Map-based cloning of a maize prostrate-growth mutant links gravitropism with inflorescence development through auxin transport, auxin signaling, and response to light.
Abstract
Auxin is a plant hormone that plays key roles in both shoot gravitropism and inflorescence development. However, these two processes appear to be parallel and to be regulated by distinct players. Here, we report that the maize (Zea mays) prostrate stem1 mutant, which is allelic to the classic mutant lazy plant1 (la1), displays prostrate growth with reduced shoot gravitropism and defective inflorescence development. Map-based cloning identified maize ZmLA1 as the functional ortholog of LAZY1 in rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana). It has a unique role in inflorescence development and displays enriched expression in reproductive organs such as tassels and ears. Transcription of ZmLA1 responds to auxin and is repressed by light. Furthermore, ZmLA1 physically interacts with a putative auxin transport regulator in the plasma membrane and a putative auxin signaling protein in the nucleus. RNA-SEQ data showed that dozens of auxin transport, auxin response, and light signaling genes were differentially expressed in la1 mutant stems. Therefore, ZmLA1 might mediate the cross talk between shoot gravitropism and inflorescence development by regulating auxin transport, auxin signaling, and probably light response in maize.
Auxin was the first phytohormone identified in plants (Pennazio, 2002). It plays a major role in several physiological and developmental processes, including tropic responses, axillary meristem initiation, apical dominance, organogenesis, flowering, root elongation, and embryogenesis (Muday and DeLong, 2001; Woodward and Bartel, 2005; Petrásek and Friml, 2009). These biological functions of auxin are achieved through the coordinated actions of auxin biosynthesis, auxin transport, and auxin signal transduction. Auxin synthesis occurs in specific tissues, including young developing leaves and flowers, cotyledons, and the apices of shoots and roots (Ljung et al., 2001). Auxin biosynthesis can occur through four Trp-dependent and one Trp-independent pathways, depending on the specific tissue, developmental stage, and species (Benjamins and Scheres, 2008). The primary Trp-dependent pathway in Arabidopsis (Arabidopsis thaliana) is mediated by two enzymatic activities, YUCCA (YUC) and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS/TRYPTOPHAN AMINOTRANSFERASE RELATED. In this pathway, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS proteins convert Trp to indole-3-pyruvate and YUCs convert indole-3-pyruvate to indole-3-acetic acid (IAA), which is the primary endogenous auxin (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Zhao, 2012). Distribution of IAA is determined by local auxin biosynthesis and by directional cell-to-cell IAA transport, known as polar auxin transport (PAT). Auxin influx and efflux carriers are required for PAT in plants. In Arabidopsis, four auxin influx genes (AUX1, LAX1, LAX2, and LAX3) and eight auxin efflux carriers (PIN-FORMED1–PIN-FORMED8) that mediate PAT have been identified and characterized (Parry et al., 2001; Zazímalová et al., 2010). PAT is essential for creating the auxin maxima and the auxin gradient, which serve as the cellular basis for localized cell division, elongation, and differentiation (Friml, 2003; Blakeslee et al., 2005; Sieberer and Leyser, 2006; Teale et al., 2006; Zazímalová et al., 2010; Peer et al., 2011).
In response to gravity, plants direct shoots upward (negative gravitropism) and roots downward (positive gravitropism). Gravitropism consists of three conserved steps: gravity sensing, gravitropic signal transduction, and asymmetric growth of responsive cells (Strohm et al., 2012). PAT plays a central role in both negative and positive gravitropic responses in higher plants (Friml et al., 2002). In tobacco (Nicotiana tabacum) shoots, auxin was found to be asymmetrically distributed in the reaction site of gravitropic growth (Li et al., 1991). In both rice (Oryza sativa) and Arabidopsis, disruption of the PAT regulator LAZY1 leads to branch-spreading phenotypes with reduced shoot gravitropism (Li et al., 2007; Yoshihara and Iino, 2007; Yoshihara et al., 2013). Many studies have shown that PAT also plays a key role in the positive gravitropism of Arabidopsis roots (Friml, 2003). The gravitropic curvature is completely lost in the PAT influx carrier aux1 mutant roots (Bennett et al., 1996), and PAT efflux carrier pin3 mutant roots show reduced gravitropism (Friml et al., 2002). ATP-binding cassette/P-glycoprotein (ABCB/PGP) transporters mediate cellular auxin transport (Geisler et al., 2005; Geisler and Murphy, 2006; Mravec et al., 2008). PGP4, which catalyzes basipetal auxin transport in Arabidopsis, induces reduced gravitropic bending in pgp4 mutant roots (Terasaka et al., 2005). Moreover, other genes, such as the GOLVEN secretory peptides and the phosphatidylinositol monophosphate5-kinase2, regulate PAT by modifying auxin efflux carriers and participate in the positive gravitropic response in Arabidopsis roots (Mei et al., 2012; Whitford et al., 2012).
PAT is required for inflorescence development owing to its fundamental role in axillary meristem initiation (Cheng and Zhao, 2007; Barazesh and McSteen, 2008a). Maize (Zea mays) has two types of inflorescences: the male inflorescence (the tassel), which is located on the shoot apex, and the female inflorescence (the ear), which is produced from the axillary meristems several nodes below the tassel. The tassel develops from four types of axillary meristems: branch meristems, spikelet pair meristems (SPMs), spikelet meristems, and floral meristems. The ear arises from SPMs, spikelet meristems, and floral meristems. In these apical and axillary meristems, the auxin efflux carrier PIN1 orthologs in maize, ZmPIN1a and ZmPIN1b, are localized in the epidermal cell layer and implicated in inflorescence morphogenesis through PAT (Carraro et al., 2006; Gallavotti et al., 2008b). BARREN INFLORESCENCE2 (BIF2), which encodes an ortholog of the Arabidopsis Ser/Thr protein kinase PINOID, has a conserved role in PAT by regulating the subcellular location of PIN proteins (Carraro et al., 2006; McSteen et al., 2007; Skirpan et al., 2009). bif2 mutants show reduced kernels in the ear and decreased numbers of branches, spikelets, and florets in the tassel (McSteen and Hake, 2001). Semidominant bif1 mutants display similar ear and tassel phenotypes to bif2, which may result from reduced auxin transport in the axillary meristems in the inflorescence (Barazesh and McSteen, 2008b). BARREN STALK1 (BA1) is a basic helix-loop-helix transcription factor that is required for axillary meristem initiation in maize. Therefore, ba1 mutants have no ears, branches, spikelets, or florets (Gallavotti et al., 2004). BA1 may act both upstream and downstream of PAT by forming an auxin local gradient at the flanks of the inflorescence meristem (Wu and McSteen, 2007; Gallavotti et al., 2008b). In addition to PAT, auxin biosynthesis also regulates inflorescence development in maize. SPARSE INFLORESCENCE1, which encodes a flavin monooxygenase that catalyzes a rate-limiting step in Trp-dependent auxin synthesis and is similar to YUC genes of Arabidopsis, is required for the initiation of axillary meristems and lateral organs during both inflorescence and vegetative development (Gallavotti et al., 2008a). VANISHING TASSEL2, another auxin biosynthesis gene, plays a significant role in axillary meristem formation in the inflorescences (Phillips et al., 2011). In fact, a genetic analysis indicated that auxin transport and auxin synthesis may act synergistically to regulate inflorescence development in maize (Gallavotti et al., 2008b).
Despite the evidence that auxin is involved in both gravitropic responses and inflorescence development in plants, few regulators, if any, have been shown to cross talk gravitropism and inflorescence development in a given species. Here, we characterized the agravitropic maize mutant lazy plant1 (la1) and map-based cloned the ZmLA1 gene. ZmLA1 encodes a functional ortholog of LAZY1 in rice and Arabidopsis, and it localizes to the plasma membrane and the nucleus. Interestingly, ZmLA1 regulates both tassel and ear development in maize, which is consistent with the high expression of ZmLA1 in reproductive organs. ZmLA1 plays opposite roles during basipetal and lateral PAT, and ZmLA1 physically interacts with both a putative auxin transport regulator and a putative auxin signaling protein. Furthermore, many auxin transport genes and auxin response genes are differentially expressed in the la1 mutant stems. Therefore, we propose that ZmLA1 is involved in shoot gravitropism and inflorescence development through the regulation of PAT and auxin signaling in maize.
RESULTS
The Maize prostrate stem1 Mutant Displays Reduced Gravitropism in the Shoot
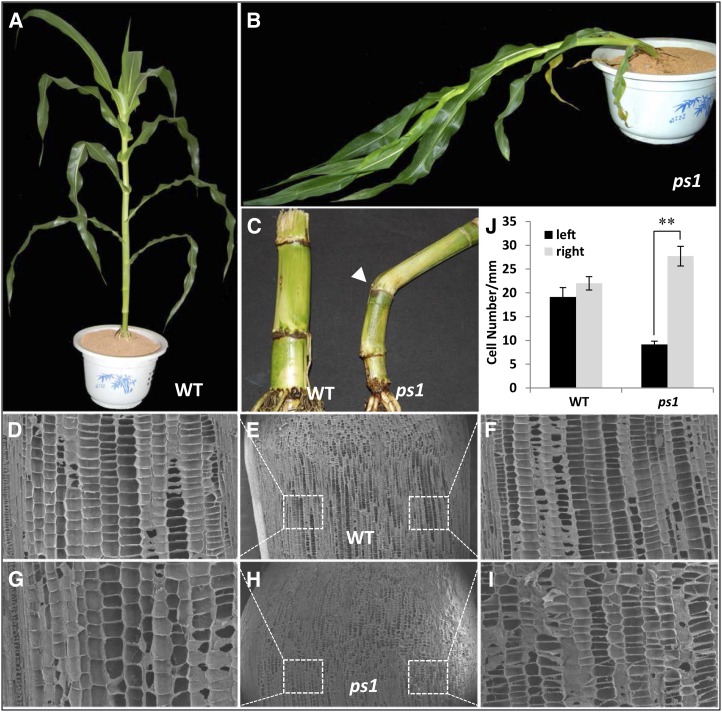
We screened the MuDR-generated mutant maize population and identified a mutant, prostrate stem1 (ps1), that exhibits a prostrate phenotype from the seven- to eight-leaf stage through maturity (Fig. 1, A and B). The bottom second through fourth nodes curved in random directions for each individual plant (Fig. 1C; Supplemental Fig. S1A). Scanning electron microscopy (SEM) analysis of the curved node showed that, unlike the equivalent cell elongation in the wild-type plants, the cell number per 1 mm of length of the near-ground side (right) was more than three times greater than that of the far-ground side (left) in the ps1 mutant (Fig. 1, D–J).
Figure 1.
Morphological characterization of maize ps1 mutant plants. A, Wild type (WT) plant at 60 d after planting segregated from ps1 × HN17 BC3F2. B, ps1 mutant plant at 60 d after planting segregated from ps1 × HN17 BC3F2. C, Near-ground stems of wild-type (left) and ps1 mutant (right) plants. The arrowhead indicates the curved node of the ps1 mutant plant. D to I, Scanning electron micrographs of longitudinal sections of the third aboveground node in wild-type (D–F) and ps1 (G–I) mutant (the curved node) plants. D and F show enlarged views of the left and right sides of E, respectively; and G and I show enlarged views of the left and right sides of H, respectively. J, Longitudinal cell numbers of the left and right sides of the third node in wild-type plants and ps1 mutants. Values are means of at least three plant measurements. For the curved node in ps1, the left and right sides refer to the far- and near-ground sides of the node, respectively. Asterisks indicates a significant difference, as indicated by P < 0.05 in Student’s t test; error bars represent the sd.
