Complex physiological responses of the brassinosteroid pathway can be explained by a biochemical model based on kinase loss-of-function alleles with brassinosteroid receptor complex occupancy as the central estimated parameter.
Abstract
Brassinosteroids (BRs) are key regulators in plant growth and development. The main BR-perceiving receptor in Arabidopsis (Arabidopsis thaliana) is BRASSINOSTEROID INSENSITIVE1 (BRI1). Seedling root growth and hypocotyl elongation can be accurately predicted using a model for BRI1 receptor activity. Genetic evidence shows that non-ligand-binding coreceptors of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) family are essential for BRI1 signal transduction. A relatively simple biochemical model based on the properties of SERK loss-of-function alleles explains complex physiological responses of the BRI1-mediated BR pathway. The model uses BRI1-BR occupancy as the central estimated parameter and includes BRI1-SERK interaction based on mass action kinetics and accurately describes wild-type root growth and hypocotyl elongation. Simulation studies suggest that the SERK coreceptors primarily act to increase the magnitude of the BRI1 signal. The model predicts that only a small number of active BRI1-SERK complexes are required to carry out BR signaling at physiological ligand concentration. Finally, when calibrated with single mutants, the model predicts that roots of the serk1serk3 double mutant are almost completely brassinolide (BL) insensitive, while the double mutant hypocotyls remain sensitive. This points to residual BRI1 signaling or to a different coreceptor requirement in shoots.
The classic biochemical model for the transduction of an extracellular signal involves ligand binding by a receptor, after which the signal is transduced to a downstream target. The main ligand-binding receptors often employ coreceptors that can enhance endocytosis (Sorkin and Von Zastrow, 2002; Molfetta et al., 2010), interactions with downstream targets, or the binding affinity between the ligand and the main receptor (Gakamsky et al., 2005; Hubbard and Miller, 2007). In plants, various signal transduction pathways in defense, development, and programmed cell death employ coreceptors to transduce an extracellular signal (Li, 2010; Calderón Villalobos et al., 2012). An important class of signaling hormones involved in growth and development are brassinosteroids (BRs), plant steroid hormones with structural similarity to animal steroid hormones (Bajguz and Tretyn, 2003). In Arabidopsis (Arabidopsis thaliana), the main ligand-perceiving receptor for BRs is the BRASSINOSTEROID INSENSITIVE1 (BRI1) receptor. Impaired BR signaling either by a reduction of BRI1 receptor level or BR availability results in severe growth and developmental phenotypes, such as a dwarfed stature, impaired photomorphogenesis, fertility defects, and impaired root growth (Clouse, 1996; Wang et al., 2001). For its signaling activity, BRI1 interacts with members of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) family that function as non-ligand-binding coreceptors (Li and Nam, 2002; Li et al., 2002; Russinova et al., 2004). In Arabidopsis, four of the five members of the SERK family are reported to have partially redundant functions and act in various signaling pathways via their interaction with different ligand-binding receptor-like kinases. SERK1 is known to be involved in male sporogenesis together with SERK2 and in brassinosteroid signaling together with SERK3 (also known as BRI1 Associated Kinase1 (BAK1)) and SERK4 (also known as BAK1-like Kinase1 (BKK1); Albrecht et al., 2008). SERK3 functions in defense (Chinchilla et al., 2007; Heese et al., 2007) and together with SERK4 in a BR-independent cell death pathway (He et al., 2007) and defense (Roux et al., 2011). While the SERK coreceptors serve both BR-dependent and -independent signaling pathways, BRI1-mediated BR signaling was recently shown to be completely dependent on three of the SERK coreceptors, SERK1, SERK3, and SERK4 (Gou et al., 2012). SERK3 is the only member of the SERK family in which the single mutant shows a BRI1-related phenotype (Albrecht et al., 2008).
A major difficulty arising from the genetic analyses of the SERK genes is that individual genes appear to participate in multiple pathways, a difficulty enhanced further in double and triple mutant combinations. For example, the strong-root phenotype observed in a serk1-1serk3-1serk4-1 triple mutant (Gou et al., 2012) is only in part the result of an impaired BR-unrelated pathway (Du et al., 2012). For these reasons, the question of how individual SERK proteins quantitatively affect the output of one specific signaling pathway, and consequently the resulting change in the physiological response, is important to help elucidate the role of these receptors. Here, we use a mathematical modeling approach in combination with experimental validation focusing on the role of SERK1 and SERK3 in the BRI1 signaling pathway. BR signaling is one of the best-understood signal transduction cascades in Arabidopsis with respect to biochemical interactions and downstream responses (Ye et al., 2011; van Esse et al., 2013; Fig. 1). In addition, quantitative data on BRI1-BL binding affinity and BRI1 and SERK receptor concentrations are available (Wang et al., 2001; van Esse et al., 2011), which is essential for modeling. Recently, we developed a quantitative model to explain and predict how root and hypocotyl growth depend on the BRI1 receptor and BL ligand concentration. With the mass action kinetics on which the biochemical part of the model is based, phenotypes of bri1 mutants could be correctly predicted (van Esse et al., 2012).
Figure 1.
