A novel subunit of the cyanobacterial NDH1 complex may mediate its coupling either to the respiratory or the photosynthetic electron flow.
Abstract
The NAD(P)H oxidoreductase or complex I (NDH1) complex participates in many processes such as respiration, cyclic electron flow, and inorganic carbon concentration in the cyanobacterial cell. Despite immense progress in our understanding of the structure-function relation of the cyanobacterial NDH1 complex, the subunits catalyzing NAD(P)H docking and oxidation are still missing. The gene sml0013 of Synechocystis 6803 encodes for a small protein of unknown function for which homologs exist in all completely known cyanobacterial genomes. The protein exhibits weak similarities to the NDH-dependent flow6 (NDF6) protein, which was reported from Arabidopsis (Arabidopsis thaliana) chloroplasts as a NDH subunit. An sml0013 inactivation mutant of Synechocystis 6803 was generated and characterized. It showed only weak differences regarding growth and pigmentation in various culture conditions; most remarkably, it exhibited a glucose-sensitive phenotype in the light. The genome-wide expression pattern of the Δsml0013::Km mutant was almost identical to the wild type when grown under high CO2 conditions as well as after shifts to low CO2 conditions. However, measurements of the photosystem I redox kinetic in cells of the Δsml0013::Km mutant revealed differences, such as a decreased capability of cyclic electron flow as well as electron flow into respiration in comparison with the wild type. These results suggest that the Sml0013 protein (named NdhP) represents a novel subunit of the cyanobacterial NDH1 complex, mediating its coupling either to the respiratory or the photosynthetic electron flow.
Cyanobacteria are the only photolithoautotrophic prokaryotes performing oxygenic photosynthesis. As in plant chloroplasts, the light reactions are situated on an internal membrane system, the thylakoids. Linear electron flow starts at PSII connected to the water-splitting center and transfers electrons via the cytochrome b6f (Cytb6f) complex and PSI to NADP+. Additionally, cyanobacteria are able to perform cyclic electron flow around PSI, producing only ATP. These light reactions allow cyanobacteria to obtain the necessary energy and reductants at varying levels in the light. In the dark, cyanobacteria also perform a respiratory electron transport to fulfill energy demands at the expense of stored carbohydrates, usually glycogen. As in heterotrophic bacteria, electrons from NAD(P)H+H+ are fed into the respiratory chain via the NAD(P)H oxidoreductase or complex I (NDH1). However, the cyanobacterial respiratory and photosynthetic electron transport chains are linked (i.e. both use several electron carriers together, such as the Cytb6f complex and mobile electron carriers). The lumenal electron carriers cytochrome c (Cytc) and plastocyanin donate electrons not only to PSI but also to the respiratory terminal cytochrome oxidase (Cytox), usually of the aa3 type, where oxygen is reduced back to water. The proton gradient generated via respiratory or photosynthetic electron transport is used by the ATPase to generate ATP (Bryant, 1994).
It has been shown that distinct, strain-dependent differences exist depending on which respiratory and photosynthetic electron flow routes are interconnected or more separated. In strains such as our model, Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803), the complete respiratory chain is localized on thylakoids, whereas in cyanobacteria such as Synechococcus elongatus PCC 7942, the respiratory chain is more separated on the cytoplasmic membrane from the thylakoid-localized photosynthetic chain (Peschek et al., 1994). To acclimate toward different environmental conditions, the cyanobacterial electron transfer network shows a relatively high degree of flexibility not only in its activity but also in its composition. For example, the preference for plastocyanin under copper-replete conditions switches to Cytc under copper-deplete conditions, while iron limitation results in a switch from the iron-containing ferredoxin to flavodoxin (Hagemann et al., 1999). The cyclic electron flow around PSI can use different routes, mainly via NDH1 but also directly to Cytb6f (Yeremenko et al., 2005). Finally, respiratory electron transport also can be connected to three different terminal oxidases depending on strain or growth conditions (Pils and Schmetterer, 2001).
Particularly high functional as well as structural diversity was shown for the cyanobacterial NDH1 complex (Zhang et al., 2004). As in other bacteria, it is involved in respiration, transferring electrons from carbohydrate catabolism into the plastoquinone (PQ) pool (Haimovich-Dayan et al., 2011). However, NDH1 also is involved in the cyclic electron flow around PSI (Yeremenko et al., 2005; Bernát et al., 2011). These two NDH1 functions are conserved in the chloroplastidial NDH complex that is phylogenetically derived from the cyanobacterial one (Ifuku et al., 2011). Moreover, it also has been established that NDH1 is essential for the CO2 conversion into HCO3− as part of the cyanobacterial inorganic carbon-concentrating mechanism (Ogawa, 1991; Shibata et al., 2001). This functional diversity is reflected in a structural diversity thought to serve these different purposes. For example, many of the smaller NDH1 subunits are encoded by multigene families (e.g. ndhF or ndhD), which are differentially expressed under changing conditions such as high or low CO2. The expression changes result in the generation of differently sized NDH1 complexes with different subunit composition, which can preferentially function in respiratory electron transport and cyclic electron transfer around PSI (called NDH1L) or in the conversion of CO2 into HCO3− (called NDH1MS; Zhang et al., 2004). Despite intensive investigations on the cyanobacterial as well as the chloroplastidial NDH1 complexes, the subunits for NAD(P)H oxidation are still unknown, making the functioning of these complexes enigmatic (for review, see Battchikova et al., 2011a). Recent isolations of functional NDH complexes from Thermosynechococcus elongatus indicated that reduced ferredoxin could possibly directly transfer electrons via ferredoxin-NADP+ oxidoreductase to NDH1 (Hu et al., 2013).
Accordingly, genome searches or proteomic analyses of isolated NDH1 complexes have often been used to gain more insights into the function of the NDH1 complex. A new NDH subunit was found in chloroplasts, named NDH-dependent flow6 (NDF6; Ishikawa et al., 2008). A protein called NdhP displaying weak similarities to NDF6 was recently copurified with active NDH1 complexes from the cyanobacterium T. elongatus (Nowaczyk et al., 2011). Here, we report on the generation and characterization of the mutant Δsml0013::Km, in which the NDF6 homolog Sml0013 of Synechocystis 6803 was inactivated.
RESULTS AND DISCUSSION
Sequence Analysis
The gene sml0013 encodes for a small protein of only 40 amino acid residues. Initial BLAST-P analysis disclosed the presence of Sml0013-like proteins in the genomes of all cyanobacteria sequenced to date, including the marine picoplanktonic cyanobacteria of the genera Prochlorococcus and Synechococcus, which are characterized by a strongly reduced genome size (Scanlan et al., 2009; Larsson et al., 2011). In all genomes, these proteins are annotated as hypothetical proteins of unknown function. Moreover, proteins with similarities to Sml0013 are also encoded in the genomes of some cyanophages, for example, the Prochlorococcus phage P-SSM2 (Supplemental Fig. S1), as was already reported by Cobley (2010). A closer look into the genome organization and sequences allowed distinguishing three groups of Sml0013-like proteins among cyanobacteria. Their distribution correlates with the three major cyanobacterial clades defined by Gupta and Mathews (2010). Among basal cyanobacteria of clade A, such as Gloeobacter violaceus PCC 7421, these proteins are found as single genes. In all genomes of clade B cyanobacteria, such as Synechocystis 6803, they are linked to a gene encoding a protein belonging to the COG1236 class with a β-lactamase fold, predicted as exonuclease involved in RNA processing. There exists a GenBank accession reporting that the mutation of this gene resulted in a changed phycobilisome content in the cyanobacterium Microchaeta diplosiphon (ABB88926.1). Finally, among the clade C cyanobacteria comprising mostly marine picoplanktonic strains, the Sml0013 homolog is encoded upstream of a gene for putative amidases/creatinases (Supplemental Fig. S1). The ubiquitous distribution and the highly conserved sequence and genome organization suggest a meaningful role of Sml0013-like proteins among cyanobacteria.