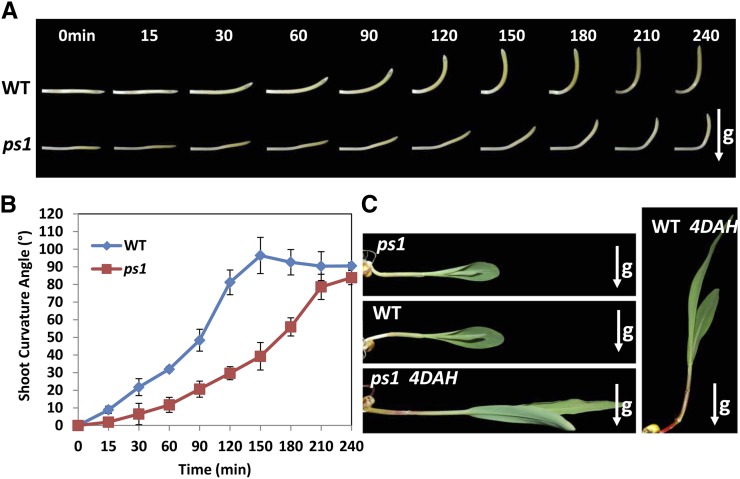
Since the prostrate phenotype is usually caused by defective gravitropism (Roberts, 1984; Friml, 2003; Li et al., 2007), we explored the gravitropic response by examining mesocotyl-coleoptiles 36 h after seed germination (Fig. 2, A and B). We placed the upright mesocotyl-coleoptiles in the horizontal direction, and the wild-type mesocotyl-coleoptiles recovered vertical growth in less than 3 h. It took more than 4 h for the ps1 mutant to recover vertical growth under the same conditions (Fig. 2, A and B). Similarly, wild-type seedlings recovered vertical growth 4 d after horizontal placement, but ps1 seedlings showed no vertical growth during the time period we observed (Fig. 2C). This suggests that the weak gravitropism in ps1 mutant mesocotyl-coleoptiles is gradually lost by the seedling stage. Notably, root gravitropic response is unaffected in ps1 mutant plants (Supplemental Fig. S1B), implying that the agravitropism phenotype is specific to shoots. Furthermore, unlike traditional gravitropic mutants (Li et al., 2007; Wu et al., 2013), the ps1 mutants displayed no noticeable changes in leaf angle compared with wild-type plants (Table I).
Figure 2.
Shoot gravitropic response is reduced in ps1 mutants. A, The response of horizontally placed mesocotyl-coleoptiles to gravity is delayed in ps1 mutants. B, Characterization of the gravitropic response rates of mesocotyl-coleoptiles in wild-type (WT) and ps1 mutant plants. C, The gravitropic response of ps1 mutant seedlings is completely lost compared with wild-type seedlings. 4DAH, Four days after horizontal placement. Arrows indicate the direction of gravity.
Table I. Phenotype analysis of maize la1-ref mutant plants.
Values are reported as means ± sd. P values are from Student’s t test. P < 0.01 indicates a significant difference.
| Genotype | No. of Tassel Branches | Silk Fresh Weight | Ear Shrank Length | Plant Height | Leaf Angle | Days to Shed Pollen | Days to Silking |
|---|---|---|---|---|---|---|---|
| g | mm | cm | ° | ||||
| Wild type | 10.28 ± 2.39 | 9.54 ± 0.88 | 85.86 ± 11.01 | 198.27 ± 17.25 | 26.91 ± 5.33 | 65.05 ± 1.56 | 67.4 ± 1.67 |
| la1-ref | 7.84 ± 1.99 | 7.13 ± 0.7 | 42.25 ± 7.27 | 165.29 ± 22.88 | 29.89 ± 7.37 | 70.77 ± 1.43 | 73.94 ± 1.61 |
| P | 9.64E-07 | 2.77E-05 | 6.23E-05 | 5.47E-24 | 1.77E-01 | 2.74E-54 | 4.43E-62 |
Map-Based Cloning of PS1 and Phylogenetic Analysis
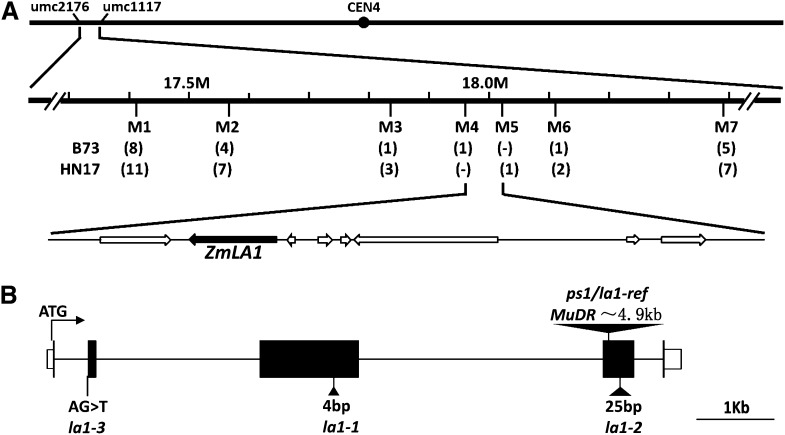
A map-based cloning strategy was used to clone the PS1 gene. We crossed the ps1 mutant with B73 or HN17 inbred lines. In each F2 population, approximately one-quarter of the individual plants displayed the prostrate phenotype (40:127 in ps1 × B73 F2 and 34:113 in ps1 × HN17 F2), which suggests that the ps1 mutant is controlled by a single recessive locus. For the ps1 × B73 F2 population, we initially mapped the PS1 gene in the region between two simple sequence repeat (SSR) markers, umc2176 and umc1117, on the short arm of chromosome 4 (Fig. 3A). We screened a large population of 609 mutant plants, and the PS1 gene interval was mapped between the molecular markers M4 and M6. At the same time, we screened the ps1 × HN17 F2 population of 678 mutant plants, and the PS1 gene was mapped between the molecular markers M3 and M5. Thus, PS1 was placed in the overlapping region between M4 and M5, a 56-kb region with eight predicted open reading frames (ORFs). Only one of these frames, GRMZM2G135019, has EST support (data from maizesequence.org). PCR analysis using overlapping primers and sequencing revealed a MuDR insertion at the 25th bp from the beginning of the fourth exon in ps1 (Fig. 3B).
Figure 3.
Map-based cloning and the gene structure of ZmLA1. A, Fine mapping of the PS1 gene. Two SSR markers (umc2176 and umc117) were used for rough mapping. Two F2 populations, which were generated from ps1 crossed with HN17 or B73, were used for fine mapping. Recombinants are indicated in parentheses below each SSR marker (M1–M7); dashes indicate a nonpolymorphic marker. The PS1 locus was mapped to a 56-kb region containing eight possible ORFs (arrows) between markers M4 and M5. The only ORF with EST support is indicated by the black arrow. B, Schematic gene structure of PS1/ZmLA1. Black boxes indicate the exons, and lines between black boxes represent introns. White boxes indicate untranslated regions. The positions of the four mutant alleles are marked and described. Insertions are represented by triangles. ps1/la1-ref is caused by an approximately 4.9-kb MuDR element insertion in the fourth exon. la1-1 and la1-2 result from a 4-bp insertion in the third exon and a 25-bp insertion in the fourth exon, respectively. These insertions caused a frame shift and a premature stop codon. la1-3 contains a G-to-T substitution, which changed the splicing acceptor in the first intron.
The classical mutant la1, first identified in the 1930s, has similar creeping growth to the ps1 mutant plant. Still, the gene conferring the la1 phenotype remains unidentified, even after decades of investigation. We obtained three allelic mutants from the Maize Genetics Cooperation Stock Center: la1-PI239110 (la1-1), la1-MTM4659 (la1-2), and la1-05HI-RnjxW22GN-333 (la1-3). Genetic complementation tests indicated that the ps1 mutant is allelic to la1. Therefore, we renamed the ps1 mutant as la1-ref and designated the PS1 gene as ZmLA1 in the remaining text. The ZmLA1 gene was sequenced in the three la1 allelic mutations, and we observed a 4-bp insertion in the third exon in la1-1 and a 25-bp insertion in the fourth exon in la1-2. Both of these insertions resulted in a frame shift and a premature stop codon. In la1-3, a single-nucleotide change at the splicing acceptor of the first splicing junction (GT-AG to GT-AT) led to inaccurate splicing (Fig. 3B). Therefore, GRMZM2G135019 was confirmed to be the ZmLA1 gene.
We isolated the 1,242-bp ZmLA1-coding DNA sequence from the stem node. ZmLA1 contained five exons spanning an approximately 6-kb genomic region with 414 predicted amino acids. Homolog proteins of ZmLA1 are widespread in land plants, and ZmLA1 homologs in Poaceae species belong to a distinct clade from those in the dicotyledon plants (Supplemental Fig. S2). Moreover, there is only one homolog in Poaceae species; generally, two or more homologs exist in eudicot species (Supplemental Fig. S2; Supplemental Data Set S1). Although no conserved domain was identified in ZmLA1, six peptide segments were relatively conserved among ZmLA1 homologs (Supplemental Fig. S3), and an ethylene-responsive element-binding factor-associated amphiphilic repression motif (LVLEL) was frequently identified at the C-terminal end of the protein (Ohta et al., 2001).
ZmLA1 Localizes to Both the Plasma Membrane and the Nucleus
The ZmLA1 protein was predicted to contain a transmembrane domain (TMD) at the N terminus and two nuclear localization signal (NLS) fragments at amino acids 275 to 298 (NLS1) and amino acids 338 to 345 (NLS2; Supplemental Fig. S3; Robbins et al., 1991). We performed subcellular localization using the constructs shown in Supplemental Figure S4A to characterize the function of these predicted domains. Similar to the ZmLA1 homologs in rice or in Arabidopsis (Li et al., 2007; Yoshihara et al., 2013), full-length ZmLA1-GFP fusion proteins localized to the plasma membrane and the nucleus of onion (Allium cepa) epidermal cells (Supplemental Fig. S4B). Upon deletion of the TMD, ZmLA1 failed to localize to the plasma membrane, suggesting that the predicted TMD is functional (Supplemental Fig. S4B). A previous study showed that the two NLSs were interdependent (Robbins et al., 1991). However, deletion of the NLS1 domain alone had no effect on localization to the nucleus. The nucleus signal of ZmLA was lost only upon deletion of both NLS1 and NLS2 (Supplemental Fig. S4B), indicating that NLS1 and NLS2 domains redundantly specify the nuclear localization of ZmLA1.
