungal and animal cells have proteins involved in acetylation that are much larger and contain domains corresponding to both RWA and TBL (Anantharaman and Aravind, 2010). When two proteins in an organism are homologous to a single fused protein in other organisms, there is a high probability that the two proteins are interacting (Marcotte et al., 1999). Therefore, we can speculate that the RWA and TBL proteins would be physically connected through protein-protein interactions in plants. According to this model, different RWA proteins may preferentially form complexes with a subset of TBL proteins. However, even in animals, it is not clear how the Cas1 protein mediates acetylation of sialic acid, and while Cas1 is definitely required, the acetylation could involve additional proteins (Arming et al., 2011).
MATERIAL AND METHODS
Plant Material and Growth
Growth condition for Arabidopsis (Arabidopsis thaliana) plants grown on soil and genotypes used have been described previously (Manabe et al., 2011). To generate the double mutants, all single mutants were crossed to one another for all the possible combinations. The F1 plants resulting from the crosses were confirmed by genotyping PCR with the primer sets previously described (Manabe et al., 2011). Then, the homozygous double mutants were selected from the segregating F2 generation. The F1 plants were crossed again to obtain triple and quadruple mutants, and the progenies were tested for genotype by PCR.
For plants grown on petri dishes, the seeds were surface sterilized first with 70% (v/v) ethanol briefly and then with sterilization solution (20% [v/v] bleach, 0.05% [v/v] Tween 20) for 20 min. Sterilized seeds were washed three times with water and stratified in water for 3 d before sowing. Seeds were sown on Murashige and Skoog (MS)-Agar medium (one-half-strength MS salt, 30 g L−1 Suc, pH 5.7) and were placed in a growth chamber (16 h of light at 22°C and 8 h of darkness at 18°C).
Complementation
The plasmid carrying RWA2-GFP fusion protein driven by double Tobacco mosaic virus 35S promoter (pMDC83-RWA2-GFP), which was also used for tobacco (Nicotiana tabacum) transient expression to examine subcellular localization (Manabe et al., 2011), was transformed into Agrobacterium tumefaciens (strain GV3101). The triple rwa1 rwa2 rwa3 mutant with heterozygous RWA4, rwa1-1 rwa2-1 rwa3-1 RWA4/rwa4-1, was transformed with the A. tumefaciens using the floral infiltration method (Clough and Bent, 1998). The T1 generation was first screened on hygromycin selection medium (one-half-strength MS salt, 30 g L−1 Suc, 50 mg L–1 hygromycin, 25 mg L–1 carbenicillin) and then transferred to nonselection medium (one-half-strength MS salt, 30 g L−1 Suc, 25 mg L–1 carbenicillin). After about 1 week, the transformants were scored for GFP fluorescence under dissection microscope. The T2 generation derived from the selected plants was tested for genotype of the RWA4 gene to obtain transformants in the quadruple mutant background.
XyG Oligosaccharide Mass Profiling
Five freshly harvested 7-d-old etiolated seedlings or 10 mg senescent stem material were ground to a fine powder using a ball mill. AIR was extracted as described previously (Gille et al., 2009). Mass profiling of XyGOs derived from the etiolated seedlings was essentially performed as described by Lerouxel et al. (2002) and shown by Günl et al. (2010) by digesting AIR with a XyG-specific endoglucanase (Pauly et al., 1999). For the senescent stem material, the AIR was suspended in water to a concentration of 10 mg mL–1, and an equivalent of 100 µg AIR (10 µL of the suspension) was used for the XEG digest as described previously (Günl et al., 2010). The spectra of the solubilized XyGOs were obtained using an Axima matrix-assisted laser desorption/ionization time-of-flight (Shimadzu) set to linear positive mode with an acceleration voltage of 20,000 V.
Xylan Oligosaccharide Mass Profiling
Fifteen plants of each genotype, rwa2 rwa1, rwa2 rwa3, rwa3 rwa4, rwa1 rwa3 rwa4, and ecotype Columbia (wild type) were grown at 23°C under 16-h photoperiod, light intensity of 150 µmol m–2 sec–1, and 70% relative humidity for 8 weeks. Basal 10-cm-long segments of inflorescence stem of five plants were used as one biological replicate, frozen in liquid N2, freeze dried, and ground using a bead mill at 30 Hz for 2 min. AIR was prepared by extracting ethanol-soluble material with 80% (v/v) ethanol containing 4 mm HEPES buffer, pH 7.5, for 10 min, recovering the insoluble material by centrifugation for 15 min at 14,000 rpm, and washing the pellet sequentially with 70% (v/v) ethanol, chloroform:methanol (1:1, v/v) mixture, and finally acetone, with each washing step followed by centrifugation as above. The pellet was then dried in a desiccator.