Extended similarity searches with entries of the entire National Center for Biotechnology Information database identified further proteins (NDF6-like) among plants displaying low degrees of similarity toward the cyanobacterial Sml0013-like proteins. The nucleus-encoded protein NDF6 was first identified in chloroplasts of Arabidopsis (Arabidopsis thaliana) and shown to be functionally associated with the chloroplastidial NDH complex (Ishikawa et al., 2008). Accordingly, the protein was named NDF6 for the NAD(P)H dehydrogenase (NDH)-dependent flow6 superfamily (PLN00180 in the Conserved Domain Database) of land plants such as Arabidopsis, rice (Oryza sativa), grape (Vitis vinifera), and Populus trichocarpa (Ishikawa et al., 2008). However, the plant NDF6-like proteins are much larger than the similar proteins from cyanobacteria (Supplemental Fig. S2). Even after removing the N-terminal transit peptide for chloroplast import, the mature proteins in chloroplasts show an approximately 60-amino acid extension at the N terminus and a 40-amino acid extension at the C terminus. Thus, only the highly conserved central part of the plant NDF6-like proteins displays the similarities to the cyanobacterial Sml0013-like proteins. This part of the proteins contains a hydrophobic stretch, which possibly forms a membrane-spanning helix (Ishikawa et al., 2008). Interestingly, the similarity of another small subunit of the NDH1 in cyanobacteria (NdhS) and chloroplasts (CRR31) also was restricted to the central part of the much larger chloroplastidial protein (Battchikova et al., 2011b), which forms in CRR31 a domain displaying some similarities to PsaE involved in ferredoxin binding at PSI (Yamamoto et al., 2011). The first evidence that the NDF6-like proteins among cyanobacteria are really associated with NDH1 was provided by Nowaczyk et al. (2011), who identified an Sml0013 homolog in purified NDH1 complexes from T. elongatus. These findings suggest that the Sml0013 protein also might be associated with the NDH1 complex. To investigate the function of this small protein, we generated the Δsml0013::Km mutant of Synechocystis 6803 (Supplemental Fig. S3).
Characterization of Growth
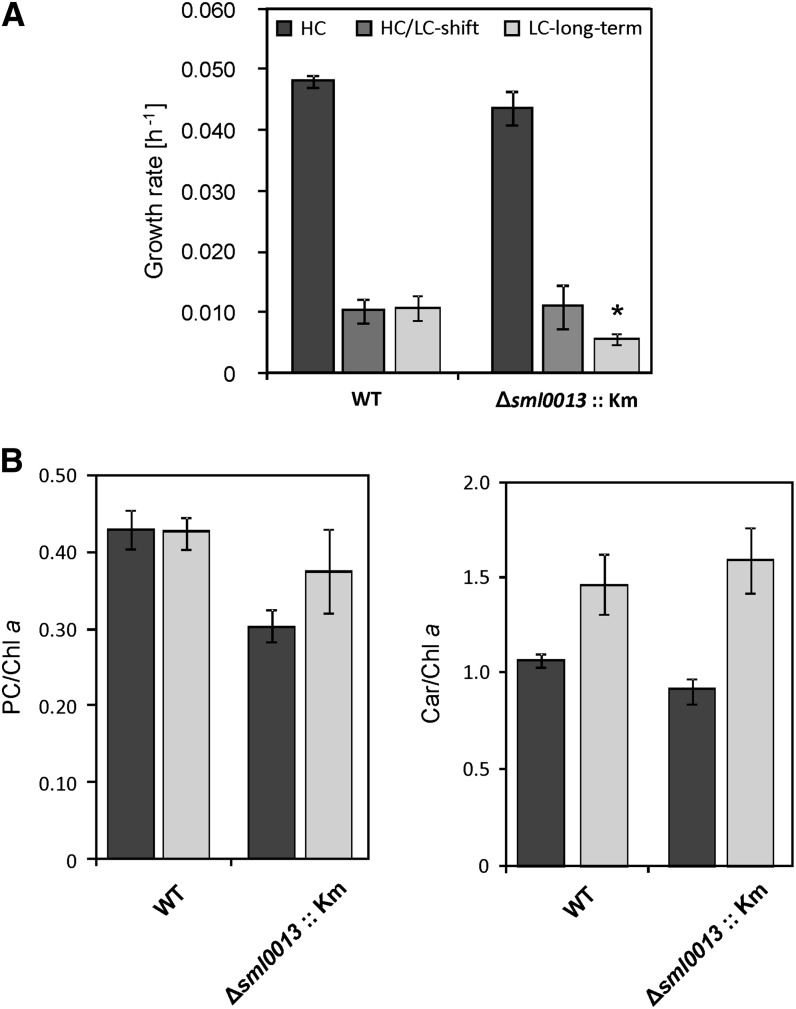
Insertion mutagenesis of sml0013 allowed the generation of many independent kanamycin (Km)-resistant clones, which showed the complete absence of the wild-type gene (Supplemental Fig. S3; Supplemental Table S1). This finding indicates that the Sml0013 protein is not essential under our standard growth conditions. To rule out polar effects, we generated an insertion mutant, Δsll0514::SmI, and a deletion mutant, Δsll0514::SmD, which were solely affected in the downstream open reading frame (ORF) sll0514 (Supplemental Fig. S4). Subsequently, the growth of the mutant clones and the wild type was compared under different conditions, including varying inorganic carbon amounts, inorganic nitrogen sources and amounts, light regimes, or temperatures. In most of the cases, wild-type and mutant cells behaved similarly (data not shown). Since it is known that the NDH1 is important for inorganic carbon uptake, we analyzed the behavior of these strains toward varying CO2 concentrations in more detail. Similar growth rates were found for the Synechocystis 6803 mutant Δsml0013::Km compared with the wild type at high CO2 (5%; HC) or 24 h after the shift to low CO2 (0.038%; LC), whereas long-term cultivation at LC decreased the growth of Δsml0013::Km cells (Fig. 1). Despite the similar growth, mutant cells were characterized by a lowered content of phycocyanin and carotenoids at HC, whereas pigmentation was not significantly different under LC conditions.
Figure 1.
Growth and pigmentation of cells cultivated under HC (5% CO2) or LC (0.035% CO2) conditions. A, Growth rates after cultivation under HC conditions, 24 h after shifting HC precultivated cells to LC, or after long-term LC acclimation. B, Pigmentation ratios of phycocyanin (PC) and carotenoids (Car) relative to chlorophyll a (Chl a) in HC-grown (dark gray columns) and long-term LC-acclimated (light gray columns) cells. Mean values and sd are shown. A statistically significant difference of mutant data in comparison with wild-type data (WT) is indicated by the asterisk (P ≤ 0.05).
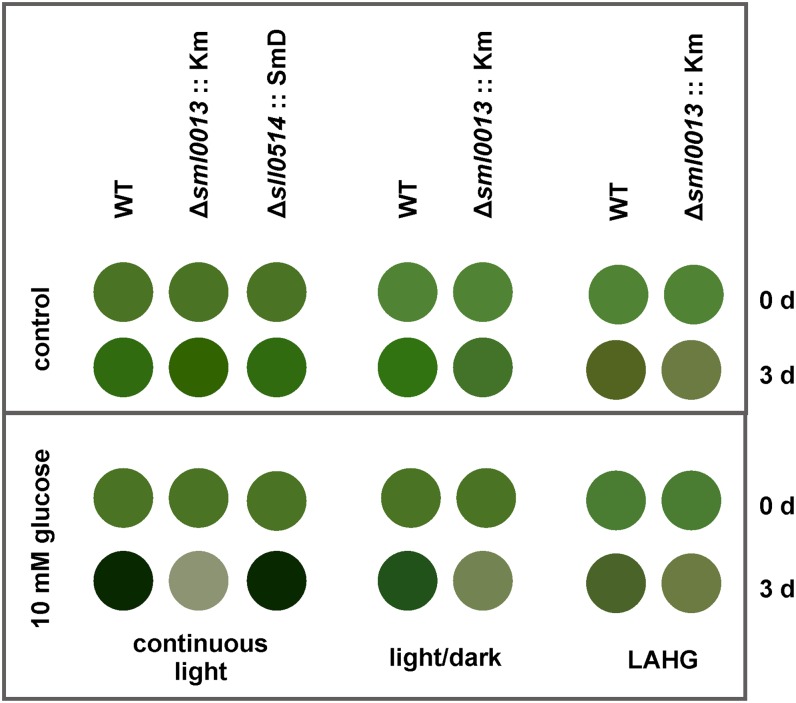
The exposure to different light regimes, continuous light, light/dark changes, or light-activated heterotrophic growth (LAHG), also did not result in any significant differences between mutant and wild-type cells (Fig. 2). In contrast, the addition of Glc to the medium revealed a Glc-sensitive phenotype of the Synechocystis 6803 mutant Δsml0013::Km when cultivated in flasks without CO2 supplementation. The strength of the phenotype became enhanced by the presence of light. Under continuous illumination, cells of the mutant Δsml0013::Km lysed 3 d after the addition of 10 mm Glc. If the Δsml0013::Km cells were cultivated with Glc under light/dark conditions, the growth became clearly diminished but Glc was not toxic, whereas no Glc effect was detected when cells were incubated under LAHG conditions. The mutants with defects in the downstream ORF sll0514 did not show the Glc-sensitive phenotype of the mutant Δsml0013::Km (Fig. 2). Moreover, precultivation at HC in the presence of Glc resulted in the release of a brownish compound into the medium by Δsml0013::Km cells, indicating stress. However, the cells survived and grew at almost the same rate as the wild type (growth rates of 0.024 ± 0.001 h−1 and 0.019 ± 0.003 h−1 for the wild type and mutant Δsml0013::Km, respectively). The Synechocystis 6803 wild-type strain used in this work is characterized by the ability to take up and utilize external Glc as a carbon resource. Accordingly, the growth of the wild type was stimulated by Glc at all light regimes (Fig. 2). Many different Glc-sensitive mutants of Synechocystis 6803 have been characterized in recent years (for review, see Haimovich-Dayan et al., 2011). Among them, mutants with defective NDH1 were found.
