Pack-MULE transposable elements selectively duplicate bona fide genes.
Abstract
The process of gene duplication followed by sequence and functional divergence is important for the generation of new genes. Pack-MULEs, nonautonomous Mutator-like elements (MULEs) that carry genic sequence(s), are potentially involved in generating new open reading frames and regulating parental gene expression. These elements are identified in many plant genomes and are most abundant in rice (Oryza sativa). Despite the abundance of Pack-MULEs, the mechanism by which parental genes are captured by Pack-MULEs remains largely unknown. In this study, we identified all MULEs in rice and examined factors likely important for sequence acquisition. Terminal inverted repeat MULEs are the predominant MULE type and account for the majority of the Pack-MULEs. In addition to genic sequences, rice MULEs capture guanine-cytosine (GC)-rich intergenic sequences, albeit at a much lower frequency. MULEs carrying nontransposon sequences have longer terminal inverted repeats and higher GC content in terminal and subterminal regions. An overrepresentation of genes with known functions and genes with orthologs among parental genes of Pack-MULEs is observed in rice, maize (Zea mays), and Arabidopsis (Arabidopsis thaliana), suggesting preferential acquisition for bona fide genes by these elements. Pack-MULEs selectively acquire/retain parental sequences through a combined effect of GC content and breadth of expression, with GC content playing a stronger role. Increased GC content and number of tissues with detectable expression result in higher chances of a gene being acquired by Pack-MULEs. Such selective acquisition/retention provides these elements greater chances of carrying functional sequences that may provide new genetic resources for the evolution of new genes or the modification of existing genes.
Transposable elements (TEs) are sequences in the genome that move from one location to another and in the process multiply in copy number. According to the transposition intermediate, TEs are classified into two major classes: class I or RNA elements, which transpose via an RNA intermediate using a copy-and-paste mechanism; and class II or DNA elements, which transpose via a DNA intermediate using a cut-and-paste mechanism. Based on their coding capacity for transposition machinery, both classes of TEs can be divided into autonomous and nonautonomous elements. Autonomous elements encode the protein products (transposase or reverse transcriptase) required for their transposition, whereas nonautonomous elements do not encode relevant proteins and rely on their cognate autonomous elements for transposition. TEs constitute over 50% of many plant genomes and as much as 85% of the maize (Zea mays) genome (Devos et al., 2005; Paterson et al., 2009; Schnable et al., 2009; Schmutz et al., 2010; Tomato Genome Consortium, 2012; Nystedt et al., 2013; Wu et al., 2013). In addition, computational and biological analyses of genomic information have revealed critical roles of transposons in gene expression, regulation, and genome evolution (Bennetzen and Kellogg, 1997; Lippman et al., 2004; Piegu et al., 2006; Ammiraju et al., 2007; Bennetzen, 2007; Slotkin and Martienssen, 2007; Zuccolo et al., 2007; Feschotte, 2008).
The Mutator superfamily is a class II/DNA TE originally discovered in maize (Robertson, 1978). Since the initial discovery of Mu1 and MuDR in maize (Robertson, 1978; Robertson et al., 1989), similar elements were later identified from the maize genome and subsequently in other organisms including plants, animals, and fungi, where they are referred to as Mutator-like elements (MULEs; Yu et al., 2000; Lisch et al., 2001; Chalvet et al., 2003; Jiang et al., 2004; Holligan et al., 2006; Marquez and Pritham, 2010). MULEs are typically characterized by an 8- to 11-bp target site duplication (TSD) flanking the element, with 9-bp TSD as the most frequent form. In addition, the majority of these elements are known for the presence of long terminal inverted repeats (TIRs), which typically range from 100 to 500 bp, a feature that largely sets them apart from other major class II TEs such as En/Spm, Helitron, PIF/Pong, and Tc1/Mariner elements. MULEs associated with long TIRs are referred to as TIR MULEs. TIR sequences appear to be important for element transposition and expression (Benito and Walbot, 1997; Raizada et al., 2001; Jiang et al., 2004). Recently, however, non-TIR MULEs have been reported in Arabidopsis (Arabidopsis thaliana), Lotus japonicus, maize, and yeast (Yarrowia lipolytica; Yu et al., 2000; Neuvéglise et al., 2005; Holligan et al., 2006; Wang and Dooner, 2006). Non-TIR MULEs refer to the MULEs with exceptionally short TIRs (less than 50 bp) and low similarity between the inverted terminal sequences. The detection of non-TIR MULEs in multiple plants suggests that extended long TIRs are dispensable for the transposition of MULEs in plants. Although elements belonging to the same TIR MULE family share an overall sequence similarity in their TIRs, they vary in their internal region. The Mu family of maize that includes multiple elements (Mu1–Mu13) share a 220-bp sequence in their TIRs, but the internal region between the TIRs may contain unique and unrelated sequences (Chomet et al., 1991; Lisch, 2002; Lisch and Jiang, 2009). The Mu4 elements, for instance, have much longer TIRs compared with other Mu elements (530 bp long), and the TIR sequence includes a fragment from a BRASSINOSTEROID INSENSITIVE1 gene (Lisch, 2002). Thus, in addition to differences in the internal sequence, elements within a MULE family vary in their TIR lengths.
Pack-MULEs are nonautonomous Mutator and MULEs that carry genes or gene fragments. Although the abundance of Pack-MULEs was not acknowledged until the availability of the entire rice (Oryza sativa) genomic sequence, the first Mutator element discovered (Mu1) was, in fact, a Pack-MULE carrying a fragment of the MRS-A gene (Talbert and Chandler, 1988), as were the other nonautonomous Mutator elements (Lisch, 2002). To date, Pack-MULEs have been characterized in both monocots and dicots, including rice, maize, L. japonicus, Arabidopsis, tomato (Solanum lycopersicum), and sacred lotus (Nelumbo nucifera; Yu et al., 2000; Jiang et al., 2004; Hoen et al., 2006; Holligan et al., 2006; Schnable et al., 2009; Ferguson and Jiang, 2012; Ming et al., 2013), suggesting their prevalence among plants. The genes from which gene sequences or fragments are captured are referred to as parental genes, and the captured fragment is referred to as the acquired sequence. Previous work identified 2,853 Pack-MULEs in rice that have transduced about 1,500 parental genes (Jiang et al., 2011). Comprehensive analyses showed that over 22% of rice Pack-MULEs are transcribed, with at least 28 elements having evidence of translation (Hanada et al., 2009). These elements often carry gene fragments from multiple loci, forming new open reading frames (ORFs). In addition to the formation of independent ORFs, Pack-MULEs can serve as part of the ORF and/or untranslated region that fuses with adjacent sequences/genes to form chimeric transcripts (Jiang et al., 2011). Pack-MULE transcripts are found in either orientation with regard to the transcription of the parental gene, with a small subset having bidirectional transcription. The formation of antisense transcripts suggests a critical role for Pack-MULE-derived transcripts in regulating the expression of parental genes through the activity of small RNAs (Hanada et al., 2009). In fact, over half of Pack-MULEs in rice are directly involved in the formation of small RNAs. Parental genes that have shared small RNAs with Pack-MULEs show lower expression levels compared with genes without an association with small RNAs (Hanada et al., 2009). Thus far, rice has remained exceptional in its Pack-MULE copy number load. Another advantage of studying Pack-MULEs in rice is the unparalleled quality of its reference genome sequence, which was accomplished using the traditional bacterial artificial chromosome (BAC-by-BAC) sequencing technology (International Rice Genome Sequencing Project, 2005).
Despite progress in MULE and Pack-MULE identification in sequenced higher eukaryotes, the process by which parental genes are captured by these elements remains to be elucidated. Thus far, two probable mechanisms have been proposed. Bennetzen and Springer (1994) suggested a model (model 1) similar to an ectopic gene conversion across a nicked-cruciform structure. Here, ectopic sequences are introduced into the internal region of the element during repair of the nick within the loop. According to this model, acquisition may or may not require the presence of transposase. The second model (model 2) proposes an aberrant gap-repair process that uses ectopic sequences as template during the repair of the empty site. In this model, an excision event is necessary, and the acquisition of new sequences occurs upon the repair of the gap at the donor site (Yamashita et al., 1999). As a result, the acquisition requires the presence of transposase but is not associated with transposition of the element. Both models predict the involvement of short stretches of homology between the broken ends and the new genomic sequence not previously associated with the element, which ultimately becomes incorporated in the internal region. Although neither of the two models has any empirical support at this time, computational analysis of Pack-MULEs in rice, maize, and Arabidopsis has shed some light on the acquisition process. A phenomenon that likely extends to all grass genomes, where significant guanine-cytosine (GC) islands and gradients persist, is the preferential acquisition of GC-rich sequences by Pack-MULEs (Jiang et al., 2011).
