A comparison of submergence-induced transcriptomes of two flooding-tolerant Rorippa species with the relatively flooding-sensitive Arabidopsis identifies gene clusters potentially associated with tolerance.
Abstract
Complete submergence represses photosynthesis and aerobic respiration, causing rapid mortality in most terrestrial plants. However, some plants have evolved traits allowing them to survive prolonged flooding, such as species of the genus Rorippa, close relatives of Arabidopsis (Arabidopsis thaliana). We studied plant survival, changes in carbohydrate and metabolite concentrations, and transcriptome responses to submergence of two species, Rorippa sylvestris and Rorippa amphibia. We exploited the close relationship between Rorippa species and the model species Arabidopsis by using Arabidopsis GeneChip microarrays for whole-genome transcript profiling of roots of young plants exposed to a 24-h submergence treatment or air. A probe mask was used based on hybridization of genomic DNA of both species to the arrays, so that weak probe signals due to Rorippa species/Arabidopsis mismatches were removed. Furthermore, we compared Rorippa species microarray results with those obtained for roots of submerged Arabidopsis plants. Both Rorippa species could tolerate deep submergence, with R. sylvestris surviving much longer than R. amphibia. Submergence resulted in the induction of genes involved in glycolysis and fermentation and the repression of many energy-consuming pathways, similar to the low-oxygen and submergence response of Arabidopsis and rice (Oryza sativa). The qualitative responses of both Rorippa species to submergence appeared roughly similar but differed quantitatively. Notably, glycolysis and fermentation genes and a gene encoding sucrose synthase were more strongly induced in the less tolerant R. amphibia than in R. sylvestris. A comparison with Arabidopsis microarray studies on submerged roots revealed some interesting differences and potential tolerance-related genes in Rorippa species.
Submergence as a result of flooding is very harmful to most plants. Restricted gas diffusion under water leads to the repression of photosynthesis and aerobic respiration, resulting in an energy and carbohydrate crisis (Drew, 1997; Voesenek et al., 2006; Bailey-Serres and Voesenek, 2008, 2010). Excessive water, either during prolonged waterlogging or complete submergence, will kill most terrestrial plants (Setter and Laureles, 1996; Drew, 1997).
Nevertheless, some specialized plant species from flood-prone areas may survive complete submergence for many months (van Eck et al., 2004; Mommer et al., 2006). Two different strategies to cope with submergence have been proposed: (1) escape from submergence by restoring air contact, characterized by rapid shoot elongation and the formation of longitudinal air channels called aerenchyma; and (2) quiescence, meaning that growth is restricted and carbohydrate reserves are preserved that can be utilized for recovery after the flood waters have subsided (Bailey-Serres and Voesenek, 2008). Even though a wide variety of plant responses have been described that can increase the survival of submergence stress, the mechanisms underlying such extreme submergence tolerance are still poorly understood, especially at the molecular level (Bailey-Serres and Voesenek, 2008).
Even many rice (Oryza sativa) varieties can be severely damaged or killed within a few weeks of complete submergence (Setter and Laureles, 1996). Lowland rice can germinate in a layer of water and flourish as long as the shoot is in contact with air. Deepwater rice, as long as it keeps up its shoot growth so that leaves remain in contact with air, is tolerant to meters of water. Recently, the genetic determinants of submergence tolerance in lowland and deepwater rice have been identified as SUB1 (Xu et al., 2006) and SNORKEL1 and SNORKEL2 (Hattori et al., 2009), respectively. These genes belong to the group VII Ethylene Response Factor (ERF) family in rice but control opposite submergence responses. The presence of the SUB1A-1 allele will suppress the growth of rice when flooded (Xu et al., 2006). Quiescence of this kind is demonstrably advantageous when paddy rice is flooded for a few weeks and the flood water is too deep to be outgrown (Fukao et al., 2006). The presence of the SNORKEL genes in deepwater rice promotes shoot elongation (Hattori et al., 2009). Recent studies in Arabidopsis (Arabidopsis thaliana) have shown that certain members of the group VII ERFs are degraded in an oxygen-dependent manner by the N-end rule pathway of protein degradation. It is suggested that this pathway functions as an oxygen-sensing mechanism in higher plants (Gibbs et al., 2011; Licausi et al., 2011). Interestingly, overexpression of one of these genes, RAP2.12, made Arabidopsis more flood tolerant (Licausi et al., 2011).
An important question that remains is what determines extreme submergence tolerance at the molecular level. Detailed understanding of the response to submergence at the level of gene expression and metabolism is mostly based on studies using maize (Zea mays), rice, and the model species Arabidopsis exposed to low oxygen levels (Sachs et al., 1996; Bailey-Serres and Voesenek, 2008). Genome-wide transcriptome studies of Arabidopsis, rice, and poplar (Populus spp.) subjected to low oxygen levels have shown that a large number of genes are differentially expressed in response to this stress, including the repression of genes involved in energy-consuming processes such as cell wall biosynthesis and the induction of genes in the glycolysis and fermentation pathways (Klok et al., 2002; Branco-Price et al., 2005, 2008; Liu et al., 2005; Loreti et al., 2005; Lasanthi-Kudahettige et al., 2007; Kreuzwieser et al., 2009; Mustroph et al., 2009; van Dongen et al., 2009). Transcriptomic studies of Arabidopsis in the past have mostly used seedlings or cell cultures exposed to low oxygen levels. However, recent studies have investigated both the molecular response and natural variation between Arabidopsis accessions to complete submergence (Lee et al., 2011; Vashisht et al., 2011) or the molecular response of Arabidopsis to root flooding (Hsu et al., 2011).
The study of wild relatives of Arabidopsis provides exciting research opportunities because it allows the use of molecular tools developed for this model and greatly extends the range of natural variation that can be studied (Mitchell-Olds, 2001; Schranz et al., 2007). Among Arabidopsis wild relatives, the genus Rorippa (yellow cress) includes the polyploid, hybridizing species Rorippa amphibia and Rorippa sylvestris, which inhabit periodically flooded river banks throughout Europe (Bleeker and Hurka, 2001; Stift et al., 2008). We have recently shown different morphological responses to submergence in R. amphibia and R. sylvestris (Stift et al., 2008; Akman et al., 2012). As will be shown here, R. amphibia is less tolerant to complete submergence than R. sylvestris, which survived over 3 months of submergence under natural light conditions. Therefore, these species provide an excellent system to address whether such variation in tolerance is associated with distinct transcriptome changes in response to submergence. In general, such effects of environmental variation on transcript profiles of contrasting genotypes or genotype-environment interactions are of particular interest in ecological genomics research (Filatov et al., 2006; Nettleton, 2006; Kammenga et al., 2007). The close relatedness of Rorippa species to Arabidopsis allowed us to use DNA microarrays developed for this model species using a genomic DNA (gDNA)-based probe selection method, shown previously to work with other members of the Brassicaceae family (Hammond et al., 2006; Broadley et al., 2008).
While underwater photosynthesis may produce some oxygen in shoots of submerged plants when light is available, oxygen is more rapidly depleted in nonphotosynthetic tissues (Voesenek and Sasidharan, 2013). Furthermore, previous molecular comparisons of submergence tolerance have been performed only on shoot tissues, related to the elongation or quiescence strategies of shoots (Xu et al., 2006; Hattori et al., 2009; Jung et al., 2010; Mustroph et al., 2010). However, very few studies have investigated root responses in relation to flooding tolerance; mostly, they investigate the effect of only hypoxia, a component of flooding stress (Klok et al., 2002; Mustroph et al., 2009; van Dongen et al., 2009). In the experimental setup used in this study, the flooding was too deep to be outgrown and the roots would experience severe hypoxia, making them important determinants of submergence tolerance. Therefore, we focused on roots to characterize the effect of submergence on whole-genome transcript profiles. Furthermore, we took advantage of recently published microarray data from roots of completely submerged Arabidopsis plants to make a direct comparison of submergence-induced transcript profiles of the extremely submergence-tolerant Rorippa species and the much less tolerant Arabidopsis.
RESULTS
R. sylvestris Has Higher Submergence Tolerance Than R. amphibia
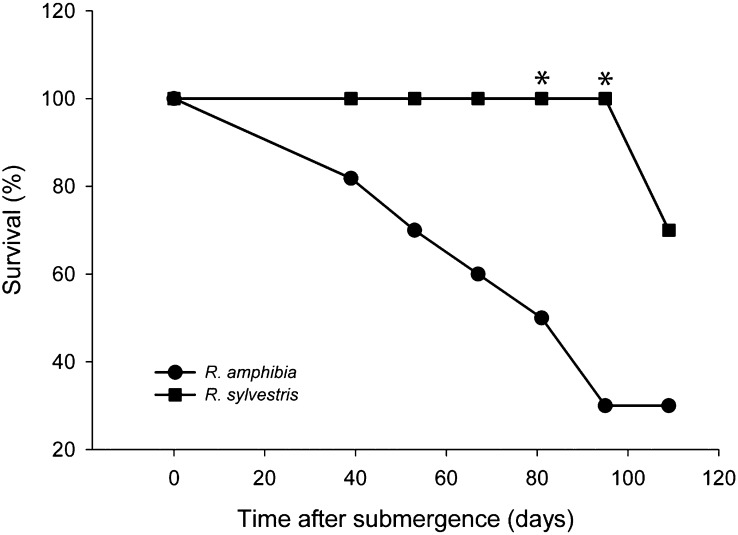
In order to confirm the submergence tolerance of Rorippa species, 52-d-old plants were subjected to complete submergence in a normal day/night rhythm. After 95 d of complete submergence in a basin, all R. sylvestris plants had survived the stress (Fig. 1). In contrast, only 30% of R. amphibia had survived at this point, with its survival gradually decreasing across the entire time course studied here. At the final time point (109 d after submergence), R. sylvestris had also suffered mortality, but the large difference between the two genotypes was maintained. Under normal growth conditions, R. amphibia produces a stem that can reach more than 1 m in length, whereas R. sylvestris does not form such a stem and reaches 0.4 m maximally (Jonsell, 1968). In accordance with previous reports, R. amphibia elongated clearly more than R. sylvestris when submerged (data not shown; Akman et al., 2012). The plants could not reach the water surface at any time because a cloth was draped over the top of the water basins. Toward the end of the experiment, most of the shoot and especially the leaves had died in both genotypes. Survival was scored as the ability to form new leaves after a recovery period of 2 weeks. The emergence of new leaves postsubmergence is probably due to the survival of the shoot meristem. It is also possible that the roots withstood complete submergence and maintained the energy reserves to sprout after desubmergence. In addition, the difference of these two Rorippa species in submergence tolerance was not restricted to older plants, as experiments with younger plants confirmed the extreme submergence tolerance of Rorippa species (data not shown). Given that most terrestrial plants cannot survive complete submergence for more than a few weeks (van Eck et al., 2004; Mommer et al., 2006; Vashisht et al., 2011), it can be stated that these Rorippa species genotypes, and in particular R. sylvestris, are extremely tolerant to submergence.