Figure 2.
Appearance of the Synechocystis 6803 wild type (WT) or mutant Δsml0013::Km as well as mutant Δsll0514::SmD after growth under different light regimes in the absence (control) or presence of 10 mm Glc. Axenic cultures of the wild type or mutants were cultivated in continuous light, light/dark changes, or LAHG under LC conditions for 3 d.
Gene Expression Changes
The genome-wide transcriptional changes in response to HC/LC shifts were compared between cells of the wild type and the mutant Δsml0013::Km. As reported before by Wang et al. (2004) and Eisenhut et al. (2007), shifts from HC into LC increased the transcript level of many genes, including those for inorganic carbon transport systems like SbtA (slr1512/slr1513 operon), BCT1 (slr0040-0044 operon), and NDH1MS (slr2006-2013 and sll1732-1736 operon; Table I). Many other genes also were found to be LC induced, for example, the flv operon (sll0217-sll0219; a graphical representation of the fold change values for each probe is provided in Supplemental File S1; the complete microarray data set is accessible in the Gene Expression Omnibus [GEO] database under accession no. GSE48415). The genes for the constitutive BicA transporter as well as subunits of the NDH1 complex showed only minor changes in both strains (Table I). Other NDH subunits also did not show any signs of an up- or down-regulation in cells of the wild type and the mutant Δsml0013::Km, which is supposed to be affected in an NDH1 subunit. Generally, the DNA microarray data set revealed that the removal of the gene sml0013 had only minor impact on the overall gene expression pattern under HC or HC/LC shift conditions in comparison with the wild type.
Table I. Selected LC-inducible genes in cells of the Synechocystis 6803 wild type or mutant Δsml0013::Km.
RNA was isolated from cells grown at HC conditions or after a shift from HC to LC for 24 h. The relative expression (fold change) of these genes is given for wild-type cells (wild-type LC/wild-type HC) and cells of the mutant (Δsml0013 LC/Δsml0013 HC). Changes of 1.87-fold or greater and 0.53-fold or less (boldface) indicate significantly increased or decreased transcript levels.
| Function | ORF | Gene Name | Wild-Type LC/Wild-Type HC | Δsml0013 LC/Δsml0013 HC |
|---|---|---|---|---|
| fold change | ||||
| Inorganic carbon transporter | ||||
| BCT1 | slr0040 | cmpA | 15.03 | 19.89 |
| slr0041 | cmpB | 14.81 | 18.21 | |
| slr0042 | porB | 12.87 | 14.93 | |
| slr0043 | cmpC | 8.13 | 10.26 | |
| slr0044 | cmpD | 9.60 | 10.93 | |
| sll0030 | cmpR | 1.41 | 1.40 | |
| SbtA | slr1512 | sbtA | 17.33 | 24.06 |
| slr1513 | sbtB | 9.56 | 11.13 | |
| NDH13 | sll1732 | ndhF3 | 23.02 | 25.71 |
| sll1733 | ndhD3 | 18.02 | 20.55 | |
| sll1734 | cupA | 11.65 | 13.25 | |
| sll1735 | 7.62 | 7.71 | ||
| sll1736 | 2.55 | 3.81 | ||
| sll1594 | ndhR | 4.24 | 2.80 | |
| NDH14 | sll0026 | ndhF4 | 0.68 | 0.68 |
| sll0027 | ndhD4 | 1.17 | 2.09 | |
| slr1302 | cupB | 0.42 | 0.33 | |
| BicA | sll0834 | bicA | 0.76 | 0.86 |
| Flavoprotein | ||||
| flv operon | sll0217 | dfa2 | 41.85 | 47.32 |
| sll0218 | 43.72 | 42.40 | ||
| sll0219 | dfa4 | 38.32 | 35.48 | |
However, a few significant gene expression changes were observed between wild-type and mutant cells. Increased mRNA levels were detected for 11 genes and decreased levels were detected for 14 genes under HC or HC/LC shift conditions (Table II). In addition, increased levels were detected for 18 noncoding and antisense RNAs and decreased levels were detected for four antisense RNAs (Supplemental Table S2). Among the latter are the antisense RNAs to flv4 (sll0217), consistent with the relatively decreased mRNA level of flv4 (Table II), supporting the control function of this antisense RNA (Eisenhut et al., 2012). Interestingly, ssr2016 was found among the genes with increased transcript levels. This gene encodes for a protein with low similarity to Pgr5, a protein known from land plants to be involved in the cyclic electron flow around PSI (Munekage et al., 2002). It has been shown that the ssr2016 mutant of Synechocystis 6803 also was affected in the antimycin A-sensitive route of cyclic electron transport, which acts independently from the NDH1 complex-mediated route (Yeremenko et al., 2005). Expression of the ATPase operon also was stimulated in mutant cells, which might indicate a decrease in the energy production of this strain. Potential imbalance in photosynthetic energy utilization in mutant cells also is mirrored by the increased expression level of ssr2595. This gene codes for HliB or ScpD, a small chlorophyll-binding protein that was found to be induced under high light, oxygen, and many other stresses affecting the growth of Synechocystis 6803 (Funk and Vermaas, 1999; Los et al., 2008; Engelken et al., 2012). In contrast to genes with elevated transcription, most of the genes characterized by a lowered expression level in mutant cells relative to the wild type code for proteins with unknown functions (Table II). The sml0013 gene, which was inactivated in the mutant Δsml0013::Km, showed the highest degree of diminished mRNA level, serving as a good control. Three of the repressed genes code for proteins that are known to be activated during nitrogen starvation (Aguirre von Wobeser et al., 2011). The NblA1 and NblA2 proteins are crucial for the degradation of phycobilisomes in the process of chlorosis after nitrogen starvation (Collier and Grossman, 1994; Baier et al., 2001), whereas the GifA protein together with GifB acts as a negative regulator for Gln synthetase, one of the main ammonia-assimilating enzymes (Garcia-Domínguez et al., 2000). Consistent with the decreased expression of gifA, an increased expression of glnA for Gln synthetase was detected (Table II). Together with the observed decreased phycocyanin/chlorophyll a ratio (Fig. 1B), the increased level of glnA mRNA and the decreased mRNA levels of nblA1, nblA2, and gifA can be taken as evidence that the nitrogen assimilation also is slightly affected in the mutant. However, as the general response to the shift from HC to LC was found to be intact, we interpret the gene expression changes between wild-type and mutant cells to be caused by the physiological response to lacking the Sml0013 protein.
Table II. Complete list of genes showing significant transcriptional changes in cells of the Synechocystis 6803 mutant Δsml0013::Km relative to the wild type.
