A novel lipid transfer pathway from anther to the microspore explains the need for plentiful lipid transfer proteins for the abundant exine.
Abstract
Lipid transfer proteins (LTPs) are small secretory proteins in plants with defined lipid-binding structures for possible lipid exocytosis. Special groups of LTPs unique to the anther tapetum are abundant, but their functions are unclear. We studied a special group of LTPs, type III LTPs, in Arabidopsis (Arabidopsis thaliana). Their transcripts were restricted to the anther tapetum, with levels peaking at the developmental stage of maximal pollen-wall exine synthesis. We constructed an LTP-Green Fluorescent Protein (LTP-GFP) plasmid, transformed it into wild-type plants, and monitored LTP-GFP in developing anthers with confocal laser scanning microscopy. LTP-GFP appeared in the tapetum and was secreted via the endoplasmic reticulum-trans-Golgi network machinery into the locule. It then moved to the microspore surface and remained as a component of exine. Immuno-transmission electron microscopy of native LTP in anthers confirmed the LTP-GFP observations. The in vivo association of LTP-GFP and exine in anthers was not observed with non-type III or structurally modified type III LTPs or in transformed exine-defective mutant plants. RNA interference knockdown of individual type III LTPs produced no observable mutant phenotypes. RNA interference knockdown of two type III LTPs produced microscopy-observable morphologic changes in the intine underneath the exine (presumably as a consequence of changes in the exine not observed by transmission electron microscopy) and pollen susceptible to dehydration damage. Overall, we reveal a novel transfer pathway of LTPs in which LTPs bound or nonbound to exine precursors are secreted from the tapetum to become microspore exine constituents; this pathway explains the need for plentiful LTPs to incorporate into the abundant exine.
Plant lipid transfer proteins (LTPs) are so termed because, in the initial studies, the proteins were shown to transfer phospholipids and fatty acids between membranes in vitro (Kader, 1996; Boutrot et al., 2008; Edstam et al., 2011). LTPs have a low molecular mass, 6 to 10 kD, and usually a high pI, 8 to 12. Each molecule has eight conserved Cys residues, which form four defined disulfide bonds. The polypeptide has three to four α-helical structures. These and other structures of the protein generate a hydrophobic cavity that could bind to different phospholipids and fatty acids. Because the proteins display nonspecific binding to different lipids, LTPs are also termed nonspecific LTPs.
Although LTPs were thought to function in lipid transfer between subcellular membranes, subsequent findings have shown that their nascent polypeptides possess an N-terminal endoplasmic reticulum (ER)-targeting signal peptide (Boutrot et al., 2008; Edstam et al., 2011). Thus, LTPs are likely secreted to the cell exterior for functioning. Numerous functions of LTPs in diverse cells or tissues have been documented or proposed. LTPs could play a role in long-distance signaling in the vascular system, pathogen defense on the organism surface, symbiotic association between plants and microbes, formation of epidermis surface lipids, seed endosperm lipid recycling, and protection of protease digestion during programmed cell death, cell wall extension, and pollen tube and style track recognition (Edstam et al., 2011).
LTPs are present, depending on the growth condition and environment, in diverse or restricted organs, tissues, and cells, including callus, maturing and germinating seeds, leaves, roots, stems, ovaries, anthers, and pollen (Boutrot et al., 2008; Edstam et al., 2011). The localization of LTP transcripts in anthers has been extensively reported, in part because anthers are essential for producing pollen and contain abundant metabolically active and secretory tapetum cells to which the transcripts could locate. In situ hybridization or LTP-promoter-GUS transformation has localized LTP transcripts or proteins in anthers and/or the tapetum of tomato (Solanum lycopersicum; TomA108 [Chen et al., 2006] and TomA5B [Aguirre and Smith, 1993]), tobacco (Nicotiana tabacum; TA32 and TA36 [Koltunow et al., 1990]), Brassica species and Arabidopsis (Arabidopsis thaliana; A9 [Paul et al., 1992; Turgut et al., 1994]), rice (Oryza sativa; YY1 [Tsuchiya et al., 1994], Osc4 [Hihara et al., 1996], and several other genes [Huang et al., 2009; Zhang et al., 2010]), sugar beet (Beta vulgaris; BvLTP1 and BvLTP2 [Matsuhira et al., 2006]), lily (Lilium longiflorum; LHM6 and LHM7 [Crossley et al., 1995]), and maize (Zea mays; MZm3-3 [Lauga et al., 2000]). However, because of the low sensitivity of detection methods, especially those used in earlier years, whether LTP transcripts are also present in lesser amounts in other organs/tissues/cells, such as the anther epidermis, is unknown. A recent study localized a rice tapetum LTP protein in the tapetum but also in the anther epidermis (Zhang et al., 2010) and perhaps in the epidermis of other organs as well. Because the tapetum is secretory in nature, the tapetum LTPs, which are also found in the anther locule, are considered to function to transfer lipid materials from the tapetum to maturing microspores. However, this function has not been defined, even though the pollen exine consists of lipid monomers and some of their synthetic enzymes are associated with the ER (Ariizumi and Toriyama, 2011; Kim and Douglas, 2013; Lallemand et al., 2013).
LTP transcripts in anthers are in high abundance. In rice, quantitative sequencing-by-synthesis transcriptomes have revealed several anther LTP transcripts in combination representing 8% of the total anther transcripts and, therefore, in an even higher proportion in the restricted tapetum (Huang et al., 2009). Similarly, LTP transcripts in anthers of Arabidopsis are abundant in the tapetum (see “Results”). Why the tapetum needs such a high amount of LTPs to carry out the proposed catalytic function of transferring lipids is an intriguing question.
In individual plant species, LTPs are encoded by several dozen paralogs. They are categorized into at least nine types or clades by comparison of LTP genes within and among species on the basis of their sequences and the presence and location of an intron in the gene, as well as their encoded proteins having a potential glycophosphatidylinositol modification site, defined number of residues between each two of the eight conserved Cys residues, and molecular mass (Boutrot et al., 2008; Edstam et al., 2011). Type I LTPs and their derivatives are the basic and ubiquitous clades. Nonplant organisms and green algae (chlorophytes and charophytes) do not have LTP genes, whereas liverworts (Marchantia spp.) do. As primitive plants evolved to higher plants, the number of LTP genes and types expanded, along with their cell expression specificity and proposed LTP functions. Arabidopsis and rice have 49 and 52 LTP paralogs, respectively, of the nine types. These paralogs have been studied on a global scale with EST databases, microarrays, transcriptome analysis, and LTP-promoter-GFP transgenic analysis (Huang et al., 2009; Edstam et al., 2011; Ng et al., 2012; Wang et al., 2012a, 2012b).
Type III LTPs (Boutrot et al., 2008), also termed type C LTPs (Edstam et al., 2011), are found only in seed plants, which produce pollen and seeds. They possess somewhat unique properties, which in combination distinguish them from other LTP types. Their genes have an intron at a DNA site not found in other LTP types (Edstam et al., 2011) and apparently express only or highly in the anther tapetum and not in the anther epidermis or other plant tissues (see “Results”). Type III LTPs have an uncharacteristic low pI of 4 in Arabidopsis (but 7 in rice), the smallest size of less than 70 residues among all LTP types, and no potential glycophosphatidylinositol modification site. Yet, the function of type III LTPs has not been examined in detail other than that, in the amphidiploid Brassica napus, the type III A3 gene showed tapetum-specific expression and its antisense knockdown had no effect on pollen viability (Paul et al., 1992; Turgut et al., 1994).
In this study, we investigated the transcripts of type III LTPs in Arabidopsis and found them abundant and highly restricted to the anther tapetum. Type III LTPs, with or without bound exine precursors, are secreted from the tapetum via the ER-Golgi system and then become components of the microspore exine. The association of type III LTPs with exine is LTP type specific and does not occur in exine-deficient mutants.
RESULTS
Several Arabidopsis Abundant LTP Transcripts Are Present Mostly in Flowers
The expression of the 49 individual LTP paralogs in Arabidopsis was diverse or restricted in various plant parts, as revealed by Affymetrix microarray data (http://affymetrix.arabidopsis.info/). LTPs encoded by paralogs exhibiting diverse expression would putatively perform a housekeeping function. LTPs encoded by paralogs exhibiting restricted expression would play a role unique to the specific plant parts. Our laboratory has been elucidating the mechanism of action of the anther tapetum. In this project, we paid attention to LTP paralogs exhibiting restricted expression in inflorescence (Table I). An inflorescence includes metabolically active tapeta, microspores, and carpels as well as petals, sepals, and various other cell types. Therefore, LTP transcripts restricted to inflorescence are not necessarily present exclusively, largely, or at all in the tapetum.
Table I. Levels of Arabidopsis LTP transcripts reported to occur almost exclusively in inflorescences.