Figure 1.
Survival upon submergence in mature Rorippa species plants. Seven-week-old R. amphibia and R. sylvestris plants were completely submerged in shaded, 1-m-deep outdoor basins. Ten individuals per genotype were taken out of the treatment at the indicated time points, and survival was scored after a 2-week recovery period as the ability to form new leaves. Asterisks denote significant differences at that time point between the two species (P < 0.05, Fischer’s exact test).
Submergence Reduced Root Sugar Content within Days
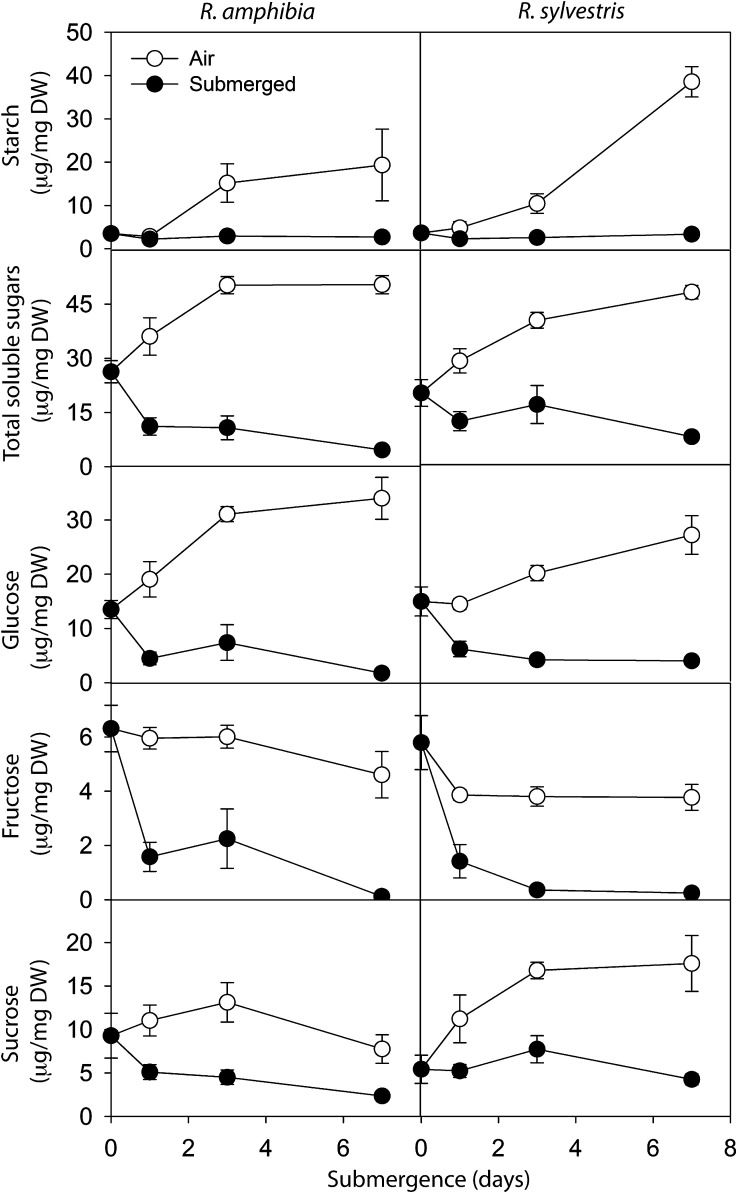
During submergence, plants rapidly have to cope with an energy crisis, because photosynthesis under water occurs at a much lower rate than above water, and glycolysis needs to be enhanced because of the low ATP yield of fermentation in comparison with oxidative respiration (Bailey-Serres and Voesenek, 2008). The adaptive responses to submergence stress that determine survival in the long run, therefore, must include efficient carbohydrate usage and energy conservation. To test the carbohydrate usage of Rorippa species, roots of young (28 d) plants submerged for up to 7 d were analyzed for sugar and starch contents (Fig. 2). Submergence did not have a significant effect on starch levels in roots of either species after 1 d of treatment (P > 0.05, Student’s t test). However, while root starch content increased in controls thereafter, it remained low in submerged roots. After 7 d of submergence, starch levels were not significantly different between the two species. The three main sugars detected were Glc, Suc, and Fru (Fig. 2). In both Rorippa species, 1 d of submergence already significantly reduced the concentration of these sugars, compared with controls in air. After 7 d, submerged R. sylvestris roots had significantly higher levels of total soluble sugars and Glc compared with R. amphibia (P < 0.05, Student’s t test; Fig. 2).
Figure 2.
The effect of submergence on starch and sugar concentrations in the roots of R. amphibia and R. sylvestris plants. Values are means ± se of four to five biological replicates. DW, Dry weight.
gDNA-Based Probe Selection Using Affymetrix Arabidopsis ATH1 GeneChip Arrays
In order to determine the molecular basis of submergence tolerance of Rorippa species, the Arabidopsis ATH1 GeneChip arrays were used for microarray experiments. To reduce possible bias in the transcriptome data due to sequence differences between Rorippa species and Arabidopsis, a probe selection method was employed based on the hybridization of gDNA of R. amphibia or R. sylvestris to the arrays. Because replicate arrays would be interrogating the same gDNA, amounting to technical but not biological replication, we analyzed the gDNA of each species once. Arabidopsis ecotype Columbia-0 gDNA was also analyzed in the same way for comparison, since it should theoretically hybridize to all perfect-match probes on the array. The control mismatch probes on this array were discarded from the analysis altogether, as they are uninformative in cross-species hybridizations. Sequence differences between Rorippa species and Arabidopsis are expected to result in reduced hybridization and, hence, a low signal of the corresponding perfect-match probes. Because every gene is interrogated by a probe set of 11 perfect-match probes on the array, discarding up to 10 poorly performing probes based on gDNA hybridization still allows measurement of the expression of the associated gene (Hammond et al., 2005, 2006). By applying increasing thresholds of gDNA hybridization signal, decreasing numbers of individual probes were retained in the resulting probe mask file, while the number of probe sets retained was reduced at a lower rate (Supplemental Fig. S1). This is consistent with previous studies using this probe selection method (Hammond et al., 2005, 2006; Broadley et al., 2008). More probes were retained at any given threshold for the Arabidopsis gDNA data set than for the Rorippa species sets, as expected. The two Rorippa species showed a similar decline in numbers of probes and probe sets with increasing thresholds. An optimal threshold was empirically determined at which the number of completely removed probe sets was minimized in order to avoid the loss of all information regarding the corresponding gene transcript. At a threshold level of the gDNA hybridization signal of 80, almost all probe sets (97%) were retained, and the number of probes was reduced by approximately two-thirds (Supplemental Fig. S1). Most probe sets contained three probes when this probe mask was used (Supplemental Fig. S2). The gDNA hybridization signal did not appear to be correlated with the number of probes per probe set, indicating that no bias had occurred in the probe selection (Supplemental Fig. S3).
Submergence-Induced Large-Scale Changes in Gene Expression
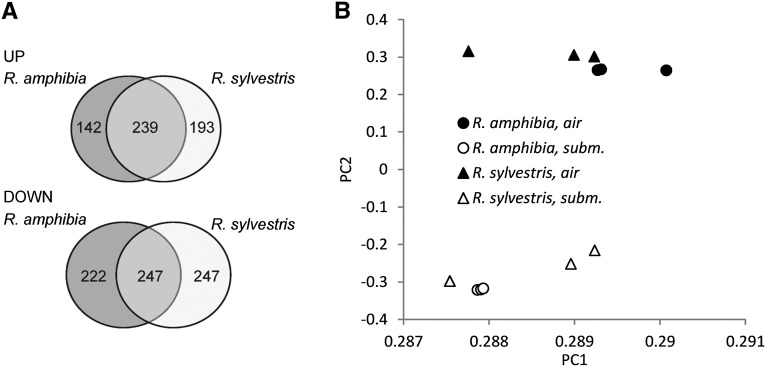
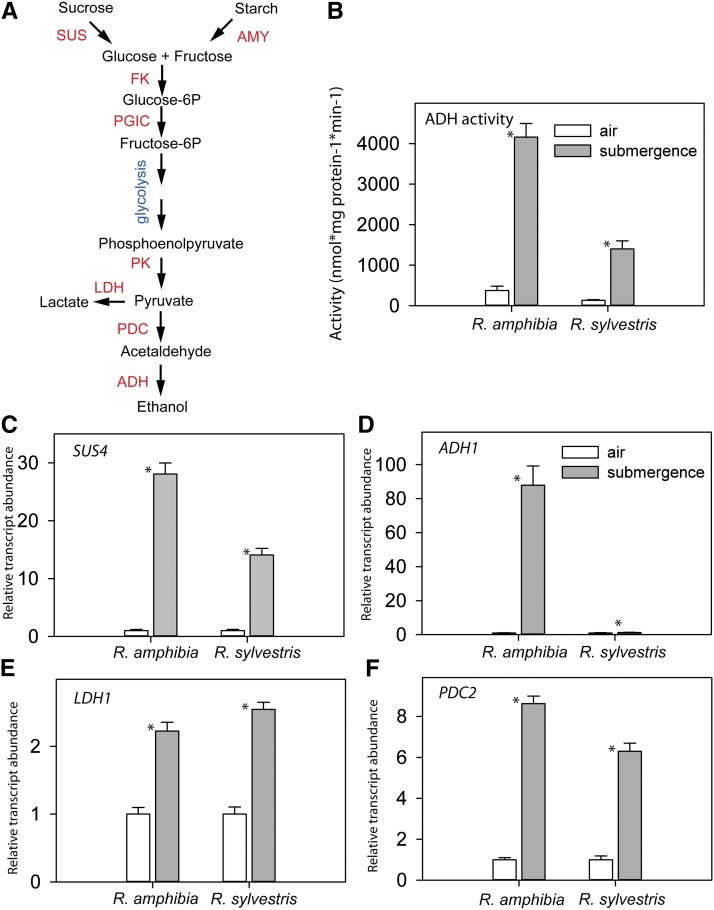
Affymetrix GeneChip arrays were used to analyze root transcript profiles of the two Rorippa species genotypes exposed to 24 h of complete submergence. Approximately 400 genes were induced and approximately 470 genes were repressed by the submergence treatment in both R. amphibia and R. sylvestris (Fig. 3A). When no probe mask was applied, less than one-half the number of significantly regulated genes were detected (data not shown), confirming previous findings that the probe filtering enhanced the sensitivity to detect changes in transcript levels (Hammond et al., 2005, 2006). About 60% of induced genes were shared between both Rorippa species, while only about 50% of reduced genes were found to be changed in both species (Fig. 3A). The commonly induced genes included well-known hypoxic genes involved in glycolysis and fermentation (alcohol dehydrogenase [ADH1], pyruvate decarboxylase [PDC1], phosphofructokinase [PFK6], and sucrose synthase [SUS4]) and transcription factors LATERAL ORGAN BOUNDARIES DOMAIN-CONTAINING protein (LBD41). A principal component analysis (PCA) of transcript levels of all interrogated genes clearly separated treated from control samples along the first and second components, explaining 94% and 3% of total variation, respectively (Fig. 3B). As expected, for each of the genotypes, the three independent replicates clustered together (Fig. 3B).