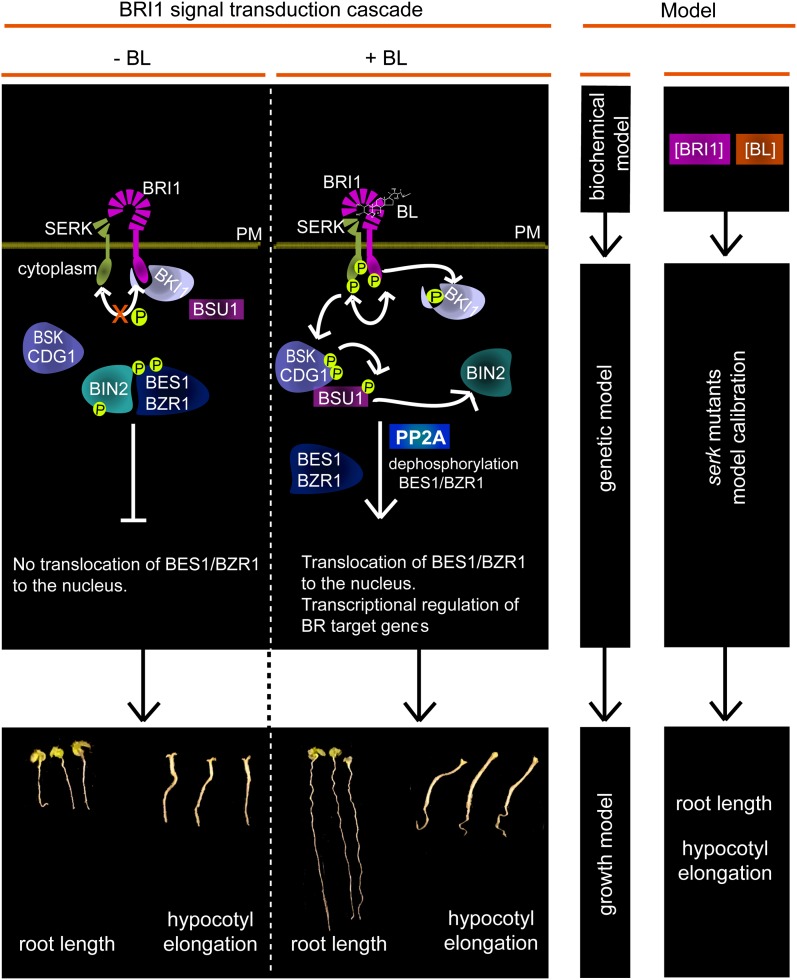
Modeling oligomeric receptor complexes. In the left panel, a brief scheme of the BRI1 pathway is shown (redrawn after Ye et al., 2011). In the absence of BL, the kinase activity of unbound BRI1 receptor is restricted by negative regulators such as BRI1 KINASE INHIBITOR1 (BKI1). BL binding to BRI1 results in transphosphorylation between BRI1 and SERKs, which leads to the phosphorylation of BSKs and CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1). The phosphorylated BSKs and CDG1 subsequently interact with BRI1 SUPPRESSOR1 (BSU1) phosphatase, promoting its interaction with BRASSINOSTEROID-INSENSITIVE2. Inactivation of BRASSINOSTEROID-INSENSITIVE2 by BSU1 results in the dephosphorylation and activation of the BR transcriptional regulators BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS SUPPRESSOR1 (BES1). The middle and right panels summarize the modeling approach. The biochemical model describes BL-dependent BRI1 activation (van Esse et al., 2012). Using genetically identified components, the model is now extended using mutated SERK1 and SERK3 coreceptors as an example. In this way, the input data from the biochemical model, modified by the coreceptors and combined into a growth model, are linked to the physiological readout in the form of root growth and hypocotyl elongation. PM, Plasma membrane; PP2A, protein phosphatase 2A. [See online article for color version of this figure.]
In this work, the model is extended to include the contribution of the coreceptors SERK1 and SERK3 to BL-dependent BRI1 activity (Fig. 1). For this, the root length and hypocotyl elongation of serk1 and serk3 single and double mutants were used as physiological readouts of BRI1 activity. The predictive power of the resulting model was tested by a leave-one-out cross validation. In silico simulations show that, in roots, the signal perceived by BRI1 is transduced by a complex consisting of BRI1 with SERK1, SERK3, or both, while in the hypocotyl, BRI1 signaling requires another SERK member, all in line with the experimental observations (Gou et al., 2012). Previously, it has been postulated that SERK3 enhances the phosphorylation of BRI1, thereby quantitatively increasing the signaling output (Wang et al., 2008). The model presented here correctly reflects this observation by predicting that the prime activity of the SERK coreceptors is to increase the physiological output.
RESULTS
Mathematical Model
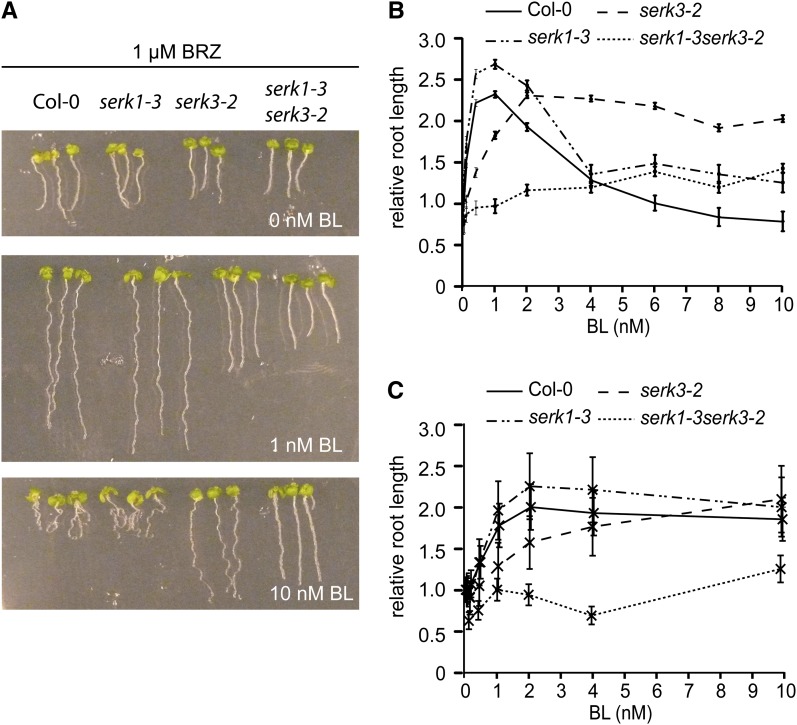
The mathematical model consists of a biochemical part and a root growth part. The growth model is largely based on phenomenological considerations and couples the output of BRI1 signaling to root growth. To determine the contribution of the SERK1 and SERK3 coreceptors to overall BRI1-mediated BR signaling, the stimulatory and inhibitory effects of brassinolide (BL) on root growth of the serk1-3 and serk3-2 single and double mutants were measured (Fig. 2). To highlight the stimulatory effect of exogenous BL, the BR biosynthesis inhibitor brassinazole (BRZ) was used throughout. The serk1-3serk3-2 mutant has only a minor stimulatory response toward exogenously applied BL (Fig. 2B). No inhibitory effect of BL on root growth was observed, even in the absence of BRZ (Supplemental Fig. S1). This indicates that roots of the serk1-3serk3-2 double mutant are almost completely insensitive toward BL. The biochemical model (Supplemental File S1A) describes three types of receptor combinations responsible for BRI1-mediated signaling in roots (i.e. [BRI1], [BRI1 SERK1], and [BRI1 SERK3]). All three only transduce the signal when BL is bound to BRI1. To incorporate the coreceptors into the existing model describing BR-activated BRI1-mediated signaling (van Esse et al., 2012), several assumptions were made. The first is that BRI1 binds to SERK proteins in a 1:1 ratio. The second is that the protein concentrations of BRI1 (62 nm), SERK1 (120 nm), and SERK3 (30 nm) in wild-type seedling roots (van Esse et al., 2011) remain constant over time. The third concerns the amount of SERK3 and SERK1 interacting with BRI1 at saturating ligand levels. While the precise stoichiometry is unknown, a 2:1:1 ratio is assumed between BRI1, SERK1, and SERK3 in functional complexes (Supplemental File S1). In wild-type seedlings, no more than 10% of the total number of SERK3 coreceptors are required in brassinosteroid signaling at a saturating ligand concentration (Albrecht et al., 2012), so at most, 3 nm SERK3, 3 nm SERK1, and 6 nm BRI1 are simultaneously active in signaling. Fourth, the affinity of BRI1 for BL remains unaltered in serk mutants. This is based on the findings of Kinoshita et al. (2005), reporting that the dissociation constant between BRI1 and BL of 7.4 nm (Wang et al., 2001) remains unaltered in the absence of BAK1 (SERK3). Finally, although Arabidopsis plasma membrane-localized proteins are relatively immobile (Martinière et al., 2012), spatial inhomogeneity is not explicitly modeled.
