An Arabidopsis gene product affects the size of the pectic polysaccharide, rhamnogalacturonan I, in seed coat mucilage.
Abstract
The function of a putative galacturonosyltransferase from Arabidopsis (Arabidopsis thaliana; At1g02720; GALACTURONOSYLTRANSFERASE-LIKE5 [AtGATL5]) was studied using a combination of molecular genetic, chemical, and immunological approaches. AtGATL5 is expressed in all plant tissues, with highest expression levels in siliques 7 DPA. Furthermore, its expression is positively regulated by several transcription factors that are known to regulate seed coat mucilage production. AtGATL5 is localized in both endoplasmic reticulum and Golgi, in comparison with marker proteins resident to these subcellular compartments. A transfer DNA insertion in the AtGATL5 gene generates seed coat epidermal cell defects both in mucilage synthesis and cell adhesion. Transformation of atgatl5-1 mutants with the wild-type AtGATL5 gene results in the complementation of all morphological phenotypes. Compositional analyses of the mucilage isolated from the atgatl5-1 mutant demonstrated that galacturonic acid and rhamnose contents are decreased significantly in atgatl5-1 compared with wild-type mucilage. No changes in structure were observed between soluble mucilage isolated from wild-type and mutant seeds, except that the molecular weight of the mutant mucilage increased 63% compared with that of the wild type. These data provide evidence that AtGATL5 might function in the regulation of the final size of the mucilage rhamnogalacturonan I.
Pectins are highly complex glycans and are made up of diverse galacturonic acid (GalA)-containing polysaccharides. They are particularly abundant in primary cell walls and in the middle lamella, the junction between adjacent cells (Mohnen, 2008). Pectin consists primarily of three polysaccharides: homogalacturonan (HG), substituted HGs, and rhamnogalacturonan I (RG-I; Willats et al., 2001b; Mohnen, 2008). HG is composed of a linear chain of α-1,4-linked GalA residues that are often methyl esterified on C6 and can be acetylated on C2 and/or C3. Substituted HGs include rhamnogalacturonan II, xylogalacturonan, and apiogalacturonan. Rhamnogalacturonan II has a galacturonan backbone and four complex but evolutionarily conserved side chains containing 12 different monosaccharides in over 20 different linkages. Xylogalacturonan is an HG substituted at O-3 with a β-linked Xyl, and apiogalacturonan is a HG substituted at O-2 or O-3 with d-apiofuranose. RG-I consists of a unique backbone with the disaccharide (α-1,4-GalA-α-1,2-Rha) as the basic repeating unit. The Rha residues are frequently substituted with chains of galactan, arabinans, or arabinogalactans. RG-I side chain structures are very complex, variable, and highly cell type and developmental stage dependent, which suggests diverse functional roles for this polysaccharide in plants (Willats et al., 2001b; Mohnen, 2008).
Pectin synthesis is catalyzed by glycosyltransferases (GTs) that transfer a glycosyl residue from a nucleotide sugar donor to an oligosaccharide or polysaccharide acceptor. Because of the complexity of pectin, more than 50 GTs are predicted to be required for pectin synthesis (Mohnen, 2008). Recently, several putative GTs involved in the biosynthesis of different pectins have been identified using mutational and/or biochemical approaches (for review, see Mohnen, 2008; Harholt et al., 2010). Among these genes, GALACTURONOSYLTRANSFERASE1 (GAUT1) has been shown to encode a functional galacturonosyltransferase (GalAT; Sterling et al., 2006) and acts in a complex with GAUT7 to synthesize the HG backbone (Atmodjo et al., 2011). QUASIMODO1 (QUA1)/GAUT8 was also implicated in HG backbone synthesis due to the observation that the qua1 mutant showed decreased activity of HG GalA transferase (Bouton et al., 2002; Orfila et al., 2005). However, there have been no studies on the synthesis of the RG-I backbone, and no candidate GTs involved in this process have yet been reported (Harholt et al., 2010). A recent report suggests a possible role for GAUT11 in seed mucilage pectin biosynthesis (Caffall et al., 2009), but further experiments are needed to verify whether GAUT11 is a candidate GT participating in RG-I backbone synthesis.
Arabidopsis (Arabidopsis thaliana) is one of the plant species that produces myxospermous seed, where the epidermal cells of the seed coat accumulate a large quantity of mucilage that is composed mainly of relatively unbranched RG-I, which is occasionally substituted with 5-arabinans and nonreducing terminal galactosyl residues at the Rha O-3 position (Penfield et al., 2001; Western et al., 2004; Dean et al., 2007). The ability of Arabidopsis mucilage to bind the antibodies JIM5 and JIM7 also suggests the presence of lowly and highly methyl-esterified HG (Western et al., 2000; Willats et al., 2001a; Macquet et al., 2007a). Small amounts of cellulose, arabinan, and galactan were also found in Arabidopsis seed mucilage (Willats et al., 2001a; Macquet et al., 2007a; Young et al., 2008). Recent studies showed that cellulose plays an adhesive role for the attachment of mucilage pectin to the seed coat epidermal cells (Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011). Variations in both mucilage composition and structure have been observed in other plants (Anderson, 1933; Muralikrishna et al., 1987; Naran et al., 2008).
Seed coat mucilage represents an excellent model system for studying pectin, and specifically RG-I, biosynthesis (Arsovski et al., 2010; Haughn and Western, 2012). A number of genes that play roles in Arabidopsis seed mucilage polysaccharide biosynthesis/metabolism have been identified. CELLULOSE SYNTHASE5 (CESA5) was found to be involved in cellulose synthesis (Sullivan et al., 2011), while MUCILAGE-MODIFIED4/RHAMNOSE SYNTHESIS2 (MUM4/RHM2) encodes a UDP-Rha synthase (Usadel et al., 2004; Western et al., 2004; Oka et al., 2007). The mum2, atsbt1.7, atbxl1, pmei6, fly1, and per36 mutants were found to be defective in pectin modification (Dean et al., 2007; Macquet et al., 2007b; Rautengarten et al., 2008; Arsovski et al., 2009; Saez-Aguayo et al., 2013; Voiniciuc et al., 2013) or in the degradation of the outer cell wall of the outer integument (Kunieda et al., 2013), all of which show a mucilage-release defect.
There are also several plant lines carrying mutations in genes encoding putative transcription factors that are affected in mucilage production, including apetala2 (ap2; Jofuku et al., 1994), transparent testa glabra1 (ttg1; Koornneef, 1981), ttg2 (Johnson et al., 2002), glabra2 (gl2; Rerie et al., 1994), ats (Léon-Kloosterziel et al., 1994), transparent testa8 (tt8; Nesi et al., 2000), enhancer of glabra3 (egl3; Zhang et al., 2003), myb domain protein61 (myb61; Penfield et al., 2001), nars1/nars2 (Kunieda et al., 2008), myb5 (Gonzalez et al., 2009; Li et al., 2009), and luh/mum1 (Bui et al., 2011; Huang et al., 2011; Walker et al., 2011). These transcription factors appear to act through at least two, or possibly three, distinct pathways to regulate mucilage biosynthesis (Huang et al., 2011). Hormones are also involved in mucilage production. Mutations in two genes, GIBBERELLIN3-OXIDASE4 (GA3OX4; Kim et al., 2005) and ABSCISIC ACID DEFICIENT1 (ABA1; Karssen et al., 1983), related to GA3 and abscisic acid metabolism, respectively, also affect mucilage synthesis.
Despite the identification of many genes that are connected to mucilage synthesis, genes involved in RG-I backbone synthesis have not yet been identified explicitly. Here, we present evidence that AtGATL5, a protein with significant sequence similarity to GAUT1 and belonging to family GT8 in the Carbohydrate-Active Enzymes classification system (http://www.cazy.org; Cantarel et al., 2009), is involved in the synthesis of the seed coat mucilage RG-I backbone in Arabidopsis.
RESULTS
AtGATL5 Encodes a Putative GT That Is Targeted to the Secretory Pathway
AtGATL5 (At1g02720) encodes a putative GT that belongs in the GATL subclade of the GT8 family (Yin et al., 2010). The coding region of AtGATL5 consists of a single exon encoding a protein with a predicted molecular mass of 41 kD. Like all other AtGATL proteins, the catalytic domain of the AtGATL5 protein contains a DxDxxxxxD motif, which is thought to be involved in nucleotide sugar binding, and also contains several conserved motifs characteristic of family GT8 GTs (Wiggins and Munro, 1998; Yin et al., 2010).
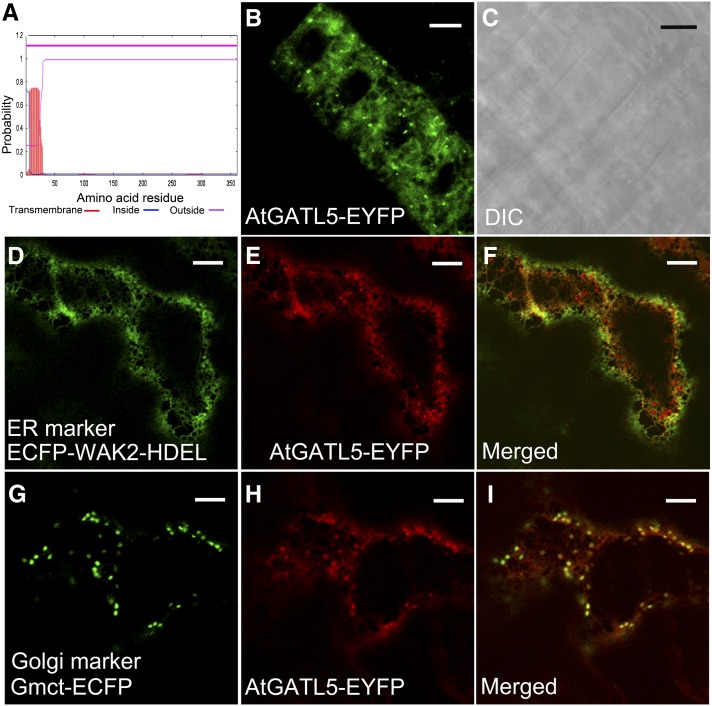
Analysis of the AtGATL5 protein sequence using the TMHMM2.0 program for the prediction of transmembrane helices in proteins (http://www.cbs.dtu.dk/services/TMHMM-2.0/) predicts that AtGATL5 has no transmembrane domain, suggesting that the AtGATL5 protein is not an integral membrane protein (Fig. 1A). To determine the actual subcellular localization of AtGATL5, the AtGATL5 gene was fused in frame at the C terminus with EYFP and transformed into wild-type Arabidopsis plants. Transgenic Arabidopsis plants exhibited both punctate and network-like fluorescence signals in the cytoplasm of root epidermal cells (Fig. 1, B and C), indicating that AtGATL5 is located in subcellular organelles. This construct also has been used to complement the reduced mucilage phenotype of the atgatl5-1 mutant (Supplemental Fig. S1), demonstrating that the fusion protein retains AtGATL5 functionality. To further explore the subcellular localization of AtGATL5, Pro-35S:AtGATL5-EYFP was cotransformed into Nicotiana benthamiana leaf epidermal cells with genes encoding either Gmct-ECFP, an enhanced cyan fluorescent protein-tagged Golgi marker (Saint-Jore-Dupas et al., 2006; Nelson et al., 2007), or ECFP-WAK2-HDEL, an endoplasmic reticulum (ER) marker (Nelson et al., 2007). As shown in Figure 1, D to I, enhanced yellow fluorescent protein (EYFP)-tagged AtGATL5 colocalizes well with both marker proteins, indicating that the AtGATL5 protein localizes to both Golgi and ER.