In this study, a comprehensive analysis of all MULEs in the rice genome, including Pack-MULEs, was performed to further understand how Pack-MULEs select and acquire parental gene sequences. The results from this study indicate that element TIR and sub-TIR properties differ between Pack-MULEs and non-Pack-MULEs and may be involved in target selection and acquisition. Analysis of the parental genes of Pack-MULEs in rice, maize, and Arabidopsis supports the role of GC content and ubiquity in the expression of the parental genes in sequence acquisition, which explains the significant preference of MULEs to duplicate genic sequences.
RESULTS
Rice MULEs Preferentially Acquire Genic Sequences
To understand the mechanism of sequence acquisition by Pack-MULEs, we compared Pack-MULEs with MULEs that do not carry non-TE genomic sequences. To this end, we established a procedure to collect all MULEs in the rice genome, which resulted in a total of 13,857 MULEs with TSDs (Tables I and II). MULEs were categorized into TIR MULE and non-TIR MULE according to a distinct similarity and length of TIRs (see “Materials and Methods”). Among the MULE elements with TSDs, 87% were TIR MULEs, suggesting that this MULE type is more predominant than the non-TIR MULEs.
Table I. Copy numbers and percentage of different classes of TIR MULEs in the rice genome.
N/A, Not applicable.
| Element Type | TIR Type | Internal Region |
Copy No.a | |
|---|---|---|---|---|
| Protein Match | Parental Copy | |||
| Pack-MULEs | 2,812 (23.21) | |||
| Pack-MULE genic | PMTIR | Known protein | Genic sequence | 2,755 |
| Pack-MULE plus intergenic | PMTIR | Known protein | Genic and intergenic sequences | 57 |
| MULE-intergenic | PMTIR | N/A | Intergenic sequence | 17 (0.14) |
| MULE-HypProt | PMTIR/non-PMTIR | Hypothetical protein | N/A | 1,196 (9.87) |
| MULE-other (non-Pack-MULEs) | PMTIR | N/A | N/A | 3,695 (30.50) |
| Non-PMTIR | N/A | N/A | 3,915 (32.32) | |
| Auto-MULEs | PMTIR/non-PMTIR | MULE transposase | N/A | 479 (3.95) |
| Total | 12,114 | |||
Numbers in parentheses represent percentage of total copy number.
Table II. Copy numbers and percentage of different classes of non-TIR MULEs in the rice genome.
N/A, Not applicable.
| Element Type | TIR Type | Internal Region |
Copy No.a | |
|---|---|---|---|---|
| Protein Match | Parental Copy | |||
| Pack-MULEs | 112 (6.42) | |||
| Pack-MULE genic | PMTIR | Known protein | Genic sequence | 106 |
| Pack-MULE plus intergenic | PMTIR | Known protein | Genic and intergenic sequences | 6 |
| MULE-intergenic | PMTIR | N/A | Intergenic sequence | 5 (0.29) |
| MULE-HypProt | PMTIR/non-PMTIR | Hypothetical protein | N/A | 119 (6.88) |
| MULE-other (non-Pack-MULEs) | PMTIR | N/A | N/A | 428 (24.54) |
| Non-PMTIR | N/A | N/A | 1,038 (59.52) | |
| Auto-MULEs | PMTIR/non-PMTIR | MULE transposase | N/A | 41 (2.35) |
| Total | 1,743 | |||
Numbers in parentheses represent percentage of total copy number.
If the internal region of a MULE has a non-TE genomic homolog, we call the genomic homolog the parental copy or parental gene (if it is from the genic region; see below). According to the internal sequence contained within the TIR, MULEs were further classified into five groups: (1) Pack-MULEs, as defined previously (Jiang et al., 2004), refers to elements containing genic sequence(s) (Supplemental Table S1); (2) MULE-intergenic refers to elements with a non-TE parental copy located in intergenic regions (Supplemental Table S2); (3) MULE-other or non-Pack-MULEs are elements whose internal sequences have no identifiable parental origin/sequence (Supplemental Table S3); (4) Auto-MULEs are elements containing sequences with homology to known Mutator/MULE transposases (Supplemental Table S4); and (5) MULE-HypProt are elements containing annotated hypothetical genes or with homology to hypothetical genes yet without a recognizable parental copy (Supplemental Table S5). MULE-HypProt could represent ancient sequence acquisitions where the internal regions are too diverged or evolved to allow the identification of the parental copies. Alternatively, it is a result of misannotation from an automated gene annotation pipeline. The non-Pack-MULEs in each MULE type were subsequently categorized into two groups based on whether the TIR family is involved in sequence acquisition. PMTIR refers to TIR families that contain or include Pack-MULEs, while non-PMTIR refers to TIR families that contain exclusively non-Pack-MULEs.
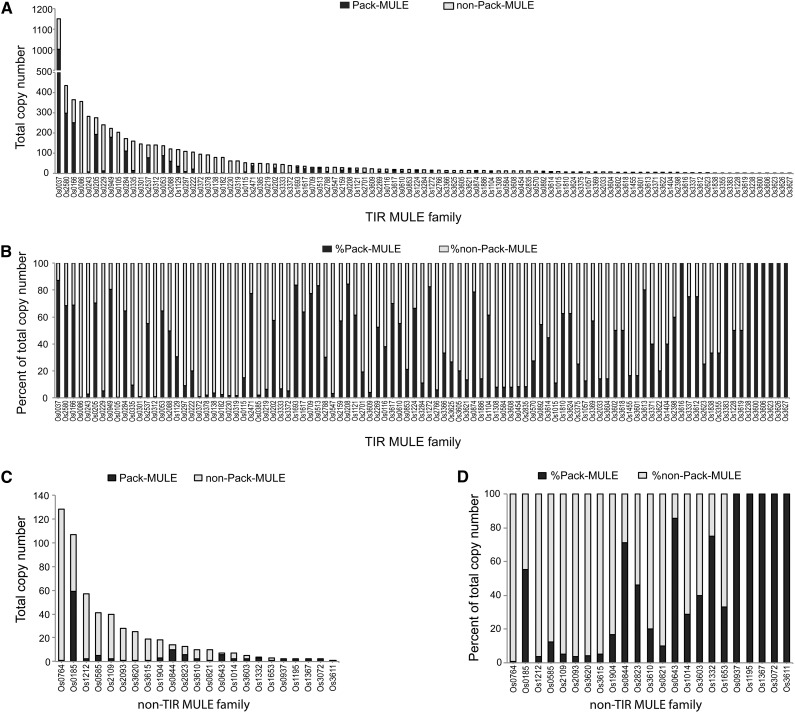
Among the 13,857 MULEs identified, 2,924 (21.1%) carry gene or gene fragments, suggesting that the majority of MULEs do not acquire genes (Tables I and II). A total of 251 TIR families were identified in the rice genome, which included 186 TIR MULEs and 65 non-TIR MULEs. Among these TIRs, 122 were associated with sequence acquisition (referred to as PMTIR). The copy numbers of Pack-MULEs range from one to 1,002 elements/copies per TIR family (Fig. 1, A and C; Supplemental Table S1). The TIR family with the most family members, Os0037, has a total of 1,151 elements, with the majority being Pack-MULEs (87%). Pack-MULEs identified are predominantly of the TIR MULE type (96.2%), suggesting that MULEs with typical long MULE TIRs are more likely to be associated with gene sequence acquisition. This is also true if the abundance of Pack-MULEs is corrected by the total copy number: 23% of the TIR MULEs are Pack-MULEs, while only 6% of the non-TIR MULEs carry gene fragments. Nevertheless, regardless of the MULE type, the composition of Pack-MULEs and non-Pack-MULEs across different MULE TIR families that vary in total copy numbers suggests that the abundance of Pack-MULEs is not correlated to the abundance of the family in the genome (Fig. 1, B and D). In other words, TIR families with high copy numbers are not more likely and frequently to acquire gene fragments than families with fewer copies. Meanwhile, 129 TIR families were devoid of Pack-MULEs (non-PMTIR), comprising a total copy number of 4,953 elements (Supplemental Fig. S1, A and B; Supplemental Table S3).
Figure 1.
Partition of Pack-MULEs and non-Pack-MULEs among TIR MULE and non-TIR MULE families in the rice genome. A, Copy number and Pack-MULE distribution in TIR MULE families associated with gene acquisition. B, Percentage of Pack-MULE and non-Pack-MULE total copy number for TIR MULE families associated with gene acquisition. C, Copy number and Pack-MULE distribution in non-TIR MULE families associated with gene acquisition. D, Percentage of Pack-MULE and non-Pack-MULE total copy number for non-TIR MULE families associated with gene acquisition.