Temporal and Spatial Expression Patterns of ZmLA1
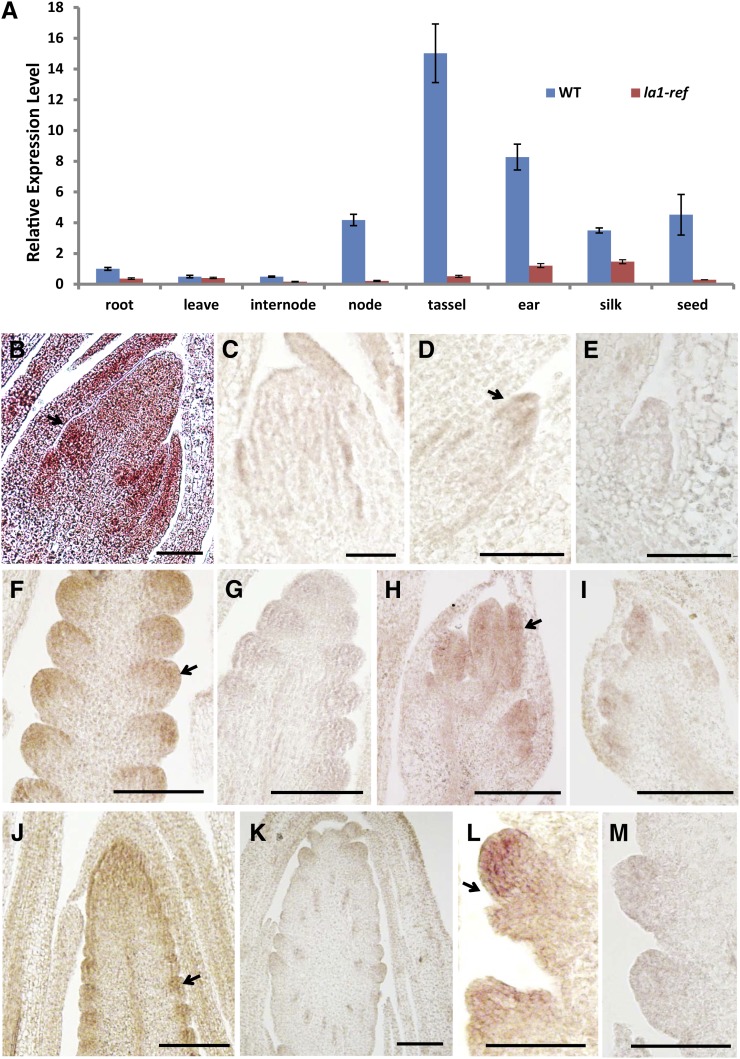
We explored the transcript accumulation of ZmLA1 via real-time quantitative reverse transcription (qRT)-PCR (Fig. 4A). Consistent with its role in shoot gravitropic response, transcripts of ZmLA1 accumulated in the node of the stem. Low levels of accumulation were evident in the internode, root, and leaves (Fig. 4A). Interestingly, ZmLA1 was abundantly expressed in reproductive organs such as the tassel and the ear (Fig. 4A). The expression of ZmLA1 in la1-ref mutant plants was greatly reduced in all the tissues we examined.
Figure 4.
Expression pattern of ZmLA1. A, qRT-PCR analysis of ZmLA1 in multiple tissues in wild-type (WT) and la1-ref plants. Three biological replicates were performed for each sample, and Actin1 (GRMZM2G126010) was used as an internal reference. Error bars indicate the se between biological replicates. B to M, In situ hybridization of ZmLA1 in wild-type (B, D, F, H, J, and L) and la1-ref mutant (C, E, G, I, K, and M) plants. All samples are 10-µm longitudinal sections: 2-week-old shoot apical meristems (B and C); axillary meristem (D and E); developing SPMs on the flank of inflorescence meristem of 34-d-old tassels (F and G); developing male flower primordia (H and I); 54-d-old ears (J and K); and developing SPMs on the flank of inflorescence meristem of 54-d-old ears (L and M). Arrows indicate the expression of ZmLA1. Bars = 50 µm.
We next investigated the expression pattern of ZmLA1 by in situ hybridization (Fig. 4, B–M). Unlike its homologs in rice or Arabidopsis, ZmLA1 was detected in the initiating leaf founder cells and young leaf primordia in seedling apices (arrow in Fig. 4B) as well as the tips of axillary meristems (arrow in Fig. 4D). Furthermore, transcripts of ZmLA1 were detected in SPMs of developing tassels (arrow in Fig. 4F), male flower primordia (arrow in Fig. 4H), and SPMs of developing ears (arrows in Fig. 4, J and L). In la1-ref mutants, the expression of ZmLA1 was dramatically reduced, but the general expression pattern remained unchanged (Fig. 4, C, E, G, I, K, and M).
ZmLA1 Regulates Both Ear and Tassel Development
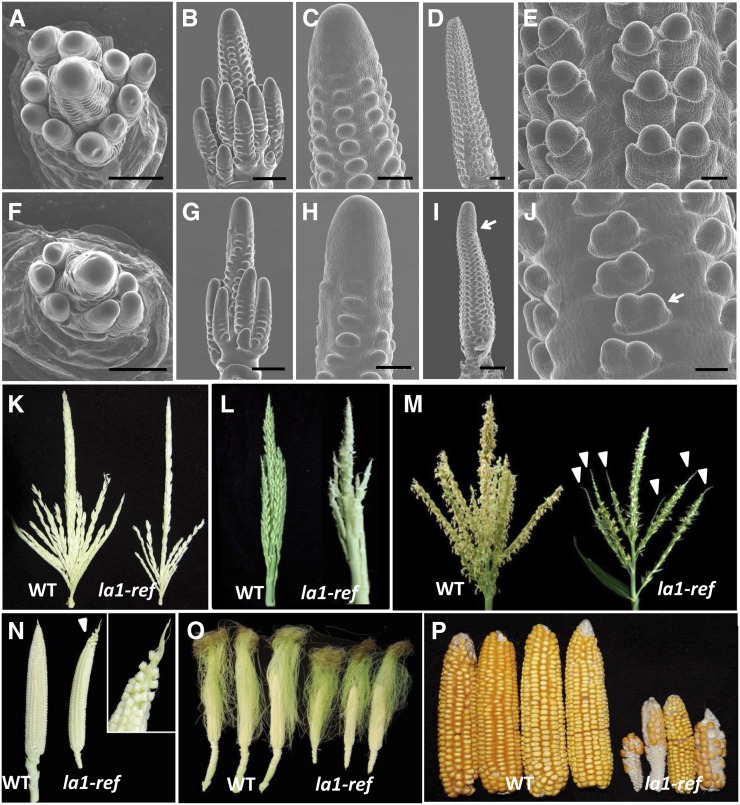
The abundant expression of ZmLA1 in inflorescences led us to explore ear and tassel development in detail. Compared with the well-organized wild-type tassels (Fig. 5, A–C) and ears (Fig. 5, D–E), la1-ref mutants have a reduced number of tassel branches (Fig. 5F), twisted rows of SPMs (Fig. 5, G and H), bent ear tips (arrow in Fig. 5I), and uneven SPMs in the ear (arrow in Fig. 5J). Statistical quantification confirmed that la1-ref mutant plants produced approximately three fewer branches than wild-type plants in the pollen-shedding stage (Fig. 5K; Table I). Subsequently, tassel spikelets were extremely arrested (Fig. 5L), pollen shedding was largely reduced, and spikelets were aborted on the tips of tassel branches (arrowheads in Fig. 5M) in la1-ref mutants. Overall, pollen shedding and silking was delayed by approximately 5 and 7 d, respectively. Mutant ears had disturbed spikelets at the tip region (Fig. 5N) and reduced ear shank and silk numbers (Fig. 5O), and the mature ears of la1-ref mutants were extremely small and disorganized (Fig. 5P). Similar inflorescence phenotypes were also observed in the allelic mutants (Supplemental Fig. S5; Supplemental Table S1), suggesting that, in addition to its role in shoot gravitropism, ZmLA1 regulates inflorescence development in maize.
Figure 5.
Comparison of inflorescences between wild-type (WT) and la1-ref mutant plants. All plants used for phenotype comparison are in the genetic background from the inbred line HN17, except in L, in which B73 is the background. A to J, SEM images of developing inflorescence in wild-type (A–E) or la1-ref mutant (F–J) plants. A and F show top view of 34-d-old tassels; B, C, G, and H show side views of 34-d-old tassels; and D, E, I, and J show side views of 54-d-old ears. Arrows in I and J indicate the bending ear and the uneven development of the ear SPM in la1-ref mutants, respectively. K to P, Development of the ear and tassel are largely disturbed in la1-ref mutants during late developmental stages. K, la1-ref mutant tassel has a reduced branch number before the heading stage. L, Spikelets are extremely arrested in la1-ref mutant tassels. M, Less pollen is shed from la1-ref tassels, and spikelets are aborted on the tips of tassel branches (arrowheads). N, Developing ears (silks were removed) show defective ear tips in la1-ref mutants; the inset shows the area to which the arrowhead is pointing. O, la1-ref mutants have reduced ear shank length and silk numbers. P, Mature ears of la1-ref mutants are much smaller and disorganized than those in wild-type plants. Bars = 500 µm except in C, H, E, and J, in which bars = 100 µm.
ZmLA1 Plays Opposite Roles during Basipetal and Lateral PAT
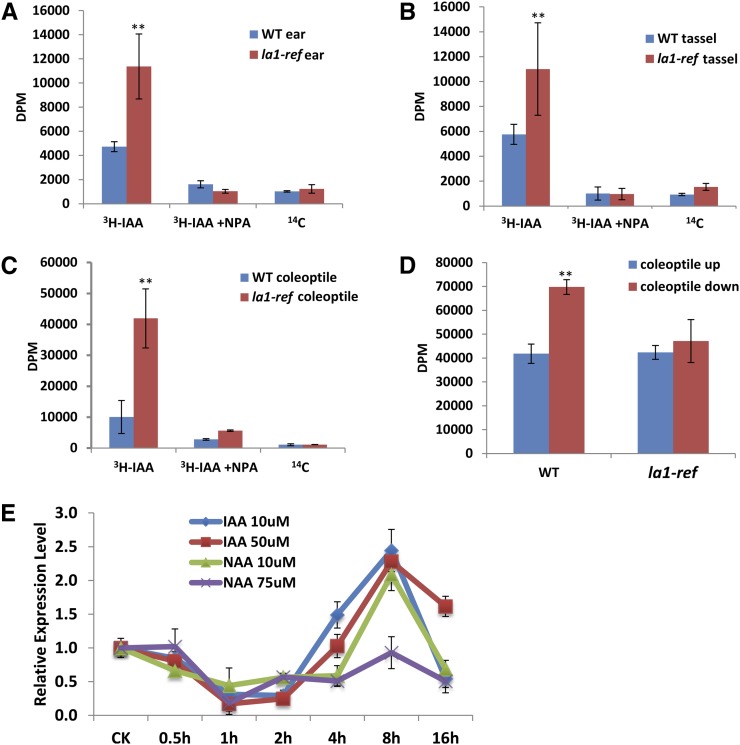
Since auxin plays an important role in gravitropism (Robert and Friml, 2009), we measured basipetal and lateral PAT using [3H]IAA. Consistent with the defective inflorescence, basipetal PAT was significantly enhanced in la1-ref ears (Fig. 6A) and tassels (Fig. 6B). Similarly, basipetal PAT was more than 4 times higher in la1-ref etiolated coleoptiles (Fig. 6C), suggesting that ZmLA1 inhibits basipetal PAT in maize. In the lateral auxin transport assay, wild-type coleoptiles accumulated more auxin in the lower side than in the upper side in response to gravity, while the amount of [3H]IAA was distributed evenly in the upper and lower halves of la1-ref coleoptiles. This finding indicates that ZmLA1 promotes lateral auxin transport (Fig. 6D). Thus, ZmLA1 plays opposing roles in the regulation of PAT, including repressing basipetal PAT while stimulating lateral auxin transport in maize.