Figure 3.
Root transcriptome profiles of Rorippa species using Arabidopsis gene chips. A, Numbers of genes regulated by submergence treatment and overlap between species. Within each genotype, expression values of submergence-induced genes show a ratio of submerged to air of greater than 2.0, and submergence-repressed genes show a ratio of submerged to air of less than 0.5 with a significant treatment effect (FDR < 0.05; t test with multiple testing correction according to Benjamini and Hochberg, 1995). B, Results of PCA applied to the expression of all the genes represented on the microarray. A probe mask with a threshold of 80 was applied to the expression data. For each of the 12 microarrays, expression data of 21,288 genes were used as input.
Comparison of Submergence-Induced Root Transcript Profiles between R. amphibia and R. sylvestris
In order to compare the two Rorippa species with varying tolerance to submergence, we first compared both lists of submergence-regulated genes (Fig. 3A). About 40% of the up-regulated genes were specific for either species. In order to get more insight into the gene expression differences, we also performed a pairwise comparison between species. Second, a complex pairwise comparison was performed to find differentially regulated genes (Supplemental Table S1). Both methods revealed some differences in submergence response between the two species. About 16% of induced genes were more strongly induced in R. amphibia as compared with R. sylvestris and vice versa. Genes with a higher induction in R. amphibia included genes involved in photosynthesis, while R. sylvestris had more genes induced that were involved in defense response. Among the most differentially induced genes in R. amphibia, there was POLYGALACTURONASE-INHIBITING PROTEIN2 (At5g06870), a WRKY transcription factor (At5g13080, AtWRKY75), and an F-box family protein (At4g23960). In R. sylvestris, the more induced genes included a nodulin MtN21 family protein (At4g28040), an unknown protein (At3g19030), and CATALASE1 (At1g20630). The PCA further emphasized that the two species were not dramatically different from each other under control and submerged conditions (Fig. 3B).
Submergence Led to the Repression of Energy-Consuming Processes and the Induction of Photosynthesis, Glycolysis, and Fermentation Genes
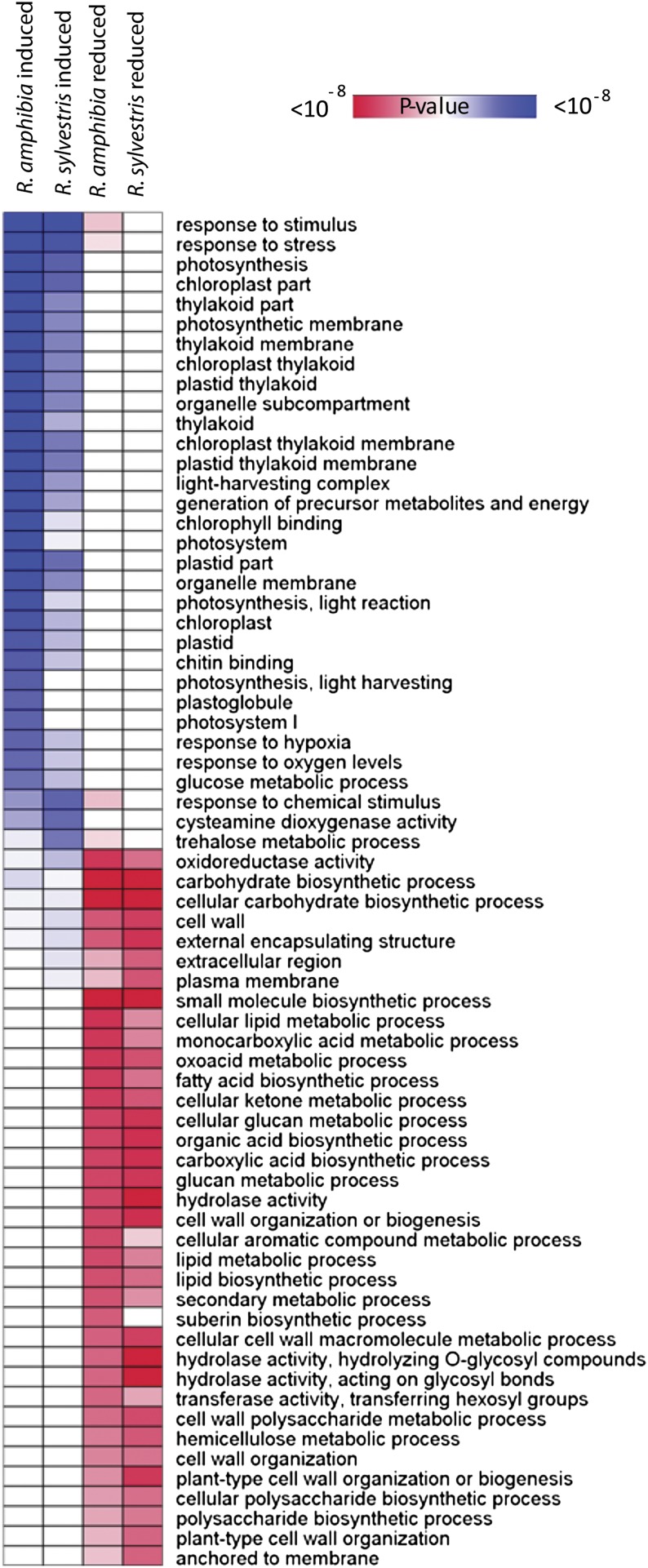
In order to get an overview of the transcriptome changes induced by the submergence treatment, overrepresentation analysis of gene categories based on the Gene Ontology (GO) database was performed using the GOHyperGAll function (Fig. 4; Horan et al., 2008). In both genotypes, this analysis showed an overrepresentation in down-regulated genes of various energy-demanding pathways, namely cell wall precursor synthesis, fatty acid biosynthesis, and amino acid synthesis (Supplemental Table S2). Glycolysis and fermentation pathways were more overrepresented in the data set of submergence up-regulated genes of R. amphibia than in R. sylvestris (i.e. GO:0006006, Glc metabolic process; adjusted P value = 4.51E-05 and 9.22E-03, respectively; Supplemental Table S2). Surprisingly, genes involved in the light reactions of photosynthesis were clearly overrepresented in the set of up-regulated genes of R. amphibia and to a lower extent in R. sylvestris (Fig. 4). Hence, the qualitative response to submergence appeared roughly similar, but genotypes did show quantitative differences.
Figure 4.
Overrepresentation analysis of Rorippa species microarray data. Enrichment of GO categories of the lists of significantly induced or reduced genes (signal log ratio > 1 or < −1, FDR < 0.05; Supplemental Table S1) was calculated by use of the GOHyperGAll function in R (Horan et al., 2008). Selected significantly enriched GO terms are shown; a full list of GO terms is found in Supplemental Table S2. Statistical significances of GO term enrichment are represented by a false-color heat map (blue = up-regulated genes, red = down-regulated genes). Categories of GO terms that were significantly overrepresented are indicated with increasing color intensity (blue or red).
Submergence Caused Stronger Induction of SUS4 and Glycolysis and Fermentation Genes in R. amphibia Than in R. sylvestris
Closer inspection of genes involved in carbohydrate catabolism revealed that several were expressed at a significantly higher level under submerged conditions in R. amphibia than in R. sylvestris (Table I). Cloning and expression analyses of selected genes using quantitative reverse transcription (qRT)-PCR confirmed that transcript levels of the homolog of the Arabidopsis gene SUS4 as well as of the genes involved in the final reactions of the fermentation pathway, PDC2 and ADH1, were higher in submerged R. amphibia roots (Fig. 5, C, D, and F). Accordingly, submerged (24 h) roots of R. amphibia also had higher ADH activity than submerged roots of R. sylvestris relative to their respective air controls (Fig. 5B). Surprisingly, submerged R. amphibia roots recorded lower ethanol content than corresponding air controls after 7 d (Supplemental Fig. S4). At other days, the values did not differ significantly. It is likely, however, that most ethanol was released into the root medium during the submergence period, as shown previously (Mustroph and Albrecht, 2007). In accordance with the expression and activity of ADH, R. amphibia roots contained more ethanol than R. sylvestris roots (Supplemental Fig. S4).
Table I. Expression values of selected genes involved in carbohydrate catabolism obtained from the microarray analysis.
Values are means ± sd; n = 3. Values within the same row followed by a different letter are significantly different (P < 0.05, Tukey’s honest significant difference test).
| Arabidopsis Genome Initiative Locus | Common Name |
R. amphibia |
R. sylvestris |
||
|---|---|---|---|---|---|
| Air | Submergence | Air | Submergence | ||
| At1g35580 | CINV1 | 185 ± 10 b | 276 ± 29 c | 82 ± 16 a | 116 ± 4 a |
| At3g43190 | SUS4 | 87 ± 19 a | 352 ± 15 b | 77 ± 9 a | 289 ± 62 b |
| At5g20830 | SUS1 | 1,232 ± 56 a | 3,358 ± 51 c | 1,110 ± 49 a | 2,005 ± 316 b |
| At2g36460 | Aldolase | 298 ± 22 a | 1,470 ± 202 b | 38 ± 7 a | 156 ± 44 a |
| At5g42740 | PGIC | 457 ± 14 b | 1,121 ± 67 d | 271 ± 15 a | 856 ± 52 c |
| At5g63680 | Pyruvate kinase, putative | 1,774 ± 89 b | 2,534 ± 183 c | 1,149 ± 88 a | 1,233 ± 56 a |
| At1g77120 | ADH1 | 202 ± 12 a | 1,788 ± 84 c | 129 ± 13 a | 865 ± 160 b |
| At4g33070 | PDC1 | 380 ± 14 a | 1,098 ± 58 b | 119 ± 15 a | 779 ± 195 b |
| At5g54960 | PDC2 | 1,215 ± 99 a | 3,944 ± 359 b | 1,924 ± 119 a | 3,367 ± 268 b |
| At4g17260 | LDH | 345 ± 35 a | 551 ± 63 b | 230 ± 9 a | 248 ± 60 a |
| At1g17290/At1g72330 | AlaAT1 | 1,038 ± 25 a | 3510 ± 6 b | 846 ± 34 a | 2,524 ± 465 c |
Figure 5.