The relative expression (fold change) of these genes is given for HC-grown cells (Δsml0013 HC/wild-type HC) and cells shifted from HC into LC (Δsml0013 LC/wild-type LC) for 24 h. Changes of 1.87-fold or greater and 0.53-fold or less indicate significantly increased or decreased transcript levels.
| ORF | Gene | Annotation | Δsml0013 HC/Wild-Type HC | Δsml0013 LC/Wild-Type LC |
|---|---|---|---|---|
| fold change | ||||
| Increased expression | ||||
| slr0798 | ziaA | Zinc-transporting P-type ATPase (zinc efflux pump) involved in zinc tolerance | 3.95 | 0.77 |
| ssr2595 | hliB | High light-inducible polypeptide HliB | 1.93 | 0.81 |
| ssr2016 | pgr5 | Hypothetical protein | 1.92 | 0.82 |
| sll1322 | atpB | ATP synthase B chain | 0.99 | 1.90 |
| sll1323 | atpG | ATP synthase G chain | 1.03 | 1.99 |
| sll1324 | atpF | ATP synthase F chain | 1.04 | 1.89 |
| ssl2615 | atpH | ATP synthase C chain | 1.09 | 2.28 |
| slr0447 | urtA | ABC-type urea transport system substrate-binding protein | 1.12 | 2.01 |
| sll1325 | atpD | ATP synthase δ-chain | 1.06 | 1.99 |
| slr1756 | glnA | Gln synthetase | 1.10 | 1.88 |
| sll1688 | Hypothetical protein | 1.92 | 0.82 | |
| Decreased expression | ||||
| slr1957 | Hypothetical protein | 0.47 | 0.68 | |
| sll1898 | ctaA | Hypothetical protein | 0.48 | 0.70 |
| sll0514 | Hypothetical protein | 0.40 | 0.44 | |
| ssr2062 | Hypothetical protein | 0.39 | 0.46 | |
| ssl1911 | gifA | Gln synthetase-inactivating factor IF7 | 0.38 | 0.89 |
| slr1634 | Hypothetical protein | 0.31 | 0.96 | |
| sll1862 | Hypothetical protein | 0.76 | 0.45 | |
| slr0442 | Hypothetical protein | 1.07 | 0.41 | |
| sll1695 | pilA2 | Pilin polypeptide PilA2 | 1.13 | 0.33 |
| sll1696 | Hypothetical protein | 1.05 | 0.42 | |
| ssl0452 | nblA1 | Phycobilisome degradation protein NblA1 | 1.39 | 0.35 |
| ssl0453 | nblA2 | Phycobilisome degradation protein NblA2 | 1.37 | 0.50 |
| sll1689 | sigE | Alternative σ-factor SigE | 0.48 | 0.73 |
| sml0013 | ndhP | Hypothetical protein | 0.17 | 0.14 |
NDH Activity Measurement via PSII Fluorescence
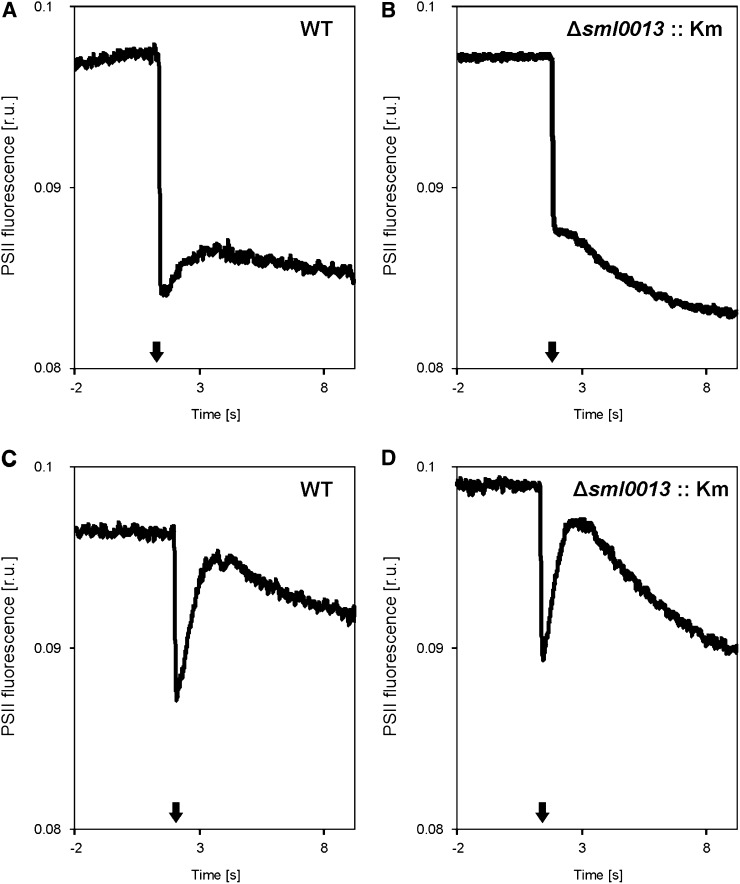
To get evidence that the Sml0013 protein is functionally associated with NDH1, we monitored the postillumination rise in chlorophyll a fluorescence. This parameter is often used as a measure for the rate of NDH1-based cyclic electron transport rates in cyanobacteria (Battchikova et al., 2011b) or in chloroplasts (Ishikawa et al., 2008). Our measurements revealed a small postillumination rise in cells of the wild type when grown under HC, which was almost absent in the mutant Δsml0013::Km under the same conditions (Fig. 3). In contrast, LC-acclimated cells of the wild type and Δsml0013::Km showed a much larger increase in PSII fluorescence after switching off the actinic light. It has been shown that acclimation to low CO2 supply increases the amount and composition of NDH1 subunits as well as the rate of PSI cyclic electron transport (Wang et al., 2004; Zhang et al., 2004; Eisenhut et al., 2007). Accordingly, our data imply that the absence of Sml0013 is mostly affecting the NDH1L complex that dominates under HC, while the NDH1MS complexes for CO2 uptake, especially under LC, are rather not affected by this mutation.
Figure 3.
Postillumination (at time point 0 s, actinic light was switched off; black arrows) rise of PSII chlorophyll a fluorescence in cells of the Synechocystis 6803 wild type (WT) or mutant Δsml0013::Km after growth at HC or LC conditions. A, Wild type at HC. B, Δsml0013::Km at HC. C, Wild type at LC. D, Δsml0013::Km at LC.
PSI Redox Kinetics
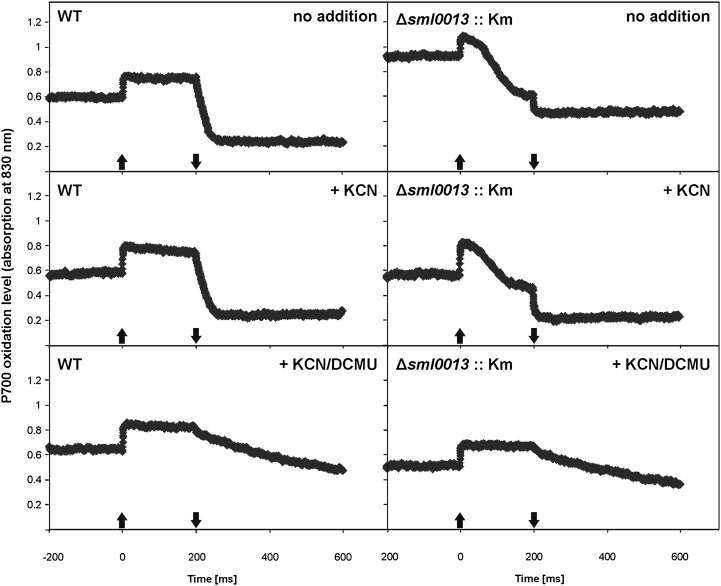
Since one main function of NDH1 is participation in the cyclic electron transport around PSI (for review, see Battchikova et al., 2011a), the redox kinetics of PSI were measured using the pulse-amplitude measurement (PAM) technique. In a first attempt, the slow kinetics of PSI oxidation in the presence of a 20-s far-red light treatment were measured with cells of both strains cultivated under HC conditions. The traces of wild-type and mutant cells were almost similar without added inhibitors (Fig. 4), as reported before for mutants of Synechocystis 6803 defective in ndhF genes (Bernát et al., 2011). The P700+ signal increased after actinic light was switched on and returned to the initial level after it was switched off. To distinguish between electron flow around PSI and electron drain to respiration via Cytox, KCN was added. The addition of KCN changed the redox kinetics of PSI in the wild type, while the traces of the Δsml0013::Km mutant cells remained almost unchanged. The PSI oxidation in wild-type cells almost lost the fast increase after far-red light illumination; instead, it showed a rather slow PSI oxidation increase (Fig. 4). Similar curves were reported by Bernát et al. (2011), where the wild type showed much slower P700 oxidation, whereas it was almost not affected by KCN addition in the ΔndhF1/3/4 triple mutant. Obviously, under KCN-free conditions, many electrons are flowing into respiration in wild-type cells; therefore, PSI oxidation was rapid, while inhibited respiration led to a prolonged oxidation time due to increased transfer of electrons toward PSI, which can be a sign for increased cyclic electron transport around PSI or an increased flow of electrons from metabolic activities [e.g. NAD(P)H+H+ produced by catabolic activities] to PSI. This slow increase is almost absent in KCN-treated mutant cells, which indicates that the absence of Sml0013 resulted rather in an impaired electron influx into the PQ pool and not in a changed electron flow to Cytox. The addition of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) to KCN-treated cells had only a small impact on the PSI oxidation traces in Δsml0013::Km and wild-type cells. Since it is known that cultivation under different CO2 amounts changes the composition of the NDH1 (Zhang et al., 2004), we made the same measurements with cell material acclimated long term to LC conditions. These measurements with LC-acclimated cells showed qualitatively the same results (Supplemental Fig. S5) as those with HC cells discussed above.