Figure 6.
Auxin transport assay and ZmLA1 expression is responsive to auxin. A to C, The basipetal auxin transport assay showed increased auxin transport in la1-ref ears (A), tassels (B), and coleoptiles (C). [3H]IAA together with the auxin transport inhibitor NPA and [14C]benzoic acid alone were used us negative controls. D, Suppressed lateral auxin transport in la1-ref coleoptiles. The amount of [3H]IAA is significantly enriched in the down (red) halves of the wild-type coleoptiles compared with the up (blue) halves; the amount of [3H]IAA is distributed evenly in the up and down halves of la1-ref coleoptiles. E, ZmLA1 expression shows similar response patterns to different concentrations of exogenous IAA or naphthylacetic acid (NAA). Asterisks indicate a significant difference, as indicated by P < 0.05 in Student’s t test; error bars represent the sd of three biological replicates. [See online article for color version of this figure.]
ZmLA1 Expression Is Responsive to Auxin and Light Signals
To further characterize the relationship between ZmLA1 and PAT, we investigated the change in ZmLA1 expression after exogenous auxin treatment. Both IAA and naphthylacetic acid application dramatically repressed the transcription of ZmLA1 within 1 h. However, the expression of ZmLA1 recovered to its original level within 4 h, and its expression more than doubled 8 h after auxin treatment (Fig. 6E). This repression-induction pattern of ZmLA1 may indicate complicated feedback systems between auxin homeostasis, PAT, and ZmLA1. A parallel experiment showed that ZmLA1 was insensitive to N-1-naphthylphthalamic acid (NPA) treatment (Supplemental Fig. S6).
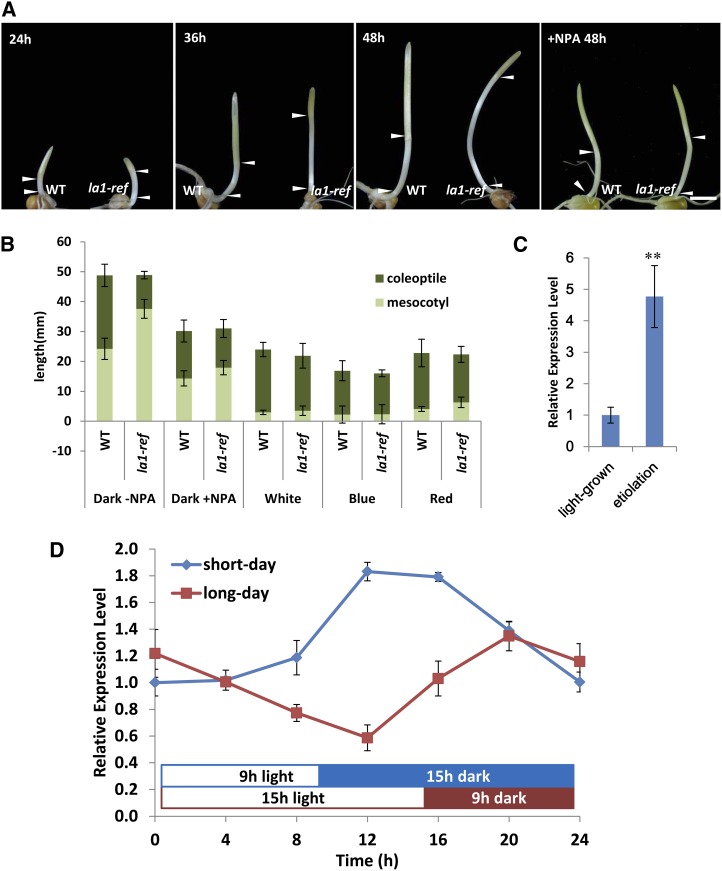
Light is one of the key environmental factors influencing the life cycle of plants, and, as such, it has been shown to mediate PAT (for review, see Sassi et al., 2013). Light prevents mesocotyl elongation, while dark enhances it (Iino, 1982; Jensen et al., 1998). In our experiments, we observed that mesocotyl elongation in la1-ref mutants was accelerated in the dark compared with wild-type plants. The corresponding coleoptile growth was inhibited, so that the whole shoot was approximately the same height as the wild-type plant (Fig. 7, A and B). Furthermore, treatment with 5 μm NPA (Katekar et al., 1981; Bailly et al., 2008; Peer et al., 2009) inhibited the acceleration of la1-ref mesocotyls in the dark, implying that the longer mesocotyls and shorter coleoptiles in la1-ref mutants are primarily due to increased basipetal PAT (Fig. 7, A and B). However, under white, blue, or red light conditions, neither mesocotyl elongation nor coleoptile growth displayed any differences between wild-type and la1-ref mutant plants (Fig. 7B). Transcript accumulation of ZmLA1 in etiolated seedlings was approximately 5 times higher than that in light-grown seedlings (Fig. 7C). Also, ZmLA1 expression was stimulated when plants were switched from light to dark in both long-day and short-day conditions (Fig. 7D). These findings suggest that the transcription of ZmLA1 is repressed by light signals.
Figure 7.
Comparison of mesocotyl elongation and response of ZmLA1 transcript to light. A, Mesocotyl elongation of wild-type (WT; left) and la1-ref (right) plants in the dark. The left three panels show plants grown for 24, 36, and 48 h after germination, respectively. The right panel indicates plants treated with 5 μm NPA for 48 h after germination. The intervals between arrowheads indicate the mesocotyls. Bar = 1 cm. B, Quantification of coleoptile and mesocotyl elongation under dark conditions without NPA treatment, dark conditions with 5 μm NPA treatment, and white, blue, or red light for 36 h after germination. The mesocotyl-coleoptile ratio is much higher in the la1-ref mutant background under dark conditions and is obviously reduced by NPA treatment. C, ZmLA1 expression in the etiolated seedling is approximately 5-fold higher than that in normal seedlings. The etiolated seedling was grown in constant dark, and the normal seedling was grown in constant light for 3 d. D, ZmLA1 expression is induced upon transition from light to dark in both long-day (15 h of light/9 h of dark) and short-day (9 h of light/15 h of dark) conditions. Asterisks indicate a significant difference, as indicated by P < 0.05 in Student’s t test; error bars represent the sd of three biological replicates.
A bioinformatics analysis using a promoter database (PlantCARE and PLACE) identified four auxin-responsive elements (two TGA box, one SAUR motif, and one AuxRR core), 14 light-responsive elements, and one circadian rhythm-related element in the ZmLA1 promoter region (1,500 bp upstream from the start codon; Supplemental Table S2), which supports the notion that ZmLA1 may participate in both auxin response and light signaling pathways.
ZmLA1 Interacts with a Putative Protein Kinase and an Auxin Signaling Protein
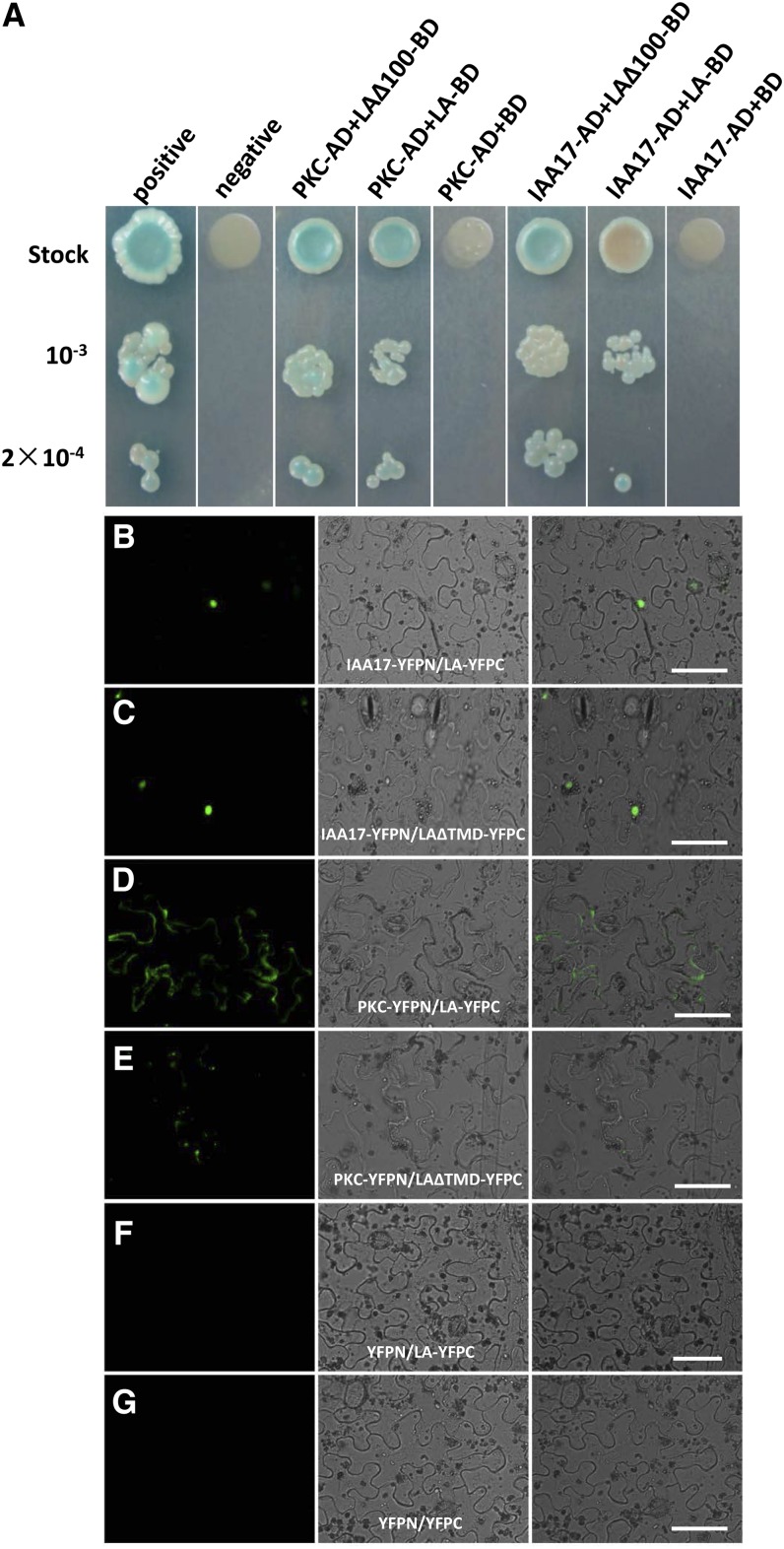
We performed a yeast two-hybrid assay to identify the proteins that interact with ZmLA1. The TMD (residues 71–90) at the N terminus of ZmLA1 may have a negative effect on the screen efficiency, so a truncated ZmLA1 with the first 100 residues deleted (LAΔ100) was used as a bait to screen a yeast two-hybrid library. We found 55 positive clones out of approximately two million transformants. Of these, 17 clones were in the reading frame that was derived from eight ORFs (Table II). We did not detect any members of an auxin transporter family, known trafficking factors, or anchoring proteins such as BIG or TRANSPORT INHIBITOR RESPONSE3, likely owing to the low efficiency of a yeast two-hybrid screen to identify membrane-localized proteins. However, two auxin-related candidates, GRMZM2G147243 and GRMZM2G026203, were identified out of the eight proteins that were likely to interact with ZmLA1 (Table II). GRMZM2G147243 (denoted as IAA17) is a predicted Aux/IAA family member that may be involved in early auxin response (Abel et al., 1994; Ulmasov et al., 1997). GRMZM2G026203 (denoted as PKC) has a protein kinase catalytic domain that plays a fundamental role in the asymmetrical localization of PIN proteins during PAT in Arabidopsis (Friml et al., 2004; Michniewicz et al., 2007; Tripathi et al., 2009; Zourelidou et al., 2009; Zhang et al., 2010; Marquès-Bueno et al., 2011) and in maize (McSteen et al., 2007); it is a member of the PKC superfamily. Both full-length ZmLA1 and LAΔ100 were confirmed to physically interact with IAA17 and PKC in yeast two-hybrid screens (Fig. 8A).
