Abstract
Introduction
There are growing concerns about the emergence of resistance to artemisinin-based combination therapies (ACTs). Since the widespread adoption of ACTs, there has been a decrease in the systematic surveillance of antimalarial drug resistance in many malaria-endemic countries. The aim of this work was to test whether data on travellers returning from Africa with malaria could serve as an additional surveillance system of local information sources for the emergence of drug resistance in endemic-countries.
Methodology
Data were collected from travellers with symptomatic Plasmodium falciparum malaria returning from Senegal (n = 1,993), Mali (n = 2,372), Cote d’Ivoire (n = 4,778) or Cameroon (n = 3,272) and recorded in the French Malaria Reference Centre during the period 1996–2011. Temporal trends of the proportion of parasite isolates that carried the mutant genotype, pfcrt 76T, a marker of resistance to chloroquine (CQ) and pfdhfr 108N, a marker of resistance to pyrimethamine, were compared for travellers and within-country surveys that were identified through a literature review in PubMed. The in vitro response to CQ was also compared between these two groups for parasites from Senegal.
Results
The trends in the proportion of parasites that carried pfcrt 76T, and pfdhfr 108N, were compared for parasites from travellers and patients within-country using the slopes of the curves over time; no significant differences in the trends were found for any of the 4 countries. These results were supported by in vitro analysis of parasites from the field in Senegal and travellers returning to France, where the trends were also not significantly different.
Conclusion
The results have not shown different trends in resistance between parasites derived from travellers or from parasites within-country. This work highlights the value of an international database of drug responses in travellers as an additional tool to assess the emergence of drug resistance in endemic areas where information is limited.
Introduction
A decline in artemisinin efficacy has recently been confirmed in several regions in Southeast Asia [1], [2], [3]. Concerns are growing about the potential for this artemisinin resistance to spread to sub-Saharan Africa, as it has previously been described for other antimalarial drugs. Indeed, resistance to chloroquine (CQ) and sulfadoxine-pyrimethamine (SP) emerged relatively quickly after their introduction and subsequently spread from Asia to Africa [4], [5]. Early detection of decreasing drug efficacy and the consequent updating of drug policies are crucial elements in the strategy to prevent the emergence or delay the spread of drug resistance [6], [7]. In recent years, considerable effort has been made to improve epidemiological antimalarial resistance surveillance in countries with limited resources. Therapeutic efficacy studies remain the gold standard for guiding drug policy, as they take into account the complex interactions between the host, parasite and drug [8]. However, many settings in endemic countries lack the financial resources necessary to maintain a sustainable, accurate and reliable antimalarial resistance surveillance system, resulting in gaps in the spatial and temporal available information.
In recent years, globalization and a substantial increase in international travel and population mobility, have provided the potential for the rapid spread of infectious diseases and antimicrobial resistance [9]. More than 900 million international journeys are undertaken annually and this figure has been consistently rising over the years (United Nations World Tourism Organization: UNWTO).
Malaria is endemic in over 100 countries and represents an important infectious disease threat for these nations. Of the 125 million people travelling to malaria endemic countries each year, approximately 10,000 malaria infections were reported worldwide in returning travellers in 2010. Under-reporting is thought to be substantial and, hence, this number may, in reality, exceed 30,000 [10]. In Europe, a 10-fold increase in imported infections was reported from 1970 to 2000 (from 1,500 to about 15,000 cases) before decreasing to about 6,000 cases in 2010 (http://data.euro.who.int/cisid); most of these cases were reported in France or the United Kingdom [11]. Travellers who return from endemic countries infected with malaria often present with low immunity against the parasites and there is no risk of re-infection, so they are a particularly valuable source of information.
In fact, historically, the emergence of CQ resistance in Africa was mainly detected through surveillance of travellers (Figure 1, Table 1). The current study was undertaken to test the idea that surveillance of parasites from travellers can be used to accurately assess the evolution of antimalarial drug resistance and provide complementary information to existing monitoring. As a proof of concept, the aim was to compare trends in molecular and in vitro markers of drug resistance observed in the imported malaria population with the trends described in field studies.
Figure 1. Map of Africa illustrating the emergence of CQ resistance in East, Central and West Africa detected through travellers’ surveillance from the late 1970s to the early 1980s.
