One of the genes coding for the first enzyme of the phosphorylated pathway of serine biosynthesis is essential for embryo and pollen development, suggesting that this pathway is an important link connecting primary metabolism with development.
Abstract
This work contributes to unraveling the role of the phosphorylated pathway of serine (Ser) biosynthesis in Arabidopsis (Arabidopsis thaliana) by functionally characterizing genes coding for the first enzyme of this pathway, 3-phosphoglycerate dehydrogenase (PGDH). We identified two Arabidopsis plastid-localized PGDH genes (3-PGDH and EMBRYO SAC DEVELOPMENT ARREST9 [EDA9]) with a high percentage of amino acid identity with a previously identified PGDH. All three genes displayed a different expression pattern indicating that they are not functionally redundant. pgdh and 3-pgdh mutants presented no drastic visual phenotypes, but eda9 displayed delayed embryo development, leading to aborted embryos that could be classified as early curled cotyledons. The embryo-lethal phenotype of eda9 was complemented with an EDA9 complementary DNA under the control of a 35S promoter (Pro-35S:EDA9). However, this construct, which is poorly expressed in the anther tapetum, did not complement mutant fertility. Microspore development in eda9.1eda9.1 Pro-35S:EDA9 was arrested at the polarized stage. Pollen from these lines lacked tryphine in the interstices of the exine layer, displayed shrunken and collapsed forms, and were unable to germinate when cultured in vitro. A metabolomic analysis of PGDH mutant and overexpressing plants revealed that all three PGDH family genes can regulate Ser homeostasis, with PGDH being quantitatively the most important in the process of Ser biosynthesis at the whole-plant level. By contrast, the essential role of EDA9 could be related to its expression in very specific cell types. We demonstrate the crucial role of EDA9 in embryo and pollen development, suggesting that the phosphorylated pathway of Ser biosynthesis is an important link connecting primary metabolism with development.
Plant primary metabolism is a complex process where many interacting pathways must be finely coordinated and integrated in order to achieve proper plant development and acclimation to the environment. An example of such complexity is the biosynthesis of the amino acid l-Ser, which takes place in at least two different organelles and by different pathways. This amino acid is essential for the synthesis of proteins and other biomolecules needed for cell proliferation, including nucleotides and Ser-derived lipids, such as phosphatidylserine and sphingolipids. Additionally, d-Ser has been attributed a signaling function in male gametophyte-pistil communication (Michard et al., 2011).
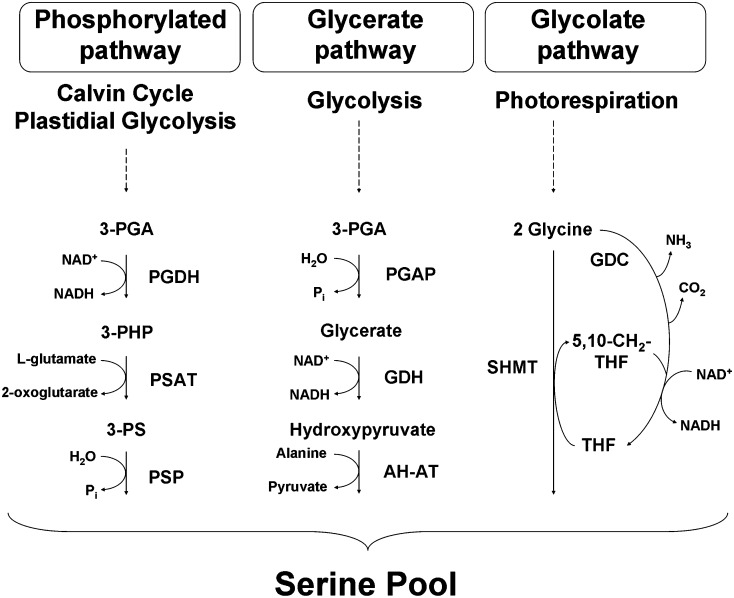
Despite the important role played by Ser in plants, the biological significance of the coexistence of several Ser biosynthetic pathways and how they interact to maintain amino acid homeostasis in cells is not yet understood. Three different Ser biosynthesis pathways have been described in plants (Kleczkowski and Givan, 1988; Ros et al., 2013; Fig. 1). One is the glycolate pathway, which takes place in mitochondria and is associated with photorespiration (Tolbert, 1980, 1997; Douce et al., 2001; Bauwe et al., 2010; Maurino and Peterhansel, 2010). In this pathway, two molecules of Gly are converted to one molecule of Ser in a reaction catalyzed by the Gly decarboxylase complex and Ser hydroxymethyltransferase (Fig. 1). Ser synthesis through the glycolate pathway is obtained in green tissues during daylight hours (Tolbert, 1980, 1985; Douce et al., 2001), suggesting that alternative Ser biosynthesis pathways may be required in the dark and/or in nonphotosynthetic organs. In this respect, Ser can be synthesized through two nonphotorespiratory pathways (Kleczkowski and Givan, 1988), the plastidial phosphorylated pathway (Ho et al., 1998, 1999a, 1999b; Ho and Saito, 2001) and the so-called glycerate pathway, which synthesizes Ser by the dephosphorylation of 3-phosphoglycerate (3-PGA; Kleczkowski and Givan, 1988; Fig. 1). This latter pathway includes the reverse sequence of the section of the oxidative photosynthetic carbon cycle linking 3-PGA to Ser (3-PGA-glycerate-hydroxypyruvate-Ser), these reactions being catalyzed by putative enzymes such as 3-PGA phosphatase, glycerate dehydrogenase, Ala-hydroxypyruvate aminotransferase, and Gly hydroxypyruvate aminotransferase. Although the existence of enzymatic activities of this pathway has been demonstrated (Kleczkowski and Givan, 1988), its functional significance is unknown and genes coding for the specific enzymes of the pathway have not been characterized to date.
Figure 1.
Schematic representation of Ser biosynthesis in plants. The enzymes participating in each Ser biosynthetic pathway are listed separately. Photorespiratory pathway (glycolate pathway): GDC, Gly decarboxylase; SHMT, Ser hydroxymethyltransferase. Glycerate pathway: PGAP, 3-phosphoglycerate phosphatase; GDH, glycerate dehydrogenase; AH-AT, Ala-hydroxypyruvate aminotransferase. Phosphorylated pathway: PSAT, 3-phosphoserine aminotransferase; PSP, 3-phosphoserine phosphatase. Abbreviations used for metabolites are as follows: 3-PHP, 3-phosphohydroxypyruvate; 3-PS, 3-phosphoserine; THF, tetrahydrofolate; 5,10-CH2-THF, 5,10-methylene-tetrahydrofolate. This figure is adapted from Cascales-Miñana et al. (2013).
The plastidial phosphorylated pathway of serine biosynthesis (PPSB; Fig. 1), which is conserved in mammals and plants, synthesizes Ser via 3-phosphoserine utilizing 3-PGA as a precursor (Kleczkowski and Givan, 1988). Evidence for the existence of this pathway in plants stems from the isolation and characterization of its enzyme activities (Handford and Davies, 1958; Slaughter and Davies, 1968; Larsson and Albertsson, 1979; Walton and Woolhouse, 1986). The PPSB involves three enzymes catalyzing sequential reactions: 3-phosphoglycerate dehydrogenase (PGDH), 3-phosphoserine aminotransferase, and 3-phosphoserine phosphatase (PSP; Fig. 1). Genes coding for some isoforms of these enzymes have been cloned and biochemically characterized in Arabidopsis (Arabidopsis thaliana; Ho et al., 1998, 1999a, 1999b; Ho and Saito, 2001).
In humans, the PPSB plays a crucial role in cell proliferation control and oncogenesis (Bachelor et al., 2011; Locasale et al., 2011; Pollari et al., 2011; Possemato et al., 2011). The functional significance of the PPSB in plants has recently been unraveled by providing evidence for the crucial role of PSP1, the last enzyme of the pathway in embryo, pollen, and root development (Cascales-Miñana et al., 2013). However, the PPSB still requires further characterization. In order to gain a complete understanding of the PPSB function in plants, precise molecular, metabolic, and genetic knowledge of all the enzymes and genes of the pathway is needed. In this work, we follow a gain- and loss-of-function approach in Arabidopsis to characterize a family of genes coding for putative isoforms of PGDH, the first enzyme of the PPSB. Here, we identify the essential gene of this family and provide evidence for its crucial function in embryo and pollen development.