Figure 2.
Root length of serk mutants in response to BL. A, Root length of wild-type seedlings (ecotype Columbia [Col-0]) and serk1-3 and serk3-2 single and double mutants at 0 nm BL (top), 1 nm BL (middle), and 10 nm BL (bottom). All assays were performed in the presence of 1 µm BRZ. B, Root growth assays of the serk1-3 and serk3-2 single and double mutants compared with the wild type at different BL concentrations. Root lengths were measured at 8 DAG and plotted relative to the BRZ-treated control. The serk1-3 single mutant does not have a clear BL-dependent phenotype, whereas the serk3-2 mutant never reaches the wild-type root length and does not show any inhibition in response to BL. The serk1-3serk3-2 double mutant is almost completely unresponsive toward BL. C, Root lengths relative to the BRZ-treated control at 4 DAG. For all measurements, n ≥ 15 roots, and error bars indicate se. [See online article for color version of this figure.]
The main hypothesis in the biochemical model is that the formation of functional complexes consisting of BRI1, SERK1, SERK3, and BL follow mass action laws. The signal (σ) induced by BL binding to BRI1 is modeled as:
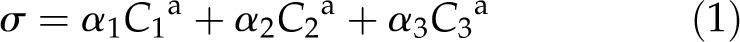 |
Here, C1a, C2a, and C3a denote [BRI1], [BRI1 SERK1], and [BRI1 SERK3]. The term C1a represents all responses toward exogenously applied BL not covered by SERK1 and SERK3. Parameters α1, α2, and α3 are proportionality constants that reflect the contribution of the different complexes to the signal strength of BRI1.
Due to the inclusion of the coreceptors, the growth model is now modeled as a combination of two separate mechanisms
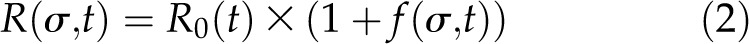 |
with R(σ,t) the root length at time t (days after germination [DAG]), f(σ,t) the root length as a result of BRI-mediated signaling, and R0(t) the root length independent of BRI1-mediated signaling or response toward BRs. Scaling of R0(σ,t) gives the root length due to BRI1 signaling relative to the root length independent of BRI1 signaling. The output of the growth model is then root length relative to the BRZ-treated control instead of absolute values (Fig. 2B). For this, the parameter R0(t) is scaled out of the equation:
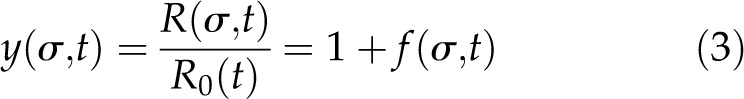 |
where y(σ,t) is the increase in root length relative to the BRZ-treated control. The function f(σ,t) is based on phenomenological considerations and consists of one activating and two inhibiting modules:
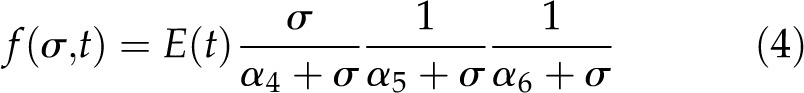 |
Here, E(t) is a time-dependent growth factor, followed by a module that represents the stimulatory effects of BR signaling on root growth and a second module for the inhibitory effects. σ denotes the signal that is induced by the binding of BL to BRI1, and α4, α5, and α6 are the half-maximum response values for the growth model (k values). The form of function E(t) is unknown and, therefore, is regarded as a free parameter that is allowed to change between different time points. The unknown model parameters such as the k values (α4, α5, α6) and E(t) were estimated by minimizing the fitting error between the model output and measured root length data. Different structures of f(σ,t) were compared using the Akaike and Bayesian information criteria and a 4-fold cross validation. The first two reflect the tradeoff between a good fit with the data and complexity, and the third measures the prediction error that is possibly due to overfitting or underfitting (Supplemental File S1, B and C). In total, 13 functions for f(σ,t) were tested, ranging from one activating or inhibiting module to three activating and three inhibiting modules and all relevant combinations in between. Model structure 1 (one activating and two inhibiting modules) scored best with respect to root growth (Supplemental Table S1) as well as hypocotyl elongation (Supplemental Table S2) and therefore was selected.
The response to BL treatment is minor at 4 DAG when compared with 6 and 8 DAG (Fig. 2, B and C). For this reason, the model cannot fit the relative root growth response curves at 4, 6, and 8 DAG simultaneously (Supplemental Fig. S2). However, growth at 4 DAG can be computed separately (Supplemental Fig. S3). Therefore, only the difference between the wild type and the serk1 and serk3 mutants in terms of relative response in root length at 6 and 8 DAG is used for model selection, calibration, and validation.