Table II. Interacting proteins identified by a yeast two-hybrid screen.
| Clone Identifier | Protein Identifier | Predicted Function |
|---|---|---|
| 6 | GRMZM2G446170_P01 | BURP domain-containing protein precursor |
| 7 | GRMZM2G026203_P01 | Ser/Thr protein kinase PKC-like superfamily |
| 26 | GRMZM2G035063_P01 | Chaperonin |
| 75 | GRMZM2G033641_P01 | Phosphatidylinositol transfer protein |
| 80, 81, 147 | GRMZM2G147243_P01 | Aux/IAA superfamily |
| 142 | GRMZM2G034005_P01 | Ankyrin repeat domain-containing protein |
| 114 | GRMZM2G324886_P01 | Calcyclin-binding protein |
Figure 8.
ZmLA1 interacts with PKC and Aux/IAA family proteins in both yeast two-hybrid assays and BiFC. A, Yeast two-hybrid assays. Two bait constructs expressed full-length ZmLA1 (LA-BD) or truncated ZmLA1 with deletion of residues 1 to 100 (LAΔ100-BD) fused with the DNA-binding domain (BD). PKC or IAA17 fused with the activation domain (AD) served as the prey constructs. pGBKT7-bait and pGADT7-prey were cotransformed into the Y2HGOLD strain, which was grown on synthetic dextrose/−Ade/−His/−Leu/−Trp/5-Bromo-4-Chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal)/Aureobasidin A plates. Colony growth required the activation of ade2, his3, and aur1-c reporter genes, and the blue-producing X-α-Gal reaction required the activation of the mel1 reporter gene to express the secreted reporter enzyme α-galactosidase. A positive interaction between T-antigen and its partner p53 and a negative interaction between T-antigen and human lamin were used as controls. Both PKC and IAA17 can interact with LA or LAΔ100 to enable strain growth into blue colony patches, whereas neither can activate these reporter genes when cotransformed with the empty plasmid pGBKT7. B to G, BiFC assay in tobacco leaf epidermis. For each group, a positive interaction is indicated by the GFP fluorescence (green) in the left panel. The tobacco cells are indicated by the bright field in the middle panel. The merged views are shown in the right panel. Testing proteins IAA17 and PKC are fused with the N terminus of YFP (IAA17-YFPN and PKC-YFPN, respectively). Full-length ZmLA and ZmLA with the TMD deletion are fused with the C terminus of YFP (LA-YFPC and LAΔTMD-YFPC, respectively). The coinfiltrated construct sets are as follows: IAA17-YFPN/LA-YFPC (B), IAA17-YFPN/LAΔTMD-YFPC (C), PKC-YFPN/LA-YFPC (D), PKC-YFPN/LAΔTMD-YFPC (E), YFPN/LA-YFPC (F), and YFPN/YFPC (G). F and G served as negative controls. The photographs were taken under the same settings, and each interaction was confirmed two times with independent infiltrations. Bars = 50 µm.
To verify the interactions between ZmLA1 and both IAA17 and PKC in vivo, bimolecular fluorescence complementation (BiFC) assays were performed in tobacco epidermis cells (Fig. 8, B–G). ZmLA1 and IAA17 interacted in the nucleus (Fig. 8B), and deletion of the TMD of ZmLA1 (LAΔTMD) did not affect this interaction (Fig. 8C). In contrast, ZmLA1 and PKC interacted on the plasma membrane (Fig. 8D), and deletion of the TMD nearly eliminated the interactions (Fig. 8E). These results suggest that ZmLA1 may regulate auxin signaling in the nucleus through IAA17 and auxin transport in the plasma membrane through PKC.
Transcriptome Analyses of la1-ref Mutant Nodes
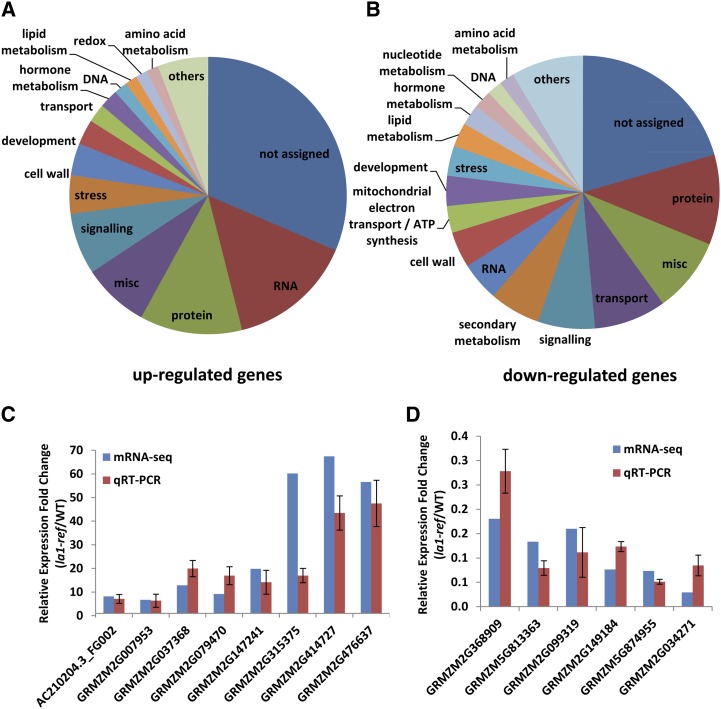
We compared the transcriptome profile of the third node in la1-ref mutants with those in wild-type plants using RNA-SEQ to examine the genome-wide effect of the ZmLA1 gene. We generated 14.6 and 36.5 million paired-end reads from two biological samples of wild-type and la1-ref mutant plants, respectively. We found 931 genes that were differentially expressed in la1-ref mutants, based on the criteria of at least a 2-fold change in expression and a false discovery rate of q < 0.05. There were more up-regulated genes (646) than down-regulated genes (285; Supplemental Table S3), implying that ZmLA1 may act as a major repressor during maize gravitropism and inflorescence development. The differentially expressed genes (DEGs) were annotated and classified into different functional categories using MapMan (Supplemental Table S4). A wide range of categories were affected in la1-ref mutant plants, including RNA regulation, transport, and hormone metabolism (Fig. 9, A and B; Supplemental Fig. S7; Supplemental Table S4). To validate the results of RNA-SEQ, we randomly selected eight up-regulated DEGs and six down-regulated DEGs for qRT-PCR analysis. The RNA-SEQ and qRT-PCR exhibited close agreement (Fig. 9, C and D; Pearson correlation coefficient, r = 0.871, P = 4.85 × 10−5), indicating the reliability of our RNA-SEQ data.
Figure 9.
Transcriptome comparison between wild-type (WT) and la1-ref mutant nodes using RNA-SEQ. A and B, Functional categories of the DEGs in la1-ref mutants using MapMan: up-regulated genes (A) and down-regulated genes (B). C and D, qRT-PCR validation of the selected DEGs: up-regulated genes (C) and down-regulated genes (D). The relative expression levels of DEGs were examined by fragments per kilobase of exon per million fragments mapped in RNA-SEQ. Three corresponding biological replicate samples were performed for the wild type and la1-ref in qRT-PCR, and three technical validations were performed in each biological replication. ZmActin1 (GRMZM2G126010) was used as an internal reference. Error bars indicate the sd between biological replicates.
Among the functional categories of DEGs, 10 genes involved in Ca2+ signaling, 21 genes involved in lipid metabolism, and seven genes involved in pH/H+ modulation or proton transport, including the maize ortholog of AtAVP1 (GRMZM2G148200; Li et al., 2005), were differentially expressed in la1-ref mutant nodes (Supplemental Table S5). These data are consistent with previous studies that reported that changes in or metabolism of cytosolic Ca2+, pH, or lipid concentration contribute to gravitropic growth and auxin transport in plants (Toyota et al., 2008; Monshausen et al., 2011; Smith et al., 2013). In addition, 12 light signaling genes were up-regulated in la1-ref (Supplemental Table S5), and transcription of ZmLA1 was repressed by light (Fig. 7, C and D), suggesting that light and ZmLA1 may form a negative feedback loop.
Furthermore, genes predicted to be involved in auxin transport or auxin response were highly enriched in the DEGs (Table III). For example, the putative auxin efflux transporter ZmPIN1c (GRMZM2G149184) was down-regulated 13.1-fold, and the maize ortholog (GRMZM5G813363) of N-MYC DOWNREGULATED-LIKE1 (NDL1), which positively regulates auxin transport in a G protein-mediated pathway (Mudgil et al., 2009), was also down-regulated 7.5-fold in la1-ref mutants (Table III). However, maize DWARF BRACHYTIC2 (BR2; GRMZM2G315375), the ortholog of ABCB1 in Arabidopsis was up-regulated 60-fold in la1-ref mutants (Table III). In addition, 13 auxin response genes were differentially expressed in la1-ref mutant nodes. Of these, 11 genes were up-regulated and two genes were down-regulated (Table III), implying that ZmLA1 may play a predominantly repressive role in auxin response.