RESULTS
Identification and Expression Analysis of Plastid-Localized PGDHs
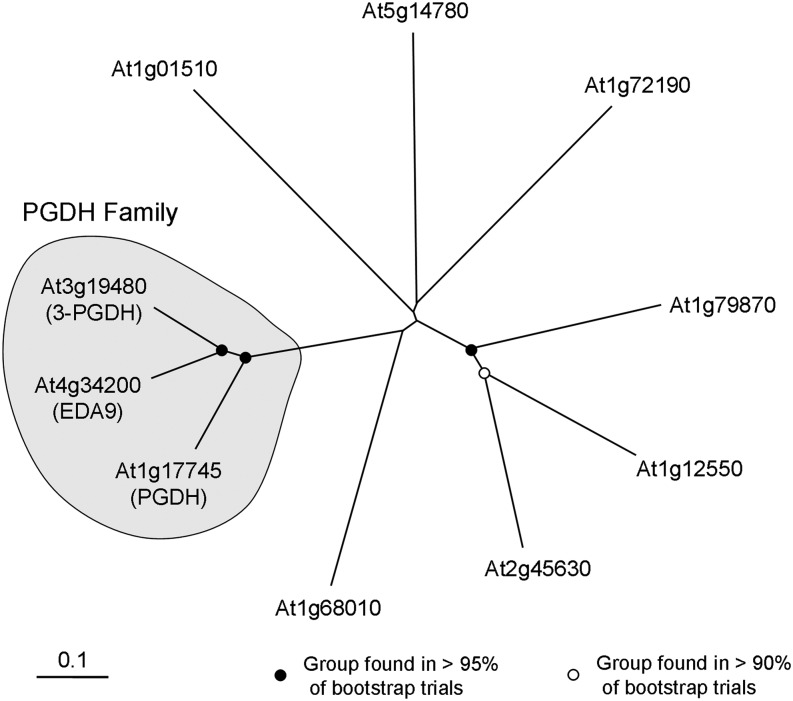
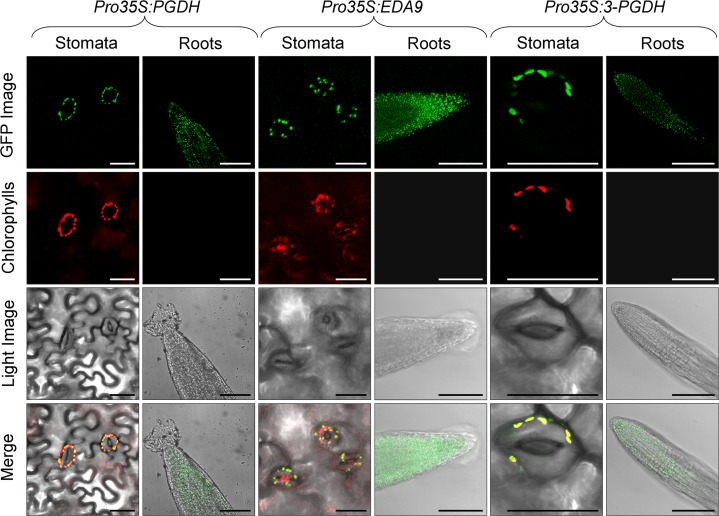
A single gene coding for a PGDH Arabidopsis isoform has previously been cloned, and it was demonstrated that the encoded protein possesses in vitro PGDH activity (Ho et al., 1999a). This gene (At1g17745) is named PGDH. By searching homologous sequences to PGDH in The Arabidopsis Information Resource database (http://www.arabidopsis.org), we identified two more genes coding for putative PGDHs (At3g19480 and At4g34200), called 3-PGDH and EMBRYO SAC DEVELOPMENT ARREST9 (EDA9), respectively. Both are described in the database as being involved in the l-Ser biosynthesis process, although there is no empirical information about it. 3-PGDH and EDA9 have 78% amino acid identity between them and show 74% and 77% amino acid identity, respectively, with PGDH. The phylogenetic tree confirmed that EDA9 and 3-PGDH are more closely related to one another than to PGDH (Fig. 2). Other genes homologous to PGDH exhibiting putative dehydrogenase activity (i.e. At1g72190, described as oxoreductase) display a much lower percentage of identity (31%) compared with PGDH, indicating that EDA9, 3-PGDH, and PGDH are the only genes of the family (Fig. 2). Accordingly, we choose the three genes for further studies and collectively refer to them as the PGDH family. PGDH is a well-known highly conserved enzyme whose crystal structure has been solved in Mycobacterium tuberculosis and human (Dey et al., 2008). The three Arabidopsis PGDH family proteins displayed 100% identity in all the residues forming the putative catalytic site and the ligand-binding domain (Supplemental Fig. S1). According to the ChloroP prediction server (http://www.cbs.dtu.dk/services/ChloroP/), the three PGDH family proteins have a putative N-terminal plastid/chloroplast localization signal. To investigate the subcellular localization of the PGDH family proteins, we stably expressed GFP fusion protein constructs in Arabidopsis. In both leaves and roots, all the PGDH family members were localized in chloroplasts and in nongreen plastids (Fig. 3), indicating that they could participate in the PPSB.
Figure 2.
Phylogenetic tree of the Arabidopsis PGDH proteins. The phylogenetic tree was constructed with deduced amino acid sequences of Arabidopsis genes producing the closest significant alignments with PGDH, as described in “Materials and Methods.” Confidence values for grouping in the tree were obtained using Bootstrap Neighbor-Joining Tree using 2,000 bootstrap trials. Tree was visualized in TreeView, and the PGDH family is grouped into a globe. Tree nodes with bootstrap values greater than 95% and greater than 90% are indicated with black and white dots, respectively. The Arabidopsis Information Resource descriptions of genes used for the alignment are as follows: At1g72190, d-isomer-specific 2-hydroxyacid dehydrogenase; At1g12550, d-isomer-specific 2-hydroxyacid dehydrogenase; At1g79870, d-isomer-specific 2-hydroxyacid dehydrogenase; At2g45630, d-isomer-specific 2-hydroxyacid dehydrogenase; At5g14780, formate dehydrogenase; At1g68010, hydroxypyruvate reductase; and At1g01510, NAD(P)-binding Rossmann-fold superfamily protein.
Figure 3.
Subcellular localization of the PGDH family isoforms by stable expression of GFP fusion proteins in Arabidopsis. Chloroplastic/plastidic localization of PGDH, EDA9, and 3-PGDH in leaves and roots is shown. GFP fluorescence, chlorophyll fluorescence, light images, and merged images are presented from top to bottom. Bars = 30 µm for stomata and 100 µm for root cell images.
The analysis of the PGDH family promoters using the promomer tool (Winter et al., 2007) revealed that the promoter regions of all three genes were significantly enriched in consensus sequences present in important genes controlling anther development, such as the floral homeotic genes AGAMOUS, AGAMOUS LIKE1, AGAMOUS LIKE2, and AGAMOUS LIKE3 (Supplemental Table S1). In addition, the promoter of EDA9 was also enriched in regulatory elements responding to abscisic acid.
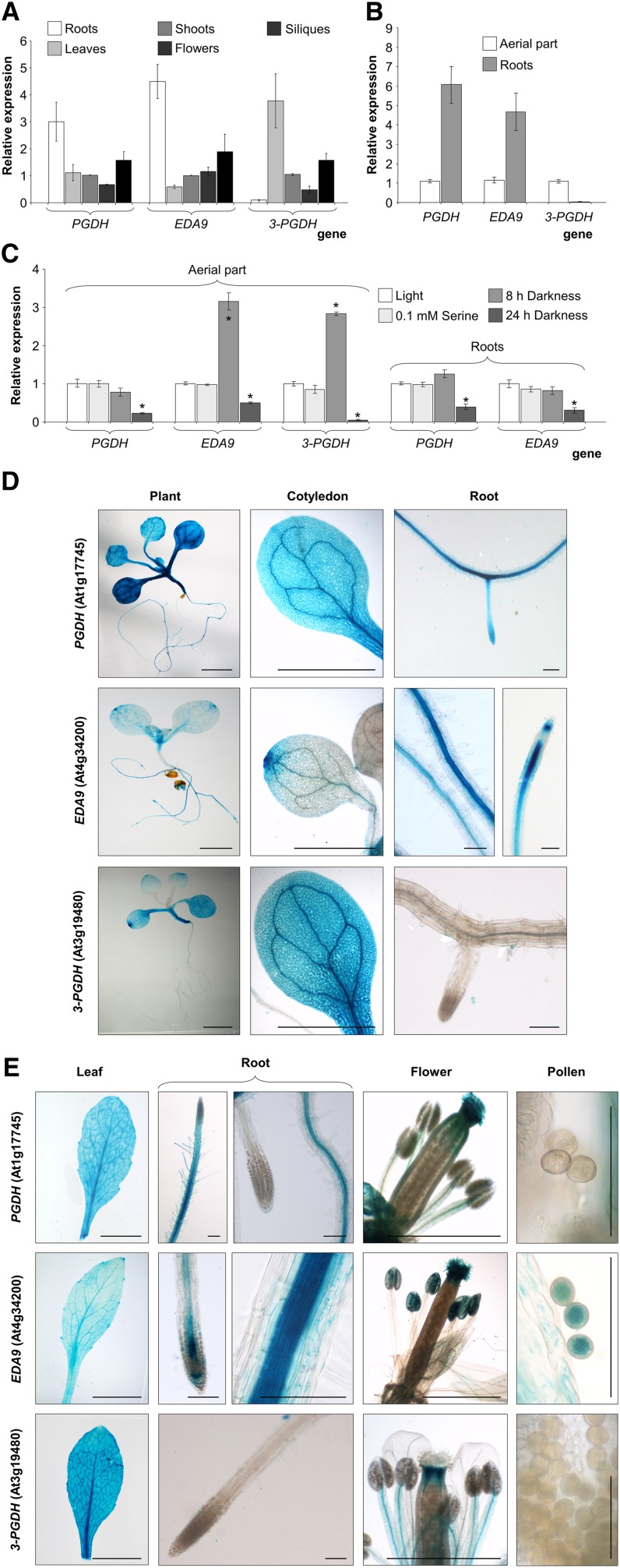
We assessed the expression patterns of the PGDH family genes by quantitative real time (RT)-PCR and by analysis of promoter-GUS. All three genes displayed different expression patterns (Fig. 4, A and B). Although expressed in all the organs studied, PGDH and EDA9 were expressed preferentially in roots at both seedling and adult stages. However, 3-PGDH was expressed mainly in the aerial parts and was not expressed, or very poorly so, in roots. When expression was studied under different dark conditions, an 8-h exposure to darkness induced EDA9 and 3-PGDH expression in the aerial parts, whereas longer exposures (24 h) repressed the expression of all the genes studied in both roots and aerial parts of the plant (Fig. 4C). Ser did not repress the expression of any of the three genes studied following a 24-h treatment.
Figure 4.
Expression analysis of PGDH family genes. A and B, RT-PCR analysis of PGDH, EDA9, and 3-PGDH in different adult (30-d-old) plant organs grown under greenhouse conditions (A) and in the aerial parts and roots of 18-d-old seedlings grown in one-fifth-strength Murashige and Skoog (MS) medium (B). C, Effects of different growth parameters on relative PGDH, EDA9, and 3-PGDH expression in roots and aerial parts of 18-d-old seedlings grown in one-fifth-strength MS medium ± 0.1 mm Ser. Values in A to C (means ± se; n = 3 independent biological replicates) are normalized to the expression in the shoots (A), aerial parts (B), and aerial parts or roots (C) under light conditions. *, Significantly different as compared with the control (P < 0.05). D and E, Expression of GUS under the control of PGDH, EDA9, and 3-PGDH promoters in seedlings (D) and adult plants (E). Bars = 2 mm (whole seedling, cotyledons, and flowers), 0.1 mm (roots and pollen), and 1 cm (leaves).
GUS expression analyses also showed a specific expression pattern for each gene (Fig. 4, D and E). Thus, PGDH was expressed mainly in the vasculature in both seedlings and adult plants. In roots, PGDH was expressed in the meristems at the seedling stage but was poorly expressed, if at all, at the adult stage. In floral organs, PGDH was expressed in the stigma and anther filaments. EDA9 was expressed mainly in the vegetative apex, in the distal zone of cotyledons and leaves, and in anthers, stigma, and pollen grains. In roots, EDA9 was expressed mainly along the root vasculature and in the meristem. 3-PGDH was expressed in the whole aerial part at both the seedling and adult stages but not in roots. It was clearly expressed in cotyledons (especially in the distal zone) and adult leaves as well as in floral organs, specifically in the stigma and anther filaments.