From the 2,924 Pack-MULEs, 1,557 unique parental genes were identified (Supplemental Table S6). Among the Pack-MULEs, 63 also contain intergenic sequences in addition to genic sequences. In addition, 22 MULE-intergenic elements were found (Supplemental Table S2), and all of them are associated with PMTIR. The intergenic components of the 63 Pack-MULEs and 22 MULE-intergenic elements are derived from a total of 60 intergenic parental sequences, suggesting that MULEs can acquire sequences other than genes, albeit at a much lower frequency. To test whether the dearth of intergenic sequence acquisition is a result of a lower proportion of the genome being the source of this type of parental sequences, we calculated the total genic and intergenic space of the rice genome. The intergenic space (79 Mb) is roughly 68% of the size of the genic space (116 Mb). However, there are about 26 times more genic parentals compared with intergenic parentals, and among Pack-MULEs, even more elements (45 times) have acquired only genes compared with those that acquired both genic and intergenic sequences.
The underrepresentation of intergenic sequences among acquisitions by MULEs suggests that genic sequences are preferentially acquired. Alternatively, this may indicate that, compared with the genic components in Pack-MULEs, the intergenic fragments in Pack-MULEs or MULE-intergenics have less selective advantage, so their retention time is shorter. If the latter was the case, one would expect to see more intergenic sequences among newer acquisition events. To test this, the age of acquisition events was roughly estimated based on the transversion rate (the amount of transversion that has occurred between the alignable length of the acquired sequence and the parental sequence). Sequence transversion rate was chosen, as it is a better indicator of age compared with either sequence similarity or transition rate. This is because the transition rate is correlated with GC content in addition to age. The median transversion rate of all acquired sequences (genic and nongenic) was used as the cutoff to classify relatively old (transversion rate > 2.75%) and recent (transversion rate ≤ 2.75%) events. The results show no significant difference in the number of intergenic acquisition events between recent and old acquisitions (2.91% versus 2.94%). Thus, a potential lack of selective advantage does not explain the dramatic underrepresentation of intergenic regions inside MULEs.
Structural Differences between Pack-MULEs and Non-Pack-MULEs
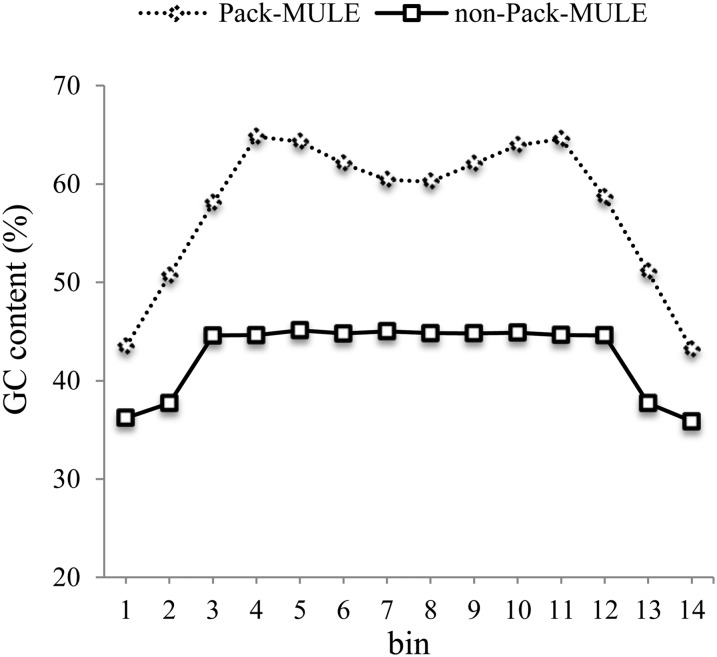
Since non-TIR MULEs do not have well-defined inverted terminal regions and only account for a minor portion of the Pack-MULEs, comparisons of structural differences were limited to elements classified as TIR MULEs. A variety of differences were observed when the sequences of Pack-MULEs were compared with those of non-Pack-MULEs. Overall, Pack-MULEs have a much higher GC content compared with non-Pack-MULEs (median, 58.2% versus 36.5%; P < 2.2 × 10−16, Wilcoxon rank-sum test [WRS]) and are much longer (1,445 versus 441 bp; P < 2.2 × 10−16, WRS). To evaluate the GC gradient along the elements, the TIR regions of each element were divided into two equal-sized bins, while the internal regions were divided into 10 equal-sized bins. As shown in Figure 2, both TIR and internal sequences of Pack-MULEs are more GC rich than those of non-Pack-MULEs. In addition, a steeper increase in GC content (15% increase) is observed from bin 1 to bin 3 of Pack-MULEs.
Figure 2.
GC content along Pack-MULEs and non-Pack-MULEs. The first two and last two bins represent TIR regions, and the internal sequence was divided into 10 equal-sized bins prior to the determination of GC content per bin.
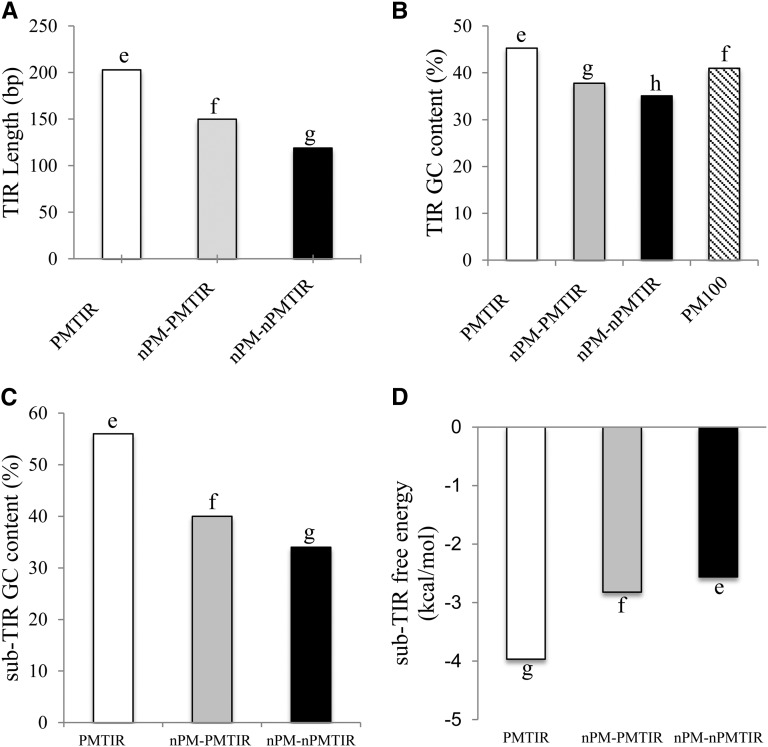
Furthermore, properties among previously deemed critical regions for sequence acquisition, TIR and sub-TIR, were compared between Pack-MULEs and non-Pack-MULEs. In our analysis, the sub-TIR was defined as the 50-bp sequence adjacent to the TIR. As shown in Figure 3, Pack-MULEs have significantly longer TIRs, higher TIR and sub-TIR GC content, and stronger sub-TIR free energy than non-Pack-MULEs (P < 2.2 × 10−16, WRS). If only elements within PMTIR families are considered, there are more non-Pack-MULEs than Pack-MULEs (Table I), yet the TIRs of Pack-MULEs are still longer, with higher GC content in TIR and sub-TIR regions (Fig. 3; P < 2.2 × 10−16, WRS). This suggests that longer TIR and higher GC content are not required or favorable for transposition but may be important in sequence acquisition. Alternatively, these differences may also be a product of a positive feedback mechanism through the acquisition of GC-rich sequences that in some cases can be converted as part of the TIR (as in the case for the Mu4 element mentioned in the introduction), therefore resulting in longer and more GC-rich TIRs and GC-rich sub-TIRs with stronger free energies. However, analysis of GC content using only the first 100-bp sequence of the Pack-MULE TIRs, a size more similar to the average TIR length of non-Pack-MULEs, shows that even the most terminal end of Pack-MULEs is more GC rich than TIRs of non-Pack-MULEs (P < 2.2 × 10−16, WRS; Fig. 3B). Since this region is distal to the internal region, it is unlikely that the higher GC content in this region in Pack-MULEs is a direct or an immediate consequence of acquisition. However, it could be an indirect consequence or result of selection if the higher GC content of TIRs promotes acquisition.
Figure 3.