Figure 4.
Slow redox kinetics of PSI measured as change in A830 after illumination with strong far-red light (710 nm) for 20 s (up and down arrows indicate switching of actinic light on and off). Cells precultured under HC conditions of the wild type (WT) and the mutant Δsml0013::Km were measured without the addition of inhibitors, after the addition of KCN to inhibit electron flow into the respiratory electron chain toward Cytc oxidase, and after combined addition of KCN and DCMU to inhibit both respiratory and photosynthetic linear electron flow from PSII.
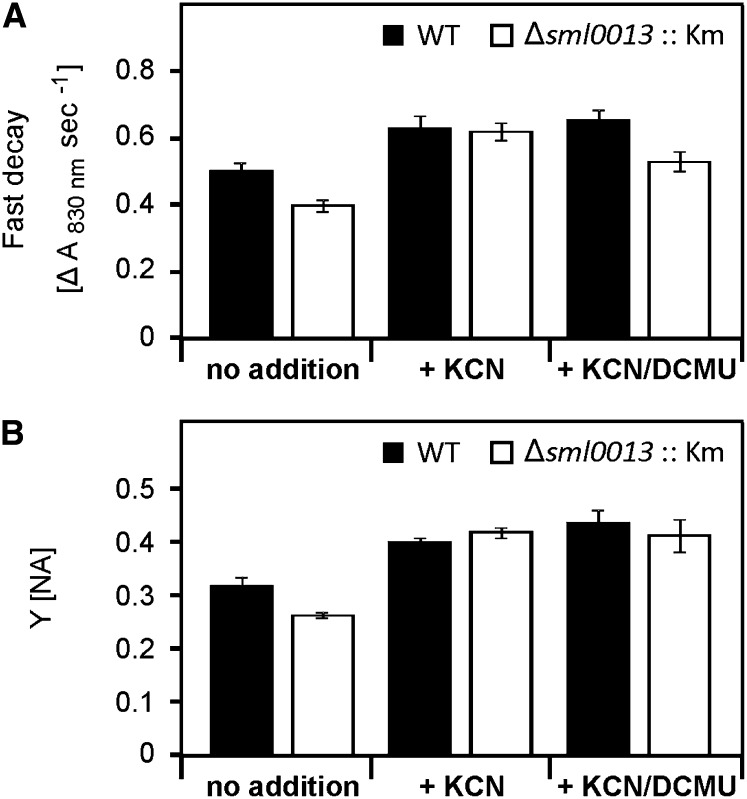
The quantitative evaluations of the slow PSI redox kinetic measurements supported the visual evaluation (Fig. 5). The decay of the P700+ signal reflecting the electron flow rate to P700+ was slower by about 20% for the mutant Δsml0013::Km compared with the wild type. This difference indicates that fewer electrons reach PSI in mutant cells, supporting the view of a diminished NDH1 function in Δsml0013::Km. The addition of KCN increased the P700+ rereduction rate in wild-type cells as well as mutant cells, because fewer electrons can escape from the cyclic electron flow around PSI to the Cytox. Interestingly, KCN addition equals the P700+ rereduction rate in both strains. KCN increased the rate almost 2-fold more in cells of Δsml0013::Km compared with the wild type, indicating that more electrons seem to be transferred toward respiration in mutant cells. Alternatively, the rate of PSI rereduction following KCN addition cannot get any faster in either of the two strains, because of a common limitation in electron transfer to PS1. An increased flow to respiration also was shown for the mutant ΔndhF1 after KCN addition (Bernát et al., 2011). The final addition of DCMU did not change this rate in the wild type, whereas cells of the mutant decreased the decay somehow, indicating that in these cells some electrons from PSII are reaching PSI even when far-red actinic light is used.
Figure 5.
Quantitative evaluation of the measurements of redox kinetics of PSI. Cells precultured under HC conditions of the wild type (WT) and the mutant Δsml0013::Km were measured without the addition of inhibitors, after the addition of KCN to inhibit electron flow into the respiratory electron chain toward Cytc oxidase, and after combined addition of KCN and DCMU to inhibit both respiratory and photosynthetic linear electron flow from PSII. From the slow kinetics, we calculated the rates of the decay phase after switching off far-red light (A). From the fast kinetics, the Y(NA) was calculated (B), reflecting the amount of PSI that can be oxidized under the different conditions. Mean values and sd from one typical experiment are shown (n = 3).
Long-term LC-acclimated cells showed similar trends for the wild type and Δsml0013::Km, albeit the rates were always slower, as in HC cells (Supplemental Fig. S6). Mutant cells always showed slower rereduction rates independent of whether inhibitors were added. KCN addition almost doubled this rate in the two strains; however, the mutant cell rate still was clearly lower than that in wild-type cells. Again, DCMU addition had a much higher effect on the A830 decay rate in Δsml0013::Km cells. The P700+ rereduction became even slower than in nontreated cells, while the wild type did not show such a high DCMU effect.
Since the addition of DCMU revealed changes in the slow kinetics of P700 oxidation with far-red light, especially with cells of Δsml0013::Km, we assumed that even under this light condition, electrons from PSII are influencing the PSI redox kinetics. Therefore, in another set of PAM experiments to measure fast PSI redox changes, we used an additional white light pulse to obtain a complete oxidation of PSI in cells preilluminated by actinic far-red light, as done before. An immediate rise in A830 was observed (Fig. 6; 0.213 ± 0.008 versus 0.135 ± 0.003 for the wild type and Δsml0013::Km when cultivated at LC; 0.221 ± 0.011 versus 0.154 ± 0.005 for the wild type and Δsml0013::Km when grown at HC). This finding indicates that the actinic far-red light was not sufficient to fully oxidize PSI, probably due to a continuous electron inflow to PSI from cyclic electron flow and/or metabolic activities. Moreover, the higher increase in wild-type cells compared with the mutant Δsml0013::Km in the absence of inhibitors implies that a higher amount of electrons from cyclic electron flow and/or metabolism reached PSI in wild-type cells. During the subsequent 200-ms white light pulse, P700+ was expected to become quickly rereduced due to electrons from PSII via linear electron flow. Interestingly, a clear rereduction of P700+ was only observed in mutant cells, while the wild type showed only a very small decrease of A830 during the short white-light period. The different behavior during the 200-ms white light pulse indicates a much better coupling of PSII to PSI in mutant compared with wild-type cells. An almost similar picture was observed after the addition of KCN. Again, cells of the mutant Δsml0013::Km showed a clear rereduction of PSI, although with a slightly slower rate, during the 200-ms white light pulse, whereas wild-type cells exhibited only a minor decrease in the PSI oxidation level (Fig. 6). In contrast to Δsml0013::Km, mutants defective in the downstream ORF sll0514 showed no difference compared with wild-type cells in measurements of fast PSI redox changes (Supplemental Fig. S4).
Figure 6.
Fast redox kinetics of PSI measured as change in A830 after preillumination with strong far-red light (710 nm) and a white light pulse of 200 ms (up arrows indicate white light on, and down arrows indicate switching off actinic far-red and white light). Cells precultured under HC conditions of the wild type (WT) and the mutant Δsml0013::Km were measured without the addition of inhibitors, after the addition of KCN to inhibit electron flow into the respiratory electron chain toward Cytc oxidase, and after combined addition of KCN and DCMU to inhibit both respiratory and photosynthetic linear electron flow from PSII.
The Y(NA), which reflects the relative proportion of acceptor-limited PSI centers (i.e. centers in the state P700 A−), was now similar in HC-grown cells of the mutant compared with the wild type (Fig. 5). This finding can be interpreted as a sign of fewer electrons being drained to Cytox in the wild type compared with the mutant. Finally, cells were treated with KCN and DCMU to block both respiratory and photosynthetic linear electron flow. This treatment abolished the fast rereduction of PSI during the 200-ms pulse in mutant cells almost completely (Fig. 6), verifying that the rereduction of PSI observed in the absence of inhibitors was due to an efficient PSII-to-PSI coupling. However, the DCMU additions had no additional effect on KCN-treated cells of the wild type as well as the mutant regarding the Y(NA) at HC (Fig. 5). The long-term acclimation to LC resulted in few alterations in the fast PSI reduction measurements. The traces as well as the Y(NA) calculations were very similar to the situation discussed above for HC cells (Supplemental Figs. S6 and S7).