Figure 1.
Subcellular localization of fluorescent protein-tagged AtGATL5 protein. Fluorescent protein-tagged AtGATL5 was expressed in Arabidopsis plants and leaf epidermal cells of N. benthamiana plants, and its subcellular localization was examined with a laser scanning confocal microscope. A, AtGATL5 protein is predicted by the TMHMM2.0 program to have only a signal peptide at its N terminus but no transmembrane domain. B and C, Fluorescent signals of Arabidopsis root epidermal cells expressing the Pro-35S:AtGATL5-EYFP construct (B) and the corresponding differential interference contrast (DIC) image (C). Note that the AtGATL5-EYFP signals show both punctate and network patterns. D to F, N. benthamiana leaf epidermal cells expressing both Pro-35S:AtGATL5-EYFP and ECFP-WAK2-HDEL constructs. D, Localization of ECFP-WAK2-HDEL ER marker protein (green). E, Localization of AtGATL5-EYFP protein (red) in the same cell as in D. F, Merged image of D and E, showing colocalization of AtGATL5-EYFP and ECFP-WAK2-HDEL proteins. G to I, N. benthamiana leaf epidermal cells expressing both Pro-35S:AtGATL5-EYFP and Gmct-ECFP constructs. G, Localization of Gmct-ECFP Golgi marker protein (green). H, Localization of AtGATL5-EYFP protein (red) in the same cell as in G. I, Merged image of G and H, showing colocalization of AtGATL5-EYFP and Gmct-ECFP proteins. Bars = 30 μm.
Expression Pattern of the AtGATL5 Gene
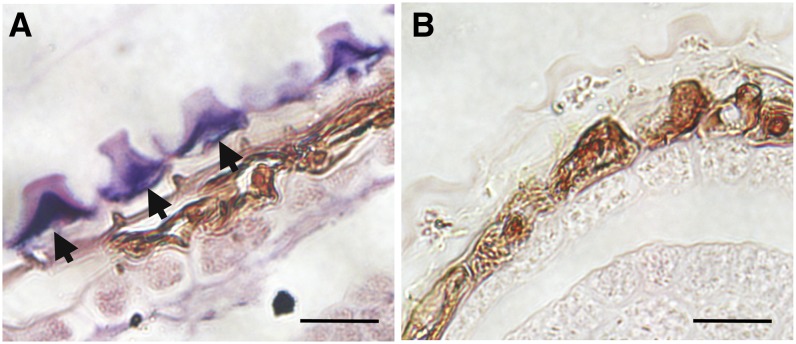
Our previous promoter:GUS expression studies showed that the AtGATL5 gene is expressed ubiquitously in all the major tissues examined in Arabidopsis (Kong et al., 2011). According to the AtGenExpress visualization tool (http://jsp.weigelworld.org/expviz/expviz.jsp; Schmid et al., 2005), the highest level of expression was found during seed development, specifically in the heart to torpedo stages of embryo development (Supplemental Fig. S2A). In addition, data available from transcriptome analyses using RNA extracted from laser-capture-dissected seed coat tissue (Le et al., 2010; http://seedgenenetwork.net/arabidopsis) indicate that AtGATL5 transcript levels are low in the seed coat during early embryogenesis and that the levels rise from the linear cotyledon stage onward (Supplemental Fig. S2B). We performed in situ hybridization to determine the specific cell type(s) in the seed in which AtGATL5 is expressed. The results demonstrate that AtGATL5 expression is confined exclusively to the epidermal cell layer of the outer integument (Fig. 2).
Figure 2.
AtGATL5 expression pattern in seed coat cells by RNA in situ hybridization. Hybridization is shown with the AtGATL5-specific antisense probe (A) and control hybridization with the sense probe (B) in the 13-DPA seed coat. Arrows indicate the hybridization signal specifically localized to the epidermal cells of the seed coat. Bars = 20 µm.
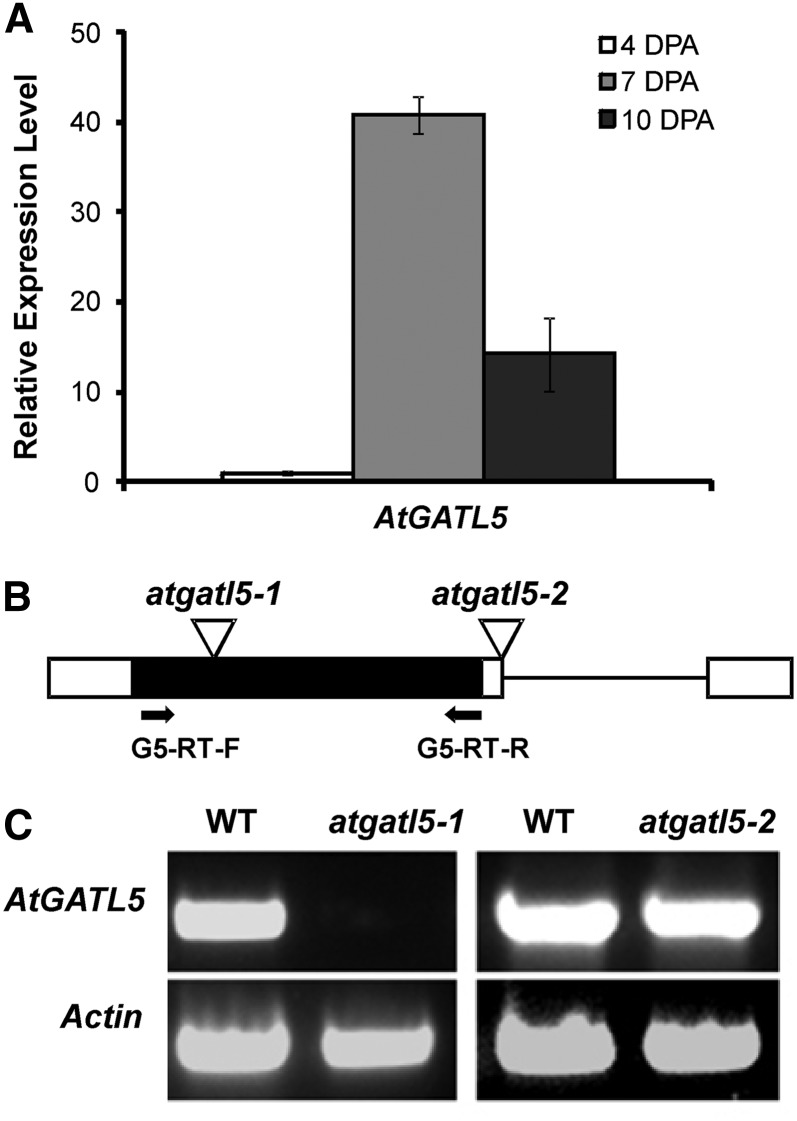
Quantitative real-time PCR was performed using RNA extracted from developing wild-type siliques at 4, 7, and 10 DPA (Fig. 3A). AtGATL5 is up-regulated approximately 41 times at 7 DPA, corresponding to seeds with late heart to torpedo stage embryos, when mucilage production in seed coats is at its peak, compared with 4-DPA seeds, when mucilage synthesis has not yet been initiated. This expression pattern is consistent with a role for AtGATL5 in mucilage synthesis.
Figure 3.
Schematic structure and steady-state RNA level analysis of the AtGATL5 gene. A, Quantitative RT-PCR analysis of AtGATL5 transcript during seed coat development. RNA was prepared from siliques at 4, 7, and 10 DPA. Data are presented as relative fold change, where the 4-DPA transcript level was set at 1.0. AtGATL5 was expressed at all stages examined, but most highly at 7 DPA. Actin2 (At3g18780) was used as an internal control. Error bars represent se of three biological replicates. B, Schematic diagrams of the locations of T-DNA inserts for the atgatl5-1 and atgatl5-2 mutant lines. The single exon and the untranslated regions are indicated as a black box and white boxes, respectively. The intron located in the 3′-untranslated region is indicated as a black line. Inverted triangles represent the sites of T-DNA insertion. Arrows indicate the positions of the two primers used for RT-PCR. C, RT-PCR showing that the AtGATL5 gene is expressed in wild-type siliques (WT) and atgatl5-2 siliques but is not expressed in siliques of the atgatl5-1 mutant.
A Knockout Mutation of AtGATL5 Results in Reduced Mucilage Production in the Seed Coat
Two independent transfer DNA (T-DNA) insertion lines, Salk_106615 and Salk_038363, were selected and screened for disruption of the AtGATL5 gene using PCR. As shown in Figure 3B, Salk_106615 has a T-DNA inserted in the exon of AtGATL5, whereas the Salk_038363 mutant carries a T-DNA insertion in the 3′ untranslated region. Plants homozygous for T-DNA insertions in AtGATL5 obtained from the independent insertion lines Salk_106615 and Salk_038363 were named atgatl5-1 and atgatl5-2, respectively (Fig. 3B). The effect of the T-DNA insertions on AtGATL5 mRNA expression levels was studied by reverse transcription (RT)-PCR amplification using primers spanning the coding region of the mRNA. No amplification was observed in the atgatl5-1 mutant, whereas a clear band corresponding to the amplification of AtGATL5 mRNA was visible in atgatl5-2 and wild-type plants (Fig. 3C). The phenotypes described in this article have been observed only in atgatl5-1 plants but not in atgatl5-2 plants.