Affymetrix microarray data, shown in arbitrary units of absolute signal intensity, are from NASC (http://affymetric.arabidopsis.info/). The LTP type, Mr (without the putative N-terminal signal peptide), and pI data are from Boutrot et al. (2008). The flower bud stages were defined according to The Arabidopsis Information Resource. Low levels of the LTP transcripts in other plant parts are shown. Type III LTP transcripts are highlighted in boldface.
| Flower Buds |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Type | Mr | pI | Stage 9 | Stage 10–11 | Stage 12 | Stage 15 | Pollen | Sepal | Petal | Seed | Seedling | Leaf | Root |
| At3g51590 | I | 9.9 | 9.6 | 1,050 | 2,275 | 403 | 5 | 12 | 17 | 3 | 1 | 6 | 8 | 7 |
| At1g66850 | II | 8 | 7.1 | 1,285 | 3,882 | 3,110 | 2 | 27 | 54 | 35 | 1 | 10 | 16 | 13 |
| At5g07230 | III | 6.9 | 4.3 | 1,595 | 879 | 1 | 1 | 5 | 7 | 1 | 5 | 1 | 2 | 7 |
| At5g62080 | III | 6.6 | 4.1 | 1,909 | 413 | 26 | 4 | 16 | 18 | 2 | 1 | 4 | 1 | 3 |
| At5g52160 | III | 6.8 | 4.6 | 474 | 25 | 6 | 2 | 9 | 2 | 1 | 5 | 5 | 3 | 2 |
| At3g07450 | IX | 8 | 12.1 | 1,349 | 156 | 8 | 9 | 14 | 12 | 5 | 1 | 15 | 12 | 8 |
| At3g52130 | IX | 10.5 | 4.4 | 1,489 | 131 | 2 | 1 | 6 | 15 | 1 | 2 | 10 | 6 | 1 |
| At4g28395 | Y | 13.4 | 5.3 | 733 | 1,409 | 17 | 2 | 30 | 7 | 2 | 6 | 2 | 8 | 2 |
Type III LTPs, with Transcripts Restricted to Inflorescence, Have Unique Features among the Nine Types of Arabidopsis LTPs
We chose type III LTPs, whose transcripts are restricted to inflorescence, for further studies (Table I). Type III LTPs have an uncharacteristic low pI of approximately 4 in Arabidopsis (but approximately 7 in rice), fewer than 70 residues (the smallest size among all LTP types), and no potential glycophosphatidylinositol modification site (Boutrot et al., 2008). Their genes possess an intron at a DNA site not found in genes of other LTP types (Edstam et al., 2011). Levels of type III LTP transcripts peaked at an early stage of flower development, designated stages 7 to 9 by the Nottingham Arabidopsis Stock Centre (NASC). In a massively parallel signature sequencing analysis of a mixture of inflorescences of stages 1 to 12, the transcript levels of two of the three type III LTPs (At5g62080 and At5g07230; data not available for At52160) combined represented 0.13% of the total transcripts per million (http://mpss.udel.edu/at/mpss_index.php). These inflorescences contain several groups of metabolically active cells/tissues, including tapeta, microspores and pollen in anthers, various cell types in the carpel, petals, and sepals, and others. Because all of the type III transcripts are restricted to the tapetum in anthers at an early developmental stage (see below), their presence in the tapetum at the peak transcript stage could be several percent of the total mRNA. The high percentage of type III LTP transcripts estimated to be in the tapetum of Arabidopsis reflects that found experimentally in the tapetum of rice (5.7% in anthers [Huang et al., 2009]), from which anthers at specific developmental stages could be obtained for more precise analysis.
Type III LTP Genes Expressed Only in the Anther Tapetum
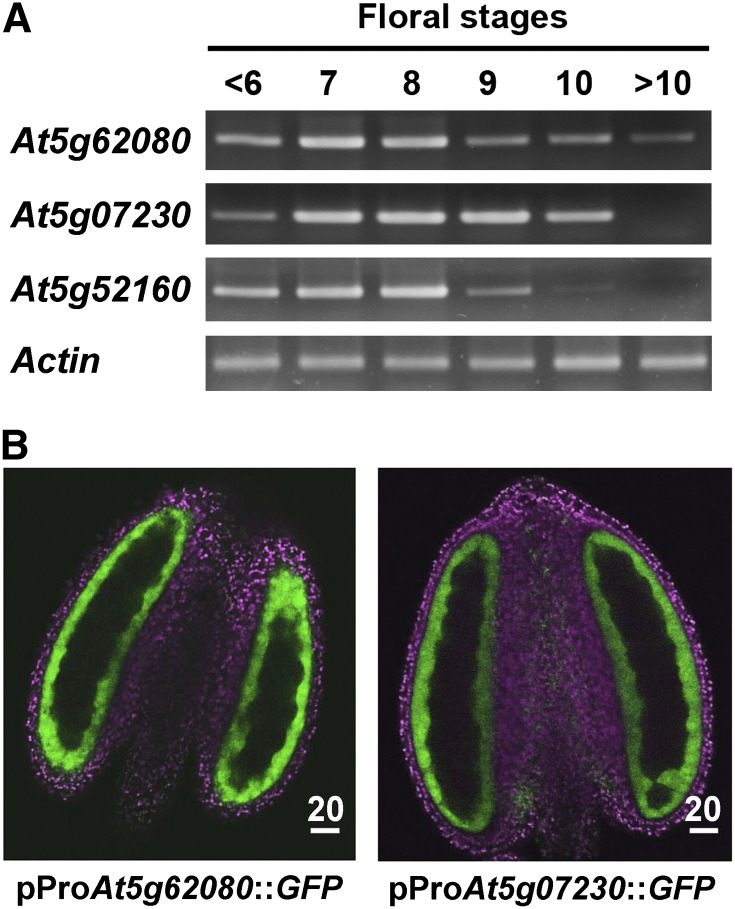
During flower development, transcript levels of At5g62080 and At5g52160 increased and peaked at stages 7 to 8 and then decreased in a similar pattern, whereas that of At5g07230 had a slightly delayed developmental timing (Fig. 1A). We focused on two type III LTPs in all subsequent studies: At5g62080, with transcripts having a developmental pattern similar to those of At5g52160 (see below), and At5g07230, with transcripts having a delayed developmental timing.
Figure 1.
Expression patterns of the three type III LTP genes in developing flowers of Arabidopsis. A, RT-PCR analysis of the transcript levels of At5g62080, At5g07230, and At5g52160 in flowers of progressive developmental stages. Actin was used as a normalization control. B, Subanther localization of At5g62080 and At5g07230 in anthers of stage 7 flowers of plants transformed with the promoter of the respective genes attached to GFP. At this developmental stage, transcripts of the two genes peaked (A). LTP-GFP (pseudocolored in green) was located in the tapetum. Chlorophyll autofluorescence (magenta) shows the other cells. The locule and its included tetrads did not show fluorescence and appeared dark. Scale bars are in μm.
We explored the subflower location of type III LTP transcripts. We attached the LTP gene promoter of approximately 2 kb to a GFP gene and transformed the DNA construct into wild-type plants. In transformed plants, the GFP signal appeared in developing anthers and peaked at stages 7 and 8 and was restricted to the tapetum of the whole flower (Fig. 1B). Other cells of the anthers, including microspores and the epidermis as well as other inflorescence cells and other cells of the whole plants, showed no GFP signal (data not shown).
Type III LTPs in the Tapetum Were Secreted to the Anther Locule via the ER-trans-Golgi Network
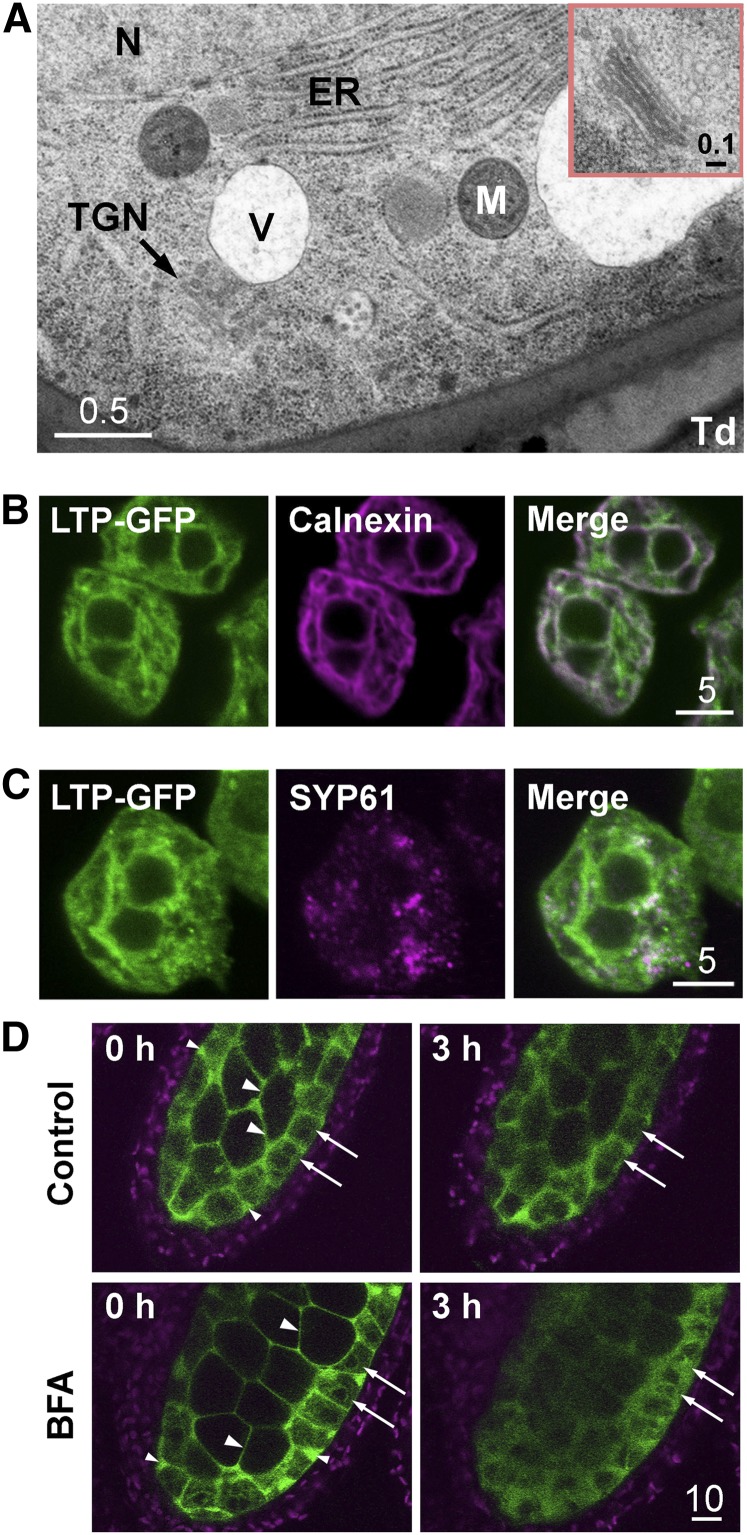
Although the tapetum in most higher plants is described as secretory, the mechanism of subcellular tapetum secretion has not been explored in much detail until recently with maize (Li et al., 2012). The mode of secretion of LTP from the tapetum has not been explored. In this study with Arabidopsis, transmission electron microscopy (TEM) revealed abundant rough ER, occasional Golgi bodies, and the trans-Golgi network (TGN; Fig. 2A). The presence of rough ER and Golgi bodies in the tapetum cells has been observed by others (Murgia et al., 1991; Platt et al., 1998; Li et al., 2012). In the Arabidopsis tapetum, the secretory organelles were distributed throughout the cell rather than concentrated in the cell portion near the locule, as in maize (Li et al., 2012).