Interestingly, the mutation of sml0013 improved the coupling of PSII and PSI, because only mutant cells showed a strong P700+ rereduction during the white light pulse independent from the CO2 content of the preculture. Similarly, the P700+ rereduction rate in measurements of the slow redox kinetics of PSI was also much more strongly influenced by DCMU in mutant than in wild-type cells. One possibility to explain this difference might be different redox levels of the PQ pool. Therefore, the PQ pool was assessed using the method described by Asada et al. (1992). However, these measurements revealed no differences in PQ reduction between wild-type and mutant cells regardless of whether they were grown under HC or LC conditions (data not shown).
CONCLUSION
Short protein-coding genes constitute an abundant, yet largely uncharacterized, fraction in bacterial genomes. The results obtained here suggest that the only 40-amino acid protein Sml0013 is a yet undefined subunit of the cyanobacterial NDH1 complex in Synechocystis 6803. It might be involved in the coupling of NDH1 to other electron transport complexes on the thylakoids, thus influencing the relative flux of electrons into the different photosynthetic or respiratory electron transport chains. As proposed before (Nowaczyk et al., 2011), we suggest to name it NdhP. The presence of NdhP homologs in the genome of all 126 cyanobacteria sequenced to date (Shih et al., 2013) indicates an important role of this protein in the natural environment of cyanobacteria. The assumption of an important role of this putative small NDH1 subunit is supported by the notion that NdhP proteins are homologous to NDF6, a subunit of the chloroplastidial NDH complex (Ishikawa et al., 2008). Thus, the chloroplastidial NDH complex, which is evolutionarily and functionally closely related to NDH1 of cyanobacteria (Shikanai, 2007; Ifuku et al., 2011), also kept this small protein subunit despite the long separate evolution.
However, the deletion of NdhP was possible in cells of Synechocystis 6803 without affecting cell viability under our standard laboratory conditions. Despite the testing of various growth conditions, only the addition of Glc to cells grown under LC conditions was lethal for the Δsml0013::Km mutant. The presence of Glc has an effect on many different cellular processes, including respiration and photosynthesis (Haimovich-Dayan et al., 2011). It has been shown that the NDH1L complex is not only involved in cyclic electron flow but also required for Glc utilization in Synechocystis 6803 (Zhang et al., 2004). Because cultivation under LAHG conditions alleviated the Glc-sensitive phenotype, it can be concluded that neither Glc nor one of its metabolites is directly toxic, but the transfer of electrons from Glc metabolism via an NDH1L complex devoid of NdhP somehow results in overreduction of the electron transfer chain on the thylakoids under light conditions. Thus, the Glc-sensitive phenotype of the Synechocystis 6803 mutant Δsml0013::Km resembles the phenotype reported for many other mutants defective in subunits of the NDH1 complex (for review, see Battchikova et al., 2011a; Haimovich-Dayan et al., 2011).
The rather minor importance of NdhP under our laboratory conditions is reflected by the finding that the overall gene expression pattern was only slightly changed in cells cultivated under HC as well as HC/LC shift conditions. None of the genes for other NDH1 subunits showed any significant difference in the expression level changes in mutant relative to wild-type cells. Interestingly, the gene for the PGR5 homolog in Synechocystis 6803 was up-regulated, which correlates with the observed changed activity of cyclic electron flow around PSI in the mutant without NdhP. Additionally, the observed alteration in the phycocyanin/chlorophyll a ratio in mutant cells also is in line with the reduced expression of the genes for proteins involved in nitrogen utilization such as glnA, nblA, and gifA.
Despite the rather minor alterations in growth and pigmentation, marked differences were detected in the PSI redox kinetics between wild-type and mutant cells. These experiments showed a decreased activity of cyclic electron flow around PSI, an increased electron transfer out of the photosynthetic into the respiratory electron transfer chain, and an enhanced coupling of PSII to PSI in cells of the mutant Δsml0013::Km. Since defects in the cyclic electron flow around PSI and coupling of the photosynthetic and respiratory electron flow also have been often reported for mutants with missing NDH1L subunits (Battchikova et al., 2011a, 2011b; Bernát et al., 2011; Bolychevtseva et al., 2011), the finding of similar alterations with cells of Δsml0013::Km support our hypothesis that NdhP is functionally associated with the NDH1 in Synechocystis 6803. Moreover, the Glc-sensitive phenotype and the absence of marked differences in the PSI redox changes between HC- and LC-acclimated cells make it most likely that NdhP is especially associated with the NDH1L complex (Zhang et al., 2004), which is known to act as part of the joint photosynthetic/respiratory electron flow in Synechocystis 6803. Our measurements of PSI redox kinetics suggest that the small NdhP protein possibly somehow improves or even mediates the coupling of PSI and NDH1L, leading to an improved cyclic electron transfer and making it less open for electron entry from PSII or the electron outflow to Cytox, two processes relatively enhanced in the ndhP mutant. Whether PSI and NDH1 supercomplexes exist in cyanobacterial cells, as has been shown for plant chloroplasts (Shikanai, 2007), is a matter of discussion that cannot be touched on by our experiments.
MATERIALS AND METHODS
Strains and Cultivation
The Glc-tolerant strain Synechocystis sp. PCC 6803 was obtained from Norio Murata (National Institute for Basic Biology) and served as the wild type. Cultures were grown on plates with BG11 medium (Rippka et al., 1979) containing 0.8% agar and buffered with 20 mm TES-KOH to pH 8.0 under constant illumination (30 µmol photons m−2 s−1) at 30°C. Cultivation of the mutant Δsml0013::Km was performed in the presence of 50 µg mL−1 kanamycin. For the physiological characterization, axenic cultures were grown under light/dark regimes: (1) continuous light at 50 µmol photons m−2 s−1; (2) diurnal light/dark cycles (12 h of light/12 h of dark); and (3) complete darkness with a 20-min light pulse at 50 µmol photons m−2 s−1 every 24 h (LAHG; Anderson and McIntosh, 1991). Growth experiments on plates or in shaken flasks were performed under controlled conditions in the climate cabinet BrightBoy GroBank Mobylux BB-XXL3+ (CLF Plant Climatics). To grow cells under photoheterotrophic or LAHG conditions, the BG11 medium was supplemented with 10 mm Glc. To test the axenic character of the cyanobacterial culture, 20 µL of the culture was dropped onto a plate with Luria-Bertani medium and cultivated at 30°C for 48 h.
To obtain higher amounts of biomass, cells were grown photoautotrophically in batch cultures using glass vessels of 3 cm diameter with 5-mm glass tubes for aeration (blubbing flow rate was 5 mL min−1) under continuous illumination of 130 µmol photons m−2 s−1 (Osram L58 W 32/3) at 29°C. For CO2 shift experiments, cells were precultivated with air enriched with 5% CO2 (defined as HC) in BG11 medium of pH 8.0. Then, cells were harvested by centrifugation (5 min at 3,000g, 20°C). The pellet was washed and resuspended in fresh BG11 medium of pH 7.0 at an optical density at 750 nm of 0.8. After 1 h of cultivation under HC conditions, CO2 limitation was set by shifting the exponentially growing cultures to bubbling with ambient air containing 0.038% CO2 (defined as LC). For long-term acclimation to HC or LC, cells were cultivated under the defined conditions for 72 h. The suspensions were daily diluted to an optical density at 750 nm of 0.8 by the corresponding medium.