Phenotypic Characterization of PGDH Family Mutants
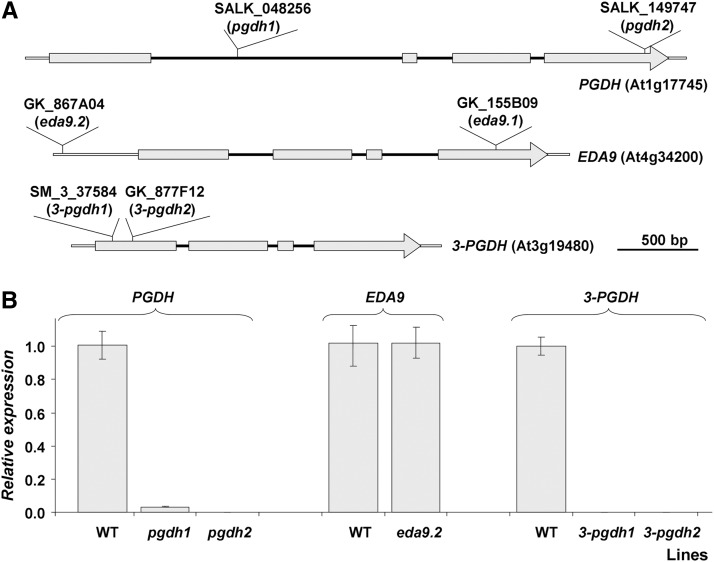
In order to shed light onto the in vivo function of PGDH family mutants, a reverse genetic approach was followed. Multiple independent transfer DNA (T-DNA) insertion lines affecting the gene family were identified in the mutant collections (SALK_048256 and SALK_149747 for PGDH, SM_337584 and GK_877F12 for 3-PGDH, and GK_867A04 and GK_155B09 for EDA9). These mutant alleles were named pgdh1 (SALK_048256), pgdh2 (SALK_149747), 3-pgdh1 (SM_337584), 3-pgdh2 (GK_877F12), eda9.1 (GK_155B09), and eda9.2 (GK_867A04). The presence and genomic locations of the T-DNA insertions were verified by PCR with genomic DNA and by sequencing of PCR products. The exact location of each T-DNA insertion is presented in Figure 5A and in Supplemental Table S2. Genotyping identified homozygous mutants for alleles pgdh1, pgdh2, 3-pgdh1, 3-pgdh2, and eda9.2. The RT-PCR analysis revealed that transcript levels of PGDH in pgdh1pgdh1 and pgdh2pgdh2 and of 3-PGDH in 3-pgdh1 3-pgdh1 and 3-pgdh2 3-pgdh2 were very low or undetectable (Fig. 5B). Despite this low gene expression level of PGDH or 3-PGDH in their respective homozygous mutants, none presented significant growth retardation under in vitro conditions (Supplemental Fig. S2). The expression level of EDA9 in eda9.2eda9.2, whose T-DNA insertion is located 471 nucleotides upstream of the start codon, was similar to that of the wild type (Fig. 5B). Consequently, this mutant did not show a growth phenotype in vitro (Supplemental Fig. S2). However, it was not possible to identify homozygous eda9.1eda9.1 individuals. An analysis of the segregation of the eda9.1 mutant allele was conducted in a population of 200 seeds obtained from heterozygous EDA9eda9.1 plants. No homozygous individuals (eda9.1eda9.1) were rescued based on PCR genotyping (Table I). The segregation analysis of the population displayed a 1:2 ratio (EDA9EDA9:EDA9eda9.1; χ2 = 2.4; P > 0.05), which is typical of a Mendelian segregation with a lethal phenotype for eda9.1eda9.1 individuals.
Figure 5.
Genomic organization and expression analysis of PGDH family T-DNA mutant lines. A, Gray boxes represent exons, and black lines represent introns. The T-DNA insertion point in each PGDH family mutant is shown. B, Detection of the PGDH, EDA9, and 3-PGDH transcripts in mutants pgdh1pgdh1, pgdh2pgdh2, eda9.2eda9.2, 3-pgdh1 3-pgdh1, and 3-pgdh2 3-pgdh2 by RT-PCR analysis. Values (means ± se; n = 3 independent biological replicates) are normalized to the expression in the wild type (WT).
Table I. Genotypes of the progeny of self-crossed EDA9eda9.1 and eda9.1eda9.1 ProEDA9:EDA9 plants.
| Self Crosses | No. of Progeny | Genotypes of Progeny (PCR) |
||
|---|---|---|---|---|
| EDA9EDA9 | EDA9eda9.1 | eda9.1eda9.1 | ||
| EDA9eda9.1 | 200 | 77 (38.5)a | 123 (61.5)a | 0 (0.0)a |
| eda9.1eda9.1 ProEDA9:EDA9 | 94 | 0 (0.0) | 0 (0.0) | 94 (100) |
Significantly different from the expected 1:2:1 ratio for normal Mendelian segregation (χ2 = 69.87; P < 0.001); not significantly different from the 1:2 ratio for eda9.1eda9.1 lethal effect (χ2 = 2.40; P > 0.05).
To corroborate these results, a second segregation analysis was conducted based on sulfadiazine resistance conferred by the T-DNA insertion in the eda9.1 mutant allele. From 516 seedlings coming from heterozygous EDA9eda9.1 plants, 65.5% were sulfadiazine resistant and 34.5% were sulfadiazine sensitive (Table II). These results do not match the expected 75% of resistant individuals for nonlethal eda9.1eda9.1 mutants. On the contrary, the observed results matched a 1:2 segregation (EDA9EDA9: EDA9eda9.1; χ2 = 0.31; P > 0.05), indicating a lethal phenotype associated with the eda9.1 mutant allele.
Table II. Antibiotic resistance in the progeny of self-crossed EDA9eda9.1 and eda9.1eda9.1 ProEDA9:EDA9 plants.
SulfS, Sensitive; SulfR, resistant
| Self-Crosses | No. of Progeny | Antibiotic Resistance |
|
|---|---|---|---|
| SulfS | SulfR | ||
| % | |||
| EDA9eda9.1 | 516 | 178 (34.5)a | 338 (65.5)a |
| eda9.1eda9.1 ProEDA9:EDA9 | 530 | 0 (0.0) | 530 (100) |
Significantly different from the expected 1:3 ratio for normal Mendelian segregation considering the antibiotic resistance phenotype (χ2 = 24,82; P < 0.001); not significantly different from the 1:2 ratio for eda9.1eda9.1 lethal effect (χ2 = 0.31; P > 0.05).
Since these data indicated that EDA9 may play a relevant role in Arabidopsis development, we next concentrated on the functional characterization of this gene.
The eda9.1eda9.1 Embryo Is Arrested at Early Developmental Stages
To investigate whether the lethality associated with the eda9.1 mutation is due to the male gametophyte, female gametophyte, or embryo defects, reciprocal out-crosses of EDA9eda9.1 plants as male/female parent (donor/recipient) and the wild type as female/male parent (recipient/donor) were performed, and the segregation of the mutant allele was studied based on the antibiotic resistance conferred by the T-DNA insertion (Table III). The results indicate that both the male and female mutant gametophytes were transmitted with a transmission efficiency of 83% and 96%, respectively, which suggest that eda9.1 triggers an embryo-lethal phenotype. EDA9eda9.1 plants were visually indistinguishable from the wild type, indicating the recessive nature of the mutant allele.
Table III. Antibiotic resistance in the progeny of reciprocal out-crosses of EDA9eda9.1 plants as male/female parent (donor/recipient) and the wild type as female/male parent (recipient/donor).
SulfS, Sensitive; SulfR, resistant
| Reciprocal Outcrosses | No. of Progeny | Antibiotic Resistance |
Transmission Efficiencya | |
|---|---|---|---|---|
| SulfS | SulfR | |||
| % | ||||
| Recipient × donor | ||||
| Wild type × EDA9eda9.1 | 392 | 214 (54.6)b | 178 (45.4)b | 83.1 |
| EDA9eda9.1 × wild type | 212 | 108 (50.94)c | 104 (49.06)c | 96.3 |
Transmission efficiency (%) = (mutant/wild type) × 100. bNot significantly different from the expected 1:1 ratio for equal transmission efficiency (χ2 = 3.31; P > 0.05). cNot significantly different from the expected 1:1 ratio for equal transmission efficiency (χ2 = 0.08; P > 0.05).