Structural differences between Pack-MULEs and non-Pack-MULEs based on TIR and sub-TIR sequences. A, Median TIR length. B, Median TIR GC content. C, Median sub-TIR GC content. D, Median sub-TIR free energy. nPM-PMTIR, Non-Pack-MULEs with Pack-MULE-associated TIRs; nPM-nPMTIRs, non-Pack-MULEs with non-Pack-MULE-exclusive TIRs; PM100, using only the first 100-bp sequence of Pack-MULE TIRs. Bars designated with different letters indicate values that are significantly different (α = 0.008 for B and α = 0.02 for A, C, and D) by WRS with Bonferroni correction.
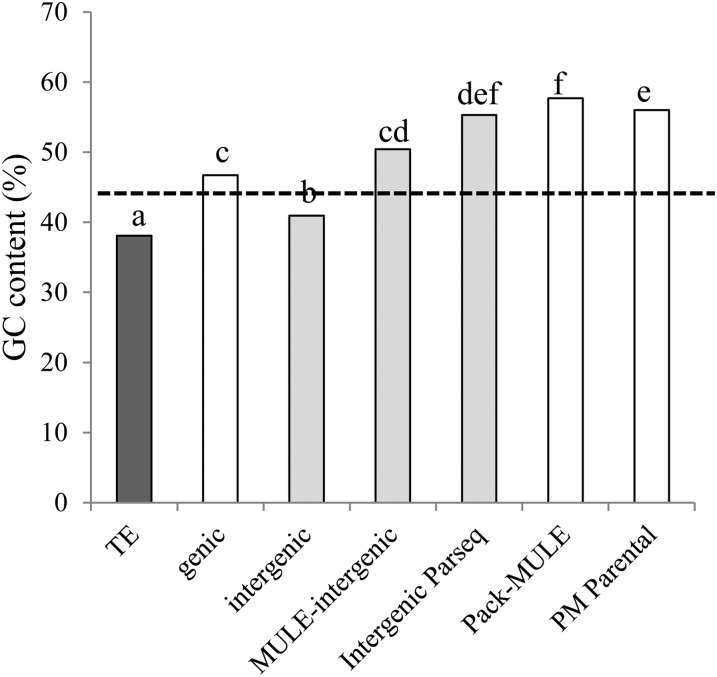
Since previous work has reported the importance of GC content in the acquisition of genic sequences by Pack-MULEs (Jiang et al., 2011), we tested the role of GC content in the acquisition of intergenic sequences by MULEs. Intergenic parental sequences of MULEs are significantly more GC rich than the overall TE and intergenic sequence of the genome (P = 1.687 × 10−15 and P = 2.59 × 10−12, respectively, WRS; Fig. 4). Similarly, Pack-MULE parental genes are significantly more GC rich than the overall genic sequence of the genome (P < 2.2 × 10−16, WRS; Fig. 4), suggesting that the preference for GC-rich sequences applies to both genic and nongenic regions.
Figure 4.
GC content of different genomic sequences in the rice genome. Genome average GC content is indicated by the dashed line. Bars designated with different letters indicate values that are significantly different (α = 0.002) by WRS with Bonferroni correction.
Underrepresentation of Genes with Unknown Function among Parental Genes
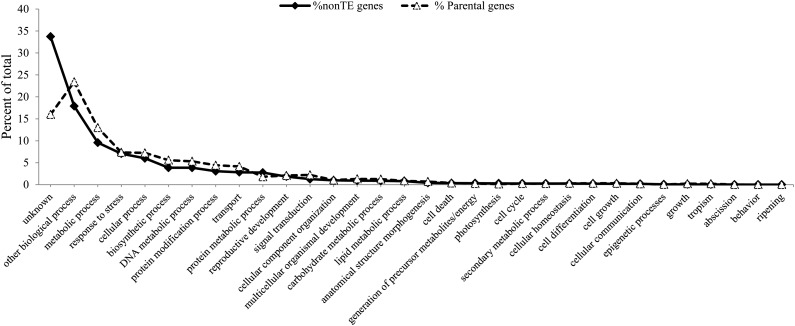
Although Pack-MULEs preferentially acquire GC-rich genes, it is not known whether they also prefer certain classes of genes or if acquisition based on gene function is random. If acquisition is random, we would expect no differences in the ratio of non-TE genes and Pack-MULE parental genes for each functional category. To test this hypothesis, the ratio of non-TE genes and rice parental genes among different GOSlim assignments of biological processes was evaluated using functional assignments and annotations made by the Rice Genome Annotation Group at Michigan State University (Kawahara et al., 2013). A total of 32 biological process categories, which includes “unknown” for genes without an assignment, were compared between Pack-MULE parental genes and non-TE genes. As shown in Figure 5, a slight overrepresentation of genes involved in biosynthetic and metabolic processes (χ2 test, P < 2.2 × 10−16) and a strong bias against genes with unknown classification among parental genes (χ2 test, P < 2.2 × 10−16) were observed. This slight preference for a few categories dissipates, however, when the unknown category is excluded from the analysis (Supplemental Fig. S2). A comparison was also conducted in maize and Arabidopsis to determine whether such a bias against genes with unknown function exists in other plant species where Pack-MULEs have been characterized. In both species, a significant underrepresentation of genes with unknown function among Pack-MULE parental genes was also found (χ2 test, maize, P < 2.2 × 10−16, Arabidopsis, P = 0.03; Tables III and IV).
Figure 5.
Percentage of GOSlim categories of Pack-MULE parental genes and all non-TE genes of rice.
Table III. GC content and expression information among Pack-MULE parental genes and non-TE genes in maize according to functional assignment.
PMPar, Pack-MULE parental gene. Numbers in each column followed by different letters are significantly different (α = 0.008 with Bonferroni adjustment).
| Gene | Total | Percentage GC Genomic | Percentage GC CDS | FPKMa | No. of Librariesa | No Expression | Percentage No Expression |
|---|---|---|---|---|---|---|---|
| Non-TE, gene unknown | 13,057 | 50.10 b | 56.50 b | 39.60 a | 11.00 a | 4,196 | 32.1 d |
| Non-TE, gene known | 26,599 | 47.50 a | 55.90 a | 70.14 b | 12.00 b | 2,873 | 10.8 b |
| PMPar, unknown | 47 | 57.20 c | 63.60 a,c | 45.43 b | 12.00 b | 2 | 4.3 a |
| PMPar, known | 188 | 55.45 c | 63.50 c | 62.71 b | 12.00 b | 7 | 3.7 a |
Median was determined only from genes that are expressed.
Table IV. GC content and expression information among Pack-MULE parental genes and non-TE genes in Arabidopsis according to functional assignment.
PMPar, Pack-MULE parental gene. Numbers in each column followed by different letters are significantly different (α = 0.008 with Bonferroni adjustment).
| Gene | Total | Percentage GC Genomic | Percentage GC CDS | FPKMa | No. of Librariesa | No Expression | Percentage No Expression |
|---|---|---|---|---|---|---|---|
| Non-TE, gene unknown | 10,239 | 39.50 b | 43.50 a | 33.83 a | 5.00 a | 4,584 | 44.8 d |
| Non-TE, gene known | 17,147 | 39.30 a | 44.40 b | 54.33 b | 6.00 b | 3,088 | 18.0 c |
| PMPar, unknown | 7 | 41.80 a,b | 43.20 a,b | 46.92 a,b | 2.50 a,b | 1 | 14.3 b |
| PMPar, known | 28 | 39.80 a,b | 44.65 a,b | 41.80 a,b | 6.00 a,b | 2 | 7.1 a |
Median was determined only from genes that are expressed.
To understand the mechanism underlying the apparent bias against genes without a known function, we compared the GC content among genes with and without a GOSlim assignment, since it is known that GC richness is favored in sequence acquisition/retention. Among non-TE genes, those without a GOSlim assignment have significantly higher GC content than counterparts with a GOSlim assignment both at the genomic and coding sequence (CDS) levels (genomic, P < 2.2 × 10−16; CDS, P < 2.2 × 10−16, WRS; Table V). Among Pack-MULE parental genes, the GC content difference between GOSlim genes and unknown genes was detectable at the genomic sequence level (P = 0.001, WRS; Table V) but not significant at the CDS level. In all four comparisons, genes with unknown category have higher or comparable GC content than those with assigned function(s). These results suggest that the gene GC content does not explain the underrepresentation of genes with unknown biological function among Pack-MULE parental genes in rice. Similarly, maize non-TE genes with unknown function have significantly higher GC content than those with known function (genomic, P < 2.2 × 10−16; CDS, P < 5.589 × 10−16, WRS; Table III). In Arabidopsis, genes with unknown function had significantly higher GC content than genes with known function only at the genomic level (P = 0.01; Table IV). These data show that acquisition bias against genes with unknown function is not species specific and supports the notion that GC content does not explain this finding.