Regulation of anaerobic metabolism. A, Schematic representation of the metabolic pathway showing sugar and starch breakdown feeding into glycolysis and fermentation. This simplified scheme is after Bailey-Serres and Voesenek (2008). B to F, Relative transcript abundance measured by qRT-PCR of Rorippa species homologs of ADH (B), SUS4 (C), ADH1 (D), PDC2 (E), and LDH1 (F) activity measured in roots of 24-h submerged Rorippa species plants. For ADH activity, data are means and sd (n = 5). For transcript abundance levels, data are means and SE (n = 4). Asterisks above the bars indicate significant differences between control and submerged samples (Student’s t test, P < 0.05). [See online article for color version of this figure.]
An exception to the differential induction of fermentation genes between the two species was LACTATE DEHYDROGENASE1 (LDH1), which was similarly up-regulated upon submergence in both species (Fig. 5E). Lactate levels in submerged roots of both species, however, were not significantly higher than the respective controls, but in general, more lactate was present in roots of R. amphibia in comparison with R. sylvestris (Supplemental Fig. S4). Other carbohydrate catabolism genes showing a stronger induction in R. amphibia than R. sylvestris included SUS1 and various glycolysis genes such as PGIC, pyruvate kinase, and aldolase (Table I).
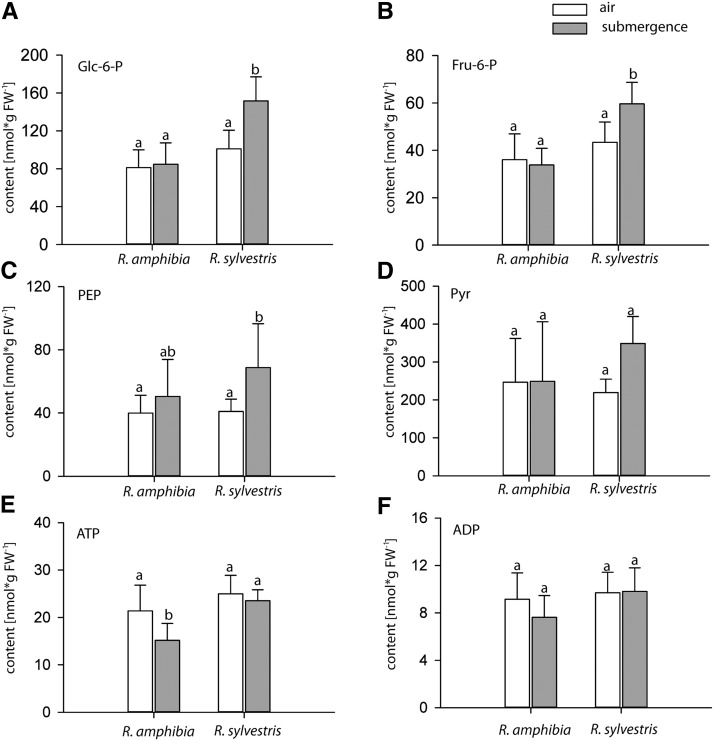
We were further interested in changes of metabolite contents during the submergence treatment and analyzed selected metabolites of primary metabolism. Interestingly, after 24 h of submergence, R. sylvestris accumulated significantly more sugar phosphates (Glc-6-P and Fru-6-P) than R. amphibia (Fig. 6). Considering that there were no significant differences in starch content of submerged roots after 24 h relative to controls (Fig. 2), an accumulation of these glycolytic intermediates likely indicates a restriction of glycolysis in R. sylvestris (Fig. 6, A and B). The levels of phosphoenolpyruvate and pyruvate were also slightly, but not significantly, higher in the more tolerant R. sylvestris (Fig. 6, C and D). These data suggested a slower glycolytic process in R. sylvestris, but despite this assumption, ATP levels did not decrease in R. sylvestris but instead dropped significantly by 30% in R. amphibia during 24 h of submergence (Fig. 6E). The ratio of ATP to ADP, however, remained unchanged in both species and treatments (Supplemental Fig. S5).
Figure 6.
Submergence-induced metabolite changes in Rorippa species. Content is shown for selected metabolites (nmol g−1 fresh weight [FW]) in roots of the Rorippa species under control conditions (air) and submergence for 24 h. A, Glc-6-P. B, Fru-6-P. C, Phosphoenolpyruvate (PEP). D, Pyruvate (Pyr). E, ATP. F, ADP. Data are means + sd (n = 8). Different letters above the bars denote statistically significant differences (P < 0.05, Tukey’s honestly significant difference test).
Comparison of Tolerant Rorippa Species with the Related But Submergence-Intolerant Arabidopsis
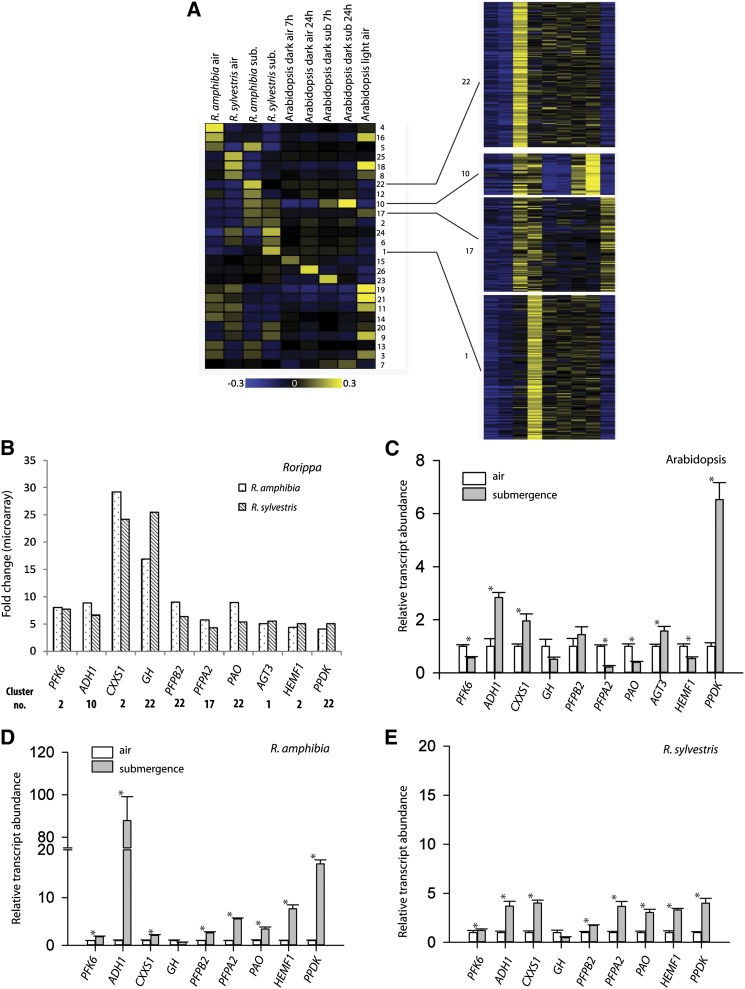
The use of Affymetrix GeneChips to monitor Rorippa species root transcriptomes allowed us to compare our data set with that of Lee et al. (2011), who studied roots and shoots of Arabidopsis plants also using a 24-h submergence treatment, although that experiment was performed in complete darkness. The gene expression data of Rorippa species were compared with root transcript profiles of submerged Arabidopsis plants, which are relatively flooding intolerant (Lee et al., 2011; Vashisht et al., 2011). This comparison was based on the Affymetrix identifiers and included a cluster analysis of the mean expression values of each sample. This analysis revealed a large variation in gene expression between the submergence-induced transcriptome profiles of Rorippa species versus Arabidopsis (Fig. 7A). Only some genes were induced in all three species at all times (cluster 10 of Supplemental Table S3), including 21 of the previously identified “core hypoxic response” genes (Mustroph et al., 2009). Among these genes were known hypoxic genes involved in primary metabolism (SUS1, SUS4, ADH1, and PDC1) and other hypoxic genes (wound-responsive protein, hemoglobin, and stearoyl-acyl-carrier-protein desaturase family protein). Many genes responded in different ways in Arabidopsis and Rorippa species. For example, genes of clusters 2 and 17 were more strongly induced in Rorippa species than in Arabidopsis. Interestingly, genes of cluster 2 were regulated preferentially by darkness in Arabidopsis. Therefore, the induction of these genes in Rorippa species could be attributed to the change in light level rather than to a submergence-associated change in oxygen concentration. Although the submergence treatment of Rorippa species plants was performed in the light at approximately 200 µmol m−2 s−1, light quantity and quality can vary in a water column, possibly leading to the observed changes in the expression of light-regulated genes, even in roots. Therefore, the relatively large number of transcriptional regulators found in cluster 2 (e.g. a basic helix loop helix transcription factor [At2g43140], WOX14 [At1g20700], and a calcium-binding elongation factor hand family protein [At5g54130]) are most likely not involved in mediating hypoxia tolerance in Rorippa species.
Figure 7.
Comparison of the submergence response of tolerant Rorippa species and sensitive Arabidopsis. A, Cluster analysis of root transcript profiles of Arabidopsis (Lee et al., 2011) and Rorippa species subjected to 24 h of submergence. RMA-normalized expression values were transformed as described in “Materials and Methods” and then clustered with the PAM function in R. The mean of each cluster is shown on the left side, and all genes of four selected clusters are shown on the right side. B, D, and E, Transcript levels of Rorippa species homologs of selected genes from indicated clusters as measured by microarray (B) and qRT-PCR (D and E) analyses in roots of 24-h submerged Rorippa species plants. Microarray data are fold change ratios of submergence versus control. C, Transcript abundance of the genes indicated measured using qRT-PCR in roots of Arabidopsis plants submerged for 24 h under conditions identical to those used for Rorippa species microarrays. Data in B to E are means of the relative transcript levels and se (n = 3 for C and n = 5 for D and E).
Genes of cluster 17 were preferentially submergence induced in Rorippa species. While these genes were not submergence induced in Arabidopsis under darkness, some of them were induced by hypoxic treatment of Arabidopsis in light [SRO5 (At5g62520), PDC family protein (At5g01320), and 2OG-Fe(II) oxygenase family protein (At1g20270); Mustroph et al., 2009], indicating that these genes could be variably induced due to the actual oxygen status and/or light levels. Other genes that were solely induced in Rorippa species and could be potential mediators of the higher tolerance of these species in comparison with Arabidopsis included a glycoside hydrolase family19 protein (At1g56680), genes of unknown function (At5g04730 and At1g12845), and some photosynthesis-related genes (At5g54270 and At1g79040). Interestingly, a pyrophosphate-Fru-6-P-phosphotransferase α-subunit (PFPA2, At1g76550) was also more highly induced in Rorippa species than in Arabidopsis (Fig. 7, C–E).