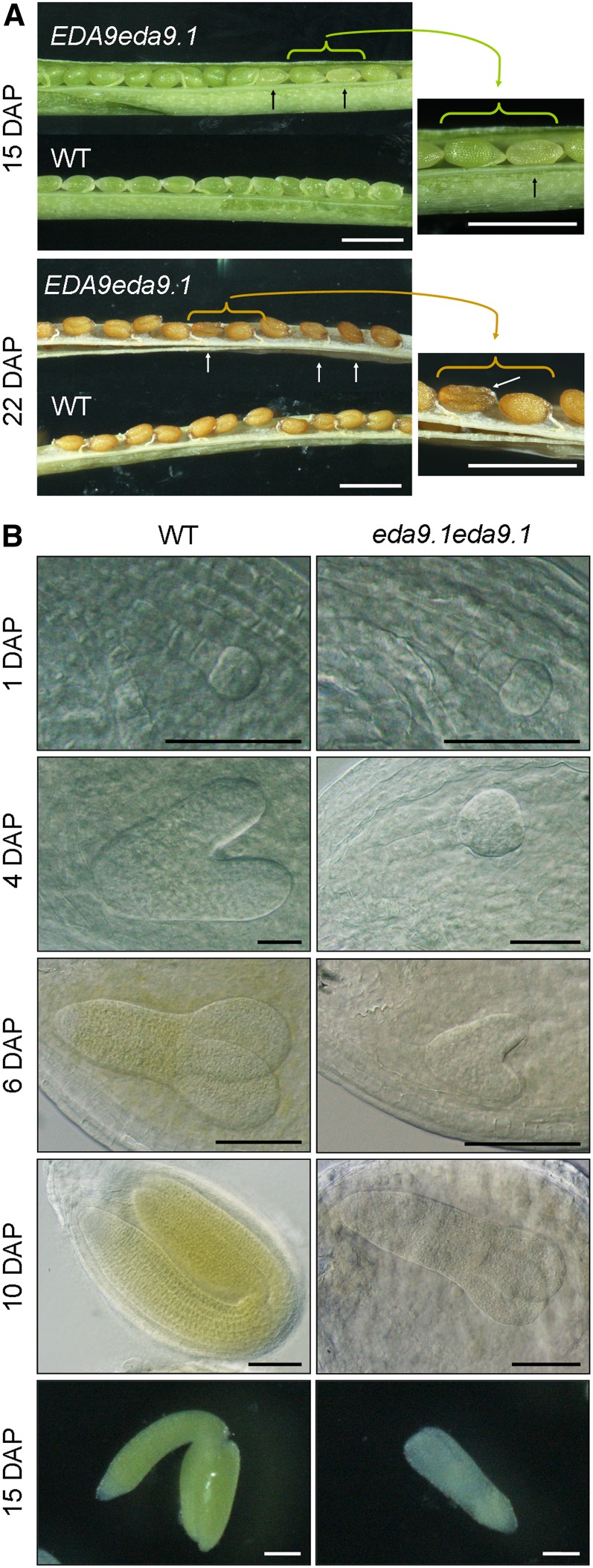
To further confirm this, siliques from heterozygous EDA9eda9.1 plants were dissected at different developmental stages. A population of abnormal seeds was observed 15 d after pollination (DAP) that was randomly distributed along the length of the silique (Fig. 6A). At this stage, mutant seeds were white and started to deflate. Mutant seeds at 22 DAP turned dark brown and were completely deflated. The segregation analysis of a population of 2,631 seeds obtained from heterozygous EDA9eda9.1 plants displayed a 1:4 ratio (20.4% mutant:79.6% normal seeds; χ2 = 0.28; P > 0.05). The percentage of eda9.1eda9.1 seeds was lower than expected, after assuming that there were no significant differences in the mutant allele transmission efficiencies. This may be related to the completely abnormal and deflated form of mutant seeds, which made it difficult to identify some of them 22 DAP and beyond as aborted seeds, and they were excluded from the segregation analysis. To characterize the nature of seed lethality, the embryos in developing siliques of heterozygous EDA9eda9.1 plants were examined (Fig. 6B). At 1 DAP, all the embryos examined reached a similar developmental stage (octant stage, according to Capron et al. [2009]). However, at 4 DAP, some of the embryos showed delayed development (early globular versus midtorpedo stage). This delayed development of embryos continued 6 DAP (heart versus early cotyledon stage) and was clearly assigned to mutant seeds 10 DAP (early cotyledon versus bent cotyledon stage). Terminal aborted embryos (15 DAP) were albino and could be classified as short cotyledons (L1) according to the SeedGenes database (http://www.seedgenes.org). PCR-based genotyping analysis revealed that the aborted embryos were eda9.1eda9.1 individuals.
Figure 6.
eda9.1 homozygous mutants are embryo lethal. A, Siliques from wild-type (WT) and heterozygous EDA9eda9.1 plants at 15 and 22 DAP observed with a binocular microscope. In the EDA9eda9.1 silique, a population of mutant seeds is marked with arrows. The right-hand images show closeups of the mutant seeds. Bars = 1 mm. B, Micrographs show the embryo development of the wild type and homozygous eda9.1eda9.1 from the same silique at different stages observed with a differential interference contrast microscope (1, 4, 6, and 10 DAP) or with a binocular microscope (15 DAP). Bars = 100 µm.
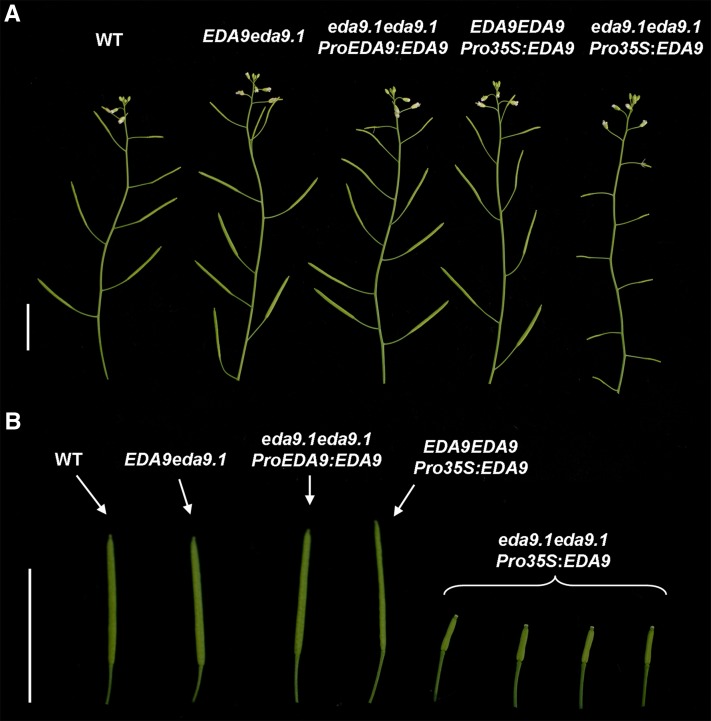
In order to corroborate the phenotype-genotype correlation of the eda9.1 mutation, we transformed EDA9eda9.1 plants with the construct ProEDA9:EDA9. We were able to complement the embryo-lethal phenotype of eda9.1eda9.1 individuals by obtaining homozygous eda9.1eda9.1 plants in the segregating population (Fig. 7). Confocal imaging of these lines confirmed that EDA9 is expressed during seed and embryo development (Fig. 8).
Figure 7.
Phenotypic characterization of eda9.1eda9.1 complemented lines. A, Morphology of adult shoots from wild-type plants (WT), heterozygous EDA9eda9.1, homozygous eda9.1eda9.1 lines transformed with a EDA9-GFP cDNA under the control of the endogenous EDA9 promoter (ProEDA9:EDA9), the wild type transformed with a EDA9-GFP cDNA under the control of the 35S promoter (EDA9EDA9 Pro-35S:EDA9), and homozygous eda9.1eda9.1 lines transformed with the 35S promoter (Pro-35S:EDA9). Bar = 1.5 cm. B, Closeups of siliques from the same lines described in A. Bar = 1.5 cm.
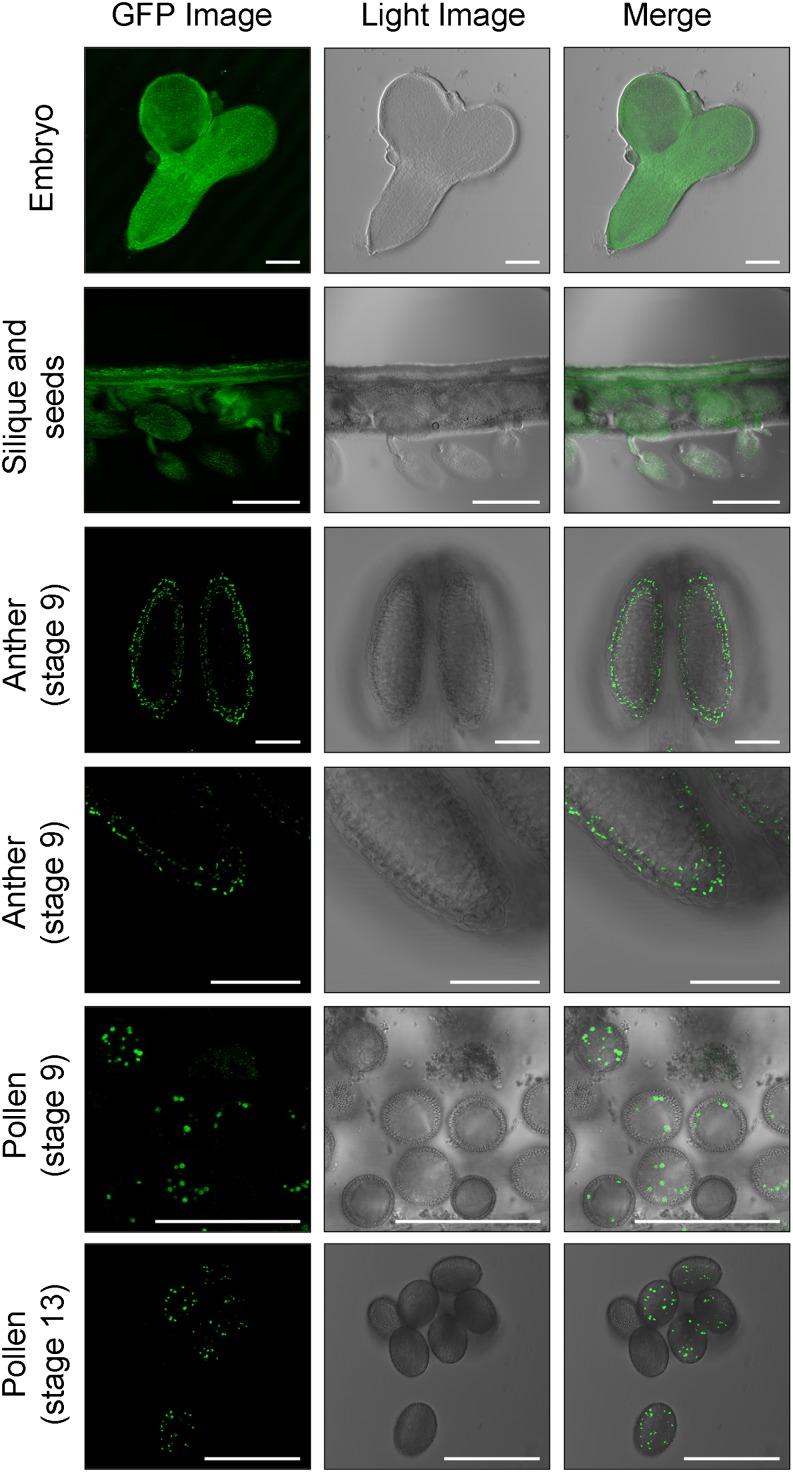
Figure 8.
EDA9 is expressed in seeds, embryo, tapetum, and pollen. EDA9 expression is shown in seeds, embryo, tapetum (stage 9), and pollen (stages 9 and 13) visualized by confocal microscopy of a EDA9-GFP fusion protein under the control of the endogenous EDA9 promoter expressed in eda9.1eda9.1. GFP fluorescence, light, and merged images are presented. Bars = 50 μm for embryos, anthers, and pollen and 500 μm for siliques and seeds.