Table V. GC content and expression information among Pack-MULE parental genes and non-TE genes in rice according to GOSlim assignment.
PMPar, Pack-MULE parental gene; NoSlim, genes without a GOSlim assignment; WithSlim, genes with a GOSlim assignment. Numbers in each column followed by different letters are significantly different (α = 0.008 with Bonferroni adjustment).
| Gene | Total | Percentage GC Genomic | Percentage GC CDS | FPKMa | No. of Librariesa | No Expression | Percentage No Expression |
|---|---|---|---|---|---|---|---|
| Non-TE, gene NoSlim | 12,010 | 51.80 b | 59.50 b | 5.81 a | 6.0 a | 6,541 | 54.46 d |
| Non-TE, gene WithSlim | 23,609 | 45.20 a | 55.00 a | 9.98 b | 8.0 c | 3,671 | 15.55 b |
| PMPar, NoSlim | 250 | 59.00 d | 68.30 c | 6.79 a | 7.0 a,b | 60 | 24.29 c |
| PMPar, WithSlim | 1,334 | 56.40 c | 67.30 c | 9.41 b | 7.0 b | 145 | 11.10 a |
Median was determined only from genes that are expressed.
Some of the genes with unknown function might be the result of misannotation. Thus, it is feasible that sequences misannotated as genes are overrepresented within the unknown group and that the apparent bias against them indicates a preference for bona fide genes. To test this, we surveyed the distribution of parental genes among non-TE genes with and without an ortholog (Schnable et al., 2009; Lin et al., 2010; Davidson et al., 2012), since genes with orthologs are more likely bona fide genes. For both rice and maize, genes with orthologs are significantly enriched among Pack-MULE parental genes (χ2 test, rice, P < 2.2 × 10−16; maize, P = 4.152 × 10−14; Supplemental Table S7). For Arabidopsis, an enrichment is observed among parental genes (91% have orthologs; Supplemental Table S7), yet this overrepresentation is not statistically significant, most likely due to the low number of parental genes.
The Effect of Gene Expression on Sequence Acquisition and Its Interaction with GC Content
Since GC content does not explain the discrepancy in gene acquisition preference mentioned above, other factors that may influence sequence acquisition were explored. The role of gene expression in rice was tested using RNA-Seq data (Michigan State University Rice Genome Annotation Group) from 10 different rice developmental stages encompassing diverse vegetative and reproductive tissues (Davidson et al., 2012). A gene was considered expressed if the fragments per kilobase of exon per one million fragments mapped (FPKM) value was 1.0 or greater in at least one expression library; otherwise, the gene was categorized as not expressed. Over one-half (54%) of non-TE genes without a GOSlim assignment were not expressed, while only 15% of non-TE genes with known functions were not expressed (Table V). Meanwhile only 24% of Pack-MULE parental genes without a GOSlim assignment and even fewer, 11%, of those with GOSlim assignments were not expressed, suggesting that gene expression may play a role in the preference for acquisition.
The role of gene expression was also evaluated in maize and Arabidopsis. Maize RNA-Seq expression data were obtained from a previous study (Davidson et al., 2011), and expression was determined using parameters similar to rice. Since similarly comprehensive RNA-Seq expression data generated from a single experiment were not readily available in Arabidopsis, we utilized the massively parallel signature sequencing data set with expression levels of genes from multiple tissues (Meyers et al., 2004a, 2004b). Using only uniquely mapping signatures, a gene was considered expressed if the transcripts per one million (TPM) value was 5.0 or greater in at least one expression library. In both species, significantly more genes with unknown function were not expressed compared with genes with known function (χ2 test, P < 2.2 × 10−16; Tables III and IV). However, the number of nonexpressed genes from either category was much lower among parental genes than the genomic average, suggesting that the underrepresentation of genes with unknown function among parental genes is connected to the lack of expression of unknown genes in all three species.
To further assess the role of gene expression, the level of expression, determined by the FPKM/TPM value, and the number of tissues with expression were compared among different groups of genes. In all three species, expressed genes with unknown function have significantly lower expression levels (P < 2.2 × 10−16, WRS) and fewer tissues with detectable expression (P < 2.2 × 10−16, WRS) than those with a GOSlim assignment (Tables III–V). The expression levels of parental genes of Pack-MULEs do not significantly differ from the genomic average, with the exception of the maize parental genes with unknown function, which showed a significantly higher expression level than the genomic average (P = 5.923 × 10−6, WRS). Interestingly, parental genes with or without a known function were expressed in similar numbers of tissues in all three species (Tables III–V), suggesting that the breadth of expression is critical to sequence acquisition. Thus, the high percentage of genes with no expression and genes with less ubiquitous expression explains why genes without a GOSlim assignment are underrepresented in the parental genes of Pack-MULEs.
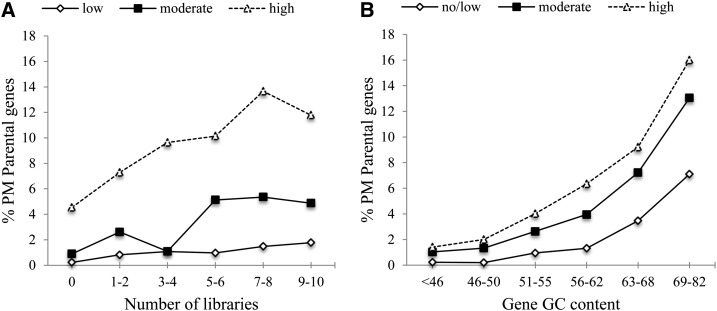
To determine whether the roles of GC content and the breadth of gene expression on sequence acquisition are independent, we categorized rice genes into different GC content groups (low, moderate, and high) as well as different expression categories (no/low, moderate, and high) based on the number of tissues with detectable expression (FPKM ≥ 1.0) and determined the proportion of Pack-MULE parental genes within each group. Although the results above suggest that expression plays a similar role in acquisition preference in maize and Arabidopsis, the analysis in this section was limited to the rice data, due to the much lower number of parental genes in the other two species. Our results in rice indicate that both GC content and the number of tissues with expression evidence play a role in sequence acquisition preference by Pack-MULEs. The ratio of parental genes among non-TE genes was used to reflect the acquisition frequency (how frequently a certain group of genes was acquired). As shown in Figure 6A, for low GC genes and moderate GC genes, a very minimal and modest increase in the proportion of parental genes, respectively, may be observed, with increase in the number of tissues with expression. In comparison, among high GC genes, a stronger increase in the ratio of parental genes occurs with more expression libraries. Meanwhile, when genes are categorized according to the number of tissues with expression, a more substantial increase in the ratio of parental genes is observed in all three expression groups as GC levels increase, and the increase is much greater among genes expressed in eight to 10 tissues (Fig. 6B). It is clear, however, that GC content plays a more dominant role than gene expression for sequence acquisition/retention. This is because variation of GC content may lead to as much as an 11-fold change in the percentage of parental genes, while that for gene expression is only 2- to 5-fold. In addition, the increase in GC content is accompanied by a boost in the percentage of parental genes, despite their expression patterns. In contrast, the effect of gene expression on the percentage of parental genes is only substantial when the genes have moderate or high GC content (Fig. 6A). It is also interesting that the effect of gene expression plateaued with expression in seven or more tissues (Fig. 6A). That explains why the median value of the number of tissues (seven tissues) with expression for parental genes with known function is slightly lower than the genomic average (eight tissues; Table V) in rice, because more ubiquitous expression (in more than seven tissues) does not confer additional advantage for acquisition.
Figure 6.
The effect of GC content and expression in gene acquisition frequency. A, Relationship between expression breadth and the ratio of parental genes among genes grouped on GC content range (low, 30%–50% GC; moderate, 51%–62% GC; high, 63%–81% GC). B, Relationship between gene GC content and the ratio of parental genes among genes grouped on number of tissue expression range (no/low, zero to one library; moderate, two to seven libraries; high, eight to 10 libraries).