Cluster 22 was enriched for genes that were more induced in R. amphibia than in R. sylvestris and not induced in Arabidopsis. Some of these genes belong to the pyrophosphate-using alternative pathway in metabolism, including the pyrophosphate-Fru-6-P-phosphotransferase β-subunit (PFPB2, At4g04040) and pyruvate-orthophosphate dikinase (PPDK, At4g15530) that could be involved in submergence tolerance. However, the analysis of in vitro PFP activities in submergence-treated Rorippa species roots did not show significant differences between the Rorippa species and Arabidopsis (Supplemental Fig. S6). Nevertheless, one cannot exclude differences in the in planta activity due to different affinities of the PFP isoforms for their substrates. This cluster was also strongly enriched in photosynthesis-related genes (Supplemental Table S3), as shown already in the direct species comparison (Fig. 4).
Genes of cluster 1 were more highly expressed in submerged R. sylvestris than R. amphibia, were not induced in Arabidopsis, and were enriched in the GO terms cobalt ion binding and antioxidant activity (i.e. At1g20630, CATALASE1). This cluster also contained genes that were related to amino acids (At2g40420, amino acid transporter family protein; At2g38400, Ala:glyoxylate transaminase). In contrast, clusters 23 and 7 contained genes that were more induced in submerged Arabidopsis roots: surprisingly, the previously identified hypoxia-responsive ERF HRE1 (Licausi et al., 2010) and a phosphate-related protein (PHI1, At1g35140). A fraction of these genes, mostly from cluster 23, were induced after 7 h, rather than after 24 h, of submergence in Arabidopsis, and this could explain the lack of induction in Rorippa species after 24 h. However, we cannot exclude that orthologs of these genes in Rorippa species can be regulated by submergence but are not represented on the ATH1 microarray.
In order to confirm this contrasting regulation between Rorippa species and Arabidopsis, the expression of a selection of these genes was measured in roots of Arabidopsis plants using qRT-PCR. Orthologs of these genes were also cloned and their expression measured in both Rorippa species. The submergence treatment for all three species (24 h) was identical to conditions used for the microarray studies on Rorippa species, including the use of similar light levels, thereby allowing a direct comparison, since the Arabidopsis microarray data were obtained from plants submerged in the dark (Lee et al., 2011). Twenty-four-hour submerged roots of both Arabidopsis and Rorippa species experienced hypoxic stress, as evidenced by a significant up-regulation of ADH1 (Fig. 7, C–E). Most of the 10 genes tested in Arabidopsis showed either no significant change (At2g43610, chitinase family protein; At4g04040, PFPB2) or significant down-regulation (At1g76550, PFPA2; At4g32840, PFK6; At3g44880, PAO, pheophorbide a oxygenase family protein; At1g03475, HEMF1, coproporphyrinogen III oxidase) of transcripts due to submergence (Fig. 7B), while orthologs of eight of these 10 genes showed significant up-regulation in both Rorippa species in accordance with the microarray results. Our expression analyses also confirmed the overall trends revealed by the cluster analyses. For example, roots of 24-h submerged Rorippa species plants had significantly higher transcript levels of PFPA2 (cluster 17) compared with air controls. In contrast, roots of Arabidopsis plants submerged under identical conditions had significantly lower PFPA2 transcript abundance relative to their nonsubmerged controls. The cluster 22 genes PAO (R. amphibia versus R. sylvestris [log fold change 1.7 versus 1.5]) and PFPB2 (R. amphibia versus R. sylvestris [log fold change 1.36 versus 0.8]) were submergence inducible only in Rorippa species, with a higher fold induction in R. amphibia.
In Arabidopsis, transcripts for CXXS1 (At1g11530), Ala:glyoxylate aminotransferase3 (At2g38400), and PPDK (At4g15530) were significantly up-regulated by submergence stress, in contrast with the results of Lee et al. (2011). Notably, that study (Lee et al., 2011) was done under dark submerged conditions and compared with dark controls, while the light control at the start of the treatment showed much lower expression values for these three genes (Supplemental Table S3), pointing to the fact that these genes are induced by a decrease in light intensity, independent of a decrease in oxygen concentrations. It can be speculated that transcript profiles during submergence, and thereby tolerance, can be considerably affected by light conditions during submergence.
DISCUSSION
The results presented here demonstrate the successful use of Arabidopsis gene chips in profiling the flooding-induced transcriptome of closely related Rorippa species. The difference between R. amphibia and R. sylvestris in the degree of submergence tolerance (Fig. 1) could be explained by some quantitative differences in gene expression in the roots after short-term submergence. Furthermore, a direct comparison of these extremely submergence-tolerant Rorippa species with the relatively intolerant Arabidopsis allowed the identification of potential tolerance-related genes.
Submergence-Regulated Genes in Rorippa Species Roots Included Many Known Hypoxia-Regulated Genes
An overview of gene regulation showed some similarity between our results obtained for Rorippa species and those found for Arabidopsis and rice (Klok et al., 2002; Branco-Price et al., 2005, 2008; Liu et al., 2005; Loreti et al., 2005; Lasanthi-Kudahettige et al., 2007; Mustroph et al., 2009; van Dongen et al., 2009). Notably, genes involved in energy-demanding processes such as cell wall precursor or fatty acid synthesis were repressed upon submergence, whereas glycolysis and fermentation genes were induced (Supplemental Tables S1 and S2). Furthermore, transcript abundance and activity of the hypoxia marker gene ADH1 were also clearly induced by the 24-h submergence treatment (Table I; Fig. 5). These data suggest that (1) submergence treatment resulted in hypoxia in the roots of the Rorippa species genotypes and (2) species widely differing in submergence tolerance up-regulate a common set of core hypoxia-responsive genes, as demonstrated previously (Christianson et al., 2010; Mustroph et al., 2010; Narsai et al., 2011). The observation that many of these genes were induced similarly in all three species despite differing tolerance (cluster 10 of Supplemental Table S3) demonstrates that the core hypoxic response, although required for plant survival, does not determine the specific tolerance of Rorippa species.
Regulation of Carbohydrate Respiration Might Underlie the Tolerance Differences between R. amphibia and R. sylvestris
Carbohydrate reserves are important for survival when plants experience an energy crisis that ensues from complete submergence, particularly when escape from (deep) floodwater is impossible and the rates of carbohydrate production under water are low, as was the case here.
Transcript levels of SUS1 and SUS4 were significantly increased after 24 h of submergence in both species (Table I; Fig. 5), and the level of gene induction was greater in R. amphibia than in R. sylvestris. It is known that SUS genes are induced by low oxygen levels in many plant species (Bailey-Serres and Voesenek, 2008), and out of the six SUS genes in Arabidopsis, it was also SUS1 and SUS4 that were found to be induced by low oxygen stress (Baud et al., 2004). Moreover, mutant studies have demonstrated that SUS genes are necessary for tolerance to soil flooding in Arabidopsis (Bieniawska et al., 2007) and tolerance to hypoxia in potato (Solanum tuberosum; Biemelt et al., 1999). The Suc degradation pathway mediated by SUS and UDP-Glc pyrophosphorylase is less energy consuming than the invertase-mediated Suc degradation pathway (Drew, 1997; Bailey-Serres and Voesenek, 2008; Huang et al., 2008). Notably, a putative invertase inhibitor (At5g62350) was more induced in R. sylvestris than in R. amphibia (Supplemental Table S1). Greater induction of SUS genes in shoots of intolerant rice varieties was associated with faster elongation and carbohydrate depletion, probably causing its more rapid mortality (Fukao et al., 2006). Similarly, it is possible that in R. amphibia, which also shows strong underwater shoot elongation (Akman et al., 2012), the greater mortality is associated with more carbohydrate use through the SUS and INVERTASE pathways. After 1 week of submergence, significantly higher amounts of total soluble sugar remained in R. sylvestris roots (Fig. 2). Greater depletion of root carbohydrates in R. amphibia could also indicate a remobilization of these reserves to fuel the shoot elongation in this escaping species. Stem growth during anaerobic conditions in pondweed (Potamogeton distinctus) was associated with enhanced starch consumption in underground turions (Harada and Ishizawa, 2003). Thus, carbohydrate translocation to stems could provide the required substrates for energy production to mediate shoot extension growth. In pondweed, elongation under anoxic conditions was accompanied by a stimulation of glycolysis (Sato et al., 2002). Genes involved in starch breakdown were also induced to a greater degree in intolerant rice varieties compared with the more tolerant SUB1 rice (Fukao et al., 2006), but we did not observe any significant treatment effects on amylase genes in our study (Supplemental Table S1). Accordingly, submergence did not affect root starch content after 24 h in either species, and both recorded similar starch contents after 7 d of submergence (Fig. 2). Under low-oxygen conditions, the breakdown products of starch and/or Suc provide substrates for glycolysis and fermentation.
Our analysis further showed that, besides carbohydrate cleavage enzymes, fermentative enzymes (ADH1 and PDC2) were also more strongly induced in R. amphibia than in R. sylvestris (Fig. 5; Table I). Accordingly, higher ADH activity was recorded in submerged R. amphibia roots than in R. sylvestris (Fig. 5B). Although levels of the fermentation end products ethanol and lactate were not higher in submerged Rorippa species roots (Supplemental Fig. S4), the loss of these metabolites into the root medium is very likely (Mustroph and Albrecht, 2007). Although mutant and overexpression studies have shown that PDC and ADH genes are important for low-oxygen stress tolerance (Ismond et al., 2003), the level of induction of these genes is not directly correlated with tolerance. Rather, it has been suggested that tolerance to low oxygen levels requires a tight balance between energy use for growth and other processes, on the one hand, and the generation of ATP through glycolysis and fermentation, on the other.
The measurements of glycolytic intermediates showed accumulations of Glc-6-P and Fru-6-P in roots of R. sylvestris after 24 h of submergence treatment, suggesting a possible inhibition of subsequent consumption processes. Although the accumulation of these metabolites could also be attributed to increased starch breakdown, this can be ruled out, considering that there were no significant changes in starch content after 24 h (Fig. 2). The intermediates phosphoenolpyruvate and pyruvate showed a similar, but not significant, trend. Remarkably, the ATP content was less reduced in R. sylvestris than in R. amphibia under submergence (Fig. 6E), despite the slower metabolism of the first species. This might indicate the higher energy demand of elongation growth in R. amphibia in comparison with R. sylvestris and, therefore, hints at the benefit for the quiescence strategy if elongation growth does not reach the oxygen-rich air zone.
Tolerant SUB1 rice, using the quiescence strategy, showed higher mRNA levels and enzyme activities of PDC and ADH during submergence than intolerant rice without SUB1, but remarkably, the level of ethanol production was lower in SUB1 rice (Fukao et al., 2006), possibly mediated by the lower induction of Suc synthases and amylases and other unknown factors. These data illustrate that mRNA levels and in vitro enzyme activities are not necessarily correlated with the rate of an enzymatic process in planta. The contrasting observations in fermentative enzyme induction of quiescent SUB1 rice and quiescent R. sylvestris could be due either to species differences and/or to different organs analyzed in the two studies. Indeed, shoots were analyzed from rice, while our study on Rorippa species was focused on roots. Both organs could have very different responses to submergence, as already suggested before for Arabidopsis (Ellis et al., 1999; Mustroph et al., 2009). Therefore, while the expression differences between the two Rorippa species, especially of SUS4, could perhaps be explained in terms of contrasting escape and quiescence strategies, it is premature to link this to differences in submergence tolerance. In addition, it is also not known how the different induction of glycolysis and fermentation genes in the two Rorippa species is regulated at the molecular level.