EDA9 Expression Is Required for Mature Pollen Development
In the adult stage, most transgenic eda9.1eda9.1 ProEDA9:EDA9 plants were fertile and visually indistinguishable from the wild type. However, when EDA9eda9.1 plants were transformed with an EDA9-GFP complementary DNA (cDNA) under the control of the 35S promoter (Pro-35S:EDA9), the resulting homozygous eda9.1eda9.1 transgenic lines in the segregating population were sterile, producing small siliques with no seeds (Fig. 7). An EDA9 overdose effect was not seen to be the cause of the observed sterility phenotype, since wild-type plants expressing Pro-35S:EDA9 were fertile (Fig. 7).
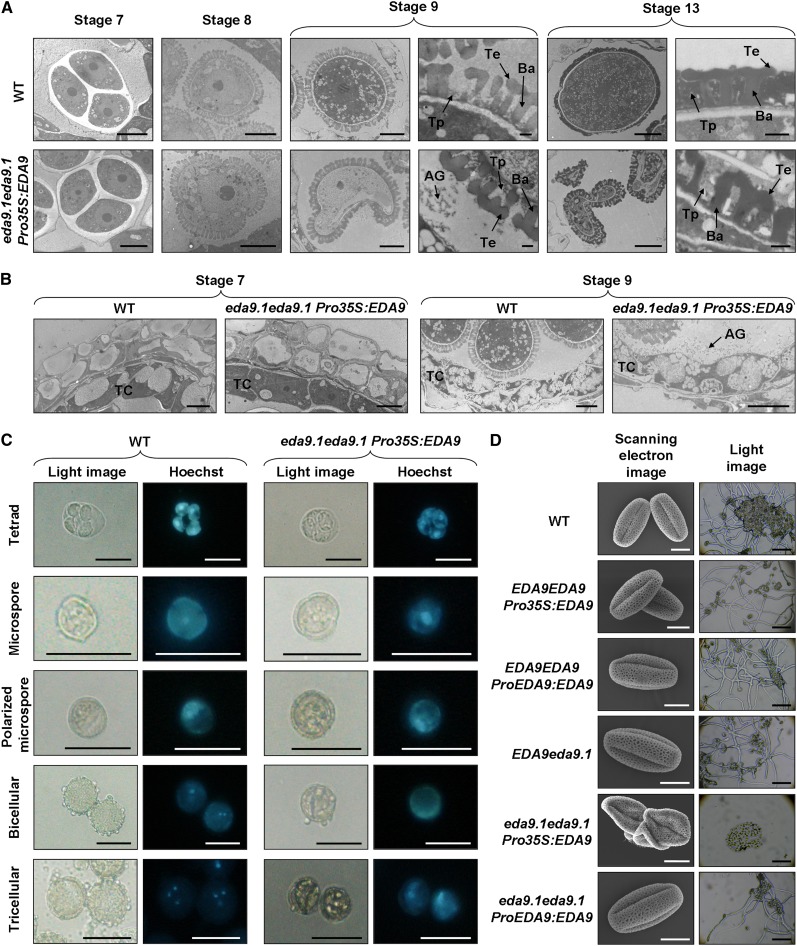
As the 35S promoter exhibits very low expression, if any, in the Arabidopsis tapetum (Grienenberger et al., 2009; Muñoz-Bertomeu et al., 2010), we performed an ontogenic serial analysis of anther and pollen development in the eda9.1eda9.1 Pro-35S:EDA9 sterile lines as compared with the wild type. For that purpose, floral buds were classified from stages 8 to 13 according to the landmark events described by Smyth et al. (1990) and analyzed by transmission electron microscopy (Fig. 9, A and B). In the eda9.1eda9.1 Pro-35S:EDA9 anthers, tetrads of microspores were formed (stage 7 according to Sanders et al. [1999]), which progressed in their development and were released from tetrads (stage 8). These initial steps of microspore development in eda9.1eda9.1 Pro-35S:EDA9 were similar to those observed in the wild type. However, eda9.1eda9.1 Pro-35S:EDA9 microspores suffered a delay in their development as compared with the wild type, which led to a degeneration process after the polarized microspore stage (from stage 9 and beyond). At the end of the maturation process (stage 13), most eda9.1eda9.1 Pro-35S:EDA9 pollen grains showed partially shrunken or completely collapsed protoplasms. The baculum and tectum of a typical exine pollen wall layer were present in the eda9.1eda9.1 Pro-35S:EDA9 gametophyte. However, the interstices of the exine pattern were not completely filled with tryphine, as was observed in the wild type.
Figure 9.
EDA9 expression is essential for proper pollen development. A, Developmental analysis of pollen (stages 7–13, according to Sanders et al. [1999]) in wild-type (WT) and homozygous eda9.1eda9.1 plants transformed with a EDA9-GFP cDNA under the control of the 35S promoter (Pro-35S:EDA9) by transmission electron microscopy. AG, Aggregated compounds; Ba, baculum; Te, tectum; Tp, tryphine. Bars = 5 μm for all images except closeups of stages 9 and 13, where bars = 0.5 μm. B, Cross sections of anthers (stages 7 and 9, according to Sanders et al. [1999]) in wild-type and homozygous eda9.1eda9.1 plants transformed with Pro-35S:EDA9 by transmission electron microscopy. TC, Tapetum cell. Bars = 5 μm. C, Developmental analysis of pollen (from tetrad to tricellular stage) in wild-type and eda9.1eda9.1 Pro-35S:EDA9 plants visualized by Hoechst staining of the microspore nucleus. Bars = 25 μm. D, Scanning electron micrographs of pollen grains (left) and light microscope images of pollen germination assays (right) from the wild type, the wild type transformed with Pro-35S:EDA9 (EDA9EDA9 Pro-35S:EDA9) and the endogenous EDA9 promoter (EDA9EDA9 ProEDA9:EDA9), and heterozygous EDA9eda9.1 and homozygous eda9.1eda9.1 plants transformed with Pro-35S:EDA9 and the endogenous EDA9 promoter (ProEDA9:EDA9). Bars = 10 μm (left column) and 200 μm (right column).
To confirm which step of gametophyte development was affected by the eda9.1 mutation, we performed Hoechst staining of microspore nucleus from eda9.1eda9.1 Pro-35S:EDA9 plants at different developmental stages (Fig. 9C). For this experiment, the most normally shaped microspores for the last stages of development were selected, since most of them were seriously malformed. Nucleus staining indicated that tetrad and polarized microspore development in eda9.1eda9.1 Pro-35S:EDA9 anthers was normal, but subsequent to the polarized microspore stage, development was arrested and no bicellular or tricellular pollen was found. These results indicate that microspores from eda9.1eda9.1 Pro-35S:EDA9 are unable to undergo the double mitosis necessary to reach the mature pollen stage. Scanning electron microscopy revealed that mature pollen from the eda9.1eda9.1 Pro-35S:EDA9 line exhibit shrunken and collapsed shapes and were unable to germinate when cultured in vitro (Fig. 9D).
The analysis of anther development indicated that the tapetal cell layer is formed in eda9.1eda9.1 Pro-35S:EDA9 anthers (Fig. 9B). We could not see any drastic morphological alteration in this eda9.1eda9.1 Pro-35S:EDA9 cell layer, as was observed in pollen. However, an accumulation of aggregated compounds in the anther locules of eda9.1eda9.1 Pro-35S:EDA9 at stage 9 was clearly observed. It is important to note that pollen in eda9.1eda9.1 Pro-35S:EDA9 anthers initiate the degeneration process at this developmental stage (Fig. 9A). In wild-type anther locules, no such compound accumulation could be seen at stage 9. The observed accumulation of aggregated compound in the eda9.1eda9.1 Pro-35S:EDA9 anther locules at stage 9 temporally correlated with the expression of EDA9 in this cell layer (Fig. 8). EDA9 was also clearly expressed in pollen, mainly in stage 9 and beyond (Fig. 8).
Metabolic Characterization of Overexpressed and Mutant Lines
Similar to the PGDH family mutants, wild-type plants overexpressing the three PGDH family genes (Oex lines) did not display a drastic alteration of the growth pattern when cultured in vitro, although some of them presented significantly reduced (Oex EDA9) growth (Supplemental Fig. S3). Metabolomic analysis, however, revealed a clearly altered metabolite content in both mutant and Oex lines as compared with the wild type (Table IV; Supplemental Table S3). The direct product of PGDH reaction, 3-phosphohydroxypyruvate, could not be determined, since phosphorylated intermediates are usually not detectable by gas chromatography-mass spectrometry. However, Ser, the final product of the PPSB, increased in both roots and aerial parts of all Oex lines. This increase was higher in roots than in the aerial parts for Oex PGDH and Oex EDA9 and similar for Oex 3-PGDH roots and aerial parts. The higher Ser increases as compared with the wild type were observed in the following order: Oex PGDH roots (4.2-fold increase) > Oex EDA9 roots (2.8-fold increase) > Oex PGDH aerial parts (2.1-fold increased) > Oex EDA9 aerial parts (1.7-fold increase) > Oex 3-PGDH aerial parts (1.45-fold increase) > Oex 3-PGDH roots (1.41-fold increase).
Table IV. Metabolite levels in the aerial parts and roots of mutants (pgdh1pgdh1 and 3-pgdh1 3-pgdh1) and overexpressing plants (Oex PGDH, Oex EDA9, and Oex 3-PGDH).