Figure 2.
Localization of At5g62080-GFP and organelle markers in early stage 7 flowers in Arabidopsis. A, TEM of a portion of a tapetum cell, revealing ER, vacuole (V), nucleus (N), mitochondrion (M), TGN, and the adjacent locule with a small portion of a tetrad (Td). The inset shows an enlarged, well-defined Golgi body. Scale bars are in μm. B, CLSM of tapetum cells of plants transformed with At5g62080-GFP, revealing fluorescence of At5g62080-GFP (in green) overlapping substantially with the ER marker calnexin (detected with antibodies; magenta), with overlapped areas in white. The twin dark spheres represent two nuclei. C, CLSM of a tapetum cell of plants transformed with At5g62080-GFP, revealing fluorescence of At5g62080-GFP (in green) overlapping partly with the TGN marker SYP61 (detected with antibodies; magenta), with overlapped areas in white. The twin dark spheres represent two nuclei. D, Treatment of anthers of At5g62080-GFP-transformed plants with BFA, an inhibitor of the TGN secretory pathway. Anthers were incubated with or without 20 μg mL−1 BFA for 3 h, and fluorescence of At5g62080-GFP (in green) was observed. The same settings (laser power and detection gain) were used for direct comparison of the anthers before and after treatment. Slight losses of fluorescence and its sharpness had occurred after the 3-h treatment. Arrows indicate the tapetum cells; large arrowheads indicate the locule space between microspore mother cells, and small arrowheads indicate the apoplastic space between tapetum cells. Chlorophyll autofluorescence from cells of the outer wall is in magenta.
Intact tapetum cells from anthers of plants transformed with an LTP-GFP construct, which included the At5g62080 promoter and coding region tagged with GFP at the 3′ terminus, were examined by confocal laser scanning microscopy (CLSM) for GFP fluorescence (shown in green) and immunoprocessed calnexin (an ER marker, in magenta; Fig. 2B). LTP and calnexin were each present as a network distributed extensively throughout the cell. The two networks overlapped substantially (overlapping area represented by white color) but not completely. In our study, 70.6% ± 4.2% of the LTP-GFP signal overlapped (see “Materials and Methods”) with the calnexin signal, and 47.8% ± 1.9% of the calnexin signal overlapped with the LTP-GFP signal. The nonoverlapping regions of LTP could represent TGN and nearby vesicles or different ER subdomains. TGN (in magenta), revealed via immunodetection of its marker SYP61 (Drakakaki et al., 2012), had a more restricted distribution within the cell; it overlapped with only 13.1% ± 2.9% of the LTP-GFP network, and 44.6% ± 11.4% of the SYP61 signal overlapped with the LTP-GFP signal (Fig. 2C).
We further tested the idea that the export of LTP-GFP was mediated by the vesicular transport system involving ER-TGN. Stage 7 anthers from transgenic plants expressing LTP-GFP were treated with brefeldin A (BFA), an inhibitor of the vesicular transport system (Nebenführ et al., 2002). Disrupting the transport pathway with BFA would block the export of LTP-GFP out of the tapetum cells and result in the accumulation of LTP-GFP inside the cells. In stage 7 anthers (Fig. 2D), LTP-GFPs were being secreted out of the tapetum cell, and the LTP-GFP signals were mostly present in the locule space between microspore mother cells (large arrowheads at 0 h) as well as in the apoplastic space between the tapetum cells (small arrowheads at 0 h). These extracellular spaces of the locule are apparently interconnected, because LTP-GFP signals can be seen to freely flow within the spaces in a live recording (data not shown). The LTP signals were relatively fewer inside the tapetum cells (arrows at 0 h). After 3 h of incubation in 20 μg mL−1 BFA, the LTP-GFP signals increased by about 60% inside the tapetum cells (Fig. 2D; see “Materials and Methods”), signifying blocking of the ER-TGN transport system. In contrast, the LTP-GFP signals in tapetum cells of anthers after incubation without BFA increased only about 7% in comparison with cells at 0 h. In both BFA-treated and untreated anthers, the overall LTP-GFP signals decreased by about 15% after the incubation, and the locule space among microspore mother cells and the tapetum became less defined (Fig. 2D), presumably because the anthers had been removed from their natural growth condition. This loss of sharpness was more pronounced in BFA-treated anthers, likely due to the effect of BFA on both the tapetum and microspore mother cells. The overall observation in Figure 2 supports the idea that LTP-GFPs were exported via the ER-TGN pathway.
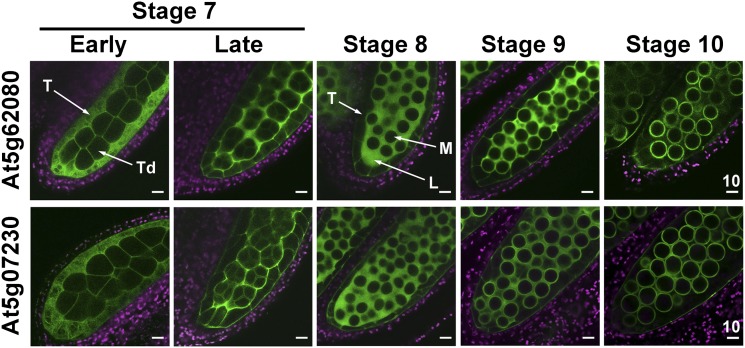
Type III LTPs Were Secreted to the Locule and Then Appeared on the Microspore Surface
We traced the locations of LTP-GFP in anthers during development in plants transformed with a type III LTP-GFP construct, which included the promoter and coding region. The LTP-GFP fluorescence signal first appeared in the tapetum at early stage 7, when microspores were in tetrads (Fig. 3). At late stage 7 and stage 8, LTP-GFP was abundant in the locule, with decreasing amounts in the still-intact tapetum. At stage 9, LTP-GFP appeared both in the locule and on the microspore surface. Eventually, at stage 10, most of the GFP was present on the microspore surface. Beginning at stage 10 and onward, the LTP-GFP fluorescence became weaker and gradually disappeared. Presumably the fluorescence properties of LTP-GFP vanished after its association with and modifications in the microspore wall.
Figure 3.
Changes in subanther locations of At5g62080-GFP and At5g07230-GFP during anther development in Arabidopsis. CLSM of the fluorescence of anthers at progressive developmental stages in plants transformed with the respective genes including the promoter and coding regions attached to GFP was performed. GFP fluorescence is shown in green, and chlorophyll autofluorescence is shown in magenta. The same settings (laser power and detection gain) were used for direct comparison of the developing anthers. At stage 10, the tapetum began undergoing programmed cell death. L, Locule; M, microspore; T, tapetum; Td, tetrad. Scale bars are in μm.