Model Calibration and Validation
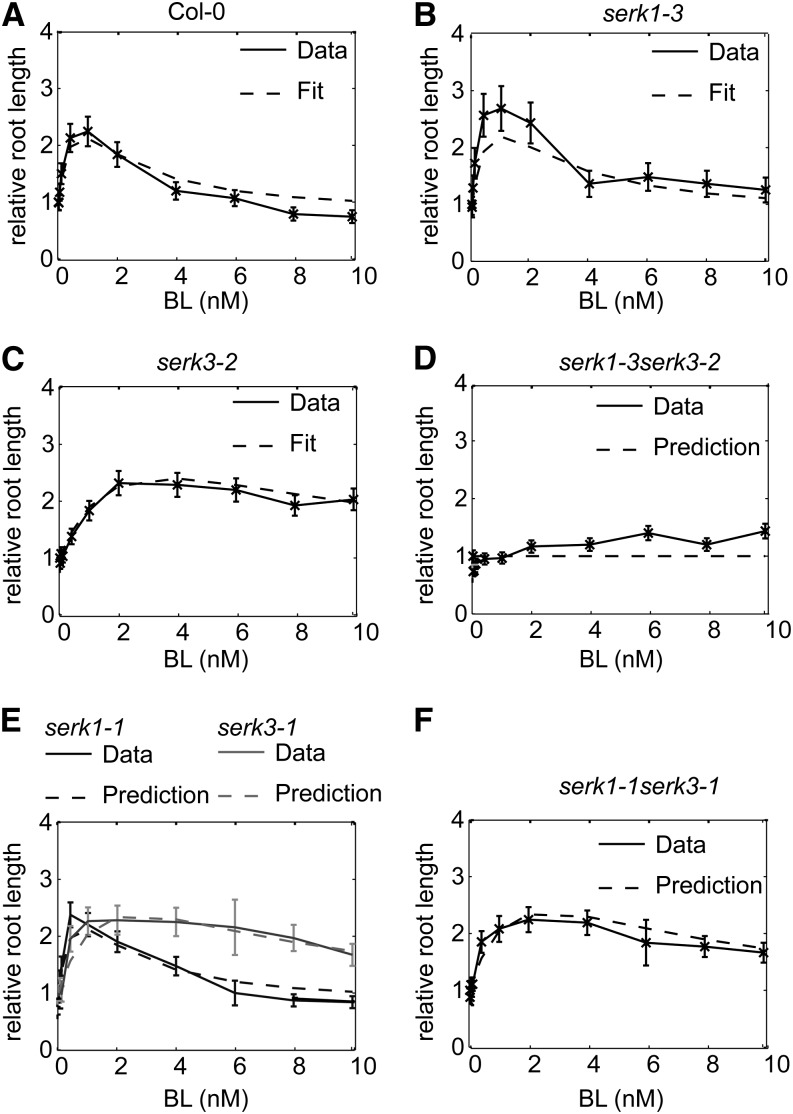
Root growth assays of the serk1-3 and serk3-2 single mutants were included in the model calibration data set to train the model on the effect of SERKs on BRI1 receptor activity. The model is able to fit the relative root growth pattern of the wild type and the serk1-3 and serk3-2 single mutants while predicting the relative root length of the serk1-3serk3-2 double mutant (Fig. 3, A–D). The serk3-2 mutant is a true null loss-of-function allele. There is a slight discrepancy between the model predictions and actual data for the root growth response of the serk1-3serk3-2 double mutant (Fig. 3D). This could be due to a minor contribution of SERK2 or SERK4 in BR signaling only occurring in the absence of SERK3 (Jeong et al., 2010; Du et al., 2012) or to trace signaling mediated by the BRI1 receptors alone. Leave-one-out cross-validation, consisting of calibration on relative root growth of the single serk mutants and the wild type, shows a small predictive error (Supplemental Fig. S4). To further test the predictive power of the model, the relative root growth response of the serk1-1 and serk3-1 single mutants and the serk1-1serk3-1 double mutant toward BL was simulated. The serk3-1 mutant is a weak mutant that still harbors low levels of SERK3 transcript (Gou et al., 2012). Small prediction errors were obtained when assuming that there is between 10% and 20% SERK3 activity left in the serk3-1 mutant background (Supplemental Fig. S5). Under this assumption, the model accurately predicts the behavior of the serk1-1serk3-1 double mutant (Fig. 3, E and F).
Figure 3.
Model fit and prediction of relative root growth response for the wild type and serk mutants. All measured root lengths are plotted relative to the BRZ-treated control. A to C, Model calibration on the relative root length of the wild type (ecotype Columbia [Col-0]), serk1-3, and serk3-2 at 6 and 8 DAG. Only results of 8 DAG are shown. D, Model validation by predicting the relative root growth response of serk1-3serk3-2 double mutant seedlings at 8 DAG. E and F, Model predictions for the relative root length of the serk1-1 and serk3-1 single mutants (E) and the serk1-1serk3-1 double mutant (F). For all experiments, n ≥ 15 roots measured in three independent replicates, and error bars indicate se; assays were performed in the presence of 1 µm BRZ.
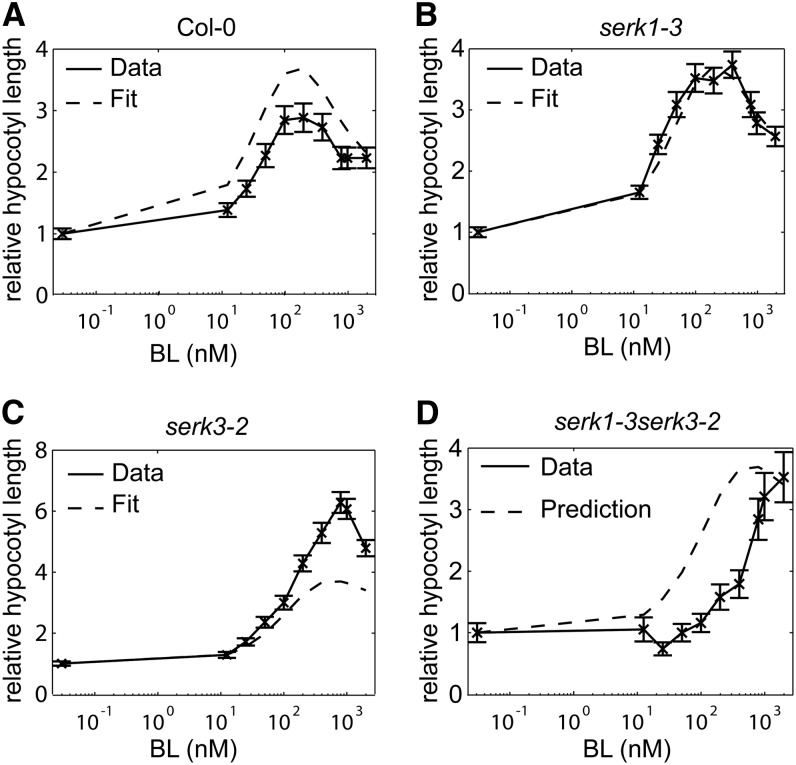
As a final validation, the model was used to predict hypocotyl elongation relative to the BRZ-treated control. The trends of the relative hypocotyl elongation of the wild type, the serk1-3 and serk3-2 single mutants, and the serk1-3serk3-2 double mutant were predicted correctly for all cross validations (Fig. 4; Supplemental Fig. S6). The model correctly predicts that in the hypocotyl there is still a marked response of the serk1-3serk3-2 double mutant toward BL. This suggests that there is either a difference in coreceptor usage by BRI1 between the roots and hypocotyl or a substantial contribution by BRI1 alone.