Submergence-Induced Induction of Photosynthesis Genes in Rorippa Species Roots
One remarkable observation was the strong induction of genes associated with the light-harvesting complexes, particularly in R. amphibia (Fig. 4), which could imply the presence and/or formation of chloroplasts in belowground tissues. Both Rorippa species are root sprouters, and in older plants, the rhizomes contain numerous buds competent to form green shoots (see “Materials and Methods”). Perhaps these tissues already possess chloroplasts, although in our experimental setup, no root sprouting was observed. It is known that several aerenchymatous species, including rice, form functional chloroplasts in the roots after prolonged illumination (Armstrong and Armstrong, 1994; Jackson and Armstrong, 1999; Rich et al., 2011). Furthermore, Arabidopsis roots contain chloroplasts and express photosynthetic genes in the root cortex (Dinneny et al., 2008). Roots may have been illuminated in our experimental setup as well, particularly near the surface of the pots, but this can be neglected because this exposure to light would be similar in both air and submerged conditions. In Arabidopsis, photomorphogenesis in roots is normally repressed, but in certain photomorphogenesis mutants, roots appear green because chloroplasts have developed (Chory et al., 1989; Oyama et al., 1997). What caused the strong induction of these photosynthesis genes after 24 h of submergence is not immediately clear. One possibility is sugar starvation, a well-known signal stimulating photosynthesis genes (Sheen, 1990; Baena-González et al., 2007). If so, this would suggest that sugar starvation was more severe in R. amphibia than in R. sylvestris. However, marker genes for energy or sugar starvation were not more strongly induced in R. amphibia than in R. sylvestris.
Comparison with Submergence-Intolerant Arabidopsis Reveals a Unique Complement of Submergence-Induced Genes in Roots
The close relatedness of Rorippa species to Arabidopsis allowed a direct comparison of root transcriptome profiles for these highly submergence-tolerant species with Arabidopsis, which is relatively sensitive to complete submergence. Arabidopsis accessions show a considerable variation in their tolerance to submergence (Vashisht et al., 2011), but even the most tolerant accession (C24) was found to have an median lethal time of only 20 d (submerged in the light). This is in contrast with median lethal time values of 75 d for R. amphibia and more than 110 d for R. sylvestris under similar conditions of submergence. Gene clusters that are differentially regulated between these two species, therefore, are potential tolerance-related genes. Here, cluster 22 and cluster 1 contain potential candidates for species differences within the Rorippa genus, while cluster 17 has genes differentially expressed between Rorippa species and Arabidopsis (Supplemental Table S3). Interestingly, these clusters contain a number of genes that are associated with the inorganic pyrophosphate (PPi)-dependent alternative pathways of phosphorylation reactions in primary metabolism (PFPA2 and PFPB2). These were discussed previously as having a potential benefit for energy metabolism in plants under low-oxygen stress (Mustroph et al., 2005; Bailey-Serres and Voesenek, 2008; Huang et al., 2008). Furthermore, rice genotypes differing in submergence tolerance [M202 versus M202(SUB1)] also express PFP genes at different levels under submerged conditions (Mustroph et al., 2010). Other differentially expressed genes are associated with photosynthesis or amino acid metabolism, but their role in low-oxygen survival remains unclear so far. Interestingly, transcription factors of the ERF group VII family that have been associated with submergence tolerance in rice (Xu et al., 2006; Hattori et al., 2009) and that are involved in low-oxygen signaling (Gibbs et al., 2011; Licausi et al., 2011) had very low expression in this data set under all conditions and were also not induced by submergence (Supplemental Table S1). However, one cannot rule out orthologs that could not be detected with the Arabidopsis microarray GeneChip and differences in posttranslational regulation. Indeed, RAP2.12 was removed from the data set during the probe selection process.
The observed differences between Rorippa species and Arabidopsis could be caused by a number of factors. First, they could be caused by species-specific adaptational response differences, the most interesting factor, which are most likely different from the core hypoxic response concept mentioned before. This could involve, for example, an energy-saving quiescence strategy, including the use of PPi-using enzymes and adaptations of the photosynthetic apparatus. Second, technical problems due to variation in Rorippa species gene sequences in comparison with Arabidopsis cannot be excluded completely and could be relevant for some genes that are hardly expressed in Rorippa species as measured by Arabidopsis ATH1 chips. Third, the expression differences of some genes in the two genera could be also attributed to differences in oxygen concentrations in the roots during the submergence treatment, which is dependent on temperature and light, on the duration of the treatment, or on variation in root and shoot anatomy (e.g. aerenchyma). Therefore, it is essential to be aware that transcriptome profiles can be influenced by factors such as the conditions of submergence (dark versus light) and the developmental stage at which plants are submerged. Since the Arabidopsis microarray data set used for comparisons here were from plants submerged in the dark, we repeated submergence experiments on Arabidopsis plants with conditions similar to those used for the Rorippa species microarrays. Gene expression measurements of selected genes from the differentially expressed cluster (Fig. 7) showed that even in the roots, light conditions during submergence affected transcript profiles to some extent. For example, the gene for PPDK, identified as being induced only in submerged Rorippa species roots, was highly induced in Arabidopsis roots as well when illuminated control conditions were used as a reference. However, these experiments confirmed that some genes (PFPA2, PFPB2, PAO, and HEMF1) that are strongly induced under submergence in Rorippa species are not induced in Arabidopsis, making them interesting candidates for future studies.
CONCLUSION
In this study, we have successfully exploited the close relatedness of wild Rorippa species with Arabidopsis in order to utilize ATH1 chips for profiling the submergence-induced transcriptome of these two species. Our results suggest that differences in submergence tolerance between R. amphibia and R. sylvestris, adapting an escape and a quiescent survival strategy, respectively, could be related to differences in the fermentation capacity and carbohydrate consumption in the roots.
We also compared the molecular responses to submergence of Rorippa species with that of Arabidopsis. The commonalities in short-term gene expression responses between Rorippa species, and their common differences from the flooding-intolerant Arabidopsis, suggest important tolerance genes and pathways, such as the use of alternative PPi-dependent pathways. Future studies of gene expression under prolonged submergence of roots and shoots and the development and analyses of transgenic plants expressing candidate genes from Rorippa species will further unravel the molecular basis of the contrasting tolerance responses between these two genera.
MATERIALS AND METHODS
Plant Material
Rorippa amphibia and Rorippa sylvestris are clonal root sprouters that can be propagated using rhizome fragments. The origin of the tetraploid genotypes used in this study has been described previously (Stift et al., 2008). An R. amphibia rhizome was collected along the Yssel River near Doesburg, The Netherlands (N: 52°01'25'', E: 06°08'42''), and an R. sylvestris rhizome was collected along the Rhine River near Millingerwaard, The Netherlands (N: 51°52'02'', E: 06°08'42''), in 2003 and maintained in a greenhouse. Rhizome fragments of each genotype were surface sterilized by soaking in 10% (v/v) commercial bleach for 5 min, rinsed twice in deionized water, and transferred to petri dishes containing 2.2 g L−1 Murashige and Skoog salts (Sigma-Aldrich) at pH 5.6 with 0.8% purified agar (Hispanagar). The rhizomes were left to sprout for 11 to 14 d in a growth cabinet (MLR-350; Sanyo) with a 16-h light period (photosynthetically active radiation [PAR] of 36 µmol m−2 s−1, 20°C) and 8 h of dark at 16°C. For each genotype, similar-sized single sprouts were cut off and transferred to pots containing sterilized sand (0.5- to 1.0-mm grain size; Filcom) and saturated with complete nutrient solution (0.1 g L−1 Peter’s Professional 20:10:20 General Purpose; Scotts Europe). Plants were grown in a glasshouse under natural daylight with additional lighting provided by 600W SON-T lamps (Philips). Plants received additional nutrients or were transferred to bigger pots as indicated below and were watered regularly. Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia-0) were germinated and grown as described by Vashisht et al. (2011).
Submergence Tolerance
R. sylvestris and R. amphibia shoots were grown in the glasshouse on mesh pots (55 mm diameter) containing sand for 2 weeks, transferred to 0.5-L pots containing potting soil, and after an additional 10 d were transferred again to 3-L pots containing potting soil. Plants were 52 d old when the submergence treatment started in July 2007. Mature plants were used, since these have the capacity to show stem elongation, whereas younger plants will not form stems. The soil in each pot was covered with a layer of sand to keep it under the water. Plants were fully submerged in outdoor concrete basins of length × width × depth = 400 cm × 100 cm × 100 cm filled with tap water for up to 3 months. Plants were randomly assigned to one of nine basins and one of 30 positions within each basin. The basins were covered with shade cloth, reducing irradiance at the plant level by approximately 65% to mimic a deep flooding event. PAR was measured continuously by an underwater light sensor connected to a data logger (LI-1400; Licor). Water was periodically pumped from the bottom of the basins using a submersible pump, passed through a UV-C filter (Velda), and released back into the basin in order to prevent the buildup of algae. Ten plants of each species were removed from the basins at six time points starting from 39 d after submergence and every 2 weeks thereafter. Survival was scored as the percentage of plants that formed leaves after a recovery period of 2 weeks in the glasshouse. Seven control plants of each genotype that were kept in air in a similar basin all survived the entire experimental period, demonstrating that mortality was the result of submergence.
Sugar and Starch Analysis
In the submergence experiments, plants were grown in pots of 8.5 cm diameter filled with river sand and containing nine grains of slow-release fertilizer (Osmocote Plus 15, Peters Professional; Scotts Europe) per pot. Individual plants were submerged in plastic opaque tubs that were disinfected, rinsed, and filled with tap water to the brim 1 d prior to the start of the experiment. Control (air) plants were placed in empty tubs. Water temperature was approximately 20°C. The water was not filtered or refreshed during the experiments. Submergence experiments were performed in climate-controlled walk-in growth chambers maintained at 20°C, 200 μmol m−2 s−1 PAR (16-h photoperiod), and a relative humidity of 70%. At specific time points after the start of the submergence treatment, plants were taken out of the tubs, and roots were quickly rinsed and separated from the shoot and immediately frozen in liquid nitrogen. Roots harvested from a single plant constituted a biological replicate. Freeze-dried, ground tissue was suspended in 500 μL of 0.83 m perchloric acid, vortexed, and then centrifuged for 15 min at 4°C and 13,000 rpm. Bicin (100 μL, 1 m) and 80 μL of potassium hydroxide (4 m) were added to the resulting supernatant followed by centrifugation for 10 min at 4°C and 13,000 rpm. The supernatant was then used for the determination of total soluble sugars (Glc, Fru, and Suc) using a commercial kit (K-SUFRG; Megazyme) according to the manufacturer’s instruction. The pellet was used for starch determination using a commercial starch determination kit (K-TSTA; Megazyme). Ethanol and lactate were determined as described by Bergmeyer (1983). Briefly, lactate was measured in 1 m Gly buffer, pH 9.5, with the addition of 0.4 m hydrazine, 5 mm EDTA, 2.5 mm NAD, and 5 units of l-LDH. Ethanol was measured in 21 mm Gly buffer, pH 8.7, containing 75 mm sodium pyrophosphate, 75 mm semicarbazid, 1 mm NAD, and 5 units of ADH.