Type III LTPs Were Present in the Exine of Microspores at a Midstage of Anther Development
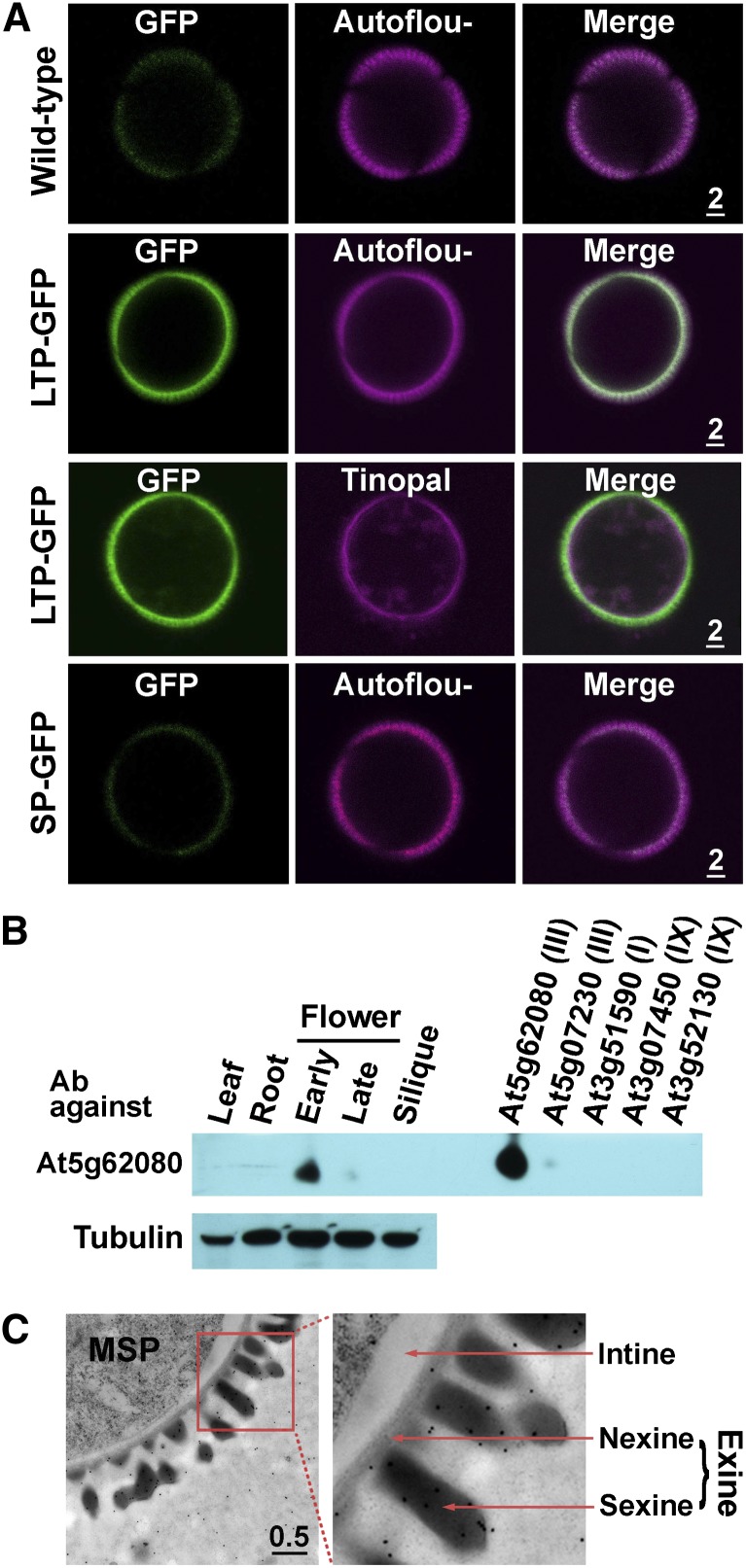
We detected the association of At5g62080 LTP-GFP with the microspore surface in stage 10 anthers of transformed plants (Fig. 3). For convenience and direct comparison, we removed the microspores from anthers and monitored the microspore surface fluorescence. In nontransformed plants, no GFP fluorescence other than faint background fluorescence was present on microspores (Fig. 4A). In transformed plants, LTP-GFP fluorescence was associated with the microspore surface, overlapping with the autofluorescence of the pollen wall. After the microspores had been stained with tinopal for the cellulosic cell wall (Regan and Moffatt, 1990), LTP-GFP fluorescence was present together with (as a white ring in Fig. 4A) and also slightly exterior to the tinopal stain. We tested whether the binding of LTP-GFP to the microspore surface required the LTP protein per se. In transformed plants with GFP attached only to the LTP signal peptide (i.e. without the mature LTP peptide), signal peptide-GFP was not found on microspores. Overall, LTP-GFP was present in the exine of microspores, and the domain required for association was the LTP per se. A similar finding, but with fewer details, of At5g07230-GFP fluorescence overlapping with pollen wall autofluorescence is shown in Supplemental Figure S1.
Figure 4.
Localization of At5g62080-GFP on microspores at a midstage (stage 10) of anther development in Arabidopsis. A, Presence of At5g62080-LTP-GFP (encoded by GFP attached to full-length LTP) and absence of At5g62080-SP-GFP (GFP attached to the LTP region encoding only the N-terminal signal peptide) on microspores of plants transformed with the respective genes. In the third row, microspores were stained with tinopal for the cellulosic cell wall. GFP fluorescence is shown in green, and exine autofluorescence (or tinopal stain) is shown in magenta. Nontransformed plants (wild-type) were used as a control. Scale bars are in μm. B, Localization of At5g62080 in flowers and other Arabidopsis organs with immunoblotting and specificity of anti-At5g62080 antibodies (Ab). At left, total proteins of equal amounts from the indicated organs were subjected to immunoblotting after SDS-PAGE with antibodies against a synthetic fragment of a unique sequence of At5g62080. Immunodetection of tubulin was used as a control. Young flower buds were stages 1 to 10; late flower buds were stages 11 to 13. At right, His-tagged recombinant protein (1 ng each) of the indicated LTPs, whose transcripts were present mostly in flowers among all organs (Table I), were subjected to the same immunodetection. The antibodies were highly specific to At5g62080 and barely recognized the closest-related At5g07230 (Supplemental Fig. S2). The antibodies recognized only one band on the SDS-PAGE blot; only this portion of the immunoblot is shown. The molecular masses of the endogenous and recombinant At5g62080 were 6.6 and 7.9 kD, respectively. C, Immuno-TEM of the outermost portion of microspores (MSP) probed with antibodies against At5g62080. The right panel is an enlarged image of the left panel. Immunogold particles were present in the locule and exine (both nexine and sexine). Quantification of the gold particles is shown in Supplemental Table S1. Scale bar is in μm.
The location of native At5g62080 LTP without the GFP tag in nontransformed plants was explored with immuno-TEM. We generated rabbit polyclonal antibodies against a synthetic peptide unique to the C-terminal region of At5g62080. Immunoblotting after SDS-PAGE revealed that the antibodies were specific for At5g62080, with a very slight reactivity toward its closest paralog protein, At5g07230 (Supplemental Fig. S2), and that At5g62080 occurred at an early stage of flower development (Fig. 4B). For immuno-TEM, anthers were treated with a freeze-fixation procedure (see “Materials and Methods”) to preserve proteins in the aqueous locule. In stage 10 anthers, with the exine just formed, immunogold particles were present largely in the nexine and sexine of exine and some in the locule (Fig. 4C). Electron-dense particles inside the microspore protoplasts were mostly larger and irregularly shaped and were not immunogold particles. At an earlier stage (stage 8) immediately before the exine appeared, most of the immunogold particles were present in the locule (Supplemental Fig. S3 shows a sequence of images of stage 8–10 anthers). Tabulation with statistical analysis of immunogold particles (Mayhew et al., 2002) in the anther locule and exine (Supplemental Table S1) indicates that there was a shift in immunogold particles from the locule to the exine from stage 8 to stage 10; this shift is consistent with the LTP-GFP fluorescence shown in Figure 3. The immunogold particles in the tapetum cells and the intracellular microspores throughout development were considered to have arisen from random labeling based on statistical analysis, reflecting the well-known high background in the relatively nonspecific immunogold detection compared with the sensitive and highly specific GFP fluorescence detection.
Structurally Modified Type III, Two Inflorescence-Specific Type IX, and One Leaf-Abundant Type I LTPs Did Not Target Microspores in Transformed Plants
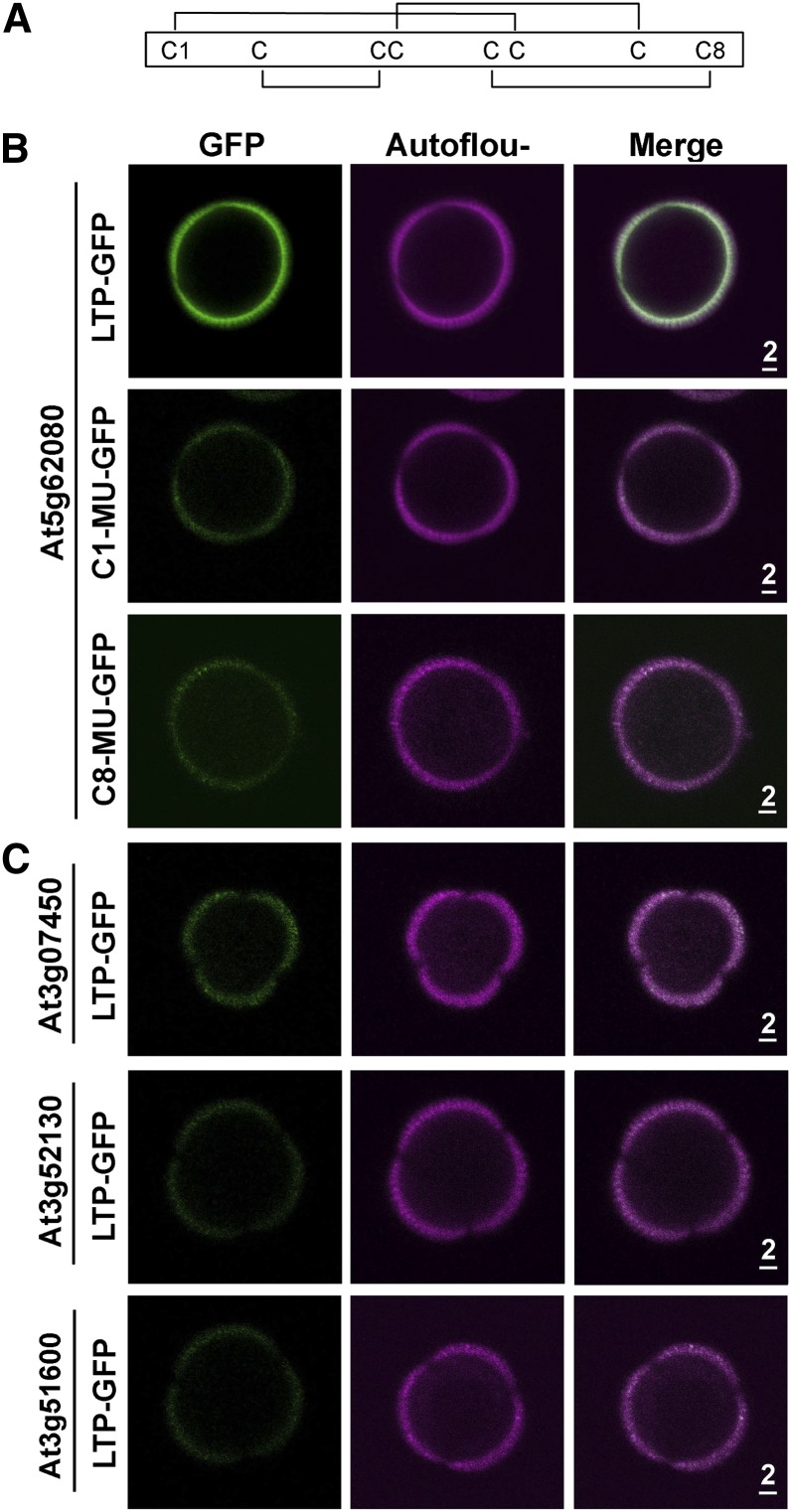
We tested whether GFP-tagged type III LTPs with modified structures would target to microspores in transformed plants. An LTP molecule has eight conserved Cys residues, which form four defined disulfide bridges for a three-dimensional structure. The four bridges in type III LTPs are shown in Figure 5A. We altered the first Cys residue (to produce C1-MU-GFP) or the eighth Cys residue (C8-MU-GFP) to an Ala residue by modifying the codon in At5g62080 and then attached GFP to the DNA construct for plant transformation. Neither of the two modified At5g62080 LTP-GFP proteins was present on microspores of transformed plants (Fig. 5B).