Figure 4.
Model fit and prediction of relative hypocotyl elongation for the wild type (ecotype Columbia [Col-0]) and serk mutants. All measured hypocotyl lengths are plotted relative to the BRZ-treated control. A to C, Calibration on the hypocotyl elongation of the serk1-3serk3-2 double mutant. All hypocotyl lengths were measured at 5 DAG, and seedlings were grown in the dark. D, Model validation by predicting the relative hypocotyl elongation of the serk1-3serk3-2 double mutant. Error bars indicate se; n ≥ 30 hypocotyls measured in three independent replicates.
Coreceptors Act by Changing the Maximum Output of BR Signaling
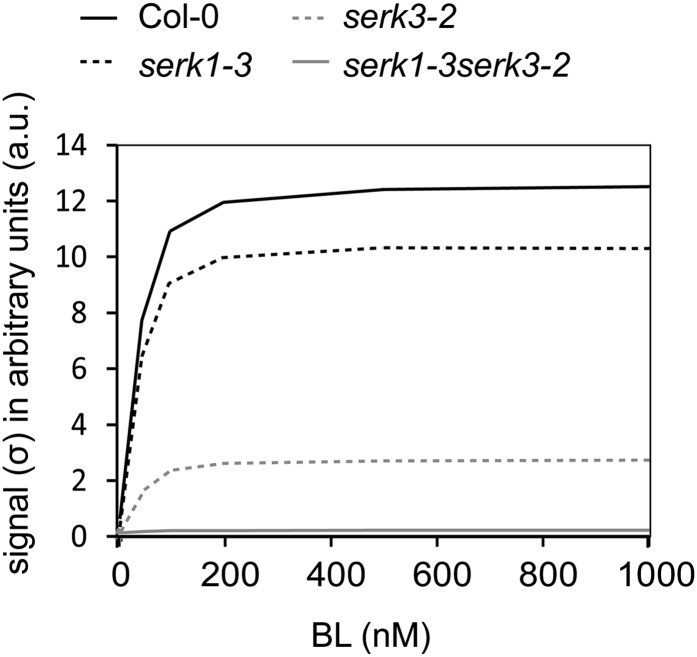
Next, the model was used to address the question of how the coreceptors affect the signal (σ) after BL binding to BRI1. We first focused on the absence of SERK3, because the serk3-2 strong loss-of-function mutant behaves as a weak loss-of-function bri1 mutant. Simulations predict that one possible explanation for the reduced σ in serk3-2 is a reduction in the concentration of BRI1 available for signaling (Supplemental Fig. S7A), while the total amount of BRI1 remains the same (see assumption 2 in the description of the mathematical model). For example, the amount of actively signaling BRI1 receptor should be about 30-fold less in the serk3-2 mutant background when compared with the wild-type background in order to result in the trend observed in the serk3-2 mutant (Supplemental Fig. S7A). However, quantitative confocal microscopy experiments demonstrate that the amount of BRI1-GFP in the plasma membrane is hardly changed in the serk3-2 background (Supplemental Fig. S7, B and C), suggesting that reduction of the total number of BRI1 receptor molecules is not a mechanism by which coreceptors attenuate the signal. It has been postulated that SERK3 acts by increasing the activity of the BRI1 receptor to achieve full signaling output (Wang et al., 2008); therefore, we tested whether our model could reflect this possibility. Indeed, predicting the behavior of σ in the absence of SERK3 shows that the σmax will drop about 6-fold and is close to zero in the double serk1-3serk3-2 mutant (Fig. 5). Therefore, we propose that in wild-type roots, SERKs primarily act by increasing the σmax of BRI1, in line with the experiments shown by Wang et al. (2008).
Figure 5.
Prediction of BRI1 signaling output in the absence of SERK coreceptors. The signal intensity of BRI1 (σ) is predicted in the presence or absence of SERK1 and SERK3. The signal transduced in serk3-2 and serk1-3 mutants is computed by plotting α3C3a (σ SERK3) and α2C2a (σ SERK1). For the signal in the serk1-3serk3-2 mutant, the term α1C1a was computed, representing the residual responses to BL. Col-0, Wild-type ecotype Columbia.
BRI1 Signaling Operates with Only a Minor Amount of Coreceptors
SERK1 and SERK3 function as coreceptors in various signaling pathways by interacting with different main ligand-perceiving receptors. If these pathways operate at the same time, this implies that the coreceptors are distributed among them. For this reason, it is of interest to know how coreceptor dosage affects BRI1-mediated BR signaling. Therefore, this possibility was incorporated into the model by the mass balance equation (Eq. 2) in Supplemental File S1A. Based on the amount of SERK3 coimmunoprecipitated by BRI1 at saturating ligand level (assumption 3 in “Results”), the model predicts that only about 0.05 nm SERK1 and SERK3 (0.04% and 0.16% of the total amount, respectively) is used in BR signaling at a physiological ligand concentration of 1 nm BL (Supplemental File S1; Supplemental Table S3). In the biosynthesis mutant det2, root growth mimics that of BRZ-treated seedlings, showing a positive effect of exogenous BL even below 1 nm BL (Supplemental Fig. S8), confirming the effect of BRZ. No attempt was made to predict det2 root growth in the models presented here, due to the previously observed correspondence in the biological effects (Nagata et al., 2000). Even after increasing the percentage of both SERK1 and SERK3 used by other pathways to 85%, the model is still able to predict BRI1-mediated root growth (Table I). This corroborates the finding that FLAGELLIN SENSITIVE2 (FLS2)-mediated flg22 signaling uses up to 70% of SERK3 without affecting BR signaling (Albrecht et al., 2012).
Table I. Effect on model prediction of changing the amount of SERK1 and SERK3 employed by non-BRI1-related pathways.