Enzyme Activity Measurements
Rorippa species and Arabidopsis plants were grown as described above for the sugar analysis, and roots were harvested after 24 h of submergence treatment. Enzymes were extracted as described by Mustroph et al. (2006), and activities of ADH, PFP, and PFK were measured as described by Mustroph and Albrecht (2003).
Submergence Experiment for Gene Expression
Rhizome segments of R. amphibia and R. sylvestris were aseptically transferred to 9-cm-diameter petri dishes on sterile 0.8% agar containing 2.2 g L−1 Murashige and Skoog salts (Sigma-Aldrich), pH 5.6, and placed for 11 d in a growth chamber (MLR-350; Sanyo) at 16 h of light, 20°C, 36 μmol quanta m−2 s−1 and 8 h of dark, 16°C. Approximately 50 meristems of each genotype were carefully excised from the rhizomes, planted on fine river sand (0.5- to 1-mm grain size; Filcom) in 8.5-cm-diameter plastic pots, and placed in a greenhouse under natural daylight. Temperature was maintained at 21°C and relative air humidity at 77%. Irradiance at the plant level during the growing period was logged continuously at half-hourly intervals with a quantum sensor (Li-1400; Licor) and was 116 μmol quanta m−2 s−1 on average, with maxima occasionally reaching 870 μmol quanta m−2 s−1. Pots remained in a layer of 1 g L−1 nutrient solution (Peat Lite Special, Peter’s Professional; Scotts Europe) for 4 d, after which they were kept in a layer of rainwater. After 16 d, when approximately six leaves were visible, a uniform selection of 18 plants of each genotype was made, half of which were randomly assigned to a control group and half of which were subjected to the submergence treatment. At noon, individual plants were placed in plastic 16-L buckets that were either empty for air controls or filled with clear rainwater for the submerged group. Water temperature was 21°C. After 24 h of treatment, the entire root system was separated from the shoot, quickly rinsed in water, and frozen in liquid N2. Groups of three randomly picked replicate samples were bulked, resulting in three independent biological replicates, and stored at −70°C until further use. A total of 12 arrays were used to interrogate the transcriptome of roots from R. amphibia and R. sylvestris plants submerged for 24 h or kept in air as a control. RNA extracted from these samples was used for qRT-PCR and microarray hybridizations. Arabidopsis plants grown according to the method of Vashisht et al. (2011) were used for submergence experiments in conditions similar to those described above. After 24 h of submergence, plants were taken out of the water and roots were quickly cleaned, separated from the shoot, and flash frozen in liquid N2.
RNA and gDNA Isolation and Microarray Hybridization
gDNA was isolated from 100 mg of fresh leaf material using the DNeasy Plant Mini Kit (Qiagen). gDNA (500 ng) was labeled and amplified by using the Bioprime DNA labeling kit (Invitrogen) and analyzed with a Nanodrop spectrophotometer and Agilent Bioanalyzer, after which 6.9 µg of labeled gDNA probe of each species was hybridized to an ATH1-121501 GeneChip (Affymetrix). Complete information about this platform can be found at the Gene Expression Omnibus Web site (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL198). Total RNA was extracted from 100 mg of tissue using the RNeasy Plant Mini Kit (Qiagen). RNA quality was assessed by electrophoresis on agarose gels, Nanodrop spectrometry, and Agilent Bioanalyzer. Complementary RNA probe generation, microarray hybridization, washing, and scanning were performed as described by the manufacturer’s protocols (Affymetrix). The data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through Gene Expression Omnibus Series accession number GSE13641 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13641).
gDNA-Based Probe Filtering and Microarray Data Analysis
The CEL files from hybridizations with gDNA were used to create custom CDF files for the different species, based on the original Arabidopsis ATH1 CDF file. According to the procedure described by Hammond et al. (2005, 2006), probe masks were created with the easy_script.pl file obtained from http://affymetrix.arabidopsis.info/xspecies/, testing different thresholds. In this manner, individual probes were removed that had lower gDNA hybridization intensity than the threshold. Out of 11 probes in a probe set interrogating each gene, up to 10 probes could be removed, or the entire set was removed when all 11 probes fell below the threshold. Using these data, plots were made showing the number of probes and probe sets retained at different gDNA hybridization thresholds (Supplemental Fig. S1). Based on these plots, a threshold was empirically determined at which the number of removed entire probe sets was minimized (because in those cases, the corresponding genes can no longer be interrogated) but the number of removed individual probes was maximized. Based on this evaluation, we decided to use a threshold of 80.
For the final CDF files for Rorippa species, the original Arabidopsis CDF file was first masked with the hybridized R. amphibia gDNA CEL file and subsequently masked with the R. sylvestris gDNA CEL file. The custom CDF file was then used to produce an R-usable library with the “makecdfenv” package in R. Subsequently, CEL files from the mRNA hybridization Affymetrix chips were processed by use of the R program and Bioconductor packages (Gentleman et al., 2004) and the custom-made CDF library. Robust multichip average (RMA) normalization was performed using the default settings of the corresponding R function (Irizarry et al., 2003). Analysis of differentially expressed genes between two different conditions was performed with the Limma package using the RMA-normalized expression values (Smyth, 2004). The Benjamini and Hochberg method was selected to adjust P values for multiple testing and to determine false discovery rates (FDRs; Benjamini and Hochberg, 1995). As a confidence threshold, an FDR of less than 0.05 and a fold change cutoff of 2.0 was used.
Furthermore, the Rorippa species expression data were compared with expression data from submerged Arabidopsis plants (Lee et al., 2011). To avoid systematic errors, we treated the original Arabidopsis microarray data exactly as the Rorippa species data by first creating a custom CDF file with the help of a CEL file derived from Arabidopsis gDNA and then following the exact same procedure as for the Rorippa species data set. To more easily compare the response to submergence between species, k-means clustering was performed with all genes that showed differential expression due to submergence as determined by the Limma analysis described above. Unlogged RMA-normalized expression values (means of replicates) were first transformed by mean centering and subsequent division by the sum of the expression values separately for each genus. These data were then used for clustering in R with the function “PAM.” After testing several numbers of clusters, we decided to use 26 clusters for the data set.
Overrepresentation analysis of GO categories was performed for specific gene lists obtained by the Limma and k-mean clustering analyses as described by Horan et al. (2008). In summary, the Arabidopsis gene-to-GO mappings from The Arabidopsis Information Resource (available at http://geneontology.org; downloaded November 13, 2010) were used for these analyses. The hypergeometric distribution was applied to test gene sets for the overrepresentation of GO terms. To perform this test, the GOHyperGAll function was used in R, which computes for a given sample population of genes the enrichment test for all nodes in the GO network and returns raw and adjusted P values. As an adjustment method for multiple testing, it uses the Bonferroni method according to Boyle et al. (2004). GO categories with an adjusted P < 0.05 were deemed significantly enriched. PCA was performed using the R software and prcomp function based on the entire data set. Microarray expression data of a selected set of genes were validated using qRT-PCR (Supplemental Table S4).
Cloning and qRT-PCR
Cloning was performed using standard protocols (Sambrook and Russell, 2001). Partial genomic sequences of orthologs of Arabidopsis genes were obtained from R. amphibia and R. sylvestris using primers designed based on the Arabidopsis sequences. Complementary DNA (cDNA) was used to PCR amplify the respective Rorippa species genes, and PCR products were directly sequenced. Since the Rorippa species genotypes used in this study are tetraploid, up to four alleles may be present. Based on the obtained sequences, primers were designed that detected all present alleles in both species and amplified approximately 150 bp for use in qRT-PCR with primer3 software (http://primer3.ut.ee). Primer sequences for the Arabidopsis and Rorippa species genes studied using real-time reverse transcription-PCR are given in Supplemental Table S5. RNA samples were treated with DNase (DNAfree; Ambion) to remove any gDNA, and cDNA was synthesized from 500 ng of RNA using 50 ng of random hexamers (Invitrogen) and 100 units of SuperScript III reverse transcriptase (Invitrogen) in a 20-µL reaction according to the manufacturer’s instructions. Quantitative PCR was performed using an ABI 7500 in a reaction mixture (20 µL) containing 10 µL of 2× SYBR Green (Platinum SYBR Green Supermix gPCR UDG; Invitrogen), 0.3 µL of 10 µm of each primer, 0.04 µL of a one-tenth dilution of 50× ROX reference dye, and 10 ng of cDNA. Relative transcript levels were corrected against ACTIN2 transcript levels (for Rorippa species genes) and AtUBQ10 (for Arabidopsis) and calculated using the ΔΔCt method (Livak and Schmittgen, 2001).
Measurement of Metabolite Content
Rorippa species plants were grown as described above, and roots were harvested after 24 h of submergence treatment. Metabolites were extracted with perchloric acid and subsequently neutralized with KOH as described previously (Mustroph et al., 2006). Adenylate concentrations in the extracts were determined with the luciferase system as described by Wulff and Döppen (1985) for ATP and by Hampp (1985) for ADP. Other metabolites were determined after Burrell et al. (1994) with modifications. Glc-6-P and Fru-6-P were measured in 100 mm HEPES, pH 7.5, with the addition of 1 mm MgCl2 and 0.4 mm NADP. The reaction was started by adding 10 units of Glucose-6-phosphate dehydrogenase for Glc-6-P and 7 units of phosphoglucose isomerase for Fru-6-P. Pyruvate and phosphoenolpyruvate were measured in 100 mm HEPES, pH 7.5, with the addition of 2 mm MgCl2, 0.2 mm ADP, and 0.1 mm NADH. The reaction was started with 10 units of LDH for pyruvate and 1 unit of pyruvate kinase for phosphoenolpyruvate. Measurements were done with eight replicates per species and treatment.