Data are relative values to the mean response calculated for the wild type. Wild-type values (included in Supplemental Table S3) were normalized to 1. Mutant data represent means ± se of seven independent determinations. Oex line data represent means ± se of 14 to 21 independent determinations corresponding to samples of two (Oex EDA9) or three (Oex PGDH and Oex 3-PGDH) independent transgenic plants. Those values that are significantly different from the wild type are shown in boldface (P < 0.05). The full data set is shown in Supplemental Table S3.
| Metabolite | Mutants |
Overexpressing Plants |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aerial Part |
Root |
Aerial Part |
Root |
|||||||
| pgdh1 | 3-pgdh1 | pgdh1 | 3-pgdh1 | Oex PGDH | Oex EDA9 | Oex 3-PGDH | Oex PGDH | Oex EDA9 | Oex 3-PGDH | |
| Amino acids | ||||||||||
| Asn | 2.04 ± 0.24 | 1.25 ± 0.13 | 1.15 ± 0.12 | 0.99 ± 0.10 | 0.66 ± 0.15 | 1.01 ± 0.33 | 0.91 ± 0.24 | 0.98 ± 0.12 | 1.83 ± 0.17 | 0.73 ± 0.14 |
| Gly | 0.66 ± 0.03 | 0.81 ± 0.03 | 0.78 ± 0.03 | 0.87 ± 0.03 | 0.77 ± 0.02 | 1.05 ± 0.04 | 1.17 ± 0.08 | 1.73 ± 0.05 | 1.25 ± 0.09 | 0.83 ± 0.03 |
| Ile | 0.82 ± 0.02 | 1.07 ± 0.03 | 1.33 ± 0.04 | 0.72 ± 0.02 | 0.97 ± 0.03 | 1.11 ± 0.18 | 0.81 ± 0.02 | 0.95 ± 0.02 | 1.11 ± 0.03 | 0.81 ± 0.02 |
| Lys | 1.35 ± 0.06 | 1.15 ± 0.03 | 1.20 ± 0.02 | 0.79 ± 0.03 | 0.81 ± 0.05 | 1.02 ± 0.07 | 0.75 ± 0.03 | 0.97 ± 0.02 | 1.19 ± 0.06 | 0.66 ± 0.02 |
| Met | 0.98 ± 0.03 | 1.33 ± 0.02 | 0.93 ± 0.02 | 0.93 ± 0.02 | 1.17 ± 0.04 | 1.33 ± 0.02 | 1.15 ± 0.03 | 1.44 ± 0.03 | 1.36 ± 0.03 | 1.00 ± 0.02 |
| Orn | 1.37 ± 0.06 | 1.29 ± 0.04 | 1.26 ± 0.13 | 0.70 ± 0.06 | 0.64 ± 0.03 | 1.16 ± 0.06 | 0.82 ± 0.04 | 0.68 ± 0.08 | 0.91 ± 0.19 | 0.56 ± 0.04 |
| Pro | 0.45 ± 0.01 | 0.85 ± 0.01 | 0.35 ± 0.01 | 0.74 ± 0.02 | 0.80 ± 0.04 | 1.28 ± 0.09 | 1.18 ± 0.09 | 0.77 ± 0.02 | 1.31 ± 0.02 | 0.76 ± 0.01 |
| Ser | 0.82 ± 0.02 | 1.33 ± 0.01 | 0.62 ± 0.02 | 0.97 ± 0.03 | 2.07 ± 0.07 | 1.66 ± 0.08 | 1.45 ± 0.05 | 4.19 ± 0.12 | 2.75 ± 0.07 | 1.41 ± 0.01 |
| Val | 0.82 ± 0.02 | 1.11 ± 0.02 | 1.17 ± 0.03 | 0.78 ± 0.02 | 0.98 ± 0.03 | 1.26 ± 0.18 | 0.96 ± 0.02 | 0.98 ± 0.01 | 1.21 ± 0.03 | 0.88 ± 0.01 |
| Organic acids | ||||||||||
| Citric acid | 0.72 ± 0.05 | 0.98 ± 0.03 | 0.49 ± 0.02 | 0.93 ± 0.02 | 0.67 ± 0.03 | 0.78 ± 0.07 | 0.55 ± 0.02 | 0.85 ± 0.02 | 0.86 ± 0.02 | 0.66 ± 0.01 |
| Glyceric acid | 0.71 ± 0.03 | 0.55 ± 0.01 | 1.00 ± 0.03 | 0.51 ± 0.01 | 0.52 ± 0.02 | 0.72 ± 0.02 | 0.46 ± 0.01 | 0.49 ± 0.01 | 0.74 ± 0.01 | 0.28 ± 0.01 |
| Malic acid | 0.54 ± 0.02 | 0.98 ± 0.02 | 0.70 ± 0.02 | 0.84 ± 0.01 | 0.87 ± 0.03 | 0.89 ± 0.05 | 0.70 ± 0.02 | 1.01 ± 0.01 | 1.02 ± 0.01 | 0.70 ± 0.01 |
| Malonic acid | 0.67 ± 0.02 | 1.26 ± 0.02 | 0.75 ± 0.04 | 1.05 ± 0.02 | ||||||
| Succinic acid | 1.00 ± 0.02 | 1.32 ± 0.02 | 0.65 ± 0.02 | 0.96 ± 0.02 | 1.74 ± 0.08 | 1.81 ± 0.13 | 1.24 ± 0.03 | 1.05 ± 0.01 | 1.23 ± 0.02 | 0.77 ± 0.01 |
| Sugars and sugar alcohols | ||||||||||
| Fru | 0.84 ± 0.03 | 0.68 ± 0.02 | 1.15 ± 0.04 | 1.03 ± 0.02 | 1.12 ± 0.04 | 1.27 ± 0.06 | 1.29 ± 0.06 | 0.34 ± 0.00 | 0.92 ± 0.01 | 0.42 ± 0.02 |
| Galactinol | 0.47 ± 0.02 | 0.59 ± 0.01 | 0.67 ± 0.01 | 0.58 ± 0.01 | 0.53 ± 0.04 | 1.11 ± 0.11 | 0.46 ± 0.02 | 0.61 ± 0.01 | 1.59 ± 0.02 | 0.86 ± 0.01 |
| Glc | 0.84 ± 0.02 | 0.90 ± 0.02 | 0.94 ± 0.09 | 0.92 ± 0.04 | 0.60 ± 0.02 | 0.73 ± 0.04 | 0.87 ± 0.03 | 1.48 ± 0.06 | 1.65 ± 0.06 | 0.87 ± 0.06 |
| Raffinose | 0.39 ± 0.03 | 0.51 ± 0.02 | 0.58 ± 0.02 | 0.67 ± 0.02 | 0.49 ± 0.03 | 0.88 ± 0.10 | 0.49 ± 0.03 | 0.55 ± 0.01 | 1.12 ± 0.02 | 0.69 ± 0.01 |
The metabolite profiles of pgdh1pgdh1 and 3-pgdh1 3-pgdh1 were nonidentical, indicating that PGDH and 3-PGDH perform different metabolic functions in the plant. For instance, while in aerial parts some metabolites changed in the same direction in both mutants (increased [Orn] or decreased [Gly, Pro, glyceric acid, Fru, Glc, galactinol, and raffinose]) as compared with controls, some others only changed significantly in one of the mutants (Asn, Met, citric acid, and succinic acid) or even changed contrapuntally in the mutants (Val and malonic acid). A similar behavior of both mutants with respect to some metabolites could also be observed in roots, changing in the same sense (Pro, citric acid, malic acid, galactinol, and raffinose). But again, some other metabolites changed in the opposite direction (Ile, Lys, and Val). Ser was significantly reduced in both roots and aerial parts of pgdh1pgdh1 but was not reduced in the roots and even increased in the aerial parts of 3-pgdh1 3-pgdh1.
DISCUSSION
Ser biosynthesis provides a good example of the complexity of plant primary metabolism. Three different Ser biosynthesis pathways have been described in plants (Kleczkowski and Givan, 1988; Ros et al., 2013). In quantitative terms, Ser production through the glycolate pathway is considered the most important, at least in photosynthetic cells (Tolbert, 1980; Douce et al., 2001). Experiments performed in the 1970s and 1980s using high CO2 concentrations and photorespiratory inhibitors provided strong evidence that nonphotorespiratory pathways could play an important role in Ser supply to plants (Johnson and Hatch, 1969; Snyder and Tolbert, 1974; Platt et al., 1977; Servaites, 1977; Morot-Gaudry, 1980). However, no genetic evidence for the physiological functions of these nonphotorespiratory pathways has been provided until very recently, when the last enzyme of the PPSB was characterized (Cascales-Miñana et al., 2013).
We identified three genes in the database coding for putative PGDH genes that we considered to constitute the PGDH family. The biochemical activity of PGDH has been demonstrated previously (Ho et al., 1999), but not that of 3-PGDH and EDA9. Due to the high percentage of identity among all three PGDH family proteins, especially the 100% identity in the amino acid residues forming the catalytic site and ligand-binding domain, we assumed that both 3-PGDH and EDA9 also have PGDH activity. This assumption would be substantiated by the significant increase in the Ser content, the final product of the PPSB in the Oex 3-PGDH and Oex EDA9 lines (an increase from 1.4- to 2.8-fold as compared with the wild type). In fact, Ser was the amino acid that increased the most in both Oex lines. Additionally, mutations in PSP1, the single gene coding for the last reaction of PPSB, produced the same embryo and pollen defects as eda9.1eda9.1, which would corroborate the participation of EDA9 in the same pathway.
According to our expression data, the PGDH family displays a very specific expression pattern at the organ, tissue, and cell levels. Both PGDH and EDA9 were preferentially expressed in roots, which supports the idea that the PPSB plays an important role in supplying Ser to this organ or to other nonphotosynthetic tissues (Ho and Saito, 2001; Cascales-Miñana et al., 2013). It has also been suggested that the PPSB could be relevant in photosynthetic organs, especially in the dark when the pathway of carbon flux from glycolate to Ser ceases to function (Kleczkowski and Givan, 1988; Ho and Saito, 2001). The induction of 3-PGDH and EDA9 expression in aerial parts of the plant under dark conditions (8-h exposure) provides experimental evidence for this idea. Finally, it has been proposed that in green tissues, the PPSB is probably of minor significance as compared with the glycolate pathway during daylight hours when photorespiration takes place (Ho and Saito, 2001). However, the specific expression of 3-PGDH in aerial parts supports a more important role of the PPSB in photosynthetic organs, even during the light period. This function might be more related to specific cell types (nonphotosynthetic) in these photosynthetic organs. A repression of all the PGDH family genes was observed in photosynthetic and nonphotosynthetic organs after 24 h of dark exposure. These opposing effects observed after different times of exposure to darkness could be related to the energetic state of cells. Long-term darkness conditions imply a reduced energy supply to cells from photosynthetic activity. Thus, a low energetic cell level could activate other mechanisms involved in the repression of PGDH family genes, short-circuiting the enhancing effect of darkness exposures. Treatments with Ser, the product of PPSB activity, did not repress the expression of any of the three PGDH family genes. However, translational or posttranslational inhibition of PGDH family expression/activity cannot be ruled out.
Our expression experiments indicated that an important transcriptional regulation of the PGDH family genes takes place, and not only at the tissue and cell levels but also by the environment (light-dark transitions). This strong transcriptional control was not seen with PSP1, which was similarly expressed in all organs studied (Cascales-Miñana et al., 2013). Thus, the presence of three genes coding for PGDH with different expression patterns suggests that the first enzyme of the PPSB might be the bottleneck regulating the pathway at the transcriptional level.