The Enrichment of GC-Rich Sequences inside Pack-MULEs Is Due to Selective Acquisition and Preferential Retention
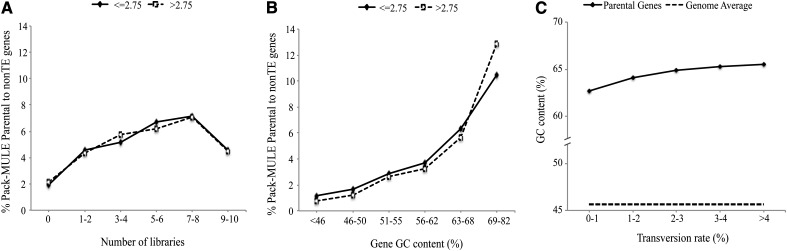
The apparent preference for higher GC content and relatively ubiquitous expression in sequence acquisition in rice, however, can be an artifact of selection, since sequences with higher GC content and more ubiquitous expression are more likely derived from coding regions and, thus, are more likely to be functional. If that is the case, one would expect the preference to be more dramatic among old than among recent acquisition events. Again, the age of acquisition events was roughly estimated through the transversion rate between the acquired sequence and the parental gene. Parental genes were separated into two groups: recent acquisitions, those with transversion rate of 2.75% or less; and old acquisitions, those with transversion rate of 2.75% or greater. As shown in Figure 7A, the two groups of parental genes show an overall similar percentage with increasing number of tissue expression, suggesting that the number of expressed tissues does not have a significant influence on the retention of their gene fragments. In contrast, there are significantly more parental genes in old acquisition events (transversion rate > 2.75%) compared with recent events among genes with a GC content of 69% to 82% (Fig. 7B), suggesting that selection may play a role in the apparent enrichment of parental genes with extremely high GC content. To further characterize the impact of gene GC content on the retention of the relevant gene fragments, we tested the relationship of GC content and transversion rate of all rice parental genes and found a low, albeit statistically significant, correlation (0.09; P = 0.0003, Spearman; Fig. 7C); that is, the GC content of parental genes progressively increases with transversion rate between Pack-MULEs and the parental genes. Again, this indicates that selection plays a role in the retention of GC-rich genes.
Figure 7.
Comparison of GC content and the breadth of expression of parental genes between recent and old acquisition events estimated by transversion rate. A, Effect of acquisition age on the breadth of gene expression. B, Effect of acquisition age on GC content. C, Relationship between the GC content of parental genes and the transversion rate.
To obtain the best possible assessment of the GC content of parental genes upon acquisition, we calculated the GC content of the 14 parental genes with a 0% transversion rate. Theoretically, these sequences represent the most recent acquisition events and have been subjected to little selection. The GC content of all of them is higher than 50%, and the average value is 66.1%, which is dramatically higher than the genome average GC content (45.6%) of non-TE genes. This fact, together with the minor increment of GC content of parental genes over evolutionary time (Fig. 7C), suggests a strong preference for GC-rich genes upon acquisition. Taken together, our results suggest that selection may play a role in the retention of fragments from different parental genes, although it is insufficient to fully explain the enrichment of GC-rich genes among parental genes of Pack-MULEs.
DISCUSSION
The process of gene duplication followed by sequence and functional divergence (neofunctionalization) is one of the most important means for the generation of new genes (Flagel and Wendel, 2009). Studies have shown that all major families of TEs are involved in gene duplication in plants (Jiang et al., 2004; Kawasaki and Nitasaka, 2004; Morgante et al., 2005; Zabala and Vodkin, 2005; Wang et al., 2006; Schnable et al., 2009). In the rice genome, over 1,500 parental genes have been transduced by Pack-MULEs, which can generate independent or chimeric transcripts when fused with nearby sequences (Jiang et al., 2011). In addition, these transcripts may regulate parental gene expression, suggesting a very important role of Pack-MULEs in novel gene formation and evolution. It was shown previously that Pack-MULEs preferentially acquire GC-rich sequences, a phenomenon only seen in grasses. Aside from that, the process by which these sequences are selected and captured by Pack-MULEs remains largely an enigma.
Our findings in this study show that rice TIR MULEs have a higher propensity to acquire genomic sequences compared with non-TIR MULEs, and this bias may be related to differences in structural properties such as TIR length and TIR GC content. It remains unclear at this stage whether the capacity of sequence acquisition among TIR MULEs contributes to the overall success of TIR MULEs versus non-TIR MULEs, since 87% of the rice MULEs belong to TIR MULEs. MULEs and Pack-MULEs in rice are capable of acquiring both genic and intergenic sequences, although the acquisition preference for genic sequences is much more pronounced compared with intergenic sequences (Table I). This may suggest that Pack-MULEs are either more competent to acquire genes or that genes are more readily acquired by Pack-MULEs over other sequences in the genome. Consistent with previous work, GC content was a factor in the preferential acquisition or retention of intergenic fragments, because the GC content of the intergenic sequences inside MULEs or Pack-MULEs is much higher than the genomic, TE, and intergenic GC contents. Interestingly, the GC-rich internal sequences of Pack-MULEs are accompanied by higher GC content of TIRs and sub-TIRs in Pack-MULEs compared with that of non-Pack-MULEs. One of the models for sequence acquisition suggests the formation of a cruciform structure during the process, with the TIRs forming the stem of the hairpin (Bennetzen and Springer, 1994). In this model, an endonucleolytic attack occurs in the single-stranded loop, aided by sequences containing homology to a parental sequence, and initiates repair through illegitimate recombination. Our results are consistent with this model in the following respects. On one hand, the GC-rich internal regions and GC-rich sub-TIRs seem to imply that sequence homology between the element and the acquisition target likely plays a role in acquisition. If this is the case, one would expect AT-rich sequences to be acquired as well if the sub-TIR sequence is also AT rich. This was not observed, since non-Pack-MULEs have relatively AT-rich sub-TIRs (Fig. 2) but they do not carry any recognizable genomic sequences. This is possibly because the pairing of AT-rich sequences is not as stable as that of GC-rich sequences to initialize the repair process. On the other hand, a long TIR would lead to a more stable cruciform that may facilitate the acquisition process. This hypothesis may also explain why MULEs are more frequently associated with sequence acquisitions than other “cut-and-paste” DNA transposons, in that most MULEs have extended long TIRs.
Although a particular functional category of genes does not seem to be more preferentially acquired by Pack-MULEs, a bias against genes with unknown function is obvious, and this was not species specific. Hypothetical genes and genes with unknown function can often result from misannotation. These genes, in most cases, are generated by gene prediction programs and, therefore, may lack supporting expression data. As a result, it is conceivable that there are more false-positive annotations within this group compared with genes with known function. Such bias may reflect the preference of Pack-MULEs for bona fide genes. In other words, Pack-MULEs might be better than gene annotation programs in distinguishing genuine genes from other sequences. The underrepresentation of genes without a known biological function prompted the analysis of expression among annotated genes, which showed that a relatively ubiquitous expression throughout development may play a role in sequence acquisition: Pack-MULEs preferentially acquire genes that are expressed in multiple tissues/developmental stages (Fig. 6A). Our analysis also shows an enrichment of genes with orthologs among parental genes (Supplemental Table S7). More importantly, our data explain the strong propensity of Pack-MULEs to transduce genes over other nongenic sequences in the genome.
Gene GC content and the ubiquity of expression show an additive effect on preferential selection and acquisition by Pack-MULEs. Interestingly, it appears that selection acts differentially on GC content and the ubiquity of expression. The preference for expression ubiquity seems to be largely at the acquisition level, since it is evenly distributed among old and recent acquisition events (Fig. 7A). In contrast, there is a detectable level of selection that favors the retention of fragments from highly GC-rich genes (Fig. 7, B and C). Such differentiation is understandable from a mechanistic point of view: once a gene fragment is acquired by a Pack-MULE, the characteristic of expression is no longer associated with the fragment. Since the acquired fragments may not be expressed in the same pattern as their parental copies, there is no basis for selection for or against the expression pattern of the parental genes. This is consistent with the fact that Pack-MULEs are often associated with different tissue specificity from their parental genes (Hanada et al., 2009). On the other hand, GC content is always associated with the fragment. High GC content could induce a series of genetic and epigenetic changes in the genome. Genetically, it may modify the 5′ end of the adjacent genes and intensify the negative GC gradient (Jiang et al., 2011). Epigenetically, GC-rich sequences offer more methylation targets that could influence the chromatin structure and expression of the nearby genes (Kalisz and Purugganan, 2004; Tatarinova et al., 2010; see below). All these features may form the basis for selection. Apparently, the high GC content is favored here, which may imply that it has provided certain benefits for the organism. Despite the possible selection for high GC content over evolutionary time, the degree of selection seems too moderate to explain the dramatic difference in the GC content between parental genes and all non-TE genes (Fig. 7C). Accordingly, it is likely that the preferential acquisition by Pack-MULEs for GC-rich genes is also responsible, or more important, for the enrichment of GC-rich genes among parental genes. In addition, we cannot rule out the possibility that variation in GC content over evolutionary time is due to a change in acquisition preference. This could occur, for example, when different MULE families have slightly different acquisition preferences and their amplification rate has not been constant in each time range. Future computational and biochemical analyses are required to test whether acquisition preference varies among different MULE families.