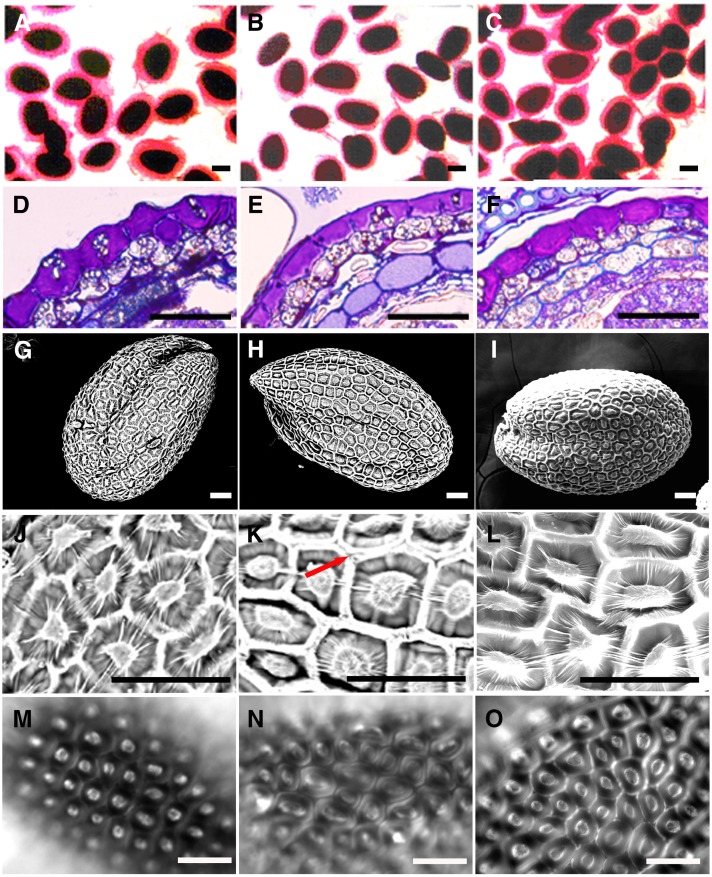
When mature hydrated seeds were stained with ruthenium red, which stains negatively charged biopolymers such as pectin and DNA (Koornneef, 1981), only a thin irregular staining pattern was observed in atgatl5-1 seeds, whereas wild-type seeds showed intense regular spherical staining (Fig. 4, A and B). Further studies were carried out on sections prepared from seeds that were stained with toluidine blue O, a polychromatic dye that stains mucilage pink (Western et al., 2000). Significant pink-staining mucilage was seen in the epidermal cells of both mutant and wild-type seeds after the completion of mucilage synthesis (10 DPA; Fig. 4, D and E).
Figure 4.
Seed coat mucilage phenotypes of wild-type and mutant plants. A to C, Ruthenium red staining of wild-type (A), atgatl5-1 (B), and complemented atgatl5-1 (C) seeds. Note that the red-stained capsule of mucilage surrounding the seeds is thinner in atgatl5-1 than in wild-type and atgatl5-1 complemented seeds. D to F, Cross sections of 10-DPA wild-type (D), atgatl5-1 (E), and complemented atgatl5-1 (F) seed coat epidermis stained with toluidine blue. G to L, Scanning electron micrographs showing whole seed and enlarged seed coat details of wild-type (G and J), atgatl5-1 (H and K), and complemented atgatl5-1 (I and L) seeds. Note that atgatl5-1 shows a lack of/less adhesion between hexagonal cell walls (K; arrow), in contrast to the wild type, which can be rescued by the expression of AtGATL5 in atgatl5-1 (L). M to O, Calcofluor white staining of wild-type (M), atgatl5-1 (N), and complemented atgatl5-1 (O) seeds showing the lack of/less adhesion between hexagonal cell walls in contrast to the wild type, which can be rescued by the expression of AtGATL5 in atgatl5-1 (O). Bars = 250 µm (A–C) and 50 µm (D–O).
To further analyze the defects present in the atgatl5-1 seed coat, seeds of the wild type and atgatl5-1 were observed by scanning electron microscopy. As shown in Figure 4, G, H, J, and K, the central volcano-shaped columella in atgatl5-1 seed coat epidermal cells is broader than that in wild-type seeds, in agreement with the reduced amount of mucilage (Fig. 4, A and B). These results were confirmed by calcofluor white staining of Arabidopsis seeds, which brightly stains the columella and the hexagonal walls of epidermal cells (Fig. 4, M and N) and also shows that atgatl5-1 mucilage secretory cells have a wider columella (Fig. 4N) than wild-type secretory cells (Fig. 4M). The increased width of the columella is consistent with the phenotype seen in other mutants with severely (mum4 and ttg1) and moderately (men4) reduced mucilage production (Western et al., 2004; Arsovski et al., 2009). Besides the mucilage defect and changes in mucilage secretory cell structure, the atgatl5-1 mutant also shows less adhesion between the hexagonal seed coat epidermal cell walls and wild-type seeds, as evidenced by the appearance of two distinct walls side by side in some areas of the seed coat, rather than a single thick wall as seen for wild-type cells, in either the scanning electron microscopy images or in the calcofluor white-stained seeds (Fig. 4, J, K, M, and N; Supplemental Fig. S1, G and H).
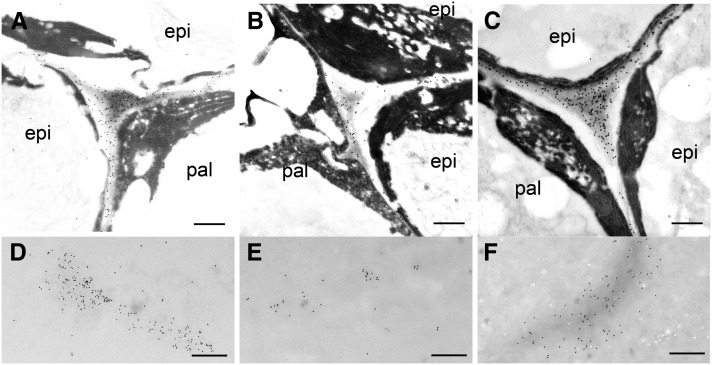
Immunogold labeling, using CCRC-M38 (a nonmethylesterified HG-directed monoclonal antibody; Pattathil et al., 2010), was employed to visualize the localization of HG epitopes in seed coat epidermal cells. Nonmethylesterified HG is evenly distributed in the middle lamella and cell corners of seed coat epidermal cells (Fig. 5, A–C). Labeling is also observed in clumps within the apoplastic space interior to the cell wall of seed coat epidermal cells, which is where the mucilage is deposited (Fig. 5, D–F). Less labeling was observed in both areas in atgatl5-1 seeds compared with the wild type and with the complemented atgatl5-1 seeds (Fig. 5).
Figure 5.
Gold immunolabeling of HG epitopes in epidermal cell walls of the seed coats of 10-DPA seeds with CCRC-M38. Images show labeling of HG epitopes in the middle lamella and cell corner of the testa epidermal cell of wild-type (A), atgatl5-1 (B), and complemented atgatl5-1 (C) seeds and in the apoplast of wild-type (D), atgatl5-1 (E), and complemented atgatl5-1 (F) seeds. epi, Epidermal cell of the first layer of the outer integument, where mucilage is synthesized; pal, palisade cell of the second layer of the outer integument. Bars = 900 nm (A–C) and 500 nm (D–F).
Homozygous atgatl5-1 mutant plants are morphologically indistinguishable from wild-type plants throughout development and reached similar sizes in their aboveground growth. No significant differences in root growth were observed when atgatl5-1 and wild-type seeds were germinated on vertical agar plates (data not shown).
Complementation of the atgatl5-1 mutant with a wild-type copy of the AtGATL5 gene was undertaken in order to verify that the mutant phenotypes result from the knockout of the AtGATL5 gene. An exogenous copy of the AtGATL5 gene driven by its native promoter was transformed into atgatl5-1 mutants. Twelve independent transformants were examined, and all showed the same characteristics. Representative results are shown in Figures 4, C, F, I, L, and O, 5, C and F, and Supplemental Figure S1, C, F, and I and demonstrate that transformation of the AtGATL5 gene into atgatl5-1 restored all observed mutant phenotypes to the wild type. These experiments show that the phenotypes observed in atgatl5-1 are the result of the mutation in the AtGATL5 gene.
Compositional and Linkage Analyses of Seed Mucilage
Arabidopsis seed mucilage has two layers. The outer layer is water soluble and is easily extracted using either water or ammonium oxalate (Macquet et al., 2007a). The inner layer is strongly attached to the seed and cannot be removed with either hot acid or alkali (Macquet et al., 2007a). The sugar composition of the water-extractable outer layer showed a reduction in the amounts of GalA and Rha, but no change in the molar ratios of these two sugars, in the atgatl5-1 mutant mucilage compared with the wild type (Supplemental Table S1). In order to obtain a more complete picture of total mucilage composition, rhamnogalacturonan hydrolase treatment was used to solubilize both layers of mucilage from atgatl5-1 and wild-type seeds, and the monosaccharide compositions of the released materials were analyzed. Rhamnogalacturonan hydrolase cleaves unbranched RG-I to oligomers with Rha at the nonreducing end (Azadi et al., 1995), and ruthenium red staining of the extracted seeds showed that, compared with water or ammonium oxalate extraction, rhamnogalacturonan hydrolase removes the mucilage more extensively than does water (Supplemental Fig. S3). Although some cell wall components from the seed coat or the embryo might also be released, compositional analysis of the enzyme-released mucilage best represents the composition of the total seed mucilage, as the molar ratio of Rha and GalA is close to 1:1 (Table I). The total amount of sugar released by the enzyme treatment was reduced approximately 35% in atgatl5-1 seeds compared with wild-type seeds (Table I). Most of this decline in total sugar released from mutant seed is accounted for by decreases in the amounts of GalA and Rha released (40% and 34%, respectively) compared with the wild type. Other monosaccharides, including Ara, Fuc, Xyl, Man, Gal, and Glc, only account for a few percent of the total monosaccharides released and did not change appreciably in atgatl5-1 (Table I; Supplemental Table S1). In the wild type and the atgatl5-1 mutant, the water-soluble mucilage fraction accounts for around 70% of the total mucilage; the other 30% is strongly attached to the seed coat. Transformation of the atgatl5-1 mutant with a wild-type copy of the AtGATL5 gene rescued the reduced GalA and Rha to wild-type levels (Table I). These results suggest that the amount of RG-I, which is the major component of mucilage, synthesized by atgatl5-1 seeds is significantly reduced. This is in agreement with microscopic observations that indicated that the atgatl5-1 mutant testa epidermal cells release lower quantities of mucilage, as measured by staining with ruthenium red (Fig. 4).
Table I. Monosaccharide composition of mucilage extracted from wild-type, atgatl5-1, and complemented atgatl5-1 seeds by mutant seeds using rhamnogalacturonan hydrolase digestion.