An amount of 3 mg of Arabidopsis AIR was incubated in 1 mL 20 mm NaOAc, pH 5.0, with 20,000 nkat GH10 endo-1,4-β-d-xylanase from Aspergillus aculeatus per gram AIR, at 40°C for 24 h. The liberated XOSs were purified using Hypersep Hypercarb Porous Graphitized Carbon columns (Thermo Scientific), according to Packer et al. (1998) and Chong et al. (2011). The acidic XOSs were eluted in 50% (v/v) acetonitrile acidified with 0.05% (v/v) trifluoroacetic acid, dried, and finally dissolved in 30 μL Milli-Q water. As the neutral XOSs without MeGlcA substituents are less informative for structural fingerprinting, only the acidic XOSs were analyzed in this work. The atmospheric pressure matrix-assisted laser desorption/ionization-ion trap mass spectrometry analysis was carried out as previously described (Chong et al., 2011). Mass lists corresponding to acidic XOS mass peaks were extracted from Bruker’s liquid chomatography/mass selective detector ion trap software 5.2, and calculation of percentage intensities for the selected mass peaks was as previously described (Chong et al., 2011). The endo-1,4-β-d-xylanase liberated two series of acidic XOSs that comprised of GlcA (1→2)-linked XOS (GlcA-XOS) and MeGlcA (1→2)-linked XOS (MeGlcA-XOS) (Chong et al., 2011). Intensities in percentage of all the identified MeGlcA-XOS and GlcA-XOS mass spectrometry peaks were analyzed in principal components analysis with SIMCA-P+ 12.0.1 (Umetric) to reveal differences between the samples.
Histology
Basal stem segments of 12-week-old rwa1 rwa2 rwa3 rwa4 and of ecotype Columbia were fixed in 4% (w/v) paraformaldehyde and embedded in London white resin (high grade) as described in Yin et al., 2011. One-micrometer-thick sections were stained with toluidine blue and observed under light microscope.
Cell Wall Preparation and Measurement of Monosaccharide and Released Acetate
Fresh tissues were frozen with liquid nitrogen and lyophilized before grinding. The senesced stems were from plants whose water supply was gradually withheld and then dried well in the growth room. The stems were then dried further in a lyophilizer for 24 h. The dried materials were ground with bead beater and AIR was extracted from them as previously described (Manabe et al., 2011). The AIR from fresh samples was destarched before the measurement, but not for the senesced stems, as they included little starch. Acetic acid release was measured as previously described (Manabe et al., 2011).
Observation of Shoot
Rosettes were observed in a dissection microscope equipped with a camera. Several shots were taken and stacked together using Photoshop CS4 (Adobe Systems) to recreate the plants that were too large and exceeded the camera frame.
Root Growth Measurement
To measure root growth, the position of the root tip was marked at the bottom of petri dishes 2 d after germination and scanned 5 d later with a flatbed scanner (ScanMaker 9800XL, Microtek Lab) at 300 dpi. The scanned images were saved as a TIF format, and root length was measured with ImageJ software as described in Buer et al. (2000).
2D-NMR Wall Analysis
2D-NMR analysis of Arabidopsis stem material (the wild type, rwa1rwa2rwa3, rwa1rwa2rwa4, rwa1rwa3rwa4, and rwa2rwa3rwa4) was performed using the method previously reported (Cheng et al., 2013). In brief, ball-milled AIR (25 mg) was dissolved in 0.75 mL deuterated dimethyl sulfoxide (99%; Cambridge Isotope Laboratories) supplemented with 10 µL deuterated 1-ethyl-3-methylimidazolium acetate. The 2D-NMR spectrum was recorded at 318 K on a Bruker Avance 600-MHz NMR spectrometer equipped with an inverse gradient 5-mm TXI 1H/13C/15N CryoProbe. The one-bond 13C-1H correction of heteronuclear single quantum coherence was determined by Bruker standard pulse sequence “hsqcetgpsisp.2”. Important parameters applied in the experiments include the following: 16-ppm wide, centered at 7.0 ppm in F2 (1H) dimension with 2,048 data points; 240-ppm wide, centered at 90 ppm in F1 (13C) dimension with 256 data points; 128 scans; two dummy scans; 1-s interscan delay; 6.0-µs prescan delay; and a receiver gain of 46,931. The spectra were calibrated by the dimethyl sulfoxide solvent peak (δC 39.9 ppm and δH 2.499 ppm), and the interpretation of cross peaks in the spectra of was based on previously published data (Kim and Ralph, 2010; Gille et al., 2011). The NMR data processing was performed using Bruker’s Topspin 3.1 software.