To our knowledge, our study is the first to elucidate the direct involvement of gene expression in sequence duplication by DNA transposons. Furthermore, our data suggest that GC richness offers a more dominant effect in this process. This is because, as discussed above, high GC content is very likely favored by both acquisition and selection. Studies to determine the relationship between GC content and expression level and the breadth of expression are conflicting, with studies reporting a strong positive correlation (Lercher et al., 2003; Kudla et al., 2006) and those reporting weak or unclear correlation (Gilbert et al., 2004; Sémon et al., 2005). Nevertheless, these and other studies established the association of GC-rich sequences with open chromatin (Vinogradov, 2003). The open chromatin provided by GC-rich sequences potentially allows these sequences to be more accessible by host enzymes during interrupted gap repair (model 2; see the introduction) or during internal strand repair of cruciform structures (model 1; see the introduction). Expression level, as measured by FPKM/TPM values, does not appear critical to the likelihood of a gene being transduced by a Pack-MULE (Tables III–V). Nevertheless, we cannot rule out the possibility that parental genes were expressed at a level higher than average prior to the formation of relevant Pack-MULEs. This is because Pack-MULEs have a negative regulatory effect on the expression level of parental genes, which may render the difference no longer detectable after the acquisition. In contrast, preferential acquisition is positively correlated with the breadth of expression when the genes are expressed in seven or fewer tissues (Fig. 6). The open chromatin configuration during active transcription may allow access to a sequence for duplication. Consequently, the greater number of tissues with detectable expression allows the gene greater chances of being transduplicated by Pack-MULEs.
CONCLUSION
The unprecedented copy number of Pack-MULEs, the massive duplication of thousands of genes in the rice genome, combined with their biased acquisition for GC-rich genes and insertion in 5′ regions of genes suggest an evolutionary importance of these elements in gene evolution and regulation. Our findings in rice show that sequence acquisition by Pack-MULEs relies on structural/sequential properties of the elements and the acquisition targets. TIR MULEs are the predominant MULE type in the rice genome and account for the majority of the Pack-MULEs. Although Pack-MULEs can duplicate both genic and intergenic sequences, a much stronger preference for genic sequences exists. Pack-MULEs exhibit a non-species-specific bias against genes with unknown function and enrichment of parental genes with orthologs, suggesting its preferential acquisition for bona fide genes. Structural properties of elements, GC content, and the breadth of expression of parental genes influence the selection and acquisition of sequences. Increased GC content and number of tissues with detectable expression results in a higher likelihood of a gene being acquired by a Pack-MULE. Moreover, GC-rich sequences acquired by Pack-MULEs are preferentially retained compared with sequences that are not so GC rich. Although the molecular mechanism for how Pack-MULEs locate and duplicate intergenic and genic sequences remains to be empirically evaluated, our study demonstrates that the activity of Pack-MULEs leads to the selective duplication/retention of CDSs, because CDSs are more GC rich and have a wider breadth of tissue expression. Such selection enables them to carry the most likely functional sequences instead of “junk” and so provide new resources for the evolution of new genes or the modification of existing genes.
MATERIALS AND METHODS
Identification of MULEs and Pack-MULEs
The sequences for rice (Oryza sativa subsp. japonica ‘Nipponbare’) pseudomolecules and gene annotation information were downloaded from the Rice Genome Annotation Project at Michigan State University (http://rice.plantbiology.msu.edu/; release 7.0). Prior to the identification of MULEs and Pack-MULEs, MULE TIRs were classified as TIR MULEs or non-TIR MULEs. MULE families whose TIRs are at least 50 bp in length with at least 60% similarity are considered as TIR MULEs. However, MULE families less than 150 bp in size (small elements) where TIR length is at least 40 bp and their terminal sequence is related to a TIR MULE were also classified as TIR MULEs, because the short TIR is due to deletion and not to phylogeny origin. All other families are considered non-TIR MULEs. The procedure for the annotation and identification of Pack-MULEs was similar to that described previously (Hanada et al., 2009). The annotation of other MULEs was similar to that of Pack-MULEs, except that there is no requirement for the internal region of MULEs to match proteins. Auto-MULEs are MULEs with matches to previously known MULE transposases. Elements with flanking 9- to 11-bp TSD with no more than two mismatches (or 1-bp mismatch plus 1-bp insertion/deletion) were accepted for further classification and analysis. For elements with 8-bp TSD, only 1-bp mismatch or 1-bp insertion/deletion was accepted. The presence of TSD for non-Pack-MULEs was detected by custom perl scripts, and a maximum 10-bp swing from the putative element ends was allowed. For all elements with parental copies, TSD was verified by manual examination of elements and flanking sequences.
The identification of the parental origin of the sequences captured by MULEs and Pack-MULEs was conducted as described previously (Jiang et al., 2011). For an individual Pack-MULE, the sequence with the highest similarity score (BLASTN, E = 1e−10) that was not associated with a MULE TIR was considered as the parental copy of the internal sequence in a Pack-MULE. Elements without matching any proteins that did not contain a recognizable parental genomic sequence were classified as MULE-other or non-Pack-MULEs. Elements with recognizable nongenic parental sequences were classified as MULE-intergenic. Elements with hits only to hypothetical proteins and without parental sequences were classified as MULE-HypProt.
TIR and Sub-TIR Analyses
Since the majority of Pack-MULEs belong to TIR-MULEs, TIR and sub-TIR analyses were performed only on the TIR-MULEs. To identify the TIR length of each individual element, the terminal 800-bp sequence (or half of the element if the element is shorter than 1,600 bp) of each element was aligned using DIALIGN2 (Morgenstern, 2004). A custom perl script was used to determine the length of the TIR on each side, whereby considerable sequence alignment falls off. The sub-TIR was defined as the 50-bp sequence immediately following the TIR, as determined previously. The GC content of each individual TIR and sub-TIR sequence was calculated using a custom perl script. Calculations of sub-TIR free energy were performed using UNAFold (Markham and Zuker, 2008), available at http://mfold.rna.albany.edu. The statistical difference between each group was examined using the R package (http://www.r-project.org). A Bonferroni correction was applied to account for multiple comparisons.
Analysis of GC Content
To calculate the GC content of MULEs and Pack-MULEs, nested TE insertions were first curated and removed from the element sequence. Determination of the GC content of parental genes was conducted after masking with the rice repeat library that excluded Pack-MULEs. To calculate the GC gradient along MULE sequences, the TIR sequences (on both ends of the elements) and the internal region (the sequences between the TIRs) were divided into two and 10 equal-sized bins, respectively. A custom perl script was used to determine the GC content of each bin. Comparisons of GC content between groups were performed using the R package (http://www.r-project.org).
Gene Functional and Expression Analyses
The biological process GOSlim assignments and RNA-Seq expression data for rice genes were downloaded from the Rice Genome Annotation Project at Michigan State University (http://rice.plantbiology.msu.edu/). GOSlim categories were calculated such that a total count of 1 was generated from each gene; that is, genes with multiple GOSlim assignments were given an equal proportion totaling to 1. To classify expressed genes, only RNA-Seq libraries with calculated FPKM values were used, to avoid misclassifying background or noise reads from expression calls made using a single read to a gene. Genes were considered expressed if the FPKM values were 1 or greater in at least one expression library.
The maize (Zea mays) filtered gene set sequence (release 5b) and functional annotation were downloaded from the maize sequencing project (http://www.maizesequence.org). The Arabidopsis (Arabidopsis thaliana) gene sequences and functional annotation were downloaded from The Arabidopsis Information Resource 10 (http://www.arabidopsis.org). The genes were classified as “known” if a functional annotation is available; otherwise, the genes were classified as unknown. Maize RNA-Seq expression data were obtained from previously published work (Davidson et al., 2011). Evaluation of the expression of maize genes was similar to rice (FPKM ≥ 1 in at least one expression library) from RNA-Seq, which includes 13 different expression libraries. Since a similarly comprehensive RNA-Seq expression library is not readily available for Arabidopsis, the MPSS data set from eight different expression libraries was downloaded (http://mpss.udel.edu/at/mpss_index.php; Meyers et al., 2004a, 2004b). To determine the expression patterns of genes in Arabidopsis, eight libraries were used, and a gene was classified as expressed if the TPM values were 5 or greater in at least one expression library. Comparisons of various expression parameters among groups were performed using the R package (http://www.r-project.org).
To determine the distribution of parental genes among non-TE genes with and without orthologs, Arabidopsis and rice gene orthologous data were downloaded from the Rice Genome Annotation Project at Michigan State University (http://rice.plantbiology.msu.edu/; Lin et al., 2010; Davidson et al., 2012). For maize genes, data were downloaded from the maize sequencing project (http://www.maizesequence.org; Schnable et al., 2009).