The amounts of sugar are presented as mean values (μg sugar mg−1 seeds) ± se of three independent samples. Significance was tested by the Mann-Whitney U test comparing atgatl5-1 with the wild type (*P < 0.05).
| Sugar | Wild Type | atgatl5-1 | Complemented atgatl5-1 |
|---|---|---|---|
| Ara | 0.07 ± 0.03 | 0.07 ± 0.04 | 0.08 ± 0.01 |
| Rha | 10.3 ± 0.27 | 6.84 ± 0.36* | 10.7 ± 0.81 |
| Fuc | 0.1 ± 0.04 | 0.13 ± 0.03 | 0.16 ± 0.10 |
| Xyl | 0.28 ± 0.16 | 0.27 ± 0.09 | 0.33 ± 0.03 |
| Man | 0.16 ± 0.15 | 0.26 ± 0.18 | 0.21 ± 0.01 |
| Gal | 0.14 ± 0.10 | 0.16 ± 0.05 | 0.21 ± 0.05 |
| Glc | 0.29 ± 0.19 | 0.38 ± 0.10 | 0.27 ± 0.04 |
| GalA | 15.7 ± 0.81 | 9.37 ± 0.67* | 16.4 ± 1.77 |
| Total | 27.0 | 17.45 | 28.3 |
| GalA/Rha (mol/mol) | 1.28 | 1.16 | 1.3 |
To investigate the structure of seed mucilage in the wild type and the atgatl5-1 mutant in greater detail, glycosyl linkage analysis was performed on the soluble polysaccharides extracted from wild-type and atgatl5-1 seed mucilage without enzyme treatment. Consistent with previous studies (Penfield et al., 2001; Dean et al., 2007), the predominant linkages in the mucilage of both the wild type and atgatl5-1 are 2-linked Rha (2-Rhap) and 4-linked GalA (4-GalAp); both of these linkages decreased slightly in atgatl5-1 mucilage compared with the wild type (Table II). Consistent with the monosaccharide analysis data in Table I, no detectable changes in the glycosyl linkage composition of other sugars were found in atgatl5-1 mucilage. These finding suggest that the reduced mucilage content in atgatl5-1 seeds is not associated with major changes in RG-I structure.
Table II. Linkage analysis of water-extractable mucilage from wild-type and atgatl5-1 mature seeds.
Values are means of two samples with variance less than 5%. Results are given as mol %. tr, Trace amounts less than 0.5%. All sugar-associated peaks in the chromatogram are accounted for by these data.
| Sugar and Linkage | Wild Type | atgatl5-1 |
|---|---|---|
| Fuc | ||
| t-Fuc | tr | tr |
| Rha | ||
| t-Rha | 14 | 15 |
| 2-Rha | 35 | 33 |
| 2,3-Rha | 1 | 1 |
| 2,4-Rha | 1 | 1 |
| Ara | ||
| 5-Araf | 1 | 1 |
| Xyl | ||
| t-Xylp | tr | tr |
| 4-Xylp | 1 | 1 |
| Man | ||
| 4-Manp | 1 | 2 |
| 4,6-Manp | 1 | 1 |
| Gal | ||
| t-Galp | tr | 1 |
| 3,6-Galp | tr | tr |
| Glc | ||
| 4-Glcp | 2 | 2 |
| 4,6-Glcp | tr | tr |
| GalA | ||
| t-GalAp | tr | 1 |
| 4-GalAp | 39 | 37 |
| 3,4-GalAp | 1 | 1 |
| 2,4-GalAp | 1 | 1 |
| 4,6-GalAp | 2 | 1 |
Immunohistochemistry of atgatl5-1 Seed Mucilage
The glycosyl composition and linkage composition analyses suggest that only the amount of RG-I is reduced in the atgatl5-1 mutant mucilage. To further examine whether the structural features of the mutant mucilage are altered or not, over 100 antibodies that recognize most major classes of cell wall polysaccharides, including pectins (HG and RG-I), hemicelluloses (xyloglucan and xylan), galactans, and arabinogalactans, were employed to probe the mutant mucilage by ELISA. As shown in Supplemental Figure S4, a total of 18 antibodies that recognize epitopes on HG (CCRC-M34, CCRC-M38, JIM5, JIM7, JIM136, and JIM137), RG-I (CCRC-M14, CCRC-M30, CCRC-M35, CCRC-M36, CCRC-M56, CCRC-M69, CCRC-M72, CCRC-M79, CCRC-M80, CCRC-M82, and CCRC-M83), and arabinogalactan (JIM19) bind strongly to mucilage. No significant differences in binding were observed between wild-type and atgatl5-1 mucilage for any of the antibodies tested, indicating that the diverse epitopes recognized by these antibodies are not altered in the mutant mucilage. These immunological observations further support the conclusion drawn from the compositional and linkage analyses above that the atgatl5-1 mucilage has not undergone a major change in polysaccharide structure, at least with respect to composition.
Macromolecular Characterization of Mucilage
The data presented above have clearly shown that the atgatl5-1 mutation leads to a discernible reduction in the amount of seed mucilage produced, yet no alterations in mucilage structure could be detected. We then determined whether the size of the mucilage was affected by the AtGATL5 mutation. The water-soluble mucilage was analyzed by high-performance size-exclusion chromatography (HP-SEC) combined with multiple-angle laser light scattering. The elution profiles indicate that most large-Mr polysaccharides in the seed mucilage preparation elute at 13 to 17 min after injection. The detection of the elution profile by both the light-scattering and refractive index concentration detectors is nearly superimposable. From the refractive index detector, there are later-eluting smaller Mr materials (Supplemental Fig. S5). Most likely, these correspond to small sugars, ions, or pigments, since there was no corresponding signal from the light-scattering detector and the extracts separated by HP-SEC were not dialyzed prior to analysis (Macquet et al., 2007a). The macromolecular characteristics of the mucilage are listed in Table III. Wild-type and complemented atgatl5-1 mucilage polysaccharides have an average molecular mass of 415 and 527 kD, respectively. In contrast, the atgatl5-1 mucilage has a molecular mass of 678 kD, which is significantly larger than that of wild-type and complemented atgatl5-1 mucilages.
Table III. Weight average and root mean square radius of seed coat mucilage.
Mn, Number-average molar mass; Mw, weight-average Mr; Mw/Mn, polydispersity coefficient; Rw, root mean square radius. Results are averages of three biological replicates. Significance was tested by the Mann-Whitney U test comparing atgatl5-1 with the wild type (*P < 0.05).
| Parameter | Wild Type | atgatl5-1 | Complemented atgatl5-1 |
|---|---|---|---|
| Mw (kD) | 415 ± 14 | 678 ±61* | 527 ± 25 |
| Mw/Mn | 1.08 | 1.47 | 1.07 |
| Rw (nm) | 41 ± 0.1 | 63 ± 5 | 52 ± 2 |
Test for HG:GalAT Activity of AtGATL5
AtGATL5 belongs to Carbohydrate-Active Enzymes family GT8 (Yin et al., 2010), several of whose members have been implicated in the synthesis of HG (Bouton et al., 2002; Leboeuf et al., 2005; Orfila et al., 2005; Sterling et al., 2006; Caffall et al., 2009; Atmodjo et al., 2011), including one functionally characterized GalAT, GAUT1 (Sterling et al., 2006). Thus, we tested whether AtGATL5 functions as a HG:GalAT. Microsomal membranes prepared from N. benthamiana leaves transiently expressing AtGATL5-EYFP were used for the HG:GalAT assay. Results from two independent experiments revealed no significant differences in the activity measured between the control and AtGATL5-EYFP-expressing plants (Supplemental Fig. S6), suggesting that AtGATL5 does not have a HG:GalAT activity comparable to GAUT1.
Maximal Accumulation of AtGATL5 Transcript Requires Upstream Transcription Factors
A number of transcription factors have been identified that are associated with mucilage synthesis in the seed coat, including AP2 (Jofuku et al., 1994), TTG1 (Koornneef, 1981), TTG2 (Johnson et al., 2002), GL2 (Rerie et al., 1994), and MYB61 (Penfield et al., 2001). AP2 and a TTG1 protein complex, which includes TTG1, EGL3, TT8, MYB5, and TT2, positively regulate GL2 and TTG2. GL2, in turn, activates transcription of the Rha synthase, MUM4 (Western et al., 2004). MYB61, however, appears to regulate mucilage production through an alternative pathway (Western et al., 2004).
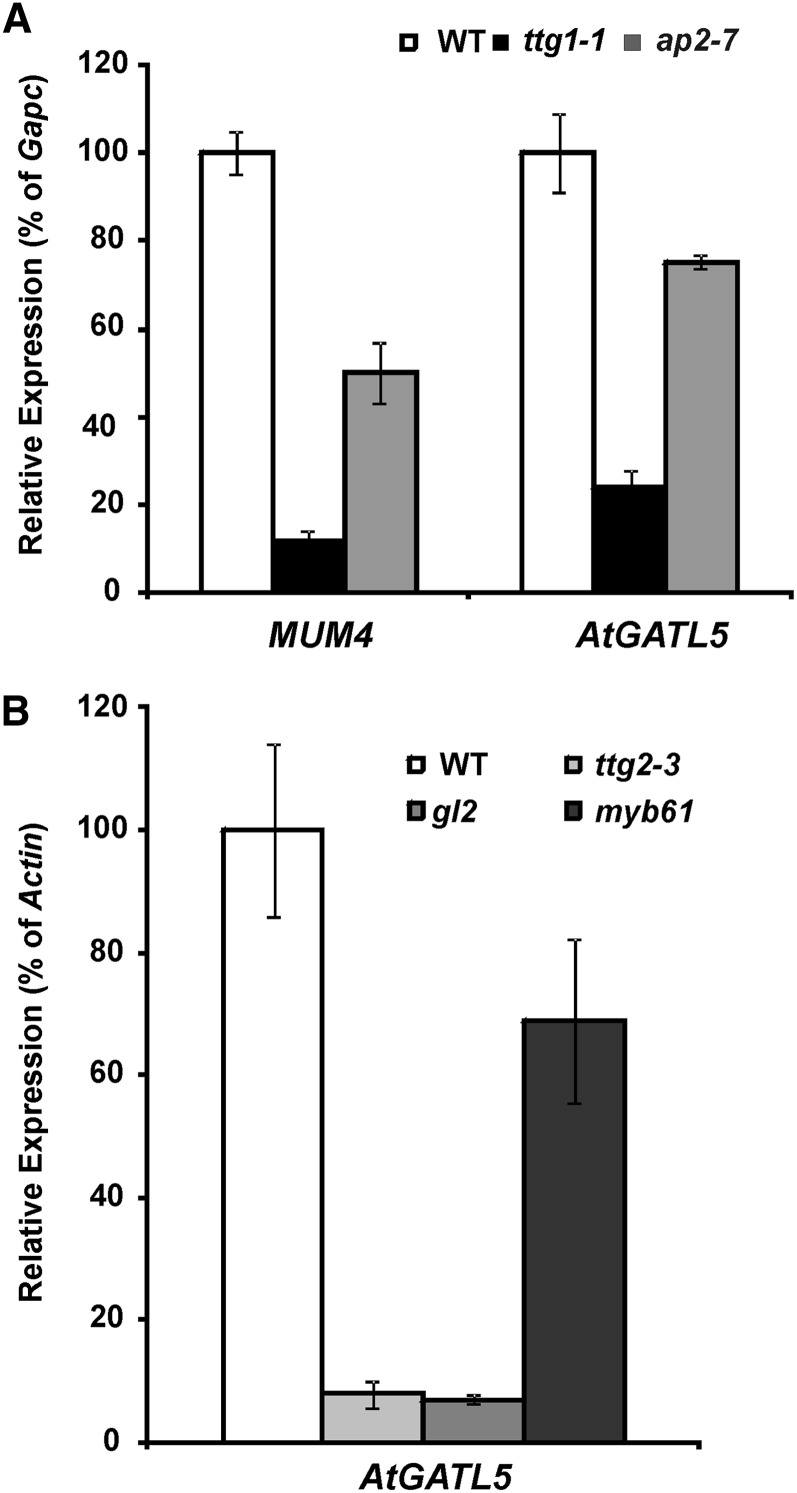
Since TTG1 and AP2 appear to act upstream of other regulators (i.e. GL2 and TTG2), microarray analysis was used to identify genes that are regulated by these two transcription factors. RNA isolated from wild-type, ap2-7, and ttg1-1 siliques at 7 DPA was used to probe Affymetrix ATH1 chips. The microarray identified 264 genes that were down-regulated in the ap2-7 mutant and 52 genes that were down-regulated in the ttg1-1 mutant. The 25 genes that are down-regulated in both ap2-7 and ttg1-1 mutants are listed in Table IV. As expected, MUM4 was one of the genes down-regulated in both mutants. Among those genes coregulated with MUM4 by TTG1 and AP2, AtGATL5 is one of the most strongly down-regulated genes in both ap2-7 and ttg1-1 mutants (Table IV). Quantitative RT-PCR analysis showed a marked decrease in AtGATL5 and MUM4 expression in ap2-7 and ttg1-1 as compared with controls (Fig. 6A). These results validate our microarray experiments. Together, they suggest that, similar to MUM4, AtGATL5 is up-regulated at the time of mucilage synthesis by a pathway that includes both AP2 and the TTG1 transcription factor complex.