Sequence data from this article can be found in the GenBank database under the following accession numbers: for R. amphibia, HB1 (KF042825), ADH1 (JQ582800), GH (KF042827), SUS4 (KF042829), PAO (KF042831), HEMF (KF042833), LDH1 (KF042836), PDC2 (KF042838), PFPB2 (KF042842), PFPA1 (KF042844), PFPA2 (KF042846), PFK6 (KF042848), and PPDK (KF042850); for R. sylvestris, HB1 (KF042826), ADH1 (JQ582801), GH (KF042827), SUS4 (KF042828), PAO (KF042832), HEMF (KF042834), CXXS1 (KF042835), LDH1 (KF042837), PDC2 (KF042839), PFPB2 (KF042843), PFPA1 (KF042845), PFPA2 (KF042847), PFK6 (KF042848), and PPDK (KF042851).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rorippa species gDNA hybridization-based probe selection of ATH1 GeneChips.
Supplemental Figure S2. Histogram of the number of probes per probe set.
Supplemental Figure S3. Box plots showing the average R. amphibia and R. sylvestris gDNA hybridization intensity by the number of probes per probe set.
Supplemental Figure S4. Submergence-induced changes in fermentation end products in Rorippa species roots.
Supplemental Figure S5. Submergence-induced changes in the ATP-ADP ratio in Rorippa species.
Supplemental Figure S6. Enzyme activities in roots of Rorippa species or Arabidopsis plants that were treated with submergence for 24 h or kept in aerated soil.
Supplemental Table S1. Microarray data for Rorippa species genes hybridized to Arabidopsis ATH1 microarray chips.
Supplemental Table S2. GO term enrichment analysis of significantly enriched gene groups identified by pairwise comparisons from Supplemental Table S1.
Supplemental Table S3. K-means cluster analysis of microarray data from Rorippa species and Arabidopsis.
Supplemental Table S4. Confirmation of microarray expression data of selected Rorippa species genes by qRT-PCR.
Supplemental Table S5. Sequences of primer combinations (5′–3′) for the Arabidopsis and Rorippa species genes studied using real-time reverse transcription-PCR.
Acknowledgments
We gratefully acknowledge Neil Graham and Sean May (Nottingham Arabidopsis Stock Centre) for assistance with the probe filtering method and comments on the manuscript. We thank Han Rauwerda (Microarray Department Amsterdam) for providing Supplemental Figures S2 and S3 and for useful discussions. We thank Floyd Wittink and other members of the Microarray Department Amsterdam for performing the microarray hybridizations. Rob Bregman, Peter Kuperus, Amit V. Bhikarie, and Ludek Tikovsky (University of Amsterdam, The Netherlands) provided excellent technical assistance.
Glossary
- ERF
Ethylene Response Factor
- gDNA
genomic DNA
- PCA
principal component analysis
- qRT
quantitative reverse transcription
- GO
Gene Ontology
- PPi
inorganic pyrophosphate
- PAR
photosynthetically active radiation
- RMA
robust multichip average
- FDR
false discovery rate
- cDNA
complementary DNA
References
- Akman M, Bhikharie AV, McLean EH, Boonman A, Visser EJ, Schranz ME, van Tienderen PH. (2012) Wait or escape? Contrasting submergence tolerance strategies of Rorippa amphibia, Rorippa sylvestris and their hybrid. Ann Bot (Lond) 109: 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J, Armstrong W. (1994) Chlorophyll development in mature lysigenous and schizogenous root aerenchymas provides evidence of continuing cortical cell viability. New Phytol 126: 493–497 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. (2010) Life in the balance: a signaling network controlling survival of flooding. Curr Opin Plant Biol 13: 489–494 [DOI] [PubMed] [Google Scholar]
- Baud S, Vaultier M-N, Rochat C. (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55: 397–409 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bergmeyer H (1983) Methods of Enzymatic Analysis. Verlag Chemie, Weinheim, Germany [Google Scholar]
- Biemelt S, Hajirezaei MR, Melzer M, Albrecht G, Sonnewald U. (1999) Sucrose synthase activity does not restrict glycolysis in roots of transgenic potato plants under hypoxic conditions. Planta 210: 41–49 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Bleeker W, Hurka H. (2001) Introgressive hybridization in Rorippa (Brassicaceae): gene flow and its consequences in natural and anthropogenic habitats. Mol Ecol 10: 2013–2022 [DOI] [PubMed] [Google Scholar]
- Boyle EI, Weng SA, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. (2004) GO:TermFinder: open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot (Lond) 96: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley MR, White PJ, Hammond JP, Graham NS, Bowen HC, Emmerson ZF, Fray RG, Iannetta PP, McNicol JW, May ST. (2008) Evidence of neutral transcriptome evolution in plants. New Phytol 180: 587–593 [DOI] [PubMed] [Google Scholar]
- Burrell MM, Mooney PJ, Blundy M, Carter D, Wilson F, Green J, Blundy KS, Ap Rees T. (1994) Genetic manipulation of 6-phosphofructokinase in potato tubers. Planta 194: 95–101 [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW. (2010) Comparisons of early transcriptome responses to low-oxygen environments in three dicotyledonous plant species. Plant Signal Behav 5: 1006–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Drew MC. (1997) Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol 48: 223–250 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov V, Dowdle J, Smirnoff N, Ford-Lloyd B, Newbury HJ, Macnair MR. (2006) Comparison of gene expression in segregating families identifies genes and genomic regions involved in a novel adaptation, zinc hyperaccumulation. Mol Ecol 15: 3045–3059 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.1–R80.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, et al. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bowen HC, White PJ, Mills V, Pyke KA, Baker AJM, Whiting SN, May ST, Broadley MR. (2006) A comparison of the Thlaspi caerulescens and Thlaspi arvense shoot transcriptomes. New Phytol 170: 239–260 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, Craigon DJ, Higgins J, Emmerson ZF, Townsend HJ, White PJ, May ST. (2005) Using genomic DNA-based probe-selection to improve the sensitivity of high-density oligonucleotide arrays when applied to heterologous species. Plant Methods 1: 10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp R (1985) ADP, AMP: luminometric method. In HU Bergmeyer, ed, Methods of Enzymatic Analysis. Verlag Chemie, Weinheim, Germany, pp 371–379 [Google Scholar]
- Harada T, Ishizawa K. (2003) Starch degradation and sucrose metabolism during anaerobic growth of pondweed (Potamogeton distinctus A. Benn.) turions. Plant Soil 253: 125–135 [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M, Girke T. (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147: 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Chou MY, Peng HP, Chou SJ, Shih MC. (2011) Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS ONE 6: e28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH. (2008) Does anoxia tolerance involve altering the energy currency towards PPi? Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismond KP, Dolferus R, de Pauw M, Dennis ES, Good AG. (2003) Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol 132: 1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB, Armstrong W. (1999) Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol 1: 274–287 [Google Scholar]
- Jonsell B. (1968) Studies in the north-west European species of Rorippa S. Str. Symb Bot Ups 19: 1–221 [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC. (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152: 1674–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga JE, Herman MA, Ouborg NJ, Johnson L, Breitling R. (2007) Microarray challenges in ecology. Trends Ecol Evol 22: 273–279 [DOI] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES. (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, Whelan J. (2009) Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol 149: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P. (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J. (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T. (2001) Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol Evol 16: 693–700 [Google Scholar]
- Mommer L, Lenssen JPM, Huber H, Visser EJW, De Kroon H. (2006) Ecophysiological determinants of plant performance under flooding: a comparative study of seven plant families. J Ecol 94: 1117–1129 [Google Scholar]
- Mustroph A, Albrecht G. (2003) Tolerance of crop plants to oxygen deficiency stress: fermentative activity and photosynthetic capacity of entire seedlings under hypoxia and anoxia. Physiol Plant 117: 508–520 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Albrecht G. (2007) Fermentation metabolism in roots of wheat seedlings after hypoxic pre-treatment in different anoxic incubation systems. J Plant Physiol 164: 394–407 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Albrecht G, Hajirezaei M, Grimm B, Biemelt S. (2005) Low levels of pyrophosphate in transgenic potato plants expressing E. coli pyrophosphatase lead to decreased vitality under oxygen deficiency. Ann Bot (Lond) 96: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Boamfa EI, Laarhoven LJ, Harren FJ, Albrecht G, Grimm B. (2006) Organ-specific analysis of the anaerobic primary metabolism in rice and wheat seedlings. I. Dark ethanol production is dominated by the shoots. Planta 225: 103–114 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Rocha M, Geigenberger P, Whelan J, van Dongen JT. (2011) Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol 190: 472–487 [DOI] [PubMed] [Google Scholar]
- Nettleton D. (2006) A discussion of statistical methods for design and analysis of microarray experiments for plant scientists. Plant Cell 18: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SM, Ludwig M, Pedersen O, Colmer TD. (2011) Aquatic adventitious roots of the wetland plant Meionectes brownii can photosynthesize: implications for root function during flooding. New Phytol 190: 311–319 [DOI] [PubMed] [Google Scholar]
- Sachs MM, Subbaiah CC, Saab IN. (1996) Anaerobic gene expression and flooding tolerance in maize. J Exp Bot 47: 1–15 [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sato T, Harada T, Ishizawa K. (2002) Stimulation of glycolysis in anaerobic elongation of pondweed (Potamogeton distinctus) turions. J Exp Bot 53: 1847–1856 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Song BH, Windsor AJ, Mitchell-Olds T. (2007) Comparative genomics in the Brassicaceae: a family-wide perspective. Curr Opin Plant Biol 10: 168–175 [DOI] [PubMed] [Google Scholar]
- Setter TL, Laureles EV. (1996) The beneficial effect of reduced elongation growth on submergence tolerance of rice. J Exp Bot 47: 1551–1559 [Google Scholar]
- Sheen J. (1990) Metabolic repression of transcription in higher plants. Plant Cell 2: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: e3. [DOI] [PubMed] [Google Scholar]
- Stift M, Luttikhuizen PC, Visser EJ, van Tienderen PH. (2008) Different flooding responses in Rorippa amphibia and Rorippa sylvestris, and their modes of expression in F1 hybrids. New Phytol 180: 229–239 [DOI] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eck WHJM, van de Steeg HM, Blom CWPM, de Kroon H. (2004) Is tolerance to summer flooding correlated with distribution patterns in river floodplains? A comparative study of 20 terrestrial grassland species. Oikos 107: 393–405 [Google Scholar]
- Vashisht D, Hesselink A, Pierik R, Ammerlaan JMH, Bailey-Serres J, Visser EJW, Pedersen O, van Zanten M, Vreugdenhil D, Jamar DCL, et al. (2011) Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol 190: 299–310 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Colmer TD, Pierik R, Millenaar FF, Peeters AJM. (2006) How plants cope with complete submergence. New Phytol 170: 213–226 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Sasidharan R. (2013) Ethylene—and oxygen signalling—drive plant survival during flooding. Plant Biol (Stuttg) 15: 426–435 [DOI] [PubMed] [Google Scholar]
- Wulff K, Döppen W (1985) ATP: luminometric method. In HU Bergmeyer, ed, Methods of Enzymatic Analysis. Verlag Chemie, Weinheim, Germany, pp 357–370 [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]