Mutants of the three PGDH family genes clearly display different phenotypes. While single pgdh and 3-pgdh mutants are viable and have no apparent phenotype under standard growth conditions, homozygous eda9.1eda9.1 mutants are unviable. This result indicates that the essential gene in this family is EDA9 and that PGDH and 3-PGDH may have compensating effects and/or specific nonessential functions under normal growth conditions. Homozygous eda9.1eda9.1 is embryo lethal, indicating that EDA9 activity is essential for embryo development. Specifically, mutants show a delay in embryo development starting at the globular stage, which finally leads to abortion. According to microarray databases, EDA9 is expressed in embryos from the globular stage onward. It is at the globular-heart transition stage when the first steps of proplastid differentiation occur (Devic, 2008). The delay in eda9.1eda9.1 mutant embryo development is probably related to the establishment of the PPSB in embryo plastids at the globular stage, as has been discussed previously (Cascales-Miñana et al., 2013).
eda9.1eda9.1 Pro-35S:EDA9 anthers displayed a male-sterile phenotype. According to microarray databases, the expression of EDA9 at the polarized microscope and bicellular stages correlates well temporally with the developmental alterations observed in pollen during maturation. However, reciprocal crossing revealed that the male mutant allele eda9.1 showed no significant difference in transmission efficiency as compared with the wild-type allele, indicating that there are no male gametophyte defects when gametes are produced by heterozygous anthers. These results suggest that EDA9 deficiency could not only affect the development of microspores but also that of other anther cellular types important for pollen development. Several of the genes controlling tapetum development have been found to affect pollen viability and/or development (Colcombet et al., 2005; Yang et al., 2007). Cells from the tapetum and the microspores develop as “communicating” neighbors. The tapetal cell layer plays an important role in the formation of the pollen wall during microgametogenesis, secreting molecules into the locule for microspore maturation. We could observe that the tapetally derived tryphine (Dickinson and Lewis, 1973) did not fill the interstices of the exine pattern in eda9.1eda9.1 Pro-35S:EDA9 pollen. An accumulation of compounds that aggregated in the anther loculus could be seen in the eda9.1eda9.1 Pro-35S:EDA9 anther at critical stages of pollen development (stage 9). The nature of the aggregated compounds in the anther locule is not known, but the proteinaceous fibrogranular nature of tryphine deposits liberated by the tapetal cells could be a good candidate. These deposits could not be seen in the wild type at the developmental stages observed in the eda9.1eda9.1 Pro-35S:EDA9 anther, which could indicate that the delivery of components from the tapetum is not properly incorporated into the pollen wall, because there is a time offset between the development of the two cell types or a problem with the component itself that leads to improper delivery of tapetum components to the pollen wall. Since we found a temporal correlation between EDA9 expression in the tapetum and microspore developmental alterations in eda9.1eda9.1 Pro-35S:EDA9, we propose that the absence of EDA9 activity is somehow able to affect the proper development of both microspores and tapetal cells, which, in turn, affects pollen maturation. Thus, our results support the essential role of EDA9 in the tapetum for proper pollen development.
eda9.1eda9.1 phenocopied the embryo and pollen defects of PSP1 mutants (Cascales-Miñana et al., 2013). The expression pattern of EDA9 is similar to that of PSP1. For instance, they are both expressed in the embryo, meristems, anthers, and pollen. We hypothesize that the essential role of these genes may be related to their expression in very specific cell types in which Ser would be supplied exclusively by the intracellular PPSB. Our results suggest that other Ser biosynthetic pathways, such as the glycerate pathway or the activity of the Gly decarboxylase complex:Ser hydroxymethyltransferase-coupled reactions, are of minor biological relevance in the tapetum, male microspores, or embryo cells. As Ser is easily transported within the phloem (Riens et al., 1991; Hunt et al., 2010), the Ser synthesized in photosynthetic cells through the photorespiratory pathway could readily be supplied to nonphotosynthetic cells. If our hypothesis is correct, some cellular types might be devoid of Ser transporters or might not be conveniently connected to the vasculature, or a combination of both of these factors may be involved.
Ser levels were significantly increased in Oex lines from all three PGDH family genes as compared with the wild type, which indicates that the three PGDH family enzymes could participate in the biosynthesis of this amino acid. Oex PGDH and Oex EDA9 lines increased the Ser level in roots more than in the aerial parts. Since PGDH and EDA9 were expressed under the control of the 35S promoter, these results would indicate a posttranscriptional/translational regulatory mechanism controlling their encoded enzyme activities.
PGDH overexpression provoked the highest increases in Ser in both aerial parts and roots as compared with 3-PGDH and EDA9. However, PGDH is not the essential gene for embryo and pollen development. These results would support that the essential function of EDA9 is related to its expression in specific cell types. The function of 3-PGDH seemed to be less important in our experimental conditions, since 3-pgdh1 3-pgdh1 did not display reduced Ser levels and the Oex 3-PGDH lines were less efficient in amino acid biosynthesis. Nevertheless, 3-PGDH could have a function under specific conditions, such as during the night in photosynthetic cells. This would be supported by its high expression level in the aerial parts and its induction under dark exposure. It is also interesting that the Ser levels were not reduced but increased in the aerial parts of 3-pgdh1 3-pgdh1. This increase may be related to the activation of the glycolate pathways by an unknown mechanism.
One question to be answered is which metabolic changes are important targets that translate into developmental modifications. In humans, it has been shown that PGDH expression alters Glc metabolism and affects cell morphology, proliferation, and oncogenesis (Bachelor et al., 2011; Locasale et al., 2011; Pollari et al., 2011; Possemato et al., 2011; Ying et al., 2012). Our results indicate that EDA9 plays an essential function in nonphotosynthetic actively dividing cells such as embryos and anthers. Thus, Ser could well be the link connecting primary metabolism with the control of cell cycle progression in specific cell types. This could be related to the role of Ser in transcriptional regulation (Timm et al., 2013).
This work corroborates that the PPSB plays an essential function in plant metabolism and development, probably because it provides Ser to specific cell types where the only supplies of the amino acid would originate from the intracellular pools. Further research is required to investigate the specific function of the PPSB in photosynthetic tissues and in a future atmosphere containing increased CO2 concentrations, where the contribution of the glycolate pathway to plant Ser supply could be compromised.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were supplied by the European Arabidopsis Stock Center (Scholl et al., 2000). Unless stated otherwise, seeds were sterilized and sown on 0.8% agar plates containing one-fifth-strength MS medium with Gamborg vitamins as described previously (Muñoz-Bertomeu et al., 2009). The selection of overexpressing plants and growth in greenhouse conditions were performed as described elsewhere (Muñoz-Bertomeu et al., 2009).
Primers
All primers used in this work are listed in Supplemental Table S4.
Mutant Isolation and Characterization
The mutant alleles of At1g17745, At3g19480, and At4g34200 (SALK_048256, SALK_149747, SM_3_37584, GK_877F12, GK_867A04, and GK_155B09) were identified in the SIGnAL Collection database at the Salk Institute (Alonso et al., 2003). Mutants were identified by PCR genotyping using gene-specific primers and left border primers of the T-DNA insertion. The T-DNA insertions were confirmed by sequencing the fragment amplified by the T-DNA internal primers and gene-specific primers (Supplemental Table S4).
Cloning and Plant Transformation
For gene promoter-reporter fusions, fragments corresponding to the native PGDH, 3-PGDH, and EDA9 promoters (1,506, 1,243, and 1,514 bp, corresponding to regions −1,500 to +6, −1,240 to +3, and −1,509 to +5 relative to the PGDH, 3-PGDH, and EDA9 translation start codons, respectively) were fused to the GUS gene in pCAMBIA1303 or pBI121. Promoter fragments of PGDH and 3-PGDH were PCR amplified from genomic DNA using the primers specified in Supplemental Table S4 to introduce HindIII and SpeI sites and cloned into pCAMBIA1303, giving ProPGDH:GUS and Pro-3-PGDH:GUS. The EDA9 promoter was PCR amplified from genomic DNA using primers specified in Supplemental Table S4 to introduce HindIII and BamHI sites and cloned into pBI121, giving ProEDA9:GUS.
The cDNAs corresponding to PGDH, 3-PGDH, and EDA9 were placed under the control of the constitutive 35S promoter (Pro-35S), giving constructs Pro-35S:PGDH, Pro-35S:3-PGDH, and Pro-35S:EDA9, respectively. PGDH and EDA9 cDNAs (U13169 and U09809 supplied by the Arabidopsis Biological Resource Center) were PCR amplified with primers specified in Supplemental Table S4 and cloned in the pCR8/GW/TOPO plasmid (Invitrogen). These cDNAs were subcloned in the plasmid pMDC83 (Curtis and Grossniklaus, 2003) using Gateway technology with Clonase II (Invitrogen). A cDNA from gene 3-PGDH was obtained by RT using total RNA as a template. This cDNA was PCR amplified using primers specified in Supplemental Table S4, cloned in the pCR8/GW/TOPO plasmid, and subcloned in the plasmid pMDC83 as described above. This plasmid allowed us to clone the PGDH family cDNAs in frame with a GFP cDNA at the C-terminal position. Pro-35S:EDA9 was used to obtain the construct ProEDA9:EDA9. The 35S promoter of Pro-35S:EDA9 (localized between the PmeI and PacI sites) was exchanged with the native EDA9 promoter previously PCR amplified from ProEDA9:GUS with primers specified in Supplemental Table S4 to introduce PmeI and PacI sites, giving the ProEDA9:EDA9 vector. All PCR-derived constructs were verified by DNA sequencing.
Arabidopsis plants were transformed with the different constructs using the floral dipping method (Clough and Bent, 1998) with Agrobacterium tumefaciens carrying pSOUP. For GUS and overexpressing lines, the wild type was used. As eda9.1eda9.1 is embryo lethal, whenever necessary, the progeny of heterozygous plants (EDA9.1 eda9.1) were transformed with the different constructs. Transformants were selected by antibiotic selection, while homozygous eda9.1eda9.1, heterozygous EDA9eda9.1, and the wild type were identified by PCR genotyping using gene-specific primers and left border primers of the T-DNA insertions (Supplemental Table S4).
At least five independent, single-insertion homozygous T3 lines were obtained for all the different constructs. After characterization by RT-PCR, two to three different lines were selected for further analysis depending on the experiment. We used both syngenic wild-type lines as well as wild-type Columbia-0 as controls for our studies. Syngenic plants were the wild-type progeny from the segregation of heterozygous mutant lines.
RT-PCR
RT-PCR was performed as described previously (Cascales-Miñana et al., 2013). Primers used are listed in Supplemental Table S4.
Pollen Germination and GUS Activity Assays
In vitro pollen germination assays were done using the optimized solid medium described by Boavida and McCormick (2007) as described previously (Muñoz-Bertomeu et al., 2010).