Figure 5.
Nontargeting of structurally modified At5g62080-GFP and GFP-tagged native non-type III LTPs to exine of stage 10 microspores of transformed Arabidopsis plants. A, Schematic diagram of the disulfide bonds between the four pairs of Cys residues in At5g62080. B, Absence of At5g62080-GFP with its first (C1-MU-GFP) or eighth (C8-MU-GFP) Cys residue modified to Ala on microspores. Nonmodified LTP-GFP was used as a positive control. GFP fluorescence is shown in green, and exine autofluorescence is shown in magenta. Scale bars are in μm. C, Absence of type IX LTP-GFP (from At3g07450 and At3g52130; expressed mostly in inflorescences in Affymetrix microarray data) and type I LTP-GFP (from At3g51600; expressed abundantly in leaves) on microspores. In transformed plants, the three LTP-GFPs were driven by the At5g62080 promoter. Scale bars are in μm.
We also tested whether GFP-tagged non-type III LTPs would target microspores in transformed plants. Transcripts of At3g07450 and At3g52130 encoding type IX LTPs were present mostly in inflorescence and appeared slightly before type III LTP transcripts during flower development (Table I), and the transcripts of At3g51600 encoding type I were abundant in leaves (NASC microarray data). In plants transformed with the LTP-GFP driven by the At5g62080 promoter, a trace of At3g07450 GFP was present, whereas the other two LTP-GFPs were absent, on microspores (Fig. 5C).
Type III LTPs Did Not Target Microspores of Mutant Plants Defective in the Production of Exine Precursors
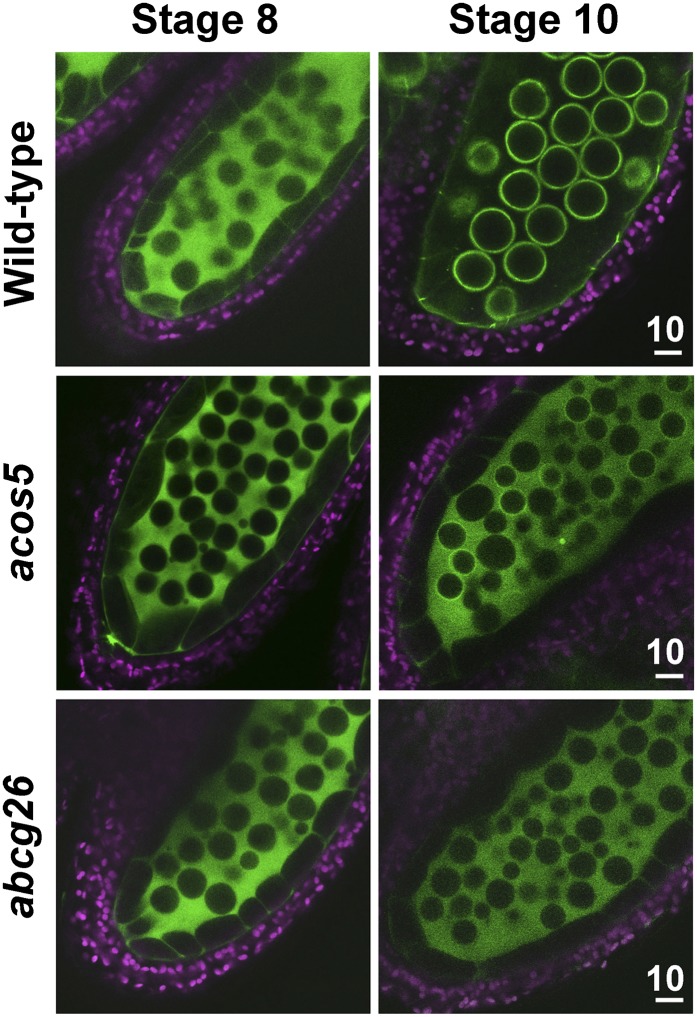
Arabidopsis knockout mutants defective in the synthesis or transport of exine precursors produce pollen with highly degenerated exine and lacking the ability to germinate. The mutant acos5 is defective in producing fatty acyl-CoA synthase in the tapetum, whose enzymic product is likely a precursor of exine (de Azevedo Souza et al., 2009). The mutant abcg26 is defective in producing the ABCG transporter in the tapetum for exine precursor export (Quilichini et al., 2010). We tested whether At5g62080 LTP-GFP would target microspores in transformed exine-defective mutants. The proteins did not target microspores in the two transformed mutants (Fig. 6). In the control wild type, LTP-GFP was present in the locule at stage 8 and on microspores at stage 10. In the two mutants, LTP-GFP was present in the locule at both stages, which indicates the successful secretion of the protein to the locule from the tapetum but the failure of the protein to bind to the microspore surface.
Figure 6.
Nontargeting of At5g62080-GFP to the microspore surface of Arabidopsis exine-defective mutants. At5g62080-GFP was transformed to the exine-defective mutants acos5 and abcg26. CLSM of the fluorescence of anthers of wild-type and transformed plants was performed. Green represents GFP fluorescence, and magenta denotes chlorophyll autofluorescence of the outer anther cells. Unlike those in the wild type, microspores in the two mutants stopped enlarging at stage 8 (de Azevedo Souza et al., 2009; Quilichini et al., 2010). Scale bars are in μm.
RNA Interference Knockdown of Two Type III LTPs Partially Impaired the Intine underneath the Exine as Well as the Viability of Pollen on Dehydration
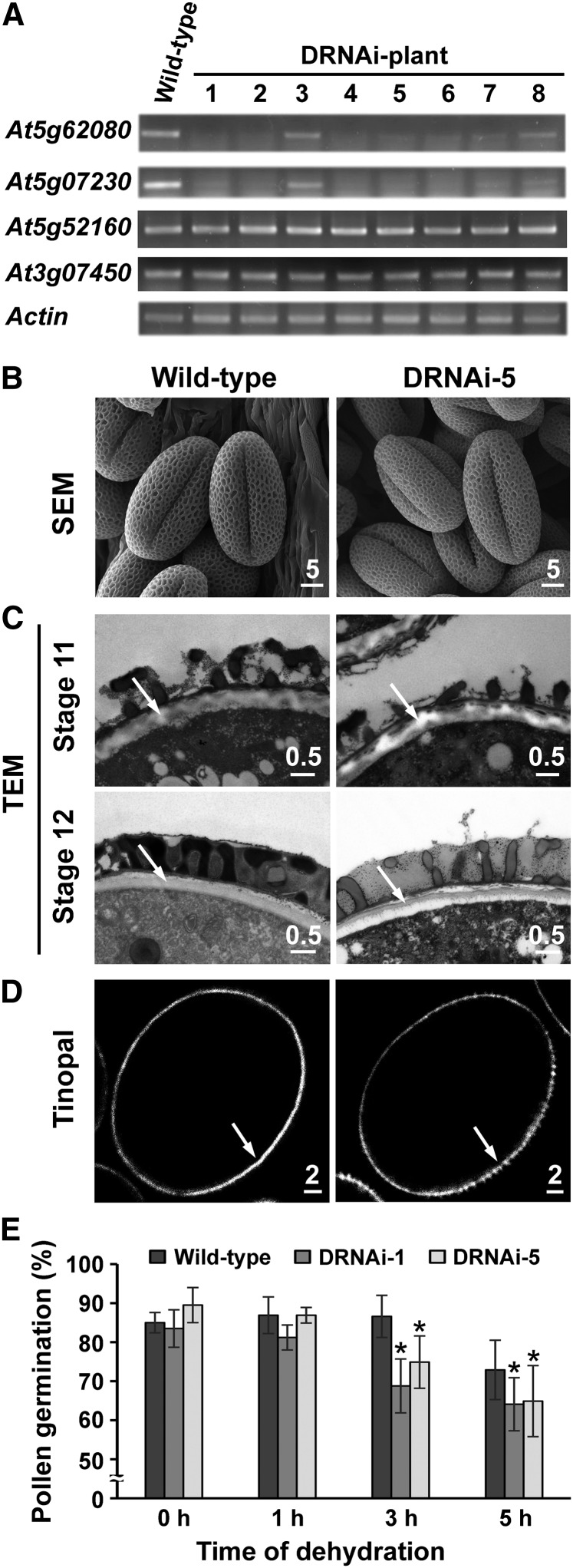
Arabidopsis transfer DNA knockout mutants of At5g62080 and At5g07230 are unavailable. Earlier, antisense knockdown of the transcripts of A9, an ortholog of At5g02730, in B. napus led to no observable mutation phenotype (Turgut et al., 1994). In our study, we generated RNA interference (RNAi) knockdown mutants of At5g62080 or At5g07230 and observed with TEM no apparent changes in the microspore surface (Supplemental Fig. S4).