In normal conditions, it is assumed that 10% of the SERK1 and SERK3 coreceptors is employed in non-BRI1-related pathways (Supplemental File S1A). This yields a low prediction error of the model (Supplemental Table S1). A prediction error deviating more than 10% from the optimal value is considered to be significantly different.
| Amount of SERK in Other Pathways | Prediction Errora |
|---|---|
| Root | |
| 10% | 63 |
| 85% | 64 |
| Hypocotyl | |
| 10% | 175 |
| 85% | 174 |
Average prediction error over four leave-one-out cross validations.
Testing Different Biochemical Model Structures
So far, biochemical model 1 was employed, assuming the occurrence of preformed BRI1-SERK complexes. Such a mechanism has been reported for several receptor-like kinases in mammalian systems (Gadella and Jovin, 1995; Lemmon and Schlessinger, 2010). Alternative mechanisms, where BRI1 binds BL before recruiting SERK3 coreceptors, have also been proposed (Wang et al., 2005). The structure of the extracellular domain of BRI1 in the presence or absence of BL does not show substantial changes and does not provide a clear indication for either mechanism (Hothorn et al., 2011). Therefore, it was tested if the root growth model is able to distinguish between the different biochemical models (Supplemental File S1A). The results show that the order of [BL BRI1 SERK] complex formation does not affect the prediction error (Table II). Next, differences in the stoichiometry were tested. A model in which the complexes consist of heterodimers (one molecule of BRI1 and one of SERK) results in a lower prediction error when compared with a model with tetrameric complexes with two BRI1 molecules and two SERKs (Table II). Changing the BRI1:SERK ratio from 1:1 to 1:4 also does not significantly alter the prediction error. Taken together, we conclude that physiological data such as growth readouts have a low predictive value for biochemical models.
Table II. Comparison of different biochemical model structures using root length as a readout.
Biochemical model 1 assumes a preassembly between BRI1, SERK1, and SERK3 in the absence of ligand, while biochemical model 2 assumes only an interaction in the presence of ligand (Supplemental File S1A). The prediction errors of biochemical model 1, biochemical model 2, and the effect of changing the stoichiometry between BRI1, SERK1, and SERK3 are shown. A deviation of 10% in prediction error from the optimal value is considered to be significantly different.
| Model | Stoichiometry, BRI1:SERK1:SERK3 | Prediction Errora |
|---|---|---|
| Biochemical model 1 | 1:1 | 63 |
| 1:2 | 67 | |
| 1:4 | 69 | |
| 2:2 | 105 | |
| Biochemical model 2 | 1:1 | 67 |
| 1:2 | 65 | |
| 1:4 | 74 | |
| 2:2 | 106 |
Average prediction error over four leave-one-out cross-validations.
DISCUSSION
In this work, we have extended a model for BR signaling mediated by the ligand-binding BRI1 receptors with the associated non-ligand-binding coreceptors of the SERK family. The resulting model can be used to incorporate downstream components of the main pathway identified by forward genetics or biochemical means. As an example, the reduced root growth and increased ligand insensitivity observed in single serk mutants were linked to the signaling output of a biochemical receptor model. The model accurately predicts relative root and hypocotyl growth in double serk mutant and wild-type seedlings and agrees well with existing experimental data presented here and elsewhere. The validated model presented here can now be used to incorporate additional BR pathway components and offers the opportunity to include quantitative elements in an otherwise genetic model.
The identification of members of the SERK family that act redundantly in different signaling pathways (He et al., 2007; Li, 2010; Du et al., 2012; Gou et al., 2012; Kim et al., 2013) has underscored the importance of a class of non-ligand-binding receptors in plant signaling pathways. In BRI1-mediated BR signaling, there is no evidence that SERK3 directly influences the ligand-binding properties (Kinoshita et al., 2005), a finding that seems to rule out a role as a classical coreceptor as used in comparable animal signaling systems. Moreover, there appears to be no change in structure of the ligand-binding domain of BRI1 after incubation with BL, but a ligand-induced platform was proposed to occur in BRI1, allowing interaction with SERK3 (Hothorn et al., 2011). Yet, the SERKs clearly modulate and are necessary for sustained BRI1-mediated signaling (Wang et al., 2005). Regardless of the precise biochemical mechanism, this would qualify them as true coreceptors. Recently, it was shown that SERK3, also functioning as a coreceptor for the flagellin-triggered FLS2 signaling pathway, is not rate limiting between both BR and flg22 pathways; in fact, most of the SERK3 proteins can be recruited by FLS2 without affecting BRI1-mediated root growth (Albrecht et al., 2012). However, it is unclear what the precise role of these receptors is. One possibility is that they act to narrow down the broad BR signal affecting a multitude of developmental events, such as cell division, cell elongation, cell death, and cell differentiation (Chinchilla et al., 2007; Heese et al., 2007; Albrecht et al., 2008; Lewis et al., 2010; González-García et al., 2011; Hacham et al., 2011), and that their effects on fertility and on immunity are downstream effects of alterations in such basic cellular pathways. However, genetic evidence does not support that option, mainly because the phenotypes encountered mimic precisely the phenotype of a loss of function in the main receptor or are weak mutants showing the entire array of phenotypes. On top of that, it becomes evident that a single member of the SERK family can simultaneously affect different pathways. In some cases, this property can be traced back to individual residues in the SERK3 protein (Schwessinger et al., 2011). For these reasons, we decided to develop a more mathematical approach to provide a better quantitative insight into the role of the SERK family.
In the complete absence of the coreceptors, the maximal signal transduced by BRI1 is significantly reduced. The most obvious explanation for this effect, therefore, is that SERKs act by transducing the signal from the ligand-binding receptor to the downstream targets. Evidence for this role is that mutual transphosphorylation between BRI1 and SERK3 was found to be essential for enhancing BR signaling in vivo (Wang et al., 2008). In FLS2-mediated signaling, SERK3 enhances the signal by interacting with BOTRYTIS-INDUCED KINASE1 (BIK1), a member of the RECEPTOR-LIKE CYTOPLASMIC KINASE superfamily and a downstream target of FLS2. In the absence of SERK3, FLS2 is unable to phosphorylate BIK1 (Lu et al., 2010). Similarly, brassinosteroid signaling kinases (BSKs), which are also members of the RECEPTOR-LIKE CYTOPLASMIC KINASE superfamily, are a substrate of BRI1 and are activated by upon BL binding to BRI1. However, in vitro kinase assays have demonstrated that BRI1 and not SERK3 phosphorylates BSKs (Tang et al., 2008). This indicates that a mechanism by which SERK3 enhances BRI1 signaling via specific phosphorylation of BSKs is not likely.