Table IV. List of overlapping differentially expressed genes identified by microarray analysis of ttg1-1 and ap2-7 mutants compared with wild-type Ler and Col, respectively.
FC, Fold change: a value of 1 means no change; a fold change of 2 or more means that the expression of the gene was induced in the mutant versus the wild type (i.e. a FC of 2 means that the expression of the gene was doubled in the mutant versus the wild type); a fold change of 0.5 or less means that the gene was repressed in the mutant versus the wild type.
| Locus | Name | Description | FC ttg1-1 versus Ler | FC ap2-7 versus Col |
|---|---|---|---|---|
| At1g02720 | AtGATL5 | Glycosyl transferase family8 protein | 0.10 | 0.16 |
| At1g09550 | Pectinacetylesterase, putative | 0.14 | 0.20 | |
| At1g17860 | Trypsin and protease inhibitor family protein | 0.17 | 0.36 | |
| At1g21070 | Low similarity to GDP-Man transporter | 0.45 | 0.20 | |
| At1g24540 | CYP86C1 | Cytochrome P450, putative | 0.19 | 2.38 |
| At1g30900 | Vacuolar sorting receptor, putative | 0.38 | 0.31 | |
| At1g53500 | MUM4 | NDP-Rha synthase (RHM2) | 0.30 | 0.31 |
| At1g61720 | BAN | Dihydroflavonol 4-reductase | 0.08 | 2.02 |
| At1g62900 | O-Methyltransferase, putative | 0.15 | 0.30 | |
| At2g38900 | Ser protease inhibitor, potato inhibitor I-type family protein | 0.03 | 2.10 | |
| At2g46720 | KCS13 | Member of the 3-ketoacyl-CoA synthase family | 0.39 | 4.66 |
| At3g04200 | Germin-like protein, putative | 0.26 | 0.13 | |
| At3g08770 | LTP6 | Lipid transfer protein6 | 0.01 | 2.91 |
| At3g13900 | Haloacid dehalogenase-like hydrolase family protein | 0.20 | 0.31 | |
| At3g52540 | ATOFP18 | Ovate family protein ATOFP18/OFP18 | 0.30 | 0.37 |
| At3g52550 | Expressed protein, similar to ATOFP15/OFP15 | 0.14 | 0.27 | |
| At4g27830 | Glycosyl hydrolase family1 protein | 0.09 | 0.29 | |
| At5g13400 | Proton-dependent oligopeptide transport family protein | 0.39 | 0.39 | |
| At5g15740 | Expressed protein | 0.34 | 0.14 | |
| At5g17220 | ATGSTF12 | Glutathione S-transferase, putative | 0.07 | 3.13 |
| At5g42800 | DFR | Dihydroflavonol 4-reductase | 0.02 | 2.80 |
| At5g45770 | Leu-rich repeat family protein | 0.16 | 0.03 | |
| At5g47330 | Palmitoyl protein thioesterase family protein | 0.48 | 0.33 | |
| At5g51950 | Glc-methanol-choline oxidoreductase family protein | 0.06 | 2.29 | |
| At5g60630 | Similar to Hyp-rich glycoprotein family protein | 0.01 | 0.37 |
Figure 6.
Quantitative RT-PCR analysis of MUM4 and AtGATL5 transcripts in the wild type (WT), ttg1-1, ap2-7, ttg2-3, gl2, and myb61. RNA was prepared from 7-DPA siliques of each plant line. Error bars represent se of three biological replicates. Relative expression levels in all samples were normalized using GapC-1 (A) or Actin2 (B) as a constitutively expressed internal control, and the expression levels of each gene in the wild type are set to 1.
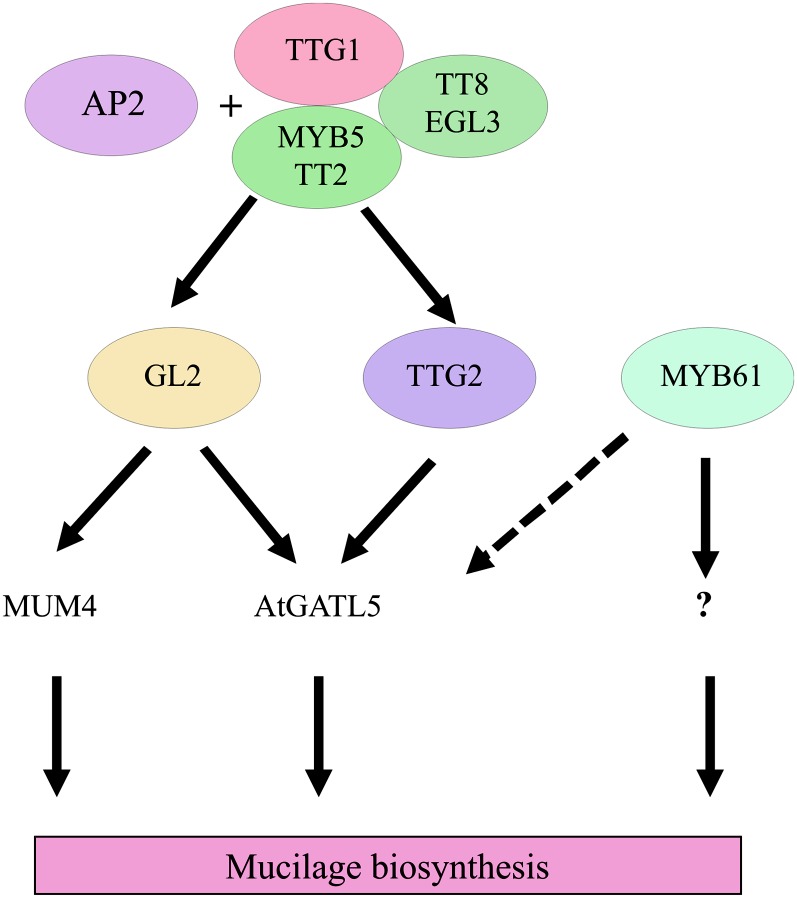
To determine if any of the other known transcription factors affecting mucilage production are involved in the regulation of AtGATL5, we performed quantitative RT-PCR using RNA extracted from 7-DPA siliques of the wild type and ttg2-3, gl2, and myb61 mutants. The level of AtGATL5 transcript was reduced dramatically in both ttg2-3 and gl2 mutant siliques compared with wild-type siliques (Fig. 6B), suggesting that, in addition to AP2 and TTG1, both TTG2 and GL2 are required for maximum levels of AtGATL5 expression at the time of mucilage production. AtGATL5 transcript levels were also down-regulated slightly in myb61 siliques compared with the wild type, although not as dramatically as in ttg2-3 and gl2 siliques. These results suggest that there might be some cross talk between the GL2, TTG2, and MYB61 pathways or that AtGATL5 may be located near the bottom of the mucilage synthesis pathway, where these regulatory pathways converge (Fig. 7).
Figure 7.
Proposed regulatory pathway for seed coat mucilage biosynthesis. AP2 and TTG1, EGL3/TT8, and MYB5/TT2 complex activate GL2 and TTG2, GL2 acts upstream of MUM4, and AtGATL5 is positively regulated by both GL2 and TTG2. MYB61 is independent from the AP2 and TTG1 pathway and positively regulates AtGATL5 slightly (dashed arrow). This pathway cartoon was adapted from Western et al. (2004), Gonzalez et al. (2009), and Li et al. (2009).
DISCUSSION
A domain structure analysis carried out previously on the AtGATL family indicated that none of the members of this family, including AtGATL5, have any apparent membrane-spanning domain but only a signal peptide (Kong et al., 2011). Furthermore, analysis of the amino acid sequences of these proteins did not indicate any other known type of membrane-anchoring domain. Nonetheless, subcellular localization experiments show that all of the AtGATL proteins examined thus far, including AtGATL1, AtGATL2, AtGATL3, AtGATL7, and AtGATL9, are localized in the Golgi and/or ER (Kong et al., 2011). In this study, the AtGATL5 protein was visualized in living Arabidopsis root cells and N. benthamiana leaf cells using a fluorescent protein-tagged AtGATL5 fusion protein, and the results show that AtGATL5 is localized in both the Golgi and the ER (Fig. 1). Considering the fact that all of the cell wall matrix polysaccharide-related GTs characterized so far are localized in the Golgi (Scheible and Pauly, 2004; Sandhu et al., 2009), the subcellular localization pattern of AtGATL5 is consistent with its possible function as a GT during cell wall synthesis.