Aging Acquisition Events
To roughly estimate the age of the genic and intergenic acquisition events, Pack-MULE and MULE-intergenic sequences was aligned to parental sequences using BLASTN (M = 5, N = −11, Q = 22, R = 11, E = 1e-10, wordmask = dust, wordmask = seg, hspsepSmax = 100, hspsepQmax = 100) to determine the boundary of the alignable region. Subsequently, each pair of alignable sequences were aligned using MUSCLE (Edgar, 2004), and the output was further processed by custom perl scripts to calculate the number of transversion events between aligned sequences as well as the transversion rate for each sequence pair. An average transversion rate was assigned for parental genes that were acquired by multiple Pack-MULEs.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Copy number of MULE TIR families not associated with gene acquisition.
Supplemental Figure S2. Percentage of GOSlim categories of Pack-MULE parental genes and non-TE genes of rice, excluding genes without a functional assignment.
Supplemental Table S1. List of rice Pack-MULEs.
Supplemental Table S2. List of rice MULEs with intergenic sequence acquisition (MULE-intergenic).
Supplemental Table S3. List of rice non-Pack-MULEs.
Supplemental Table S4. List of rice autonomous MULEs.
Supplemental Table S5. List of rice MULE-HypProt.
Supplemental Table S6. List of parental genes of Pack-MULEs.
Supplemental Table S7. Distribution of non-TE and parental genes among genes with and without orthologs.
Supplemental Sequence File S1. Terminal sequences of TIR-MULE families.
Supplemental Sequence File S2. Terminal sequences of non-TIR-MULE families.
Supplemental Sequence File S3. Element sequences of small MULEs (less than 150 bp).
Supplemental Sequence File S4. Sequences of intergenic parental copies.
Acknowledgments
We thank Kevin Childs and Robin Buell (both Michigan State University) for assistance in using the rice genome database and discussions related to the study. We thank Stefan Cerbin (Michigan State University) for critical reading of the manuscript.
Glossary
- TE
transposable element
- TSD
target site duplication
- TIR
terminal inverted repeat
- ORF
open reading frame
- PMTIR
TIR families that contain or include Pack-MULEs
- non-PMTIR
TIR families that contain exclusively non-Pack-MULEs
- WRS
Wilcoxon rank-sum test
- CDS
coding sequence
- FPKM
fragments per kilobase of exon per one million fragments mapped
- TPM
transcripts per one million
- MULE
Mutator-like element
References
- Ammiraju JS, Zuccolo A, Yu Y, Song X, Piegu B, Chevalier F, Walling JG, Ma J, Talag J, Brar DS, et al. (2007) Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J 52: 342–351 [DOI] [PubMed] [Google Scholar]
- Benito MI, Walbot V. (1997) Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol Cell Biol 17: 5165–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. (2007) Patterns in grass genome evolution. Curr Opin Plant Biol 10: 176–181 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Kellogg EA. (1997) Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Springer PS. (1994) The generation of Mutator transposable element subfamilies in maize. Theor Appl Genet 87: 657–667 [DOI] [PubMed] [Google Scholar]
- Chalvet F, Grimaldi C, Kaper F, Langin T, Daboussi MJ. (2003) Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol Biol Evol 20: 1362–1375 [DOI] [PubMed] [Google Scholar]
- Chomet P, Lisch D, Hardeman KJ, Chandler VL, Freeling M. (1991) Identification of a regulatory transposon that controls the Mutator transposable element system in maize. Genetics 129: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu SH, Jiang N, Buell CR. (2012) Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J 71: 492–502 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, Childs K, Lin H, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM, Jiang N, et al. (2011) Utility of RNA-seq for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- Devos KM, Ma J, Pontaroli AC, Pratt LH, Bennetzen JL. (2005) Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc Natl Acad Sci USA 102: 19243–19248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AA, Jiang N. (2012) Mutator-like elements with multiple long terminal inverted repeats in plants. Comp Funct Genomics 2012: 695827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183: 557–564 [DOI] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. (2004) Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118: 555–566 [DOI] [PubMed] [Google Scholar]
- Hanada K, Vallejo V, Nobuta K, Slotkin RK, Lisch D, Meyers BC, Shiu SH, Jiang N. (2009) The functional role of pack-MULEs in rice inferred from purifying selection and expression profile. Plant Cell 21: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen DR, Park KC, Elrouby N, Yu Z, Mohabir N, Cowan RK, Bureau TE. (2006) Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol Biol Evol 23: 1254–1268 [DOI] [PubMed] [Google Scholar]
- Holligan D, Zhang X, Jiang N, Pritham EJ, Wessler SR. (2006) The transposable element landscape of the model legume Lotus japonicus. Genetics 174: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Jiang N, Ferguson AA, Slotkin RK, Lisch D. (2011) Pack-Mutator-like transposable elements (Pack-MULEs) induce directional modification of genes through biased insertion and DNA acquisition. Proc Natl Acad Sci USA 108: 1537–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisz S, Purugganan MD. (2004) Epialleles via DNA methylation: consequences for plant evolution. Trends Ecol Evol 19: 309–314 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Nitasaka E. (2004) Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol 45: 933–944 [DOI] [PubMed] [Google Scholar]
- Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. (2006) High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol 4: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Pavlícek A, Hurst LD. (2003) A unification of mosaic structures in the human genome. Hum Mol Genet 12: 2411–2415 [DOI] [PubMed] [Google Scholar]
- Lin H, Moghe G, Ouyang S, Iezzoni A, Shiu SH, Gu X, Buell CR. (2010) Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol Biol 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lisch D. (2002) Mutator transposons. Trends Plant Sci 7: 498–504 [DOI] [PubMed] [Google Scholar]
- Lisch D, Jiang N (2009) Mutator and MULE transposons. In JL Bennetzen, S Hake, eds, Handbook of Maize: Genetics and Genomics. Springer, New York, pp 277–306 [Google Scholar]
- Lisch DR, Freeling M, Langham RJ, Choy MY. (2001) Mutator transposase is widespread in the grasses. Plant Physiol 125: 1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M. (2008) UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 453: 3–31 [DOI] [PubMed] [Google Scholar]
- Marquez CP, Pritham EJ. (2010) Phantom, a new subclass of Mutator DNA transposons found in insect viruses and widely distributed in animals. Genetics 185: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD. (2004a) Arabidopsis MPSS: an online resource for quantitative expression analysis. Plant Physiol 135: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. (2004b) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14: 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Vanburen R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M, et al. (2013) Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol 14: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. (2005) Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet 37: 997–1002 [DOI] [PubMed] [Google Scholar]
- Morgenstern B. (2004) DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res 32: W33–W36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvéglise C, Chalvet F, Wincker P, Gaillardin C, Casaregola S. (2005) Mutator-like element in the yeast Yarrowia lipolytica displays multiple alternative splicings. Eukaryot Cell 4: 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al. (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16: 1262–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada MN, Benito MI, Walbot V. (2001) The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J 25: 79–91 [DOI] [PubMed] [Google Scholar]
- Robertson D, Woessner JP, Gillham NW, Boynton JE. (1989) Molecular characterization of two point mutants in the chloroplast atpB gene of the green alga Chlamydomonas reinhardtii defective in assembly of the ATP synthase complex. J Biol Chem 264: 2331–2337 [PubMed] [Google Scholar]
- Robertson DS. (1978) Characterization of a Mutator system in maize. Mutat Res 51: 21–28 [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sémon M, Mouchiroud D, Duret L. (2005) Relationship between gene expression and GC-content in mammals: statistical significance and biological relevance. Hum Mol Genet 14: 421–427 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Talbert LE, Chandler VL. (1988) Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol Biol Evol 5: 519–529 [DOI] [PubMed] [Google Scholar]
- Tatarinova TV, Alexandrov NN, Bouck JB, Feldmann KA. (2010) GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics 11: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. (2003) DNA helix: the importance of being GC-rich. Nucleic Acids Res 31: 1838–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Dooner HK. (2006) Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci USA 103: 17644–17649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zheng H, Fan C, Li J, Shi J, Cai Z, Zhang G, Liu D, Zhang J, Vang S, et al. (2006) High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18: 1791–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, et al. (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23: 396–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Takano-Shimizu T, Kitamura K, Mikami T, Kishima Y. (1999) Resistance to gap repair of the transposon Tam3 in Antirrhinum majus: a role of the end regions. Genetics 153: 1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Wright SI, Bureau TE. (2000) Mutator-like elements in Arabidopsis thaliana: structure, diversity and evolution. Genetics 156: 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala G, Vodkin LO. (2005) The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell 17: 2619–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo A, Sebastian A, Talag J, Yu Y, Kim H, Collura K, Kudrna D, Wing RA. (2007) Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol 7: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]