BRI1 signaling is regulated by the presence of positive (BSKs) and negative (BRI1-ASSOCIATED KINASE1) regulators. The protein phosphatase 2A stimulates BRI1 signaling by dephosphorylating the downstream BRI1 transcriptional regulator BRASSINAZOLE-RESISTANT1 but is also required for the degradation of BRI1 via dephosphorylation of the activated receptor (Di Rubbo et al., 2011; Tang et al., 2011; Wu et al., 2011). Attenuation of the BRI1 signal is regulated by the phosphorylation of specific Ser and Thr residues (Oh et al., 2012). Thus, there appears to be evidence for a role of the SERKs as amplifiers of the entire signaling pathway, rather than having a specific effect on only one element. The modeling presented here is indeed able to predict the alterations described for the role of the SERK receptors in BR signaling. Although not addressed in this study, the model could also be useful for other pathways that use different main ligand-perceiving receptors employing SERK proteins. Simulations also demonstrate that in roots, the signal transduced by BRI1 is mainly affected by BRI1 in complex with SERK1 or SERK3. However, when simulating hypocotyl elongation in the absence of SERK1 and SERK3, the model predicts correctly that there is still a significant BL-induced increase in hypocotyl elongation. This implies that in the hypocotyl, there is either a larger contribution of BRI1 alone or, alternatively, another SERK member compensates for the loss of SERK1 and SERK3 in hypocotyl tissue. In Arabidopsis seedlings, signal transduction by BRI1 is completely abolished in the absence of SERK1, SERK3, and SERK4 (Gou et al., 2012), making that member a likely candidate. In that respect, the interaction of BRI1 with different coreceptors might reflect a mechanism that regulates the sensitivity of a cell or tissue for BRs depending on the family members present.
However, there are clear limitations to what can be understood about the underlying biochemical model predicted by the model based on physiological readout. Our modeling results emphasize that one type of coreceptor can be employed for various pathways simultaneously. This was based on the fact that remarkably low numbers of the SERK proteins are needed to sustain BR signaling. This predicts that a high level of separation must exist between the different receptor complexes active in the same cell. SERK3 has been implicated as a means to provide cross talk between different pathways (Vert and Chory, 2011). In one study, it appeared that SERK3 was rate limiting between BRI1- and FLS-mediated signaling (Belkhadir et al., 2012), while we showed that in wild-type conditions, this does not appear to be the case. Therefore, a role of SERK proteins as direct mediators in receptor cross talk seems unlikely (Albrecht et al., 2012).
In general, there are several mechanisms by which coreceptors can alter the activity of the main receptor. In mammalian systems, for example, the T-cell coreceptors CDC4 and CDC8 are positive regulators that enhance the interaction between the T-cell receptor and its ligand (Gao et al., 2002; Berg and Sewell, 2011), whereas the non-ligand-binding glucocorticoid receptor isoform β is a dominant negative regulator of the ligand-perceiving glucocorticoid receptor isoform α (Kino et al., 2009). Similar mechanisms have been reported for several other steroid receptors, such as the estrogen receptor β, peroxisome proliferator-activated receptors (PPARα and PPARγ), the thyroid hormone receptor α, and the epidermal growth factor receptor (EGFR). In plants, coreceptors are also employed to alter the signaling properties of the main ligand-perceiving receptor. Recently, it has been shown that efficient binding of the auxin receptor TRANSPORT INHIBITOR RESPONSE1 to its ligand requires coreceptors (Calderón Villalobos et al., 2012). In this respect, it is proposed that combinations of TRANSPORT INHIBITOR RESPONSE1 with different coreceptors result in ligand-perceiving complexes with distinct auxin-binding affinities. Such a mechanism would enhance the concentration range of the hormone, thereby contributing to the complexity of the auxin response (Calderón Villalobos et al., 2012). Similarly, overexpression of v-erb-b2 avian erythroblastic leukemia viral oncogene homolog2 (ERBB2), the non-ligand-binding coreceptor of EGFR, does not result in receptor hyperautophosphorylation. Instead, ERBB2 regulates the ubiquitin ligase c-Cbl through altering the phosphorylation patterns on the EGFR-HER2 heterodimer, thereby protecting the heterodimer from ligand-induced ubiquitination and degradation (Hartman et al., 2013). As a consequence, the EGFR-ERBB2 heterodimer is stabilized, resulting in sustained signaling.
In line with these animal receptor models, we propose that the action of the SERK coreceptors in BRI1-mediated BR signaling is to transiently stabilize ligand-induced active receptor complexes as a means to increase signaling strength. Whether this same action applies to other SERK-employing pathways remains to be determined. We believe that the modeling approach as described here can help to distinguish between the action of SERK members in the various pathways and to provide a more comprehensive framework of the multitude of signaling occurring simultaneously in plant cells.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type plants (ecotype Columbia), serk1-3 (GABI-KAT line 448E10) and serk3-2 or bak1-4 (SALK_116202) single mutants, and the serk1-3serk3-2 double mutant, all in the Columbia background, were used in all growth assays. For model validation, the serk1-1 (SALK_544330) and serk3-1 or bak1-3 (SALK_034523) single and double mutants were used. For fluorescence microscopy, wild-type seedlings expressing BRI1-GFP under the control of its own promoter (Geldner et al., 2007) were crossed with the serk3-2 mutant. Root growth and hypocotyl elongation assays were performed as described previously (van Esse et al., 2012).
Software and Modeling
All computational and modeling procedures were performed within the Matlab environment (Supplemental File S1; van Esse et al., 2012). Out of the different models tested, throughout this paper biochemical model 1 and physiological model 1 are used unless specified otherwise. For all simulations, root lengths relative to the BRZ-treated control were used.
Analysis of the Fluorescence Distribution of BRI1-GFP in the Wild Type versus the serk3-2 Background
Roots of 3- to 5-d-old seedlings containing BRI1-GFP in the wild-type or serk3-2 background were imaged using a CONFOCOR2/LSM510 confocal microscope (Zeiss) equipped with a 40× water objective (numerical aperture 1.2) and an argon ion laser. Imaging was performed as described previously (van Esse et al., 2011). Briefly, the argon laser was used for excitation of GFP at 488 nm, after which GFP fluorescence emission was detected with a band-pass filter (505–550 nm), and the pinhole diameter was adjusted to a slice thickness of approximately 0.9 µm. All images were taken within 45 s from the moment the root was exposed to the laser to limit bleaching effects. Images were analyzed using Image proplus software (MediaCybernetics; http://www.mediacy.com). The mean intensities of about seven roots measuring five cells per root were measured in two independent replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root length assays of serk1-3 and serk3-2 mutants in the absence of BRZ.