The null mutant of AtGATL5 has a significant reduction in the total amount of mucilage, and this reduction mainly results from the reduction of both Rha and GalA. No changes in the structure of the atgatl5-1 mucilage were observed except that the molecular mass increased to about 1.6 times that of wild-type mucilage (Table III), suggesting that AtGATL5 plays some role in regulating the size of the RG-I in the mucilage. In this context, an attractively simple hypothesis for AtGATL5 function in mucilage biosynthesis is that AtGATL5 is a glycosylhydrolase that controls the final size of the mucilage. The larger mucilage made when AtGATL5 is mutated would then invoke a negative regulatory mechanism that results in the decreased mucilage production observed in atgatl5-1 seed. The idea of a hydrolase functioning in plant cell wall polysaccharide synthesis has been proposed before. KORRIGAN (KOR), an endo-1,4-β-glucanase, cannot only degrade cellulose but also appears to play a role in plant cellulose synthesis, as evidenced by the reduction in cellulose content observed in the kor mutant (Nicol et al., 1998; Lane et al., 2001). However, glycosylhydrolases involved in cell wall polysaccharide reconstruction and modification after biosynthesis that have been identified to date are located in the cell wall or in the plasma membrane (Minic and Jouanin, 2006). So the localization of AtGATL5 in the endomembrane system makes the hydrolase hypothesis unlikely.
Since mutation of AtGATL5 results only in an increase in RG-I size, it appears unlikely that AtGATL5 plays a role in RG-I backbone synthesis. On the other hand, the reduction in nonmethylesterified HG in seed coat cell walls and mucilage (Fig. 5) supports the idea that AtGATL5 could function as a GalAT in the synthesis of HG. A slight reduction in the molar ratio of GalA to Rha was also observed in the total (enzyme-released) mucilage of atgatl5-1 (Table I), perhaps indicating a reduction in a HG component of the inner mucilage; no change in this ratio was observed in the outer (water-extractable) mucilage. Pectins are thought to act as adhesion molecules during plant growth, and several pectin mutants have been shown to have cell adhesion defects, including the qua1 mutant, which has been shown to have reduced HG biosynthesis activity (Willats et al., 2001c; Bouton et al., 2002; Iwai et al., 2002; Lord and Mollet, 2002; Orfila et al., 2005). The reduced cell adhesion found in atgatl5-1 seed coat epidermal cells is also consistent with the possible role of AtGATL5 in the synthesis of pectin.
RG-I is generally thought to be glycosidically attached to HG domains (Willats et al., 2001b; Caffall and Mohnen, 2009), and oligosaccharides appropriate to such a linkage have been isolated (Nakamura et al., 2002; Coenen et al., 2007). Thus, AtGATL5 could function in the synthesis of a HG primer or terminator important for RG-I synthesis, and mutational inactivation of AtGATL5 would then result in lower synthesis and/or abnormal size of the mucilage, both of which are observed in the atgatl5-1 seed. The idea of a terminal oligosaccharide playing a role in the regulation of polysaccharide size has been proposed for glucuronoxylan synthesis in Arabidopsis (York and O’Neill, 2008). It was found that a normal chain length of glucuronoxylan requires the presence of a GalA-containing tetrasaccharide sequence that is located at the reducing end of the polysaccharide, and mutation of the GAUT12/IRREGLULAR XYLEM8 (IRX8) gene results in lower levels of xylan synthesis and altered polymer length (Peña et al., 2007; Persson et al., 2007). If such a primer/terminator exists on RG-I, it would only account for a small percentage of the total mass of the mucilage, and its absence or alteration would be difficult to detect by any of the methods applied in this study. Further in-depth structural analysis of the mucilage secreted by the atgatl5-1 mutant will be necessary to determine if specific but subtle structural changes have occurred in the polysaccharide as a result of the knockout of AtGATL5.
Heterologous expression studies in N. benthamiana (Supplemental Fig. S6) showed that AtGATL5 does not have a GalAT activity comparable to GAUT1 (Sterling et al., 2006). Our studies, however, cannot exclude other possible enzymatic activities using different and at present unknown acceptors and/or donors. Furthermore, as discussed below, it appears that AtGATL5 is likely to interact with other proteins within a biosynthetic complex and, thus, may be inactive in the absence of those protein partners.
Another hypothesis for how AtGATL5 functions in mucilage biosynthesis is that the protein plays a regulatory rather than a catalytic role as a component of a protein complex that synthesizes RG-I. The AtGATL5 protein is localized to the endomembrane system (Fig. 4), yet the amino acid sequence analysis (Fig. 1) shows no transmembrane domain or other endomembrane-targeting sequences. Thus, it is highly likely that AtGATL5 is retained in the endomembrane system by virtue of interactions with at least one other protein that is membrane anchored/targeted. GAUT11, another protein in family GT8 that has a transmembrane domain (Yin et al., 2010), is an attractive candidate for the luminal enzyme that associates with AtGATL5, as mutations in GAUT11 also result in reduced seed mucilage synthesis (Caffall et al., 2009). There is literature precedence for the idea that proteins in GT families can serve regulatory, rather than catalytic, functions. For example, GAUT7, another protein in the GT8 family, is thought to play a noncatalytic role in HG biosynthesis in association with GAUT1 (Sterling et al., 2006; Atmodjo et al., 2011). Recent studies (Atmodjo et al., 2011) demonstrate that the transmembrane domain of GAUT1 is cleaved off posttranslationally and show that the mature GAUT1 protein is retained in the Golgi by association with GAUT7, which retains its transmembrane domain, as part of a large biosynthetic protein complex. A further example where a GT has been suggested to play a noncatalytic role in plant cell wall polysaccharide synthesis is in xyloglucan biosynthesis (Zabotina et al., 2012). Mutation of the XLYLOGLUCAN XYLOSYLTRANSFERASE5 (XXT5) gene results in a more dramatic decrease in xyloglucan content and more dramatic changes in xyloglucan structure than do mutations in either XXT1 or XXT2 (Zabotina et al., 2008, 2012). Yet, no catalytic activity for XXT5 could be demonstrated to date (Zabotina et al., 2008), and the presence of functional XXT5 alone in the xxt1 xxt2 double mutant does not result in any xyloglucan biosynthesis (Cavalier et al., 2008; Zabotina et al., 2012). Furthermore, XXT1, XXT2, and XXT5 appear to be part of a membrane protein complex that synthesizes xyloglucan (Chou et al., 2012). Thus, we suggest that the identification and investigation of proteins that interact with AtGATL5 may be potentially informative regarding AtGATL5 function.
Analysis of the expression patterns for AtGATL5 showed that its expression is highest in 7-DPA siliques, when seed mucilage synthesis is at its peak. AtGATL5 expression in siliques is very similar to that observed previously for MUM4, which encodes a Rha synthase that has been implicated in seed mucilage production (Western et al., 2004). It has been proposed that there are three pathways that control mucilage biosynthesis in the developing seed, which include the GL2, TTG2, and MYB61 transcription factors (Western, 2012). It was reported that MUM4 belongs to the GL2 regulatory pathway and that MUM4 up-regulation at the time of the mucilage production does not require either TTG2 or MYB61 (Western et al., 2004). In contrast, our data show that AtGATL5 is regulated not only by GL2 but also by TTG2 and MYB61 (Fig. 7), which suggests that cross talk may occur among different regulatory pathways controlling mucilage biosynthesis. Our microarray analysis shows that AtGATL5 is the only GT gene identified that is regulated by the master regulators AP2 and TTG1. Clearly, additional GTs must be involved in mucilage synthesis, and these could be regulated by other unidentified pathways. Recently, LEUNIG_HOMOLOG (LUH)/MUM1 was identified as a regulator of mucilage pectin modification (Bui et al., 2011; Huang et al., 2011; Walker et al., 2011). It would be interesting to check whether this transcription factor plays any role in regulating AtGATL5 that is related to mucilage backbone synthesis.
In addition to its expression in the seed coat, our previous studies demonstrated that AtGATL5 is also expressed in nonseed tissues. Notably, expression was observed in the secondary xylem of stems and hypocotyls (Kong et al., 2011), suggesting that AtGATL5 could also be involved in RG-I biosynthesis in these secondary cell wall-enriched tissues. MYB61, the transcription factor that is related to mucilage deposition and extrusion, was also expressed in developing vascular tissues (Penfield et al., 2001). Comparison of the GUS-promoter fusion expression patterns shows that both AtGATL5 and MYB61 have similar expression patterns in root cap and stem vascular tissue (Penfield et al., 2001; Kong et al., 2011), which suggests that MYB61 might also regulate pectin synthesis in these tissues through AtGATL5 and other related genes.
In summary, we have demonstrated that AtGATL5 is involved in seed mucilage RG-I biosynthesis in Arabidopsis. Our work also further demonstrates the utility of the Arabidopsis seed coat as a good model system for the identification of GTs involved in pectin, and particularly RG-I, biosynthesis. The usefulness of this model system has been enhanced recently by the establishment of global expression data sets covering seed coat development in Arabidopsis (Le et al., 2010; Dean et al., 2011). Although the exact role of AtGATL5 in the synthesis of RG-I remains to be determined, the work reported here provides impetus for future research using both polysaccharide structural approaches and biochemical studies of possible biosynthetic complexes to further elucidate the functions of AtGATL5 in RG-I synthesis.
MATERIALS AND METHODS
Plant Material and Genetic Analysis
Seeds of Arabidopsis (Arabidopsis thaliana) were sterilized, cold treated for 48 h, and germinated on one-half-strength Murashige and Skoog medium in an environmental chamber under a 16-h-light/8-h-dark cycle at 19°C during the light period and 15°C during the dark period. The light intensity was 120 µmol m–2 s–1, and the relative humidity was maintained at 70%. Seedlings were then transferred to soil and grown in greenhouse chambers under the same conditions.
Insertion lines for atgatl5 (Salk_106615 and Salk_038363), gl2 (Salk_039825), ttg2-3 (Salk_148838), myb61 (Salk_106556), as well as seeds for ap2-7 (Columbia [Col] ecotype; Kunst et al., 1989) and ttg1-1 (Landsberg erecta [Ler]; Koornneef, 1981), were obtained from the Arabidopsis Biological Resource Center (http://arabidopsis.org). PCR was performed, with primer sequences generated against the genomic regions flanking the insert and a standard primer for the 3′ end of the insertion sequence, to obtain homozygous insertion lines (primers are listed in Supplemental Table S2).
Complementation of atgatl5-1 Mutants
A 4.5-kb genomic DNA fragment containing the AtGATL5 gene (including a 2.1-kb 5′ upstream region, the entire exon sequence, and a 1.3-kb 3′ downstream region) were amplified by PCR using a pfx50 DNA polymerase (Invitrogen) with gene-specific primers G5F-PstI and G5R-XbaI (Supplemental Table S2), confirmed by sequencing, and cloned into the vector pCambia1300. The constructs were introduced into the atgatl5-1 mutant by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). Transgenic plants were selected by growth on nutrient agar containing hygromycin and used for analysis.