GUS activity assays were performed as described previously (Cascales-Miñana et al., 2013). At least three independent transgenic lines showed identical GUS staining patterns, which differed only in the expression level of GUS. Images were acquired with a Leica DM1000 microscope and a Leica DC350 digital camera.
Microscopy
For differential interference contrast microscopy, siliques from heterozygous EDA9 eda9.1 plants were dissected longitudinally at 1, 4, 6, and 10 DAP. Ovules from individual siliques were morphologically classified and processed as described previously (Cascales-Miñana et al., 2013). For transmission electron microscopy of anther and pollen, inflorescences containing buds at different developmental stages from wild-type and eda9.1eda9.1 Pro-35S:EDA9 plants were collected and classified into different development groups according to Smyth et al. (1990). Buds from the same group were removed from plants and processed as described previously (Cascales-Miñana et al., 2013).
For microspore nucleus staining with Hoechst, inflorescences containing buds at different developmental stages from wild-type and eda9.1eda9.1 Pro-35S:EDA9 plants were collected and classified in different groups as described for transmission electron microscopy. Pollen grains released from anthers were stained with 10 µg mL−1 Hoechst dye and observed as described (Cascales-Miñana et al., 2013).
For scanning electron microscopy, pollen was mounted on standard stubs and coated with gold-palladium prior to observation with a field emission microscope (Hitachi S-4100). GFP fluorescence was observed with a confocal microscope (Leica TCS-SP).
Metabolite Determination
Aerial parts and roots of 15-d-old wild-type, mutant, and Oex line plants (two to three different lines for each gene) grown on one-fifth-strength MS plates were used to determine metabolite content in derivatized methanol extracts by gas chromatography-mass spectrometry using the protocol defined by Lisec et al. (2006). Plants were grown in a growth chamber in a 22°C, 16-h-day/16°C, 8-h-night photoperiod, with light at 100 μmol m−2 s−1, and were sampled for metabolite analysis after 4 to 5 h in the light.
Bioinformatics and Statistics
PGDH genes were initially identified in The Arabidopsis Information Resource (http://www.arabidopsis.org/). The percentage of identity between different PGDHs was obtained by aligning pair sequences using bl2seq at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequences were aligned using the ClustalX program (Thompson et al., 1997), version 1.83, and the GENEDOC program. Phylogenetic analyses were performed according to the neighbor-joining method (Saitou and Nei, 1987). Bootstrapping was performed with 2,000 replicates to obtain support values for each internal branch, and the representation of the calculated consensus tree was drawn using the TreeView program (Page, 1996). Putative chloroplast/plastid localization sequences were identified using the ChloroP prediction server (Emanuelsson et al., 1999).
Experimental values represent mean values and se, and n represents the number of independent samples. P values were calculated with Student’s t test (two-tailed) using Microsoft Excel. The level of significance was fixed at 5% (0.05), representing the probability of error if the hypothesis of a significant difference between mean values was accepted. In the segregation analysis, data were tested to evaluate if the observed values agreed with Mendelian proportions using the χ2 test. P values higher than 0.05 indicate that the observed values are not significantly different from the expected ratio. P values lower than 0.001 indicate that the observed values differ significantly from the expected ratio.
Arabidopsis Genome Initiative locus identifiers for the Arabidopsis genes used in this article are as follows: At1g17745 (PGDH), At3g19480 (3-PGDH), and At4g34200 (EDA9).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignment of Arabidopsis PGDH family proteins using the GENEDOC program.
Supplemental Figure S2. Relative fresh weight (aerial parts and roots) of wild-type and PGDH family mutant seedlings grown in one-fifth-strength MS medium for 18 d.
Supplemental Figure S3. Relative fresh weight (aerial parts and roots) of wild-type and PGDH family overexpressing seedlings grown in one-fifth-strength MS medium for 18 d.
Supplemental Table S1. Genomic organization of PGDH family T-DNA mutant lines.
Supplemental Table S2. Some of the significantly enriched consensus sequences in the promoter regions of PGDH family genes.
Supplemental Table S3. Metabolite levels in the aerial parts and roots of wild-type, mutant (pgdh1pgdh1 and 3-pgdh1 3-pgdh1), and overexpressing (Oex PGDH, Oex EDA9, and Oex 3-PGDH) plants.
Supplemental Table S4. List of primers used in this work.
Acknowledgments
We thank Servei Central de Suport a la Investigació Experimental and Unitat Central de Investigació de Medicina of the Universitat de València for technical assistance. We also thank Hellen Warburton for the language review.
Glossary
- 3-PGA
3-phosphoglycerate
- PPSB
phosphorylated pathway of serine biosynthesis
- RT
reverse transcription
- T-DNA
transfer DNA
- DAP
days after pollination
- cDNA
complementary DNA
- Oex line
wild-type plants overexpressing the three PGDH family genes
- MS 1/5
one-fifth-strength Murashige and Skoog
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bachelor MA, Lu Y, Owens DM. (2011) L-3-Phosphoserine phosphatase (PSPH) regulates cutaneous squamous cell carcinoma proliferation independent of L-serine biosynthesis. J Dermatol Sci 63: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR. (2010) Photorespiration: players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S. (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Capron A, Chatfield S, Provart N, Berleth T. (2009) Embryogenesis: pattern formation from a single cell. The Arabidopsis Book 7: e0126., doi/10.1199/tab.0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales-Miñana B, Muñoz-Bertomeu J, Flores-Tornero M, Anoman AD, Pertusa J, Alaiz M, Osorio S, Fernie AR, Segura J, Ros R. (2013) The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25: 2084–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17: 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M. (2008) The importance of being essential: EMBRYO-DEFECTIVE genes in Arabidopsis. C R Biol 331: 726–736 [DOI] [PubMed] [Google Scholar]
- Dey S, Burton RL, Grant GA, Sacchettini JC. (2008) Structural analysis of substrate and effector binding in Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase. Biochemistry 47: 8271–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson HG, Lewis FRS. (1973) The formation of the tryphine coating the pollen grains of Raphanus, and its properties relating to the self-incompatibility system. Proc R Soc Lond B Biol Sci 184: 149–165 [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rébeillé F. (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M. (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Handford J, Davies DD. (1958) Formation of phosphoserine from 3-phospho-glycerate in higher plants. Nature 182: 532–533 [Google Scholar]
- Ho CL, Noji M, Saito K. (1999a) Plastidic pathway of serine biosynthesis: molecular cloning and expression of 3-phosphoserine phosphatase from Arabidopsis thaliana. J Biol Chem 274: 11007–11012 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Saito K. (1999b) Regulation of serine biosynthesis in Arabidopsis: crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J Biol Chem 274: 397–402 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Yamazaki M, Saito K. (1998) Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis. Plant J 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Ho CL, Saito K. (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20: 243–259 [DOI] [PubMed] [Google Scholar]
- Hunt E, Gattolin S, Newbury HJ, Bale JS, Tseng HM, Barrett DA, Pritchard J. (2010) A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J Exp Bot 61: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HS, Hatch MD. (1969) The C4-dicarboxylic acid pathway of photosynthesis: identification of intermediates and products and quantitative evidence for the route of carbon flow. Biochem J 114: 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Givan CV. (1988) Serine formation in leaves by mechanisms other than the glycolate pathway. J Plant Physiol 132: 641–652 [Google Scholar]
- Larsson C, Albertsson E. (1979) Enzymes related to serine synthesis in spinach chloroplasts. Physiol Plant 45: 7–10 [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. (2011) Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurino VG, Peterhansel C. (2010) Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol 13: 249–256 [DOI] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, Feijó JA. (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil d-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Morot-Gaudry JF. (1980) Oxygen effect on photosynthetic and glycolate pathways in young maize leaves. Plant Physiol 66: 1079–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Irles-Segura A, Mateu I, Nunes-Nesi A, Fernie AR, Segura J, Ros R. (2010) The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol 152: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernández E, Pozueta-Romero J, Kuhn JM, Segura J, Ros R. (2009) Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Platt SG, Plaut Z, Bassham JA. (1977) Steady-state photosynthesis in alfalfa leaflets: effects of carbon dioxide concentration. Plant Physiol 60: 230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollari S, Käkönen SM, Edgren H, Wolf M, Kohonen P, Sara H, Guise T, Nees M, Kallioniemi O. (2011) Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat 125: 421–430 [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. (2011) Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476: 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riens B, Lohaus G, Heineke D, Heldt HW. (1991) Amino acid and sucrose content determined in the cytosolic, chloroplastic, and vacuolar compartments and in the phloem sap of spinach leaves. Plant Physiol 97: 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R, Cascales-Miñana B, Segura J, Anoman AD, Toujani W, Flores-Tornero M, Rosa-Tellez S, Muñoz-Bertomeu J. (2013) Serine biosynthesis by photorespiratory and non-photorespiratory pathways: an interesting interplay with unknown regulatory networks. Plant Biol (Stuttg) 15: 707–712 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TB, Goldberg RB. (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Scholl RL, May ST, Ware DH. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servaites JC. (1977) Chemical inhibition of the glycolate pathway in soybean leaf cells. Plant Physiol 60: 461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter JC, Davies DD. (1968) The isolation and characterization of 3-phosphoglycerate dehydrogenase from peas. Biochem J 109: 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder FW, Tolbert NE. (1974) Effect of CO2 concentration on glycine and serine formation during photorespiration. Plant Physiol 53: 514–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Florian A, Wittmiß M, Jahnke K, Hagemann M, Fernie AR, Bauwe H. (2013) Serine acts as a metabolic signal for the transcriptional control of photorespiration-related genes in Arabidopsis. Plant Physiol 162: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert NE. (1980) Photorespiration. In DD Davies, ed, The Biochemistry of Plants, Vol 2. Academic Press, New York, pp 487–523 [Google Scholar]
- Tolbert NE. (1985) The oxidative photosynthetic carbon cycle and peroxisomal glycolate metabolism. In PW Ludden, JE Burris, eds, Nitrogen Fixation and CO2 Metabolism. Elsevier, New York, pp 333–341 [Google Scholar]
- Tolbert NE. (1997) The C2 oxidative photosynthetic carbon cycle. Annu Rev Plant Physiol Plant Mol Biol 48: 1–25 [DOI] [PubMed] [Google Scholar]
- Walton NJ, Woolhouse HW. (1986) Enzymes of serine and glycine metabolism in leaves and nonphotosynthetic tissues of Pisum sativum L. Planta 167: 119–128 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. (2012) Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149: 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]