Table III. Examples of the DEGs in la1-ref mutant nodes that are predicted to be involved in auxin transport or auxin response.
| Gene Identifier in Maize | Putative Ortholog in Arabidopsis | Description | Change (log2-fold), la1-ref/Wild Type |
|---|---|---|---|
| Auxin transport-related genes | |||
| GRMZM2G109527 | AT1G71960.1 (ABCG25) | ATP-binding cassette family G25 | 5.15 |
| GRMZM2G479906 | AT1G15520.1 (PDR12) | ATPase, coupled to transmembrane movement of substances | −2.82 |
| GRMZM5G874955 | AT1G15520.1 (ABCG40, PDR12) | Pleiotropic drug resistance12 | −3.77 |
| GRMZM2G315375 | AT2G36910.1 (ABCB1) | ATP-binding cassette subfamily B1 | 5.91 |
| GRMZM2G004161 | AT3G48360.1 (BT2) | BTB and TAZ domain protein2 | 5.78 |
| GRMZM2G051005 | AT5G48930.1 (HCT) | Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase | 4.00 |
| GRMZM2G097900 | AT5G37770.1 (CML24, TCH2) | EF hand calcium-binding protein family | 6.38 |
| GRMZM2G138248 | AT4G39950.1 (CYP79B2) | Cytochrome P450/family 79/subfamily B/polypeptide 2 | −3.78 |
| GRMZM2G156296 | AT5G48930.1 (HCT) | Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase | 4.06 |
| GRMZM2G148200 | AT1G15690.1 (AVP-3, AVP1) | Inorganic H pyrophosphatase family protein | −3.11 |
| GRMZM2G149184 | AT1G73590.1 (PIN1) | Auxin efflux carrier family protein | −3.71 |
| GRMZM2G429940 | AT4G30960.1 (CIPK6, SIP3, SNRK3.14) | SOS3-interacting protein3 | 4.93 |
| GRMZM5G813363 | AT5G56750.1 | Ndr family protein | −2.91 |
| GRMZM2G465595 | AT2G01940.1 (SGR5, ATIDD15) | Nucleic acid binding/transcription factor/zinc ion binding | 4.17 |
| Auxin response genes | |||
| GRMZM2G037368 | AT4G29080.1 (IAA27, PAP2) | Phytochrome-associated protein2 | 3.68 |
| GRMZM2G163848 | AT2G22670.4 (IAA8) | IAA-induced protein8 | 5.89 |
| GRMZM2G301642 | AT1G49010.1 | Duplicated homeodomain-like superfamily protein | 4.47 |
| GRMZM2G405094 | AT4G09460.1 (MYB6) | Myb domain protein6 | 5.55 |
| GRMZM5G803308 | AT4G37260.1 (MYB73) | Myb domain protein73 | 7.14 |
| GRMZM2G414727 | AT2G46690.1 | SAUR-like auxin-responsive protein family | 6.08 |
| GRMZM2G009103 | AT5G64700.1 | Nodulin MtN21/EamA-like transporter family protein | −5.62 |
| GRMZM2G136567 | AT5G07050.1 | Nodulin MtN21/EamA-like transporter family protein | −7.23 |
| GRMZM2G066219 | AT5G47530.1 | Auxin-responsive protein | 3.13 |
| GRMZM2G097989 | AT2G29480.1 (ATGSTU2, GST20) | Auxin-responsive GST | 5.36 |
| GRMZM2G331638 | AT2G04850.1 | Auxin-responsive protein related | 3.67 |
| GRMZM5G804575 | AT1G75500.1 (WAT1) | Auxin-responsive nodulin MtN21-like transporter family | 3.08 |
| GRMZM2G007953 | AT1G75500.1 (WAT1) | Auxin-responsive nodulin MtN21-like transporter family | 2.74 |
DISCUSSION
The Function of ZmLA1 Is Partially Conserved in Shoot Gravitropism
In response to gravity, the shoot perceives and transduces the gravity signal in the shoot endodermal cells. The root responds in the same manner in the columella cells of the root cap (Blancaflor et al., 1998). Some regulators are shared between shoot and root gravitropism, while others are specific to the shoot gravity response (Masson et al., 2002; Morita, 2010; Strohm et al., 2012; Wu et al., 2013). The classical shoot gravitropism mutant phenotype “lazy” has been identified in many plants, including lazy-1 and lazy-2 in tomato (Solanum lycopersicum; Roberts, 1984; Gaiser and Lomax, 1993), lazy in rice (Jones and Adair, 1938), lazy serpentina in barley (Hordeum vulgare; Suge and Türkan, 1991), lazy in Brachypodium distachyon (Derbyshire and Byrne, 2013), and la1 in maize (Jenkins and Gerhardt, 1931; Van Overbeek, 1936, 1938). In this study, we isolated the ps1 mutant, which is a phenocopy of la1 in maize (Fig. 1). Map-based cloning and allelism tests revealed that ps1 is a new allele of la1 in maize that is caused by an insertion in the ZmLA1 gene. ZmLA1 is an ortholog of LAZY1 in rice (Li et al., 2007; Yoshihara and Iino, 2007) and AtLAZY1 in Arabidopsis (Yoshihara et al., 2013). Rice LAZY1 mediates shoot gravitropism through negative regulation of PAT, and the lazy1 mutant displays a tillers-spreading phenotype with reduced shoot gravitropism (Li et al., 2007; Yoshihara and Iino, 2007). Similarly, the Atlazy1 mutant in Arabidopsis exhibits enlarged branch angles with agravitropic shoots. Maize hardly produces tillers or branches due to domestication from teosinte (Zea mays parviglumis; Doebley et al., 1997), so the prostrate growth of maize la1 mutants was similar to the tillers-spreading phenotype in rice and the branch-angle phenotype in Arabidopsis. However, disruption of ZmLA1 caused more severe defects of shoot gravitropism in maize. The prostrate growth of maize la1 mutants can be viewed as the “largest branch angle” or “most spreading tiller,” and the maize la1 seedling completely lost its gravitropic response (Fig. 2C). This is not observed in rice or in Arabidopsis mutant plants. These data suggest that the function of ZmLA1 is partially conserved during shoot gravitropism, but, unlike its orthologs in rice or in Arabidopsis, the role of ZmLA1 is more significant during the maize shoot gravity response.
Unique Roles of ZmLA1 in the Development of Maize Inflorescence
ZmLA1 was highly expressed in the reproductive tissues of maize, such as the tassel and the ear (Fig. 4A). mRNA in situ hybridization further showed that transcripts of ZmLA1 accumulated in SPMs of developing tassels (Fig. 4F), the inner domains of male flower primordia (Fig. 4H), and SPMs of developing ears (Fig. 4, J and L). Consistent with this expression pattern, la1-ref mutant plants displayed defective tassel and ear development. During the early stage of inflorescence development, rows of SPMs were twisted in the tassels and SPMs in the ears were not coordinated (Fig. 5, F–J). Subsequently, mature la1-ref plants showed a reduced number of tassel branches, defective spikelets and pollen shedding, and small ears with a decreased number of kernels (Fig. 5, K–P). Allelic mutants la1-1 and la1-2 showed similar inflorescence defects (Supplemental Fig. S5; Supplemental Table S1). Notably, no inflorescence phenotype has been observed in the lazy1 mutant of rice or Arabidopsis. Considering that ZmLA1 belongs to a distinct clade with LAZY1 and AtLAZY1 and that maize has a different type of inflorescence, ZmLA1 may have evolved a unique role in tassel and ear development in maize.
ZmLA1 May Mediate the Cross Talk between Shoot Gravitropism and Inflorescence Development by Regulating Both Auxin Transport and Auxin Signaling
Mutation of ZmLA1 increased basipetal PAT and decreased lateral PAT in maize (Fig. 6), suggesting that ZmLA1 represses basipetal PAT while promoting lateral PAT. We further showed that auxin transport-related genes, such as ZmPIN1c and ZmNDL1, were greatly down-regulated, while the maize auxin transporter BR2 (ZmABCB1/PGP1) was significantly up-regulated approximately 60-fold in la1-ref mutant stems (Table III). PIN and BR2 can regulate both rootward PAT and lateral PAT (Blakeslee et al., 2005; Knöller et al., 2010), while PIN and PGP proteins may act in antagonistic and synergistic fashions during asymmetric distribution of auxin (Luschnig et al., 1998; Noh et al., 2003; Bouchard et al., 2006; Mravec et al., 2008). Therefore, the enhanced basipetal PAT and reduced lateral PAT in la1-ref mutants may be due to the synergistic effects of the up-regulation of BR2 and the down-regulation of ZmPIN1c and ZmNDL1.
Disruption of ZmLA1 resulted in an agravitropic shoot response (Figs. 1 and 2) and defective tassel and ear development in maize (Fig. 5). Furthermore, basipetal PAT was increased in mutant inflorescences (Fig. 6), implying that ZmLA1 may regulate tassel and ear development through auxin transport. Biochemical analyses indicated that ZmLA1 physically interacts with the auxin signaling protein IAA17 in the nucleus and the auxin transport regulator PKC in the plasma membrane (Fig. 8). A large number of auxin transport and auxin response genes were also differentially expressed in la1-ref mutant stems (Table III). These data suggest that ZmLA1 may mediate the cross talk between shoot gravitropism and inflorescence development by regulating auxin transport and auxin signaling. Given that a plant-specific AGCVIII kinase subgroup such as PINOID (Michniewicz et al., 2007) and D6 Protein Kinase (Zourelidou et al., 2009; Willige et al., 2013) have been shown to regulate auxin transport through the phosphorylation of PINs, it will be interesting to explore whether PKC can phosphorylate maize PINs in the future.
ZmLA1 May Mediate the Cross Talk between Auxin and Light
For decades, auxin has been known to play an essential role in plant directional growth toward light (phototropism; Muday, 2001; Halliday et al., 2009; Sassi et al., 2013). Light signaling components can provide feedback on auxin homeostasis and response. For example, photoreceptor phytochromes and cryptochromes are critical regulatory players that control auxin transport in response to light. Mutants of phytochromes (phya and phyb) or cryptochromes (cry1 and cry2) display abnormal hypocotyl elongation with disrupted auxin transport (Canamero et al., 2006; Salisbury et al., 2007; Nagashima et al., 2008). Phytochrome Interacting Factor5 directly activates WAG2, which regulates local auxin maxima through the phosphorylation of PIN proteins (Willige et al., 2012). In addition, light stimulates the transcript accumulation of polar auxin transporters such as PIN1 (Sassi et al., 2012).
Here, we observed that mesocotyl elongation in la1-ref mutants was accelerated in dark, but not in light, conditions and that NPA treatment inhibited such mesocotyl acceleration (Fig. 7, A and B). Furthermore, expression of ZmLA1 was induced in the dark and repressed in the light (Fig. 7, C and D). These findings can be explained by several factors. Specifically, under light conditions, repression of ZmLA1 leads to a similar up-regulation of basipetal PAT in wild-type plants and la1-ref mutants. There is no detectable difference in mesocotyl elongation between wild-type plants and la1-ref mutants. However, under dark conditions, ZmLA1 transcription is induced in wild-type plants but not in la1-ref mutants, which results in repressed basipetal PAT and, consequently, reduced mesocotyl elongation in wild-type plants. In addition, four auxin-responsive elements and 14 light-responsive elements were identified in the ZmLA1 promoter region (Supplemental Table S1). Twelve light signaling genes and 11 auxin response genes were differentially up-regulated in la1-ref stems (Table III; Supplemental Table S4), suggesting that ZmLA1 may mediate the cross talk between light and auxin by acting as a negative regulator for both light signaling and auxin response.
Taken together, ZmLA1 is one of the few regulators that functions in both gravitropic response and inflorescence development in maize. Furthermore, ZmLA1 may be involved in auxin transport, auxin signaling, and light response. The membrane-localized ZmLA1 may function in shoot gravitropism by regulating the polar localization of auxin transporters, such as ZmPINs, likely through an interaction with PKC. The nucleus-localized ZmLA1 may play a role in inflorescence development by acting as a transcriptional repressor for auxin transporters such as BR2, auxin signaling genes, likely through an interaction with IAA17, and light-response genes. Future research is needed to identify additional players and downstream targets that cross talk shoot gravitropism and inflorescence development. Such findings will provide useful information for obtaining maize varieties with optimal plant architecture and maximum grain yield.
MATERIALS AND METHODS
Plant Materials
The ps1 mutant of maize (Zea mays) was separated from an M2 population generated by the cross between the inbred line Zong31 and the MuDR-active line. la1-PI239110, la1-MTM4659, and la1-05HI-RnjxW22GN-333 lines were obtained from the Maize Genetics Cooperation Stock Center. We cultivated two generations of plants every year, growing the summer generation in an experimental field of the China Agricultural University in Beijing and the winter generation in an experimental field in Sanya, Hainan. The ps1 mutant was crossed into the inbred line HN17 for three generations and then self-fertilized to obtain the BC3F2 population. Morphological comparisons were performed within siblings from the same family.