RNA Isolation, RT-PCR, and Quantitative RT-PCR
RNA isolation and analysis of gene expression by quantitative RT-PCR were performed as described previously (Kong et al., 2009). Actin2 (At3g18780) and Gapc-1 (At3g18780) were used as reference genes. Each reaction was repeated three times using complementary DNAs (cDNAs) generated from independently isolated RNA preparations. Gene-specific primers are listed in Supplemental Table S2. To determine AtGATL5 transcript levels in the homozygous mutant lines, total RNAs were isolated from 7-DPA siliques of the wild type and homozygous atgatl5-1 mutant lines, and RT-PCR was performed as described previously (Kong et al., 2011).
In Situ Hybridization
The protocol performed here is a modification of that described previously (Zhou et al., 2006). To generate digoxigenin-labeled probes, a 292-bp fragment of AtGATL5 cDNA was first subcloned into the pGM-T (Tiangen) vector using primers GATL5-Probe-F and GATL5-Probe-R (Supplemental Table S2). The DNA fragment between the Sp6 and T7 primer sequences in the construct was then amplified by PCR. Digoxigenin-labeled sense and antisense RNA probes were prepared using an in vitro transcription kit (Roche). Wild-type Arabidopsis seeds were fixed in formaldehyde:acetic acid:water (90:5:5, v/v/v) solution overnight under vacuum at 4°C and then embedded in paraffin after dehydration in a graded ethanol series of 25%, 50%, 75%, 95% (v/v), and 100%. The tissue was incubated for 1 h at room temperature in each step, and the final incubation in 100% ethanol was repeated three times. Sections (8 µm) were cut on a rotary RM 6025 microtome (Leica Microsystems), mounted onto Superfrost Plus glass slides (Thermo Fisher), and hybridized with the digoxigenin-labeled AtGATL5 antisense and sense RNA probes. The hybridization signals were detected by incubating with alkaline phosphatase-conjugated antibodies against digoxigenin and subsequent color development with alkaline phosphatase substrates. The images were collected with an Olympus X51 light microscope and processed with Photoshop version 7.0 (Adobe Systems).
Microarray Analysis
For each sample, three biological replicates were prepared. RNA was extracted from 20 siliques at 4 and 7 DPA using the RNeasy Plant Maxi Kit (Qiagen) according to the manufacturer’s protocol with the following modification: ground samples were initially resuspended in 26.8 mL of RLT-PVP-40-β buffer (23.9 mL of RLT plus 2.6 mL of 10% [w/v] polyvinylpyrrolidone-40 and 0.3 mL of β-mercaptoethanol). The quality of the RNA was determined using the 2100 Bioanalyzer (Agilent Technologies). Microarray analysis was performed using the GeneChip Arabidopsis ATH1 Genome Array (Affymetrix). Standard RNA processing, hybridization, and scanning protocols were followed as recommended by the GeneChip Expression Analysis Technical Manual (Affymetrix). Hybridization and scanning were performed at the McGill University Cell Imaging and Analysis Network facility. The robust multiarray average procedure (Irizarry et al., 2003) was performed to normalize data using the AffylmGUI R software package from Bioconductor (http://www.bioconductor.org/; Gentleman et al., 2004). Analysis of differential expression was done using a moderated Student’s t test with empirical Bayes smoothing (Smyth, 2004). A cutoff of 2-fold change and P < 0.05 were adopted to identify the differentially expressed genes.
Microscopic Investigations of Seed Coats
The seed coat expression pattern of the AtGATL5 gene was studied using the GUS reporter gene as described previously (Kong et al., 2011). For sections, developing siliques were fixed in 2.5% (v/v) glutaraldehyde buffered with 0.05 m phosphate buffer (pH 6.8) at 4°C for 24 h. After fixation, the tissues were dehydrated using an ethanol series and embedded in LR White resin (Ted Pella) as described (Freshour et al., 1996). Sections (1 µm thick) were cut using an Ultracut E ultramicrotome (Reichert-Jung) and stained with 0.1% (w/v) toluidine blue solution for light microscopy, carried out on an Eclipse80i microscope (Nikon). Images were captured with a Nikon DS-Ril camera head using NIS-Elements Basic Research software, and images were assembled using Adobe Photoshop 7.0.
Immunogold labeling was done as described previously (Freshour et al., 1996). In brief, 85-nm-thick sections were incubated with the CCRC-M38 antibody and then with gold-conjugated goat anti-mouse IgG (18 nm; Jackson ImmunoResearch Laboratories). The gold-conjugated secondary antibody was used at a dilution of 1:10 (v/v). The immunogold-labeled sections were visualized with an EM 902A transmission electron microscope (Carl Zeiss).
Subcellular Localization of AtGATL5
The AtGATL5 cDNA was PCR amplified, confirmed by sequencing, and then fused in frame with an EYFP gene under the control of the 35S promoter in a pCAMBIA-based binary vector to generate the fusion construct Pro-35S:AtGATL5-EYFP. Primers used are listed in Supplemental Table S2. The Pro-35S:AtGATL5-EYFP construct was introduced into wild-type Arabidopsis plants by A. tumefaciens-mediated transformation. Transgenic plants were selected on one-half-strength Murashige and Skoog medium with 15 mg L−1 hygromycin, and the T2 progeny were used for EYFP localization. The EYFP signals from roots of 3-d-old transgenic seedlings were viewed with a TCs SP2 spectral confocal microscope (Leica Microsystems). Images were saved and processed with Adobe Photoshop version 7.0.
The colocalization of fluorescent protein-tagged AtGATL5 with the Golgi and ER markers was carried out in Nicotiana benthamiana leaf cells as described (Kong et al., 2009). In brief, the Pro-35S:AtGATL5-EYFP construct was individually cotransformed into fully expanded leaf of N. benthamiana plants (approximately 8-week-old seedlings grown at 22°C) together with constructs encoding either the ECFP-tagged Golgi marker (Gmct-ECFP) or the ECFP-tagged ER marker (ECFP-WAK2-HDEL). At 3 or 4 d post infection, the transfected leaf in the injected area was cut and subsequently examined for yellow and cyan fluorescent signals using a Leica TCs SP2 confocal microscope.
Seed Staining
Ruthenium red staining of seeds was performed by placing the seeds in a 0.01% (w/v) ruthenium red solution and shaking them for 10 min. Seeds were removed from the imbibition solution and observed with a stereoscopic microscope (Olympus SZH-ILLD). Calcofluor white staining was performed by staining seeds for 5 min in 1 mg mL−1 calcofluor white in 2 mm sodium hydroxide, followed by two rinses in deionized water. Seeds were examined using a Leica DM6000B epifluorescence microscope with UV illumination, and images were captured with a Qimaging Retiga CCD camera operated through Openlab.
Scanning Electron Microscopy
Dry mature seeds were mounted on stubs, coated with gold, and observed using a Zeiss 1450EP scanning electron microscope at an accelerating voltage of 20 kV.
Immunological Analyses
Glycome profiling by ELISA was performed as described (Pattathil et al., 2012). Monoclonal antibodies were obtained as hybridoma cell culture supernatants from laboratory stocks (CCRC series and JIM series antibodies are available from CarboSource [http://www.carbosource/net]). Detailed information about all monoclonal antibodies included in this study can be found at WallMabDB (http://www.wallmabdb.net), an online database that provides information about the immunogens used to develop the antibodies, their isotype, the cell wall polysaccharide class they primarily recognize, and detailed epitope information where available (Pattathil et al., 2010). Whole-seed immunolabeling was conducted as described (Sullivan et al., 2011).
Cell Wall Preparation and Monosaccharide Composition Analysis
Total mucilage for monosaccharide composition analysis was extracted from mature seeds (approximately 5 mg) by shaking them in distilled water or in water containing 0.01 mg mL−1 rhamnogalacturonan hydrolase (Novozymes) for 6 h at room temperature (20°C); 50 µg of inositol was added as an internal standard. Ruthenium red staining was performed to check that total mucilage was released from the seeds. Insoluble material was removed by centrifugation, and the supernatant containing the released mucilage material was lyophilized. The GalA and Rha content was determined using trimethyl silyl ethers of methyl glycosides as described by Caffall et al. (2009), and other neutral sugar content was determined using the alditol acetate method as described by Kong et al. (2009). All analyses were carried out on three independently extracted mucilage preparations. Statistical analysis was performed using Student’s t test.
Glycosyl Linkage Composition Analysis
Mucilage used for linkage analysis was extracted by shaking intact seeds in distilled water for 5 h. The supernatant containing the released mucilage material was frozen and lyophilyzed. For linkage analysis, the sample was permethylated, depolymerized, reduced, and acetylated; the resultant partially methylated alditol acetates were analyzed by gas chromatography-mass spectrometry as described (York et al., 1985). Initially, an aliquot of the sample after dialysis was suspended in about 200 µL of dimethyl sulfoxide and placed on a magnetic stirrer for 2 d. A total of 0.6 mL of potassium dimsylate (3.6 m) was added to each sample. After 7 h at room temperature on the stirrer, the reaction mixture was cooled to 0°C, excess methyl iodide (0.7 mL) was added, and the tube was sealed. The incubation was then continued overnight at room temperature. After workup, the permethylated samples were reduced by superdeuteride. Following the workup after reduction, the samples were then repermethylated by treatment with sodium hydroxide and methyl iodide in dry dimethyl sulfoxide (Ciucanu and Kerek, 1984). The permethylated material was hydrolyzed using 2 m trifluoroacetic acid (2 h in sealed tube at 121°C), reduced with NaBD4, and acetylated using acetic anhydride/trifluoroacetic acid. The resulting partially methylated alditol acetates were analyzed on a HP 5890 gas chromatograph interfaced to a 5970 mass selective detector (electron impact ionization mode); separation was performed on a 30m Supelco 2330 bonded phase fused silica capillary column.
Microsome Preparation and GalAT Activity Assay
N. benthamiana leaves infiltrated with either Pro-35S:AtGATL5-EYFP or EYFP vector were harvested on day 3 after infiltration. Tissues were ground in homogenization buffer containing 50 mm HEPES-KOH, pH 7.3, 0.25 mm MnCl2, 25 mm KCl, 50% (v/v) glycerol, 0.1% (v/v) β-mercaptoethanol, and protease inhibitors (Roche Complete protease inhibitor tablets). The homogenate was filtered through two layers of Miracloth and centrifuged at 3,000g for 10 min, and then the supernatant was centrifuged at 280,000g for 1 h. The pellet was resuspended in the homogenization buffer that contained 25% (v/v) glycerol. Microsomal protein was quantified using the Protein Assay (Bio-Rad). HG:GalAT activity was measured essentially as described (Sterling et al., 2006) in 30-μL reactions containing 50 μg of microsomes, 50 mm HEPES, pH 7.3, 0.2 m Suc, 0.05% (w/v) bovine serum albumin, 25 mm KCl, 80 μg of oligogalacturonide mixture (DP 7-23), 6.9 μm UDP-[14C]GalA (specific activity, 180.3 mCi mmol−1; 1 Ci = 37 GBq), 1.25 mm MnCl2, and 0.5% (v/v) Triton X-100. Reactions were incubated for 3 h at 30°C and terminated by the addition of 5 μL of 400 mm NaOH. Reaction products were measured using a filter assay method (Sterling et al., 2006).