We further generated an RNAi vector driven by the At5g62080 promoter, which could create double knockdown mutants of At5g62080 and At5g07230 (Supplemental Fig. S5). Reverse transcription (RT)-PCR showed that in the studied transgenic lines, the transcript levels of At5g62080 and At5g07230 were specifically and substantially reduced (Fig. 7A). Nonetheless, these lines all produced fertile pollen. We chose two lines, DRNAi-5 and DRNAi-1, which had the fewest transcripts of At5g62080 and At5g07230 (Fig. 7A), for further study. Scanning electron microscopy of mature pollen revealed no apparent difference in the surface structure between the mutants and the wild type (Fig. 7B). TEM also revealed no appreciable and consistent difference in the exine and coat materials between the mutants and the wild type (Fig. 7C). However, the intine of microspores at stage 11 was consistently less electron dense in DRNAi-5 than in the wild type and appeared to have separated from the exine and the microspore plasma membrane (Fig. 7C). We consider it unlikely that the observed morphological defect of the intine in the mutant compared with the wild type was a TEM fixation artifact, because the two types of anthers were fixed and processed for TEM in the same experiment, and six different anthers of each type showed similar intine disparity. The disparity could be related to changes unobserved by TEM in the exine of the mutant, which would affect (1) the penetration of fixatives from the microspore surface to the underneath intine or (2) physically the proper formation of the underlying intine by microspores. Regardless, the disparity in the intine of the mutant and the wild type was also manifested with intine-specific tinopal staining: the stain was fragmented in the mutant but continuous in the wild type (Fig. 7D). Finally, we tested whether the mutants, as a consequence of unobserved deficiency in the exine trickling down to the observed deficiency in the intine, would produce pollen that was more susceptible to dehydration. Pollen was placed at 30% relative humidity and 37°C for 1 to 5 h and then allowed to germinate on an agar medium. Pollen of the mutants germinated as well as that of the wild type before dehydration treatment but became less viable after 3 to 5 h of dehydration (Fig. 7E). The lack of a drastic pollen deficiency in the mutants could reflect that At5g62080 and At5g07230 perform a redundant function, that the knockdowns were insufficient, and that the third type III LTP (At5g52160; Table I) alone would be sufficient to carry out the function.
Figure 7.
Effect of double RNAi knockdown of At5g62080 and At5g07230 on morphologic features of the pollen wall in Arabidopsis. A, RT-PCR of individual type III RNAi transgenic plants. At5g52160 (type III, much less active than the other type III genes) and At3g07450 (type IX) were both expressed mostly in flowers (Table I; Fig. 1). Actin was used as a control, whose expression was not affected by RNAi. B, Scanning electron microscopy (SEM) of pollen of the wild type and an RNAi mutant (RNAi plant 5 in A, termed DRNAi-5). Scale bars are in μm. C, TEM of stage 11 and stage 12 microspores of the wild type and DRNAi-5. Arrows indicate intine. Scale bars are in μm. D, Tinopal-stained stage 12 microspores of the wild type and DRNAi-5. Arrows indicate intine. Scale bars are in μm. E, Dehydration tolerance of pollen from the wild type and RNAi mutants. Pollens were placed at 30% relative humidity and 37°C for 1 to 5 h and allowed to germinate on solid pollen germination medium. Significant differences were calculated from the germination percentage of each mutant compared with the wild type (Student’s t test, *P < 0.01).
DISCUSSION
For two decades, transcripts of specific LTPs have been found to be present exclusively in anthers and/or tapeta of flowers. Recently available quantitative genomic data for rice (Huang et al., 2009) and Arabidopsis (this report) show that these LTP transcripts are abundant, representing as high as or higher than 10% of the total mRNA in the tapetum at a developmental stage of peak LTP occurrence. Yet, little is known about the function of tapetum LTPs other than the logical speculation based on their nascent polypeptide possessing an N-terminal signal peptide for secretion and their lipid-binding properties. Accordingly, tapetum LTPs are expected to be secreted to the locule, carrying with them exine lipid precursors for assembly on microspores (Ariizumi and Toriyama, 2011). A recent study has shown that a rice non-type III LTP is present in the locule and the anther epidermis (Zhang et al., 2010). Our findings represent a scientific advancement in that type III LTPs are secreted from the tapetum to become microspore exine constituents. Exine is an abundant morphological feature of the pollen wall, and this abundance requires plentiful LTPs as a component. Were LTPs to solely carry out a catalytic function of transporting lipids, the need for the protein amount would be minimal, especially if the LTPs were to be recycled from the locule back to the tapetum. The recycling would be logical; otherwise, the cell would have to produce an LTP molecule of 8 kD to transport a lipid molecule of 0.8 kD just once. Alternatively, as suggested by our findings, an LTP molecule would move either alone or together with a lipid molecule to the microspore surface, where the molecules would associate with other ingredients to form exine. The mode of association of the LTPs and the exine materials remains to be elucidated. The fact that LTPs are very stable (Lindorff-Larsen and Winther, 2001; Wijesinha-Bettoni et al., 2010), presumably even after their association with and modifications in exine, could reflect the bicentury-old observation that pollen exine is highly resistant to degradation.
The in vivo association of LTP with the forming exine on microspores is highly specific, requiring the native structure of type III LTP and possibly also the bound exine precursor. LTPs of other types, even the tapetum-specific type IX, cannot form the association. A non-type III LTP (rice Osc6) was detected in the locule but was not found on microspores (Zhang et al., 2010). We tried but were unable to obtain unambiguous results on the exine precursors that could bind to type III LTPs. In our studies, we used LTP-GFP for convenient and direct comparisons of the LTP locations in the tapetum and the locule as well as on microspores under different experimental settings. The GFP fluorescence of the LTP-GFP in the locule or association with the forming exine was easily detected, with no apparent loss of fluorescence intensity (Fig. 3); this detection was made in anthers at stages 7 to 9 during the formation of exine. After stage 9, the GFP signal gradually disappeared, concomitant with the completion of exine formation. This disappearance is likely due to the unique and to-be-determined mode of molecular association of LTP-GFP with other components in mature exine, such that the fluorescent property of the GFP was lost. We used immuno-TEM and detected the native LTP without the GFP tag in the anthers during development and the apparent shift of LTP from the locule to the exine from stages 8 to 10 (Fig. 4C; Supplemental Fig. S3; Supplemental Table S1).
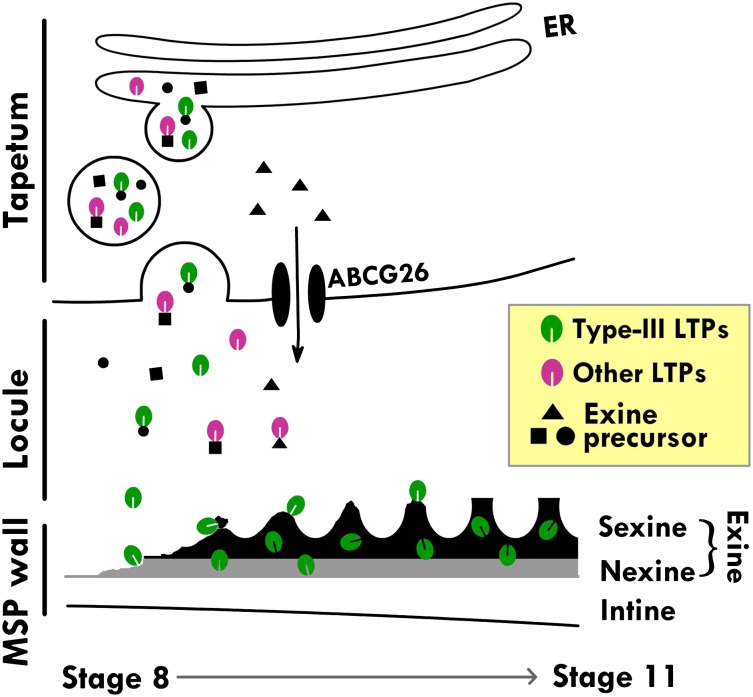
Pollen exine is a polymer collectively termed sporopollenin, which consists of unbranched and branched aliphatic and aromatic monomers that are hydroxylated or oxygenated (Ariizumi and Toriyama, 2011; Kim and Douglas, 2013; Lallemand et al., 2013). These monomers are synthesized by ER-associated or cytosolic enzymes in the tapetum. It has been assumed that the monomers before or after preliminary polymerization are secreted to the locule via the secretory pathway or the plasma membrane ABC transporter and reach the microspore surface for the final assembly into exine. With this information in mind and with our findings here, we propose a model of tapetum action in the synthesis of exine and its assembly on microspores (Fig. 8). Type III LTPs bound or nonbound to exine precursors in the ER lumen are secreted to the locule via the ER-TGN system. In the locule, type III LTPs either alone or associated with exine precursors move to the microspore surface, where they are assembled with exine precursors into exine. Other tapetum-specific but non-type III LTPs likely move via similar paths but, per se, will not be a component of mature exine. Other exine precursors are synthesized in the tapetum cytosol and are transported to the locule via the ABC transporter on the plasma membrane (Quilichini et al., 2010). Thus, diverse exine precursors produced in the tapetum are transported to the microspore surface via different routes. Simultaneously, cellulosic precursors are produced in the microspore interior and transported to the microspore surface for the assembly of the cellulosic intine (Jiang et al., 2013).
Figure 8.
Model of the action of type III (At5g62080) and non-type III LTPs in exine formation. Exine begins to appear at stage 8, and the formation is complete at stage 11. In tapetum cells, secretory vesicles derived from the ER-TGN system contain LTPs bound or unbound to ER-produced exine precursors. LTPs and exine precursors are released into the locule, where unbound LTPs may or may not bind to free exine precursors. LTPs and exine precursors, unbound or bound, move to the microspore (MSP) surface, where exine is assembled. Type III LTPs, but not other LTPs, remain in exine as permanent constituents. Other exine precursors produced by cytosolic enzymes in tapetum cells are transported to the locule via plasma membrane ABCG26 and, with or without binding to LTPs in the locule, move to the exine for assembly.