Phenotypic Analysis
Field-grown plants were used for the quantification of plant height, leaf angle, ear shank length, and tassel branches. Plant height and ear shank length were measured at maturity, while leaf angle and tassel branches were measured during the flowering time. Plant height was measured from the base of the stem to the base of the first tassel branch, and ear shank length was measured from the base of the ear to the joint where the branch originated from the stem. Leaf angle was measured at the first leaf above the ear, and tassel branches were counted using only the primary branches.
Different (white, red, and blue) light treatments were performed with uniform light intensity at 200 μmol m−2 s−1. The lengths of the mesocotyls and coleoptiles were measured 48 h after germination. Statistical analysis was performed using Student’s t test.
Gravitropism Analysis
Sterilized seeds were sown in Murashige and Skoog (MS) medium (pH 5.8) with 3% phytagel and grown in the dark at 28°C. When the vertical shoots were approximately 4 cm in length, the seedlings were placed horizontally, and the shoot curvature was measured as the shoot gravitropic response. Similar experiments were performed with 3-cm primary roots to evaluate the root gravity response.
Map-Based Cloning
Two F2 mapping populations were obtained by crossing the ps1 mutant with inbred line B73 or HN17 respectively, and position cloning was carried out with both populations. Genomic DNA was extracted from seedling leaves according to the cetyl-trimethyl-ammonium bromide method and used for bulked segregant analysis (Michelmore et al., 1991) with available SSR markers (www.maizegdb.org). Additional SSR markers, which flank the rough mapping regions, were exploited using the SSRHunter Simple Sequence Repeat Search tool (Li and Wan, 2005) based on the B73 genome sequence (www.maizesequence.org).
Real-Time qRT-PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). A sample of 2 μg of total RNA was treated with DNase I (Promega), and reverse transcription was conducted using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT)18 primers. Reverse transcription-PCR was performed using SYBR Green PCR master mix (Takara). Three biological replicates and three technical replicates were performed for each reverse transcription-PCR procedure. ZmActin1 was used as the internal reference to normalize the expression data. Relative expression levels were calculated according to the 2-2ΔΔCT (cycle threshold) method (Livak and Schmittgen, 2001), and the sd was calculated among the three biological replicates. The primers are listed in Supplemental Table S6.
Phylogenetic Analysis
ZmLA1-like proteins in other species were obtained using BLAST searches at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), Phytozome (http://www.phytozome.org), PlantGDB (http://www.plantgdb.org), Grameme (http://www.gramene.org/), and The Institute for Genomic Research (http://blast.jcvi.org/euk-blast/plantta_blast.cgi). The proposed protein sequences were aligned using ClustalW, and the neighbor-joining phylogenetic tree was produced using the bootstrap analysis with 1,000 replications in the MEGA5 software package (Tamura et al., 2011).
SEM
SEM of stems was performed with plants grown in the greenhouse (16 h of light at 30°C/8 h of dark at 24°C) for 4 weeks. The third aboveground nodes were cut longitudinally and fixed in 50% ethanol, 5% glacial acetic acid, and 3.7% formaldehyde overnight and then dehydrated with an ethanol series. Samples were critical-point dried and sputter coated with gold palladium for 60 s and viewed by SEM (Hitachi S-4700) using a 2-kV accelerating voltage. Young tassels and ears were dissected from the plants grown in the field for 5 and 8 weeks, respectively. Dissected fresh samples were directly viewed by SEM without fixation or dehydration (Whipple et al., 2010). Images were taken as soon as possible to minimize sample damage from electron beams using a 5-kV accelerating voltage and 10- to 20-mm working distance.
Subcellular Localization
The TMD was predicted with the TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and Phobius (http://phobius.sbc.su.se/). The NLSs were predicted with PSORT WWW Server (http://psort.hgc.jp/) and NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/; Nguyen Ba et al., 2009). Five vectors, LA-GFP, LAΔTMD-GFP, LAΔNLS1-GFP, LAΔNLS1NLS2-GFP, and LAΔNLS1-to-end-GFP, were constructed for subcellular localization. LA-GFP represents the full-length LA protein sequence fused with GFP, while the other vectors represent truncated LA proteins fused with GFP. Specifically, LAΔTMD-GFP indicates deletion of the proposed TMD (amino acid residues 71–90), LAΔNLS1-GFP indicates deletion of the proposed NLS1 domain (amino acid residues 275–298), LAΔNLS1NLS2-GFP indicates deletion of both of the proposed NLS1 and NLS2 domains (amino acid residues 275–298 and 338–345, respectively), and LAΔNLS1-to-end-GFP indicates deletion of the entire fragment from the proposed NLS1 domain to the C-terminal end (amino acid residues 275 to the end). Before bombardment, 2 μg of each vector plasmid was coated with tungsten particles as described previously (Varagona et al., 1992). Onion (Allium cepa) epidermal cells were peeled and placed inside-up on MS medium. Next, 10 μL of each coated vector was bombarded under a vacuum of 28 inches of mercury and 1,350 p.s.i. of helium pressure (Bio-Rad PDS-1000/He). Bombarded tissues were incubated in a dark room at 22°C for approximately 18 h, incubated in 15 μm FM4-64 for 20 min, and rinsed twice before observation using a confocal laser-scanning microscope (Carl Zeiss LSM 510) excited at 488 nm for GFP and 543 nm for FM4-64.
In Situ Hybridization
Shoot apices and young tassels and ears were dissected from plants of 2-week-old seedlings and 34-d and 54-d field-grown plants, respectively. Tissue fixation and in situ hybridization were performed as described previously (Zhang et al., 2007). The gene-specific primers are listed in Supplemental Table S6.
PAT Assays
Auxin transport assays were performed using a method modified from Haga and Iino (1998). Young ears and tassels at stage v15 and dark-grown coleoptile segments were used in the PAT assay. The ears were stripped from the husks and resected to the ear tip. We harvested 2 cm of ear and basal segments of tassel branches and 3 cm of coleoptile segments. We rinsed the samples in one-half-strength MS liquid medium for 2.5 h to remove endogenous IAA. For basipetal transport assays, the apical tips of the segments were submerged vertically in 10 μL of one-half-strength MS phytagel medium containing 500 nm [3H]IAA and 500 nm IAA and then placed in the dark for 3 h. NPA was applied to the medium as a control to inhibit PAT. [14C]Benzoic acid was used as another control to exclude the effect of free distribution of weak acid. The unsubmerged ends (0.5 cm for the ear and tassel and 1.5 cm for the coleoptile) were excised and quickly rinsed two times in one-half-strength MS liquid medium. After incubation in 1 mL of scintillation liquid for approximately 20 h, the radioactivity of each segment was detected by a liquid scintillation counter (1450 MicroBeta TriLux; Perkin-Elmer). For the lateral auxin transport assay, similar procedures were performed, except that the submerged segments were placed horizontally for 3 h in the dark and then evenly split into halves (up and down) for radioactivity measurement.
Yeast Two-Hybrid Library and BiFC
Construction of a yeast two-hybrid library was performed following the manufacturer’s instructions for the Make Your Own “Mate & Plate” Library System (Clontech). Total RNA was extracted from 4-d-old maize coleoptiles and served as the template to make the complementary DNA (cDNA) library using the cDNA Amplification Kit (Clontech). Each cDNA was cloned into plasmid pGADT7 and transformed into a Y187 yeast strain (the prey library). The truncated ZmLA protein that was missing 100 amino acid residues from the start of the N terminus (LAΔ100) was cloned into the pGBKT7 (pGBKT7-TLA) plasmid and then transformed into the Y2HGOLD yeast strain (bait). The bait and prey vectors were transformed according to the manufacturer’s instructions for the Matchmaker Gold Yeast Two-Hybrid System (Clontech). Approximately 2.1 × 106 yeast clones were screened on the selective medium synthetic dextrose/−Ade/−His/−Leu/−Trp/X-α-Gal/Aureobasidin A. Positive prey clones were isolated and sequenced. Then, the resultant full-length positive prey proteins were fused with the activation domain, and their interactions with ZmLA were confirmed using both the full-length ZmLA protein (LA-BD) and the truncated ZmLA protein without the TMD (LAΔ100-BD). For the BiFC experiment, proteins that were proposed to interact with ZmLA, IAA17 and PKC, were fused with the N terminus of yellow fluorescent protein (YFP; IAA17-YFPN and PKC-YFPN, respectively), while full-length ZmLA or ZmLA with the TMD deletion was fused with the C terminus of YFP (LA-YFPC and LAΔTMD-YFPC, respectively). Each construct set with testing interaction was cotransformed in tobacco (Nicotiana tabacum) leaves through Agrobacterium tumefaciens infiltration as described previously (Waadt and Kudla, 2008). Infiltrated tobacco leaves were incubated in the greenhouse for 4 d, and YFP fluorescence was observed using a confocal laser-scanning microscope (Carl Zeiss LSM 510) under an excitation wavelength of 488 nm.
RNA-SEQ Analysis
The third aboveground nodes were manually dissected from 4-week-old wild-type and la1-ref mutant plant siblings segregated from the HN17 background. Samples from 11 siblings were pooled as one biological repeat; two biological repeats were performed for each genotype. Total RNA was extracted with TRIzol reagent (Invitrogen). Library construction was performed according to Illumina instructions and sequenced on a HiSEquation 2000 sequencer. All the paired-end reads were mapped to the maize B73 genome using TopHat2 (Langmead et al., 2009; Trapnell et al., 2009; Kim and Salzberg, 2011). RNA-SEQ data were normalized as fragments per kilobase of exon per million fragments mapped, since the sensitivity of RNA-SEQ depends on the transcript length. Significant DEGs were identified using the Cufflinks program (Trapnell et al., 2010) as genes with at least a 2-fold change in expression and q ≤ 0.05%. The DEGs were assigned functional categories based on the MapMan annotation (Thimm et al., 2004).
Sequence data from this article can be found in the National Center for Biotechnology Information Short Read Archive sequence database under accession number SRP022165.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ps1 mutant plants have defective shoot gravitropism but not root gravitropism.
Supplemental Figure S2. Phylogenetic analysis of ZmLA1 and related homologs.
Supplemental Figure S3. Sequence alignment of ZmLA1.
Supplemental Figure S4. Subcellular localization of ZmLA1 in onion epidermis cells.
Supplemental Figure S5. Inflorescence phenotype of la1-1 and la1-2 alleles.
Supplemental Figure S6. The expression of ZmLA1 was insensitive to PAT inhibitor NPA treatment.
Supplemental Figure S7. Overview of cell functions of DEGs.
Supplemental Table S1. Phenotype analysis of la1-1 and la1-2 mutants.
Supplemental Table S2. Putative motifs in the ZmLA1 promoter (1,500 bp upstream from the start codon).
Supplemental Table S3. Transcriptome analysis identified 931 DEGs in la1-ref mutants.
Supplemental Table S4. Functional categories of the DEGs in la1-ref mutants using MapMan software.
Supplemental Table S5. Selected DEGs involved in proton transport, lipid metabolism and signaling, calcium signaling, and light signaling.
Supplemental Table S6. Primers used in this study.
Supplemental Data Set S1. Text file of the sequences and alignment used for the phylogenetic analysis shown in Supplemental Figure S2.