Mass Distribution Analysis
Destarched AIR were extracted overnight in 1 m NaOH with 2.5 mg mL−1 NaBH4. The extract was neutralized using HCl, centrifuged for 10 min at 15,000g at room temperature, and filtered through 0.44-µm filter (Q-Max PTFE). Filtered extract was analyzed on a Viskotek fitted with triple detector for light scatter, viscometry, and refractive index detection. Column used was a Gram, 3,000 Å, 8 × 300 mm (Polymer Standards Service), eluted with dimethyl sulfoxide and 5 mg mL−1 LiBr at 0.4 mL min–1 and 70°C. Molecular mass was determined using Omnisec 4.7 software and 100-kD pullulan standard. Equal amount of sugar was loaded in all runs.
The accession numbers for the genes used in this study are as follows: RWA1, AT5g46340; RWA2, At3g06550; RWA3, At2g34410; and RWA4, At1g29890. The mutant lines used for the crosses were rwa1-1 (SAIL_205_F09), rwa2-1 (GABI_571F07), rwa3-1 (SALK_133630), and rwa4-1 (SAIL_205_D08) according to the allele nomenclature introduced in Manabe et al. (2011); see also Supplemental Table S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype of 6-week-old plants.
Supplemental Figure S2. 2D-NMR spectrum of Arabidopsis stem material highlighting the region for estimation of the degree of O-acetylation of xylan and (gluco)mannan.
Supplemental Figure S3. Acidic XOSs identified in the GH10 digest of AIR from the wild type and rwa mutants.
Supplemental Figure S4. No difference in mass distribution of extracted xylan could be detected in rwa triple mutants when compared with the wild type.
Supplemental Table S1. Summary of organs, developmental stages, and polysaccharides analyzed in the study.
Supplemental Table S2. Nomenclature of Arabidopsis rwa mutants.
Acknowledgments
We thank Alicia Elizondo and Sherry Chan (Joint BioEnergy Institute) for assistance in experimentation and maintenance of plants.
Glossary
- AIR
alcohol insoluble residue
- XyG
xyloglucan
- OLIMP
oligosaccharide mass profiling
- XyGO
xyloglucan oligosaccharide
- 2D-NMR
two-dimensional NMR
- XOS
xylo-oligosaccharide
- Xylp
xylopyranosyl
- MeGlcA
4-O-methyl-α-d-GlcA
- MS
Murashige and Skoog
References
- Alvarez-Buylla ER, Benítez M, Corvera-Poiré A, Chaos Cador A, de Folter S, Gamboa de Buen A, Garay-Arroyo A, García-Ponce B, Jaimes-Miranda F, Pérez-Ruiz RV, et al. (2010) Flower development. Arabidopsis Book 8: e0127, doi/10.1199/tab.0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. (2010) Novel eukaryotic enzymes modifying cell-surface biopolymers. Biol Direct 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arming S, Wipfler D, Mayr J, Merling A, Vilas U, Schauer R, Schwartz-Albiez R, Vlasak R. (2011) The human Cas1 protein: a sialic acid-specific O-acetyltransferase? Glycobiology 21: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Masle J, Wasteneys GO. (2000) Growth conditions modulate root-wave phenotypes in Arabidopsis. Plant Cell Physiol 41: 1164–1170 [DOI] [PubMed] [Google Scholar]
- Chen X, Shekiro J, Franden MA, Wang W, Zhang M, Kuhn E, Johnson DK, Tucker MP. (2012) The impacts of deacetylation prior to dilute acid pretreatment on the bioethanol process. Biotechnol Biofuels 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Sorek H, Zimmermann H, Wemmer DE, Pauly M. (2013) Solution-state 2D NMR spectroscopy of plant cell walls enabled by a dimethylsulfoxide-d6/1-ethyl-3-methylimidazolium acetate solvent. Anal Chem 85: 3213–3221 [DOI] [PubMed] [Google Scholar]
- Chong SL, Koutaniemi S, Virkki L, Pynnönen H, Tuomainen P, Tenkanen M. (2013) Quantitation of 4-O-methylglucuronic acid from plant cell walls. Carbohydr Polym 91: 626–630 [DOI] [PubMed] [Google Scholar]