MATERIALS AND METHODS
Plant Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) were germinated on Murashige and Skoog medium on plates after sterilization and vernalization. Transfer DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center. For GFP and RNAi transgenic plants, the Murashige and Skoog medium contained 30 μg mL−1 hygromycin and 50 μg mL−1 kanamycin, respectively. After germination for 6 d, the seedlings were transplanted to soil and grown in a growth chamber at 22°C in a daily cycle of 16 h of light/8 h of dark.
Mining of Microarray Data
The microarray data of Arabidopsis were downloaded from the NASC International Affymetric Service (http://affymetrix.arabidopsis.info/) and analyzed with the use of Perl programming language. We selected LTPs with expression 10-fold higher in inflorescences than other plant parts (Table I).
RNA Extraction and RT-PCR Analysis
In the collection of developing flowers, the flower buds were harvested 10 to 18 d after the beginning of bolting. The flower stages were judged by size selection (0.6, 0.7, 0.8, and 1.0 mm for stages 7, 8, 9, and 10, respectively) with a dissecting microscope. Tissues were collected and ground to a fine powder in liquid nitrogen with the use of a mortar. Total RNA was isolated with the use of the Illustra RNAspin mini kit (GE Healthcare). First-strand cDNA was synthesized from 2.5 μg of total RNA with the use of Thermoscript III reverse transcriptase (Invitrogen) for RT-PCR. Gene-specific primer pairs were designed according to the sequences of specific genes (Supplemental Table S2). Amplified DNA fragments were analyzed on 1% agarose gels.
Generation of His-Tagged LTP
A cDNA fragment encoding LTP without an N-terminal signal peptide was conjugated with 6-His-tag DNA by three-step PCR. First, LTP fragments were generated from cDNA by the use of a gene-specific reverse primer and a partial His-tag DNA sequence as a forward primer. DNA encoding 6-His-tag and MscI sites was added to the 5′ terminus after the second and third steps of PCR by the use of a BamHI site-containing reverse primer paired with the primers pET32-6His-F and pET32-6His-MscI-F, respectively (Supplemental Table S2). After BamHI digestion, the PCR product was cloned into the MscI and BamHI sites of pET32a and then transformed into Escherichia coli strain Origami. Transformed E. coli was induced by 1 mm isopropyl-β-d-1-thiogalactopyranoside treatment, and the Trx-6His-LTP was purified with a nickel-nitrilotriacetic acid agarose column. The Trx-6His-LTP was subjected to Factor Xa protease treatment for the release of thioredoxin.
Construction of Vectors for the Transformation of Arabidopsis
For constructing GFP expression vectors, the promoter region (approximately 2 kb) was amplified from genomic DNA with gene-specific primers for At5g62080 (primers At5g62060pro-SalI and At5g62080pro-NcoI) and At5g07230 (At5g07230pro-SacI and At5g07230pro-NcoI). The sequences of all primers are shown in Supplemental Table S2. After enzyme digestion, the PCR fragments were ligated into pCAMBIA1302, and the construct was digested with the same enzyme to generate pAt5g62080pro::GFP and pAt5g07230pro::GFP. The cDNA encoding regions of At5g62080, At5g07230, At3g07450, At3g52130, and At3g51600 as well as the Cys mutant fragment of At5g62080 were amplified by RT-PCR with primer pairs containing specific restriction sites. The cDNA fragments were cloned into the NcoI/SpeI sites of pAt5g62080pro::GFP or pAt5g07230pro::GFP to generate pAt5g62080pro::LTP-GFP or pAt5g07230pro::LTP-GFP. For At5g62080, At3g07450, and At3g52130, the PCR fragments were cloned into the NcoI/SpeI sites by sticky-end PCR because of the presence of NcoI sites in the coding region. All constructs were confirmed by sequencing.
For the RNAi expression vector, the 35S promoter of pHELLSGATE (Wesley et al., 2001) was replaced with the At5g62080 promoter. First, the 2-kb promoter DNA of At5g62080 and a negative selectable marker (ccdB) were amplified by PCR with primers containing SacI/HpaI and HpaI/KpnI sites (Supplemental Table S2), respectively. The two PCR fragments were digested with SacI and KpnI, respectively, fused by T4 ligase, and cloned into KpnI/SacI-digested pHELLSGATE to replace the 35S::ccdB cassette. The product was denoted pAt5g62080pro-HELLSGATE.
cDNA fragments encoded by At5g62080 and At5g07230 were ligated and conjugated with the attB recombination site by PCR. First, PCR was used to generate a megaprimer from At5g07230 cDNA with the primers 080-230-linker-F and At5g07230-attB2-R. A second-round PCR was performed to create the At5g62080-At5g07230 fusion, with the megaprimer used as the reverse primer and the forward primer At5g62080-attB1-F. After creation of the At5g62080-At5g07230 fusion, PCR was used to add the aatB site with primers attB1-F and attB2-R. The attB1-At5g62080-At5g07230-attB2 fragment was sequenced for confirmation and cloned into pAt5g62080pro-HELLSGATE by the use of BP clonase (Invitrogen) to produce the pDouble-RNAi (Supplemental Fig. S5).
All constructs were transformed into Arabidopsis by the floral dip method (Clough and Bent, 1998).
Antibody Preparation, SDS-PAGE, and Immunoblotting
The putative amino acid sequence was analyzed by the use of IEDB antibody epitope prediction software (http://tools.immuneepitope.org/tools/bcell/iedb_input). The peptide RAATSLPSLCNLPPVDCGINA was selected. The peptide was chemically synthesized (Minotope) and conjugated to ovalbumin. The conjugate was injected into rabbits, and antibodies were purified from serum. The antibodies were used for immuno-TEM and immunoblotting after SDS-PAGE. Antibodies against tubulin were from Sigma-Aldrich. Anti-calnexin against the castor bean (Ricinus communis) protein was a gift from Dr. Sean Coughlan, and anti-SYP61 was a gift from Prof. L.W. Jiang.
Proteins in samples were separated by 15% (w/v) SDS-PAGE (Huang et al., 2009) and stained with Coomassie blue or subjected to immunoblotting with rabbit antibodies against At5g62080 (Huang et al., 2009).
CLSM and Immunofluorescence Localization
All images were captured on a Zeiss LSM 510Meta confocal microscope with a 40× or 63× (numerical aperture 1.2) water-immersion objective and Zeiss LSM 510 version 3.2 software. GFP and chlorophyll in samples were excited with the 488-nm argon laser line, and emission was detected with 500- to 530-nm and 650- to 710-nm band-pass filters, respectively. To distinguish GFP from the autofluorescence of the microspore exine, microspores were excited with the 488-nm argon laser line and captured with the lambda mode of Zeiss LSM 510 Meta. Images for GFP were generated by extracting channels between 505 and 526 nm, and images for exine autofluorescence were generated by extracting channels between 526 and 644 nm.
For intine staining, microspores were incubated in 10 μg mL−1 Tinapol LPW (also known as Fluorescent Brightener 28; Sigma-Aldrich) for 5 min. Tinapol stain was excited with a Coherent Chameleon two-photon laser at 740 nm, and emission was detected at 435 to 485 nm.
For immunofluorescence, anthers were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) containing 0.15 m Suc for 1 to 2 h at 24°C. Fixed anthers were cut horizontally with a razor into pieces on poly-Lys-coated slides. The gentle pressure from the cutting action pushed the loosely associated microspore mother cells, young microspores, and tapetum cells away from the anther wall cells. Then, the samples were allowed to stand for 10 min for the cells to attach to the poly-Lys-coated surface. The samples were mounted under a coverslip, which was sealed by Vaseline on two sides to generate a microperfusion chamber. The subsequent reagents were exchanged via perfusion. The samples were treated with 0.1% Triton X-100 in PBS to facilitate permeability and blocked in 0.15% normal donkey serum in PBS. The cells were then incubated sequentially with anti-calnexin or anti-SYP61, both at 1:100 dilution for 2 h at 37°C, and cyanine 3-conjugated anti-rabbit IgG (1:400; Jackson ImmunoResearch) for 45 min at 37°C, with three washes in PBS between incubations. Cyanine 3 was excited with the 543-nm helium-neon laser line, and emission was detected at 560 to 615 nm.
Colocalization of GFP and immunoreacted ER or TGN markers, as well as quantification of GFP intensity in tapetum cells and the locule of BFA-treated anthers, were analyzed with the use of the image-analysis software MetaMorph (version 7.7.4.0). In colocalization analysis, the signals corresponding to the two separate channels of each CLSM image were highlighted in images derived from the respective channel with the “Threshold” function. The percentage of overlapping between the highlighted areas of the two images corresponding to the respective channels was calculated using the “Measure Colocalization” function. The presented percentage of overlap was the average of measurements from at least five separate cells. In quantification of the GFP fluorescence intensity in tapetum cells and the locule of anthers before and after BFA (or control) treatment, fluorescence intensities in anthers were captured with the same setting (laser power and detection gain). The intracellular region of individual tapetum cells was selected manually with the “Region” tool, and the average intensity of each selected region was generated using the “Region Measurement” command. The average intensity of each region, after subtraction from background intensity, was normalized to the average intensity of the whole anther lobe to correct for the overall intensity change. Ten tapetum cells from two separate anther lobes were measured.