Acknowledgments
We thank members of the Jin laboratory for discussion and technical assistance, Dr. Adrienne Roeder (Cornell University) for critical reading and comments related to the manuscript, Ziding Zhang (China Agricultural University) for assistance with the prediction of protein structure, and Lei Zhao and Lubin Tan (China Agricultural University) for technical assistance with the PAT assay.
Glossary
- IAA
indole-3-acetic acid
- PAT
polar auxin transport
- SPM
spikelet pair meristem
- SEM
scanning electron microscopy
- SSR
simple sequence repeat
- ORF
open reading frame
- NLS
nuclear localization signal
- TMD
transmembrane domain
- qRT
quantitative reverse transcription
- NPA
N-1-naphthylphthalamic acid
- BiFC
bimolecular fluorescence complementation
- DEGs
differentially expressed genes
- MS
Murashige and Skoog
- cDNA
complementary DNA
- YFP
yellow fluorescent protein
References
- Abel S, Oeller PW, Theologis A. (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA 91: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M. (2008) Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J Biol Chem 283: 21817–21826 [DOI] [PubMed] [Google Scholar]
- Barazesh S, McSteen P. (2008a) Hormonal control of grass inflorescence development. Trends Plant Sci 13: 656–662 [DOI] [PubMed] [Google Scholar]
- Barazesh S, McSteen P. (2008b) Barren inflorescence1 functions in organogenesis during vegetative and inflorescence development in maize. Genetics 179: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Peer WA, Murphy AS. (2005) Auxin transport. Curr Opin Plant Biol 8: 494–500 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Fasano JM, Gilroy S. (1998) Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol 116: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, et al. (2006) Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem 281: 30603–30612 [DOI] [PubMed] [Google Scholar]
- Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, Habricot Y, Ahmad M. (2006) Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224: 995–1003 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S. (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhao Y. (2007) A role for auxin in flower development. J Integr Plant Biol 49: 99–104 [Google Scholar]
- Derbyshire P, Byrne ME. (2013) MORE SPIKELETS1 is required for spikelet fate in the inflorescence of Brachypodium. Plant Physiol 161: 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Friml J. (2003) Auxin transport: shaping the plant. Curr Opin Plant Biol 6: 7–12 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Gaiser JC, Lomax TL. (1993) The altered gravitropic response of the lazy-2 mutant of tomato is phytochrome regulated. Plant Physiol 102: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. (2008a) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D. (2008b) The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pè ME, Schmidt RJ. (2004) The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. (2006) The ABC of auxin transport: the role of P-glycoproteins in plant development. FEBS Lett 580: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Haga K, Iino M. (1998) Auxin-growth relationships in maize coleoptiles and pea internodes and control by auxin of the tissue sensitivity to auxin. Plant Physiol 117: 1473–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Martínez-García JF, Josse EM. (2009) Integration of light and auxin signaling. Cold Spring Harb Perspect Biol 1: a001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino M. (1982) Inhibitory action of red light on the growth of the maize mesocotyl: evaluation of the auxin hypothesis. Planta 156: 388–395 [DOI] [PubMed] [Google Scholar]
- Jenkins MT, Gerhardt F (1931) A Gene Influencing the Composition of the Culm in Maize. Agricultural Experiment Station, Iowa State College of Agriculture and Mechanic Arts, Ames, Iowa [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M. (1998) Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol 116: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Adair CR. (1938) A “lazy” mutation in rice. J Hered 29: 315–318 [Google Scholar]
- Katekar GF, Navé JF, Geissler AE. (1981) Phytotropins. III. Naphthylphthalamic acid binding sites on maize coleoptile membranes as possible receptor sites for phytotropin action. Plant Physiol 68: 1460–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Salzberg SL. (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöller AS, Blakeslee JJ, Richards EL, Peer WA, Murphy AS. (2010) Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J Exp Bot 61: 3689–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J. (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17: 402–410 [DOI] [PubMed] [Google Scholar]
- Li Q, Wan JM. (2005) [SSRHunter: development of a local searching software for SSR sites]. Yi Chuan 27: 808–810 [PubMed] [Google Scholar]
- Li Y, Hagen G, Guilfoyle TJ. (1991) An auxin-responsive promoter is differentially induced by auxin gradients during tropisms. Plant Cell 3: 1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G. (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28: 465–474 [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquès-Bueno MM, Moreno-Romero J, Abas L, De Michele R, Martínez MC. (2011) A dominant negative mutant of protein kinase CK2 exhibits altered auxin responses in Arabidopsis. Plant J 67: 169–180 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson PH, Tasaka M, Morita MT, Guan C, Chen R, Boonsirichai K. (2002) Arabidopsis thaliana: a model for the study of root and shoot gravitropism. The Arabidopsis Book 1: e0043, doi/10.1199/tab.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Hake S. (2001) barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S. (2007) barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Jia WJ, Chu YJ, Xue HW. (2012) Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res 22: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Miller ND, Murphy AS, Gilroy S. (2011) Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J 65: 309–318 [DOI] [PubMed] [Google Scholar]
- Morita MT. (2010) Directional gravity sensing in gravitropism. Annu Rev Plant Biol 61: 705–720 [DOI] [PubMed] [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrásek J, Skůpa P, Chand S, Benková E, Zazímalová E, Friml J. (2008) Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135: 3345–3354 [DOI] [PubMed] [Google Scholar]
- Muday GK. (2001) Auxins and tropisms. J Plant Growth Regul 20: 226–243 [DOI] [PubMed] [Google Scholar]
- Muday GK, DeLong A. (2001) Polar auxin transport: controlling where and how much. Trends Plant Sci 6: 535–542 [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Zhou J, Temple B, Jiang K, Jones AM. (2009) Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. Plant Cell 21: 3591–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Suzuki G, Uehara Y, Saji K, Furukawa T, Koshiba T, Sekimoto M, Fujioka S, Kuroha T, Kojima M, et al. (2008) Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. Plant J 53: 516–529 [DOI] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. (2009) NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics 10: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. (2003) Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423: 999–1002 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Marchant A, May S, Swarup R, Swarup K, James N, Graham N, Allen T, Martucci T, Yemm A, et al. (2001) Quick on the uptake: characterization of a family of plant auxin influx carriers. J Plant Growth Regul 20: 217–225 [Google Scholar]
- Peer WA, Blakeslee JJ, Yang H, Murphy AS. (2011) Seven things we think we know about auxin transport. Mol Plant 4: 487–504 [DOI] [PubMed] [Google Scholar]
- Peer WA, Hosein FN, Bandyopadhyay A, Makam SN, Otegui MS, Lee GJ, Blakeslee JJ, Cheng Y, Titapiwatanakun B, Yakubov B, et al. (2009) Mutation of the membrane-associated M1 protease APM1 results in distinct embryonic and seedling developmental defects in Arabidopsis. Plant Cell 21: 1693–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennazio S. (2002) The discovery of the chemical nature of the plant hormone auxin. Riv Biol 95: 289–308 [PubMed] [Google Scholar]
- Petrásek J, Friml J. (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64: 615–623 [DOI] [PubMed] [Google Scholar]
- Robert HS, Friml J. (2009) Auxin and other signals on the move in plants. Nat Chem Biol 5: 325–332 [DOI] [PubMed] [Google Scholar]
- Roberts JA. (1984) Tropic responses of hypocotyls from normal tomato plants and the gravitropic mutant Lazy-1. Plant Cell Environ 7: 515–520 [Google Scholar]
- Salisbury FJ, Hall A, Grierson CS, Halliday KJ. (2007) Phytochrome coordinates Arabidopsis shoot and root development. Plant J 50: 429–438 [DOI] [PubMed] [Google Scholar]
- Sassi M, Lu Y, Zhang Y, Wang J, Dhonukshe P, Blilou I, Dai M, Li J, Gong X, Jaillais Y, et al. (2012) COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139: 3402–3412 [DOI] [PubMed] [Google Scholar]
- Sassi M, Wang J, Ruberti I, Vernoux T, Xu J. (2013) Shedding light on auxin movement: light-regulation of polar auxin transport in the photocontrol of plant development. Plant Signal Behav 8: e23355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer T, Leyser O. (2006) Auxin transport, but in which direction? Science 312: 858–860 [DOI] [PubMed] [Google Scholar]
- Skirpan A, Culler AH, Gallavotti A, Jackson D, Cohen JD, McSteen P. (2009) BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol 50: 652–657 [DOI] [PubMed] [Google Scholar]
- Smith CM, Desai M, Land ES, Perera IY. (2013) A role for lipid-mediated signaling in plant gravitropism. Am J Bot 100: 153–160 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM. (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohm AK, Baldwin KL, Masson PH. (2012) Multiple roles for membrane-associated protein trafficking and signaling in gravitropism. Front Plant Sci 3: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suge H, Türkan I. (1991) Can plants normally produce seeds under microgravity in space? Jpn J Crop Sci 60: 427–433 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, et al. (2005) PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17: 2922–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Toyota M, Furuichi T, Tatsumi H, Sokabe M. (2008) Cytoplasmic calcium increases in response to changes in the gravity vector in hypocotyls and petioles of Arabidopsis seedlings. Plant Physiol 146: 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Syed N, Laxmi A, Chattopadhyay D. (2009) Role of CIPK6 in root growth and auxin transport. Plant Signal Behav 4: 663–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Overbeek J. (1936) “Lazy,” an A-geotropic form of maize “gravitational indifference” rather than structural weakness accounts for prostrate growth-habit of this form. J Hered 27: 93–96 [Google Scholar]
- Van Overbeek J. (1938) “Laziness” in maize due to abnormal distribution of growth hormone. J Hered 29: 339–341 [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4: 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Kudla J. (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 2008: t4995. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Hall DH, DeBlasio S, Taguchi-Shiobara F, Schmidt RJ, Jackson DP. (2010) A conserved mechanism of bract suppression in the grass family. Plant Cell 22: 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, Pérez AC, Kleine-Vehn J, Vanneste S, Drozdzecki A, Leitner J, Abas L, Aerts M, et al. (2012) GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell 22: 678–685 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ahlers S, Zourelidou M, Barbosa ICR, Demarsy E, Trevisan M, Davis PA, Roelfsema MRG, Hangarter R, Fankhauser C, et al. (2013) D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25: 1674–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ogiso-Tanaka E, Zourelidou M, Schwechheimer C. (2012) WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development 139: 4020–4028 [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES of ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, McSteen P. (2007) The role of auxin transport during inflorescence development in maize (Zea mays, Poaceae). Am J Bot 94: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Wu X, Tang D, Li M, Wang K, Cheng Z. (2013) Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T, Iino M. (2007) Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol 48: 678–688 [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Spalding EP, Iino M. (2013) AtLAZY1 is a signaling component required for gravitropism of the Arabidopsis thaliana inflorescence. Plant J 74: 267–279 [DOI] [PubMed] [Google Scholar]
- Zazímalová E, Murphy AS, Yang H, Hoyerová K, Hosek P. (2010) Auxin transporters: why so many? Cold Spring Harb Perspect Biol 2: a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nodzynski T, Pencík A, Rolcík J, Friml J. (2010) PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Madi S, Borsuk L, Nettleton D, Elshire RJ, Buckner B, Janick-Buckner D, Beck J, Timmermans M, Schnable PS, et al. (2007) Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Müller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C. (2009) The polarly localized D6 protein kinase is required for efficient auxin transport in Arabidopsis thaliana. Development 136: 627–636 [DOI] [PubMed] [Google Scholar]