- Chong SL, Nissilä T, Ketola RA, Koutaniemi S, Derba-Maceluch M, Mellerowicz EJ, Tenkanen M, Tuomainen P. (2011) Feasibility of using atmospheric pressure matrix-assisted laser desorption/ionization with ion trap mass spectrometry in the analysis of acetylated xylooligosaccharides derived from hardwoods and Arabidopsis thaliana. Anal Bioanal Chem 401: 2995–3009 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M. (2011) O-Acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23: 4041–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M. (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Pauly M. (2012) O-Acetylation of plant cell wall polysaccharides. Front Plant Sci 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Miller LM, Hou G, Yu XH, Chen XY, Liu CJ. (2012) Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 24: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Gille S, Pauly M. (2010) OLIgo mass profiling (OLIMP) of extracellular polysaccharides. J Vis Exp 40: 2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J. (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Ye ZH. (2011) The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 52: 1289–1301 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Yuan Y, Haghighat M, Ye ZH. (2012) Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-O-methylation of glucuronic acid on xylan. Plant Cell Physiol 53: 1934–1949 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Séveno M, Usadel B, Faye L, Lerouge P, Pauly M. (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe Y, Nafisi M, Verhertbruggen Y, Orfila C, Gille S, Rautengarten C, Cherk C, Marcus SE, Somerville S, Pauly M, et al. (2011) Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol 155: 1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. (1999) Detecting protein function and protein-protein interactions from genome sequences. Science 285: 751–753 [DOI] [PubMed] [Google Scholar]
- Obel N, Erben V, Schwarz T, Kühnel S, Fodor A, Pauly M. (2009) Microanalysis of plant cell wall polysaccharides. Mol Plant 2: 922–932 [DOI] [PubMed] [Google Scholar]
- Orfila C, Dal Degan F, Jørgensen B, Scheller HV, Ray PM, Ulvskov P. (2012) Expression of mung bean pectin acetyl esterase in potato tubers: effect on acetylation of cell wall polymers and tuber mechanical properties. Planta 236: 185–196 [DOI] [PubMed] [Google Scholar]
- Packer NH, Lawson MA, Jardine DR, Redmond JW. (1998) A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj J 15: 737–747 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A. (1999) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Pawar PMA, Koutaniemi S, Tenkanen M, Mellerowicz EJ. (2013) Acetylation of woody lignocellulose: significance and regulation. Front Plant Sci 4: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong RQ, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye ZH. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelko G, Lionetti V, Fursova O, Sundaram RM, Qi M, Whitham SA, Bogdanove AJ, Bellincampi D, Zabotina OA. (2013) Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol 162: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig MJ, Vinzant TB, Himmel ME, Decker SR. (2009) The effect of lignin removal by alkaline peroxide pretreatment on the susceptibility of corn stover to purified cellulolytic and xylanolytic enzymes. Appl Biochem Biotechnol 155: 397–406 [DOI] [PubMed] [Google Scholar]
- Shao M, Zheng H, Hu Y, Liu D, Jang JC, Ma H, Huang H. (2004) The GAOLAOZHUANGREN1 gene encodes a putative glycosyltransferase that is critical for normal development and carbohydrate metabolism. Plant Cell Physiol 45: 1453–1460 [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, Foston M, Li H, O’Neill MA, Ragauskas AJ, et al. (2012) 4-O-Methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc Natl Acad Sci USA 109: 14253–14258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A. (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A. (2009) The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57: 718–731 [DOI] [PubMed] [Google Scholar]
- Xiong G, Cheng K, Pauly M. (2013) Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol Plant 6: 1373–1375 [DOI] [PubMed] [Google Scholar]
- Yin L, Verhertbruggen Y, Oikawa A, Manisseri C, Knierim B, Prak L, Jensen JK, Knox JP, Auer M, Willats WG, et al. (2011) The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol Plant 4: 1024–1037 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Teng Q, Zhong R, Ye ZH. (2013) The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol 54: 1186–1199 [DOI] [PubMed] [Google Scholar]