Generation of Mutants
Total DNA from Synechocystis 6803 was isolated according to Hagemann et al. (1997). All other DNA techniques, such as ligation, plasmid isolation, restriction analysis, and transformation of Escherichia coli, were standard methods (Sambrook et al., 1989). To delete the gene sml0013, the encoding sequence together with approximately 750 bp of upstream or downstream sequences were amplified using specific oligonucleotide primers (Supplemental Table S1) via PCR. PCR was carried out using the Taq-PCR Master Mix (Qiagen) and additionally Elongase Enzyme Mix (Invitrogen). The fragments were separately cloned into the pGEMT (Promega) vector, resulting in pGEMT-sml0013-up as well as pGEMT-sml0013-down. To combine upstream and downstream sequences, first the pGEMT-sml0013-down vector was cut with NheI and NdeI. Then, the pGEMT-sml0013-up vector was cut with XbaI and NdeI to obtain the sml0013 upstream genome area. This fragment was inserted into the pGEMT-sml0013-down vector. The coding sequence of sml0013 was replaced by the aphII gene cartridge (for aminoglycoside phosphotransferase II, conferring kanamycin resistance), which was excised from pUC4K (Pharmacia) with StuI and BamHI. This kanamycin resistance cartridge was inserted into the ClaI cutpGEMT-sml0013-up-down vector (carrying the upstream and downstream sml0013-localized genome area). To obtain compatible ends for ligation, 5′ overhangs were filled in using the Klenow enzyme (Fermentas). The final insert was verified by DNA sequencing (Seqlab). The transformation of wild-type Synechocystis 6803 with the generated mutation was done as described by Hagemann and Zuther (1992). The genotype and complete segregation of the Synechocystis 6803 mutant Δsml0013::Km were checked by PCR using the primers sml0013_fw and sml0013_rev (Supplemental Table S1). Only DNA fragments with sizes corresponding to the mutated genes were detected with DNA from mutant clones, whereas fragments of wild-type gene sizes were completely absent (Supplemental Fig. S3).
To rule out polar effects, mutants defective in sll0514, which represents the ORF downstream of sml0013, were generated. The wild-type fragment of sll0514 was generated by PCR and cloned into pGEMT (Promega). The mutant Δsll0514::SmI was generated by insertion of the spectinomycin resistance cartridge obtained from pUC4S into the uncial BamHI site of sll0514. The mutant Δsll0514::SmD was generated by deletion of the 671-bp HpaI/SmaI fragment from sll0514 and its subsequent replacement by the spectinomycin resistance cartridge. The genotype and complete segregation of the Synechocystis 6803 mutants Δsll0514::SmI and Δsll0514::SmD were checked by PCR using the primers sll0514_fw and sll0514_rev (Supplemental Table S1). Only DNA fragments with sizes corresponding to the mutated genes were detected with DNA from mutant clones, whereas fragments of wild-type gene sizes were completely absent (Supplemental Fig. S4).
Sequence comparisons were done using the BLAST algorithm (Altschul et al., 1997) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Cyanobacterial genome sequences were analyzed and extracted from CyanoBase (http://genome.microbedb.jp/cyanobase).
DNA Microarray to Characterize Gene Expression Changes
Cells from 5 mL of culture were harvested by centrifugation at 4,000 rpm for 5 min at 4°C and were immediately frozen at −80°C. Total RNA was extracted after pretreatment with hot phenol and chloroform by the High PureRNA isolation kit (Roche Diagnostics). Transcriptome analysis was done using custom-made 4x44K Agilent RNA microarrays. In addition to probes for protein-coding genes, the microarray contains probes for noncoding RNAs and untranslated regions of the Synechocystis 6803 chromosome (GenBank accession no. NC_000911.1) and plasmid pSYSA (NC_005230) as identified by Mitschke et al. (2011). Each probe has an internal technical duplicate. The detailed description and probe sequences are deposited in the GEO database under accession number GPL15867.
Hybridization of the microarrays was done according to Georg et al. (2009). Two micrograms of RNA was directly labeled with Cy3 using the Kreatech “ULS labeling kit for Agilent gene expression arrays.” Then, 1.65 µg of the labeled RNA was hybridized to each microarray following the Agilent protocol for single-color microarrays. The microarrays were digitalized with an Agilent G2565CA Microarray Scanner using the Agilent Feature Extraction Software 10.7.3.1 and protocol GE1_107_Sep09. The raw data were quantile normalized with the R package limma. The probe sets for the different RNA features were averaged before the summarization of the biological replicates and contrast extraction by limma. Each experiment was performed with RNA from two biological replicates. The microarray data are accessible in the GEO database under accession number GSE48415.
PAM to Characterize NDH1 and PSI Redox Kinetics
The postillumination rise in chlorophyll a fluorescence after actinic light had been switched off was measured according to Ma and Mi (2005) by means of a Dual PAM (Walz). After dark acclimation for 30 min, samples were exposed to actinic red light (approximately 630 nm, 95 µmol photons m−2 s−1) for 30 s, and the kinetic of PSII chlorophyll fluorescence after switching off actinic illumination was recorded as a measure of NDH activity (Battchikova et al., 2011b). Measurements were done at low measuring light (7 µmol photons m−2 s−1) and low measuring frequency (2,000 Hz) in order to achieve undisturbed postillumination signals.
For measurements of the redox kinetics of PSI, we used the Dual PAM (Walz). Cell suspensions precultivated to either HC or LC conditions were directly taken from the culture vessels at a density of approximately 5 µg chlorophyll a mL−1. P700 redox kinetics (called “slow redox kinetics” hereafter) were recorded in a stirred 4-mL cuvette as described by Ma et al. (2008) after 20 min of preincubation in darkness. A far-red illumination (more than 705 nm, 75 W m−2) was provided for 20 s, oxidizing P700 to a steady state. Because far-red light excites almost exclusively PSI, electron donation by PSII can be neglected. Therefore, the rate of P700+ formation after the onset of far-red illumination (“initial fast increase”) provides information about the electron transport through the PQ pool and Cytb6f; the addition of KCN (final concentration, 1 mm) allowed for discrimination between PSI- and Cytox-directed electron transport. After the initial fast increase, caused by rapid emptying of both PQ pool- as well as Cytb6f-stored electrons, a second, slow increase in P700+ could be observed (“slow increase”) until reaching a steady-state level of P700+ (Pm′; Klughammer and Schreiber, 2008). After switching off the far-red actinic light, the P700+ rereduction kinetics were followed and quantitatively evaluated.
Since the far-red light-induced Pm′ does not reflect full P700 oxidation level because of remaining PSII activity, cyclic electron transport, and electron flow from metabolites to PQ via respiratory enzymes (Klughammer and Schreiber, 1994, 2008; Bernát et al., 2011), processes contributing to the slow increase, a strong, white light illumination pulse (18,000 µmol photons m−2 s−1 for 200 ms) was applied to allow the determination of maximum P700+ level (Pm), which can be derived by the extrapolation routine of the Dual-PAM software as described by Klughammer and Schreiber (2008). After onset of the saturation pulse, the P700+ level rises to a maximum followed by a biphasic decrease. Whereas the second phase, starting about 30 ms after the onset of illumination, is believed to reflect the rereduction of P700+ by electrons generated by PSII (Klughammer and Schreiber, 1994), the first phase indicates the existence of a rapid type of cyclic PSI flow (Klughammer and Schreiber, 1994, 2008). The addition of DCMU (final concentration, 0.1 mm) and KCN (final concentration, 1 mm) blocked any remaining contribution from PSII as well as the loss of electrons to the respiratory terminal electron acceptor. Therefore, the P700+ reduction kinetics in the presence of both inhibitors reflects the donation of electrons stored in cyclic pathways and electrons generated de novo by respiratory enzymes. From these measurements, we calculated the Y(NA) as (Pm − Pm′)/(Pm − P0) (P0–P700 oxidation level in darkness; Klughammer and Schreiber, 2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple alignment of cyanobacterial Sml0013-like proteins.
Supplemental Figure S2. Multiple alignment of NDF6-like proteins of plants, cyanobacteria, and a cyanophage.
Supplemental Figure S3. Deletion of the gene sml0013 in Synechocystis 6803 to generate the mutant Δsml0013::Km.
Supplemental Figure S4. Deletion of the gene sll0514 in Synechocystis 6803 to generate the mutant Δsml0514::Sm.
Supplemental Figure S5. Traces of the slow redox kinetics of PSI of long-term LC-acclimated cells.
Supplemental Figure S6. Quantitative evaluation of the measurements of the redox kinetics of PSI with long-term LC-acclimated cells.
Supplemental Figure S7. Traces of the fast redox kinetics of PSI of long-term LC-acclimated cells.
Supplemental Table S1. Primers used to generate the Synechocystis 6803 mutants.
Supplemental Table S2. List of noncoding or antisense transcripts showing significant transcriptional changes in cells of the Synechocystis 6803 mutant Δsml0013::Km.
Supplemental File S1. Graphical representation of the entire genome of Synechocystis 6803 and the fold change values for each microarray probe.
Acknowledgments
The excellent technical assistance of Klaudia Michl is highly appreciated. We also thank Teruo Ogawa, who introduced M.H. to the various functions of cyanobacterial NDH1.