Supplemental Figure S2. Model calibration on relative root lengths of seedlings at 4, 6, and 8 DAG simultaneously.
Supplemental Figure S3. Model calibration on relative root lengths of seedlings at 4 DAG.
Supplemental Figure S4. Model predictions of the leave-one-out cross validation.
Supplemental Figure S5. The model is robust against alterations in the amount of SERK3 transcript in the serk3-1 background.
Supplemental Figure S6. Leave-one-out cross validation for modeling hypocotyl elongation.
Supplemental Figure S7. The trend observed in the serk3-2 mutant is not due to a reduced BRI1 level.
Supplemental Figure S8. det2 roots mimic BRZ-treated wild-type roots.
Supplemental Table S1. Comparison of different physiological model structures using root length as a readout.
Supplemental Table S2. Comparison of different physiological model structures using hypocotyl elongation as a readout.
Supplemental Table S3. Predicted amount of BRI1 interacting with SERK in the wild-type background.
Supplemental File S1. Mathematical model description.
Acknowledgments
We thank Niko Geldner for providing the BRI1-GFP line in the wild-type background, the Arabidopsis Biological Resource Center for providing us the serk1 and serk3 mutant seeds, and Eric Boer and Jaap Molenaar for fruitful scientific discussions on the manuscript.
Glossary
- BR
brassinosteroid
- BRZ
brassinazole
- DAG
days after germination
- BSK
brassinosteroid signaling kinase
- EGFR
epidermal growth factor receptor
References
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. (2008) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol 148: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajguz A, Tretyn A. (2003) The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62: 1027–1046 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H, Sewell A (2011) Dynamic tuning of T cell receptor specificity by co-receptors and costimulation. In C Molina-París, G Lythe, eds, Mathematical Models and Immune Cell Biology. Springer, New York, pp 47–73 [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Russinova E. (2011) PP2A phosphatases: the “on-off” regulatory switches of brassinosteroid signaling. Sci Signal 4: pe25. [DOI] [PubMed] [Google Scholar]
- Du J, Yin H, Zhang S, Wei Z, Zhao B, Zhang J, Gou X, Lin H, Li J. (2012) Somatic embryogenesis receptor kinases control root development mainly via brassinosteroid-independent actions in Arabidopsis thaliana. J Integr Plant Biol 54: 388–399 [DOI] [PubMed] [Google Scholar]
- Gadella TW, Jr, Jovin TM. (1995) Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy: a stereochemical model for tyrosine kinase receptor activation. J Cell Biol 129: 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakamsky DM, Luescher IF, Pramanik A, Kopito RB, Lemonnier F, Vogel H, Rigler R, Pecht I. (2005) CD8 kinetically promotes ligand binding to the T-cell antigen receptor. Biophys J 89: 2121–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GF, Rao Z, Bell JI. (2002) Molecular coordination of alphabeta T-cell receptors and coreceptors CD8 and CD4 in their recognition of peptide-MHC ligands. Trends Immunol 23: 408–413 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI. (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J. (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman Z, Zhao H, Agazie YM. (2013) HER2 stabilizes EGFR and itself by altering autophosphorylation patterns in a manner that overcomes regulatory mechanisms and promotes proliferative and transformation signaling. Oncogene 32: 4169–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. (2007) BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol 17: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT. (2007) Receptor tyrosine kinases: mechanisms of activation and signaling. Curr Opin Cell Biol 19: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Shang Y, Kim BH, Kim SY, Song JH, Lee JS, Lee MM, Li J, Nam KH. (2010) BAK7 displays unequal genetic redundancy with BAK1 in brassinosteroid signaling and early senescence in Arabidopsis. Mol Cells 29: 259–266 [DOI] [PubMed] [Google Scholar]
- Kim BH, Kim SY, Nam KH. (2013) Assessing the diverse functions of BAK1 and its homologs in Arabidopsis, beyond BR signaling and PTI responses. Mol Cells 35: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Su YA, Chrousos GP. (2009) Human glucocorticoid receptor isoform beta: recent understanding of its potential implications in physiology and pathophysiology. Cell Mol Life Sci 66: 3435–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Fulcher EH, Darnielle L, Healy PN, Youn JY, Liljegren SJ. (2010) The SERK1 receptor-like kinase regulates organ separation in Arabidopsis flowers. Plant J 62: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. (2010) Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin Plant Biol 13: 509–514 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta-Peyret L, Luu DT, Botchway SW, Webb SE, Mongrand S, Maurel C, et al. (2012) Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci USA 109: 12805–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molfetta R, Gasparrini F, Santoni A, Paolini R. (2010) Ubiquitination and endocytosis of the high affinity receptor for IgE. Mol Immunol 47: 2427–2434 [DOI] [PubMed] [Google Scholar]
- Nagata N, Min YK, Nakano T, Asami T, Yoshida S. (2000) Treatment of dark-grown Arabidopsis thaliana with a brassinosteroid-biosynthesis inhibitor, brassinazole, induces some characteristics of light-grown plants. Planta 211: 781–790 [DOI] [PubMed] [Google Scholar]
- Oh MH, Wang X, Clouse SD, Huber SC. (2012) Deactivation of the Arabidopsis BRASSINOSTEROID INSENSITIVE 1 (BRI1) receptor kinase by autophosphorylation within the glycine-rich loop. Proc Natl Acad Sci USA 109: 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Von Zastrow M. (2002) Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol 3: 600–614 [DOI] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse GW, Harter K, de Vries SC. (2013) Computational modelling of the BRI1 receptor system. Plant Cell Environ 36: 1728–1737 [DOI] [PubMed] [Google Scholar]
- van Esse GW, van Mourik S, Stigter H, ten Hove CA, Molenaar J, de Vries SC. (2012) A mathematical model for BRASSINOSTEROID INSENSITIVE1-mediated signaling in root growth and hypocotyl elongation. Plant Physiol 160: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse GW, Westphal AH, Surendran RP, Albrecht C, van Veen B, Borst JW, de Vries SC. (2011) Quantification of the brassinosteroid insensitive1 receptor in planta. Plant Physiol 156: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. (2011) Crosstalk in cellular signaling: background noise or the real thing? Dev Cell 21: 985–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J. (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Li L, Yin Y. (2011) Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol 53: 455–468 [DOI] [PubMed] [Google Scholar]