Electron Microscopy
Scanning electron microscopy involved the use of a Quanta 200 microscope equipped with a cryo system (Quorum PP2000TR; FEI). Samples were freshly prepared and loaded on a stub. After treatment with liquid nitrogen, samples were transferred to a preparation chamber at −160°C and then etched at −85°C for 10 to 15 min. After coating with gold at −130°C, samples were transferred to the cryo stage in the scanning electron microscope chamber and examined at −160°C and 20 kV.
For TEM, anthers were fixed via a high-pressure freezing or chemical fixation procedure. For the former, anthers were fixed in a high-pressure freezer (Leica EM PACT2) and then subjected to freeze substitution in ethanol (containing 0.2% glutaraldehyde and 0.1% uranyl acetate) with the Leica Automatic Freeze-Substitution System and embedded in LR Gold resin (Structural Probe). For chemical fixation, anthers were fixed with 2.5% glutaraldehyde, 4% paraformaldehyde, and 0.1 m potassium phosphate (pH 7.0) at 4°C for 24 h. The materials were washed with 0.1 m potassium phosphate buffer (pH 7.0) for 10 min twice and then treated with 1% OsO4 in 0.1 m potassium phosphate (pH 7.0) at 24°C for 4 h. The fixed materials were rinsed with 0.1 m potassium phosphate buffer (pH 7.0), dehydrated through an acetone series, and embedded in Spurr resin. Ultrathin sections (70–90 nm) were stained with uranyl acetate and lead citrate and examined with the use of a Philips CM 100 transmission electron microscope at 80 kV.
For immunogold labeling, microtome sections were incubated with 3% bovine serum albumin for 1 h and then with primary antibodies at a preoptimized concentration at 25°C for 2 h. After six washes with Tris-buffered saline containing 1% Tween 20 (TBST) and once with TBST containing 1% bovine serum albumin, sections were incubated with colloidal gold-conjugated secondary antibodies at 25°C for 2 h. Sections were washed six times with TBST and three times with deionized water, stained with 4% uranyl acetate/0.4% lead citrate, and examined with the use of a Philips CM 100 transmission electron microscope at 80 kV.
Pollen Germination
Mature flowers of wild-type and RNAi plants were placed in a 37°C incubator with 30% relative humidity for 1 to 5 h. Pollen grains from flowers were placed on solid pollen germination medium [18% Suc, 0.01% boric acid, 1 mm CaCl2, 1 mm Ca(NO3)2, 1 mm MgSO4, and 0.5% Noble agar] for 5 h at 28°C. Germination of pollen grains of at least 300 per sample was recorded by light microscopy.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Localization of At5g07230-GFP on microspores at a midstage (stage 10) of anther development in transformed plants.
Supplemental Figure S2. A phylogenetic tree of floral-specific LTPs from Arabidopsis, rice, and other studied species.
Supplemental Figure S3. Immuno-TEM localization of native At5g62080 in the anther locule.
Supplemental Figure S4. No apparent phenotype of the surface of stage 12 microspores in At5g62080 and At5g07230 single RNAi mutants.
Supplemental Figure S5. Schematic diagram of the RNAi construct for producing double knockdown of At5g62080 and At5g07230.
Supplemental Table S1. Quantification of immunogold labeling by anti-At5g62080 in anthers at indicated developmental stages.
Supplemental Table S2. Primers used for DNA constructs and RT-PCR.
Acknowledgments
We greatly appreciate the assistance of Dr. Wann-Neng Jane (Academia Sinica) for electron microscopy, Ms. Mei-Jane Fang (Academia Sinica) for CLSM, and Mr. An Yan (University of California at Riverside) for pollen germination analysis and the gifts from Dr. Sean Coughlan (DuPont; anti-calnexin antibodies) and Dr. L.W. Jiang (Chinese University of Hong Kong; anti-SYP61 antibodies).
Glossary
- LTP
lipid transfer protein
- ER
endoplasmic reticulum
- NASC
Nottingham Arabidopsis Stock Centre
- TEM
transmission electron microscopy
- TGN
trans-Golgi network
- CLSM
confocal laser scanning microscopy
- BFA
brefeldin A
- RNAi
RNA interference
- RT
reverse transcription
- PBS
phosphate-buffered saline
- TBST
Tris-buffered saline containing 1% Tween 20
References
- Aguirre PJ, Smith AG. (1993) Molecular characterization of a gene encoding a cysteine-rich protein preferentially expressed in anthers of Lycopersicon esculentum. Plant Mol Biol 23: 477–487 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K. (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Boutrot F, Chantret N, Gautier MF. (2008) Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RD, Zimmermann E, Xu SX, Liu GS, Smith AG. (2006) Characterization of an anther- and tapetum-specific gene and its highly specific promoter isolated from tomato. Plant Cell Rep 25: 231–240 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crossley SJ, Greenland AJ, Dickinson HG. (1995) The characterisation of tapetum-specific cDNAs isolated from a Lilium henryi L. meiocyte subtractive cDNA library. Planta 196: 523–529 [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ. (2009) A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, van de Ven W, Pan S, Miao Y, Wang J, Keinath NF, Weatherly B, Jiang L, Schumacher K, Hicks G, et al. (2012) Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res 22: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstam MM, Viitanen L, Salminen TA, Edqvist J. (2011) Evolutionary history of the non-specific lipid transfer proteins. Mol Plant 4: 947–964 [DOI] [PubMed] [Google Scholar]
- Hihara Y, Hara C, Uchimiya H. (1996) Isolation and characterization of two cDNA clones for mRNAs that are abundantly expressed in immature anthers of rice (Oryza sativa L.). Plant Mol Biol 30: 1181–1193 [DOI] [PubMed] [Google Scholar]
- Huang MD, Wei FJ, Wu CC, Hsing YI, Huang AH. (2009) Analyses of advanced rice anther transcriptomes reveal global tapetum secretory functions and potential proteins for lipid exine formation. Plant Physiol 149: 694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhang Z, Cao J. (2013) Pollen wall development: the associated enzymes and metabolic pathways. Plant Biol (Stuttg) 15: 249–263 [DOI] [PubMed] [Google Scholar]
- Kader JC. (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654 [DOI] [PubMed] [Google Scholar]
- Kim SS, Douglas CJ. (2013) Sporopollenin monomer biosynthesis in Arabidopsis. J Plant Biol 56: 1–6 [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2: 1201–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand B, Erhardt M, Heitz T, Legrand M. (2013) Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol 162: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauga B, Charbonnel-Campaa L, Combes D. (2000) Characterization of MZm3-3, a Zea mays tapetum-specific transcript. Plant Sci 157: 65–75 [DOI] [PubMed] [Google Scholar]
- Li Y, Suen DF, Huang CY, Kung SY, Huang AH. (2012) The maize tapetum employs diverse mechanisms to synthesize and store proteins and flavonoids and transfer them to the pollen surface. Plant Physiol 158: 1548–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Winther JR. (2001) Surprisingly high stability of barley lipid transfer protein, LTP1, towards denaturant, heat and proteases. FEBS Lett 488: 145–148 [DOI] [PubMed] [Google Scholar]
- Matsuhira H, Shinada H, Yui-Kurino R, Hamato N, Umeda M, Mikami T, Kubo T. (2006) An anther-specific lipid transfer protein gene in sugar beet: its expression is strongly reduced in male-sterile plants with Owen cytoplasm. Physiol Plant 129: 407–414 [Google Scholar]
- Mayhew TM, Lucocq JM, Griffiths G. (2002) Relative labelling index: a novel stereological approach to test for non-random immunogold labelling of organelles and membranes on transmission electron microscopy thin sections. J Microsc 205: 153–164 [DOI] [PubMed] [Google Scholar]
- Murgia M, Charzynska M, Rougier M, Cresti M. (1991) Secretory tapetum of Brassica oleracea L.: polarity and ultrastructural features. Sex Plant Reprod 4: 28–35 [Google Scholar]
- Nebenführ A, Ritzenthaler C, Robinson DG. (2002) Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol 130: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TB, Cheung RC, Wong JH, Ye X. (2012) Lipid-transfer proteins. Biopolymers 98: 268–279 [DOI] [PubMed] [Google Scholar]
- Paul W, Hodge R, Smartt S, Draper J, Scott R. (1992) The isolation and characterisation of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol Biol 19: 611–622 [DOI] [PubMed] [Google Scholar]
- Platt KA, Huang AHC, Thomson WW. (1998) Ultrastructural study of lipid accumulation in tapetal cells of Brassica napus L. cv. Westar during microsporogenesis. Int J Plant Sci 195: 724–737 [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ. (2010) ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan SM, Moffatt BA. (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Toriyama K, Ejiri S, Hinata K. (1994) Molecular characterization of rice genes specifically expressed in the anther tapetum. Plant Mol Biol 26: 1737–1746 [DOI] [PubMed] [Google Scholar]
- Turgut K, Barsby T, Craze M, Freeman J, Hodge R, Paul W, Scott R. (1994) The highly expressed tapetum-specific A9 gene is not required for male fertility in Brassica napus. Plant Mol Biol 24: 97–104 [DOI] [PubMed] [Google Scholar]
- Wang HW, Hwang SG, Karuppanapandian T, Liu A, Kim W, Jang CS. (2012a) Insight into the molecular evolution of non-specific lipid transfer proteins via comparative analysis between rice and sorghum. DNA Res 19: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NJ, Lee CC, Cheng CS, Lo WC, Yang YF, Chen MN, Lyu PC. (2012b) Construction and analysis of a plant non-specific lipid transfer protein database (nsLTPDB). BMC Genomics (Suppl 1) 13: S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wijesinha-Bettoni R, Alexeev Y, Johnson P, Marsh J, Sancho AI, Abdullah SU, Mackie AR, Shewry PR, Smith LJ, Mills EN. (2010) The structural characteristics of nonspecific lipid transfer proteins explain their resistance to gastroduodenal proteolysis. Biochemistry 49: 2130–2139 [DOI] [PubMed] [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D. (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]