Glossary
- Cytb6f
cytochrome b6f
- Cytc
cytochrome c
- Cytox
cytochrome oxidase
- ORF
open reading frame
- HC
high CO2
- LC
low CO2
- LAHG
light-activated heterotrophic growth
- GEO
Gene Expression Omnibus
- PAM
pulse-amplitude measurement
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- PQ
plastoquinone
- Y(NA)
yield of nonphotochemical energy dissipation of reaction centers limited due to acceptor side limitation
- Pm′
steady-state level of P700+
References
- Aguirre von Wobeser E, Ibelings BW, Bok J, Krasikov V, Huisman J, Matthijs HCP. (2011) Concerted changes in gene expression and cell physiology of the cyanobacterium Synechocystis sp. strain PCC 6803 during transitions between nitrogen and light-limited growth. Plant Physiol 155: 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, McIntosh L. (1991) Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J Bacteriol 173: 2761–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K, Heber U, Schreiber U. (1992) Pool size of electrons that can be donated to P700+ as determined in intact leaves: donation to P700+ from stromal components via the intersystem chain. Plant Cell Physiol 33: 927–932 [Google Scholar]
- Baier K, Nicklisch S, Grundner C, Reinecke J, Lockau W. (2001) Expression of two nblA-homologous genes is required for phycobilisome degradation in nitrogen-starved Synechocystis sp. PCC6803. FEMS Microbiol Lett 195: 35–39 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Eisenhut M, Aro EM. (2011a) Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta 1807: 935–944 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Wei L, Du L, Bersanini L, Aro EM, Ma W. (2011b) Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J Biol Chem 286: 36992–37001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernát G, Appel J, Ogawa T, Rögner M. (2011) Distinct roles of multiple NDH-1 complexes in the cyanobacterial electron transport network as revealed by kinetic analysis of P700+ reduction in various Ndh-deficient mutants of Synechocystis sp. strain PCC6803. J Bacteriol 193: 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolychevtseva YV, Elanskaya IV, Karapetyan NV. (2011) Regulation of cyclic electron transport through photosystem I in cyanobacterium Synechocystis sp. PCC 6803 mutants deficient in respiratory dehydrogenases. Biochemistry (Mosc) 76: 427–437 [DOI] [PubMed] [Google Scholar]
- Bryant DA (1994) The Molecular Biology of Cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- Cobley J (2010) A gene required for photoregulation of phycobilisome abundances and for heterotrophic growth in Fremyella diplosiphon is present in all cyanobacterial genomes and in the genome of some cyanophages. In C Kerfeld, D Schluchter, eds, Abstract at the 10th Cyanobacterial Molecular Biology Workshop, Lake Arrowhead, California, USA, p 38 [Google Scholar]
- Collier JL, Grossman AR. (1994) A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J 13: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Aguirre von Wobeser E, Jonas L, Schubert H, Ibelings BW, Bauwe H, Matthijs HC, Hagemann M. (2007) Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol 144: 1946–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Georg J, Klähn S, Sakurai I, Mustila H, Zhang P, Hess WR, Aro EM. (2012) The antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J Biol Chem 287: 33153–33162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelken J, Funk C, Adamska I (2012) The extended light-harvesting complex (LHC) protein superfamily: classification and evolutionary dynamics. In RL Burnap, WFJ Vermaas, eds, Functional Genomics and Evolution of Photosynthetic Systems: Advances in Photosynthesis and Respiration, Vol 33. Springer, Dordrecht, The Netherlands, pp 265–284 [Google Scholar]
- Funk C, Vermaas WFJ. (1999) A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38: 9397–9404 [DOI] [PubMed] [Google Scholar]
- García-Domínguez M, Reyes JC, Florencio FJ. (2000) NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol Microbiol 35: 1192–1201 [DOI] [PubMed] [Google Scholar]
- Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. (2009) Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol 5: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Mathews DW. (2010) Signature proteins for the major clades of cyanobacteria. BMC Evol Biol 10: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M, Jeanjean R, Fulda S, Havaux M, Erdmann N. (1999) Flavodoxin accumulation contributes to enhanced cyclic electron flow around photosystem I in salt-stressed cells of Synechocystis sp. PCC 6803. Physiol Plant 105: 670–678 [Google Scholar]
- Hagemann M, Schoor A, Jeanjean R, Zuther E, Joset F. (1997) The stpA gene from Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J Bacteriol 179: 1727–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M, Zuther E. (1992) Selection and characterization of mutants of the cyanobacterium Synechocystis sp. PCC 6803 unable to tolerate high salt concentrations. Arch Microbiol 158: 429–434 [Google Scholar]
- Haimovich-Dayan M, Kahlon S, Hihara Y, Hagemann M, Ogawa T, Ohad I, Lieman-Hurwitz J, Kaplan A. (2011) Cross-talk between photomixotrophic growth and CO2 -concentrating mechanism in Synechocystis sp. strain PCC 6803. Environ Microbiol 13: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Hu P, Lv J, Fu P, Hualing M. (2013) Enzymatic characterization of an active NDH complex from Thermosynechococcus elongatus. FEBS Lett 587: 2340–2345 [DOI] [PubMed] [Google Scholar]
- Ifuku K, Endo T, Shikanai T, Aro EM. (2011) Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol 52: 1560–1568 [DOI] [PubMed] [Google Scholar]
- Ishikawa N, Takabayashi A, Ishida S, Hano Y, Endo T, Sato F. (2008) NDF6: a thylakoid protein specific to terrestrial plants is essential for activity of chloroplastic NAD(P)H dehydrogenase in Arabidopsis. Plant Cell Physiol 49: 1066–1073 [DOI] [PubMed] [Google Scholar]
- Klughammer C, Schreiber U. (1994) An improved method, using saturating light pulses, for the determination of photosystem I quantum yield via P700+-absorbance changes at 830 nm. Planta 192: 261–268 [Google Scholar]
- Klughammer C, Schreiber U. (2008) Saturation pulse method for assessment of energy conversion in PSI. PAM Application Notes 1: 11–14 [Google Scholar]
- Larsson J, Nylander JA, Bergman B. (2011) Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol 11: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los DA, Suzuki I, Zinchenko VV, Murata N (2008) Stress responses in Synechocystis: regulated genes and regulatory systems. In A Herrero, E Flores, eds, The Cyanobacteria: Molecular Biology, Genomics and Evolution. Caister Academic Press, Norfolk, UK, pp 117–158 [Google Scholar]
- Ma WM, Mi HL. (2005) Expression and activity of type 1 NAD(P)H dehydrogenase at different growth phases of the cyanobacterium Synechocystis PCC 6803. Physiol Plant 125: 135–140 [Google Scholar]
- Ma WM, Wei LZ, Wang QX. (2008) The response of electron transport mediated by active NADPH dehydrogenase complexes to heat stress in the cyanobacterium Synechocystis 6803. Sci China C Life Sci 51: 1082–1087 [DOI] [PubMed] [Google Scholar]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, et al. (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 108: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T. (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Nowaczyk MM, Wulfhorst H, Ryan CM, Souda P, Zhang H, Cramer WA, Whitelegge JP. (2011) NdhP and NdhQ: two novel small subunits of the cyanobacterial NDH-1 complex. Biochemistry 50: 1121–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. (1991) A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc Natl Acad Sci USA 88: 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek GA, Obinger C, Fromwald S, Bergman B. (1994) Correlation between immuno-gold labels and activities of the cytochrome-c oxidase (aa3-type) in membranes of salt stressed cyanobacteria. FEMS Microbiol Lett 124: 431–437 [Google Scholar]
- Pils D, Schmetterer G. (2001) Characterization of three bioenergetically active respiratory terminal oxidases in the cyanobacterium Synechocystis sp. strain PCC 6803. FEMS Microbiol Lett 203: 217–222 [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111: 1–61 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. (2009) Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T. (2001) Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci USA 98: 11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, Calteau A, Cai F, Tandeau de Marsac N, Rippka R, et al. (2013) Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA 110: 1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2007) Cyclic electron transport around photosystem I: genetic approaches. Annu Rev Plant Biol 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Wang HL, Postier BL, Burnap RL. (2004) Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J Biol Chem 279: 5739–5751 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Peng L, Fukao Y, Shikanai T. (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeremenko N, Jeanjean R, Prommeenate P, Krasikov V, Nixon PJ, Vermaas WFJ, Havaux M, Matthijs HCP. (2005) Open reading frame ssr2016 is required for antimycin A-sensitive photosystem I-driven cyclic electron flow in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 46: 1433–1436 [DOI] [PubMed] [Google Scholar]
- Zhang P, Battchikova N, Jansen T, Appel J, Ogawa T, Aro EM. (2004) Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]