HP-SEC Combined with Multiple-Angle Laser Light Scattering
The HP-SEC multiple-angle laser light scattering system consisted of a Waters P515 pump with an in-line degasser (Waters) and in-line filters of 0.2 and 0.1 μm. The solvent used for separation was 100 mm sodium nitrate and 10 mm phosphate buffer. Columns used were guard and separating columns of PL-Aquagel-OH linear mix (8-µm pore size; Polymer Laboratories). Samples were filtered through disposable syringe filters (0.45 μm, PES w/GMF; Whatman) before injection. Samples were injected with a 50-μL loop in a Spectrasystem autosampler. Two detectors were present in line, an 18-angle light-scattering detector and a refractive index detector (Wyatt Technologies). Data for molar mass determinations were analyzed using Astra software version 4.74.03 (Wyatt Technologies) taking the differential index of refraction as 0.146 (Macquet et al., 2007a). Chromatograms presented are representative of three independent replications.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_202015.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Pro35S:AtGATL5-EYFP complements the phenotypes of atgatl5-1.
Supplemental Figure S2. Expression profile of AtGATL5.
Supplemental Figure S3. Ruthenium red staining of wild-type seeds after mucilage extraction.
Supplemental Figure S4. ELISAs of water-extracted mucilage from wild-type and atgatl5-1 seeds using plant glycan-directed monoclonal antibodies.
Supplemental Figure S5. Size exclusion chromatography of the water-extracted mucilage from wild-type, atgatl5-1, and complemented atgatl5-1 seeds.
Supplemental Figure S6. HG:GalAT assay of microsomal proteins from N. benthamiana transformed with Pro35S:AtGATL5-EYFP.
Supplemental Table S1. Monosaccharide composition of water-extractable mucilage from wild-type and atgatl5-1 seeds.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We gratefully acknowledge the assistance of Utku Avci (University of Georgia) with fixation of seeds and siliques for gold immunolabeling, Malcolm O’Neill (University of Georgia) and Radnaa Naran (University of Georgia) for useful suggestions and assistance with mucilage structural analysis, Liangjiao Xue (University of Georgia) for assistance with statistical analyses, and Fanfang Fu (University of Georgia) for critical review of the manuscript.
Glossary
- GalA
GalUA
- HG
homogalacturonan
- RG-I
rhamnogalacturonan I
- GT
glycosyltransferase
- ER
endoplasmic reticulum
- ECFP
enhanced cyan fluorescent protein
- EYFP
enhanced yellow fluorescent protein
- T-DNA
transfer DNA
- RT
reverse transcription
- HP-SEC
high-performance size-exclusion chromatography
- Col
Columbia
- Ler
Landsberg erecta
- cDNA
complementary DNA
References
- Anderson E. (1933) The preparation of l-galactose from flaxseed mucilage. J Biol Chem 100: 249–253 [Google Scholar]
- Arsovski AA, Haughn GW, Western TL. (2010) Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research. Plant Signal Behav 5: 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL. (2009) AtBXL1 encodes a bifunctional β-d-xylosidase/α-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, III, Orlando R, Scheller HV, Mohnen D. (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azadi P, O’Neill MA, Bergmann C, Darvill AG, Albersheim P. (1995) The backbone of the pectic polysaccharide rhamnogalacturonan I is cleaved by an endohydrolase and an endolyase. Glycobiology 5: 783–789 [DOI] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker M-T, Talbotec J, Granier F, Lahaye M, Höfte H, Truong H-N. (2002) QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14: 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Lim N, Sijacic P, Liu ZC. (2011) LEUNIG_HOMOLOG and LEUNIG regulate seed mucilage extrusion in Arabidopsis. J Integr Plant Biol 53: 399–408 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D. (2009) Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant 2: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. (2009) The Carbohydrate-Active Enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YH, Pogorelko G, Zabotina OA. (2012) Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol 159: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res 131: 209–217 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coenen GJ, Bakx EJ, Verhoef RP, Schols HA, Voragen AGJ. (2007) Identification of the connecting linkage between homo- or xylogalacturonan and rhamnogalacturonan type I. Carbohydr Polym 70: 224–235 [Google Scholar]
- Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G. (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4: 1074–1091 [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG. (1996) Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325: 412–421 [DOI] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul A, Vibe Scheller H. (2010) Biosynthesis of pectin. Plant Physiol 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ. (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Western TL. (2012) Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front Plant Sci 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, DeBowles D, Esfandiari E, Dean G, Carpita NC, Haughn GW. (2011) The Arabidopsis transcription factor LUH/MUM1 is required for extrusion of seed coat mucilage. Plant Physiol 156: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Iwai H, Masaoka N, Ishii T, Satoh S. (2002) A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc Natl Acad Sci USA 99: 16319–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DLC, Breekland AE, Koornneef M. (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Kim YC, Nakajima M, Nakayama A, Yamaguchi I. (2005) Contribution of gibberellins to the formation of Arabidopsis seed coat through starch degradation. Plant Cell Physiol 46: 1317–1325 [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhou G, Avci U, Gu X, Jones C, Yin Y, Xu Y, Hahn MG. (2009) Two poplar glycosyltransferase genes, PdGATL1.1 and PdGATL1.2, are functional orthologs to PARVUS/AtGATL1 in Arabidopsis. Mol Plant 2: 1040–1050 [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhou G, Yin Y, Xu Y, Pattathil S, Hahn MG. (2011) Molecular analysis of a family of Arabidopsis genes related to galacturonosyltransferases. Plant Physiol 155: 1791–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M. (1981) The complex syndrome of the ttg mutants. Arabidopsis Inf Serv 18: 45–51 [Google Scholar]
- Kunieda T, Mitsuda N, Ohme-Takagi M, Takeda S, Aida M, Tasaka M, Kondo M, Nishimura M, Hara-Nishimura I. (2008) NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 20: 2631–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunieda T, Shimada T, Kondo M, Nishimura M, Nishitani K, Hara-Nishimura I. (2013) Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 25: 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW. (1989) AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng LC, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al. (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboeuf E, Guillon F, Thoiron S, Lahaye M. (2005) Biochemical and immunohistochemical analysis of pectic polysaccharides in the cell walls of Arabidopsis mutant QUASIMODO 1 suspension-cultured cells: implications for cell adhesion. J Exp Bot 56: 3171–3182 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Keijzer CJ, Koornneef M. (1994) A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell 6: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW. (2009) The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord EM, Mollet JC. (2002) Plant cell adhesion: a bioassay facilitates discovery of the first pectin biosynthetic gene. Proc Natl Acad Sci USA 99: 15843–15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM. (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendu V, Griffiths JS, Persson S, Stork J, Downie AB, Voiniciuc C, Haughn GW, DeBolt S. (2011) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z, Jouanin L. (2006) Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol Biochem 44: 435–449 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Muralikrishna G, Salimath PV, Tharanathan RN. (1987) Structural features of an arabinoxylan and a rhamnogalacturonan derived from linseed mucilage. Carbohydr Res 161: 265–271 [Google Scholar]
- Nakamura A, Furuta H, Maeda H, Takao T, Nagamatsu Y. (2002) Analysis of the molecular construction of xylogalacturonan isolated from soluble soybean polysaccharides. Biosci Biotechnol Biochem 66: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Naran R, Chen G, Carpita NC. (2008) Novel rhamnogalacturonan I and arabinoxylan polysaccharides of flax seed mucilage. Plant Physiol 148: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. (1998) A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Nemoto T, Jigami Y. (2007) Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J Biol Chem 282: 5389–5403 [DOI] [PubMed] [Google Scholar]
- Orfila C, Sørensen SO, Harholt J, Geshi N, Crombie H, Truong HN, Reid JSG, Knox JP, Scheller HV. (2005) QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta 222: 613–622 [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Miller JS, Hahn MG (2012) Immunological approaches to plant cell wall and biomass characterization: glycome profiling. In M Himmel, ed, Biomass Conversion: Methods and Protocols, Vol 908. Springer Science + Business Media, New York, pp 61–72 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye ZH. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C. (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM. (2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye ML, Plasson C, Hawes C, Driouich A, Faye L, Gomord V. (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu APS, Randhawa GS, Dhugga KS. (2009) Plant cell wall matrix polysaccharide biosynthesis. Mol Plant 2: 840–850 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Pauly M. (2004) Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr Opin Plant Biol 7: 285–295 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: e3. [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D. (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM. (2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M. (2004) RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol 134: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW. (2013) Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Tehseen M, Doblin MS, Pettolino FA, Wilson SM, Bacic A, Golz JF. (2011) The transcriptional regulator LEUNIG_HOMOLOG regulates mucilage release from the Arabidopsis testa. Plant Physiol 156: 46–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL. (2012) The sticky tale of seed coat mucilages: production, genetics, and role in seed germination and dispersal. Seed Sci Res 22: 1–25 [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW. (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins CAR, Munro S. (1998) Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA 95: 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Knox JP. (2001a) In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44 [DOI] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Mackie W, Knox JP. (2001b) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47: 9–27 [PubMed] [Google Scholar]
- Willats WGT, Orfila C, Limberg G, Buchholt HC, van Alebeek GJWM, Voragen AGJ, Marcus SE, Christensen TMIE, Mikkelsen JD, Murray BS, et al. (2001c) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls: implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Yin Y, Chen H, Hahn MG, Mohnen D, Xu Y. (2010) Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol 153: 1729–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. (1985) Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol 118: 3–40 [Google Scholar]
- York WS, O’Neill MA. (2008) Biochemical control of xylan biosynthesis: which end is up? Curr Opin Plant Biol 11: 258–265 [DOI] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL. (2008) Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG. (2012) Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol 159: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, van de Ven WTG, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV. (2008) Arabidopsis XXT5 gene encodes a putative α-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhou GK, Zhong R, Richardson EA, Morrison WH, III, Nairn CJ, Wood-Jones A, Ye ZH. (2006) The poplar glycosyltransferase GT47C is functionally conserved with Arabidopsis Fragile fiber8. Plant Cell Physiol 47: 1229–1240 [DOI] [PubMed] [Google Scholar]