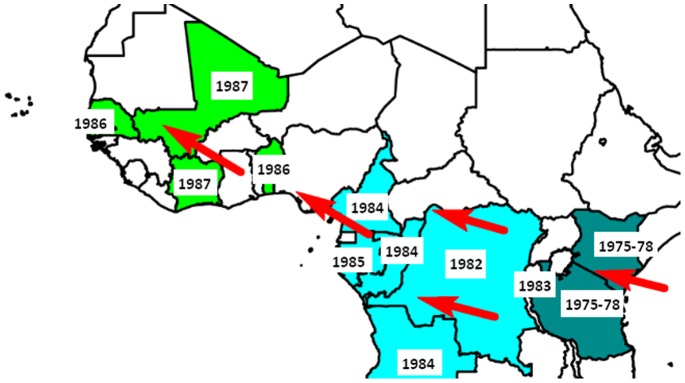
The dates of detection of index cases are displayed. The red arrows show the spread of antimalarial resistance from East Africa to West Africa.
Table 1. First published cases of CQ resistance in Africa through travellers’ surveillance*.
| Country | Date case | Date published | Country of detection | Reference |
| Kenya | 1978 | 1979 | Denmark | [29] |
| East Africa (Kenya-Tanzania) | 1975 | 1979 | United States | [30] |
| Democratic Republic of the Congo (Zaire) | 1982 | 1983 | United States | [31] |
| Burundi | 1983 | 1984 | France | [32] |
| Republic of the Congo | 1984 | 1985 | France | [33] |
| Cameroon | 1984 | 1985 | France | [34] |
| Angola | 1984 | 1984 | Denmark | [35] |
| Gabon | 1985 | 1986 | United States | [36] |
| Benin | 1986 | 1986 | France | [37] |
| Senegal | 1986 | 1987 | Sweden | [38] |
| Cote d’Ivoire | 1987 | 1988 | France | [39] |
| Mali | 1987 | 1988 | France | [40] |
The within-country studies which detected the emergence of CQ resistance in other parts of Africa during this time period are not shown here.
Materials and Methods
Data Collection
The studies were conducted by the National Reference Centre for Malaria (CNR), Paris, France and investigators from the four endemic countries, in collaboration with the WorldWide Antimalarial Resistance Network (WWARN).
Data from travellers
Data were collected from travellers with symptomatic Plasmodium falciparum malaria returning from malaria-endemic countries during the period 1996–2011. These cases were reported to the French CNR by one of the 80 hospitals participating in the sentinel network for malaria which covers about half of the cases diagnosed in France. All the travellers included in this study must have visited a malaria-endemic African country in the two months prior to diagnosis and presented with a P. falciparum infection biologically confirmed by thin and thick blood smear. Basic demographic, epidemiologic, clinical, and parasitological information as well as response to treatment, previous malaria infection and travel history information were systematically reported. Blood samples were only collected from hospitals which document anti-malarial drug resistance on a systematic basis in all Plasmodium positive diagnosis, before treatment, for molecular and in vitro analyses.
Molecular markers associated with resistance to CQ and pyrimethamine and CQ susceptibility in vitro were the tools used to compare antimalarial drug resistance trends between travellers and field studies.
No informed consent was required for this study as the procedures described here were part of the French national surveillance system of malaria. In 2006, the Commission Nationale de l’Informatique et des Libertés (CNIL), France, approved the electronic-based utilisation of patient-level data collected through the CNR’s questionnaire. We did not receive ethical approval or waiver to perform this secondary research on samples collected as a part of government surveillance for communicable diseases (Loi n° 2004-806 art.16, 21, 9 August 2004, Journal Officiel 11 August 2004). However, ethical aspects have been respected according to the French regulation (article L.1211-2, L.1111-7, L.1413-4 and L.1413-5 Code de la Santé Publique). Also an information note, which explained that the collected blood samples could be used for further research analyses, was provided to the patients who had the possibility to refuse; and the samples were anonymised for this study.
Data within-country
A literature review was performed in PubMed for publications on malaria from African endemic countries during the period 1996–2011. The search terms [country name+(pfcrt OR chloroquine resistance)] and [country name+(DHFR OR sulfadoxine-pyrimethamine resistance OR sulfadoxine pyrimethamine resistance)] were used.
After performing a sample size calculation (see below sample size calculation section), four African countries, Senegal, Mali, Cote d’Ivoire and Cameroon were included in this study. They had sufficiently large numbers of both travellers and field molecular data, from 1996–2011, for a meaningful comparison.
The collaboration of investigators within the four targeted countries, facilitated by WWARN, enabled the collation and standardisation of published field data and the identification and standardisation of unpublished field data. The field studies used for the analyses are summarized in Table 2.
Table 2. Summary of the molecular and in vitro field studies in the four endemic countries included in the analysis (both published and unpublished).
| Country | Site | Age population | Year of study | Reference | |
| Molecular marker analysis | |||||
| Pfcrt 76 | Senegal | Pikine | ≥5 ya | 2000 | [41] |
| Pikine | ≥18ya | 2001 | [42] | ||
| Thiadiaye | Pregnant women | 2002 | [43] | ||
| Dakar | 3–65ya | 2002 | [44] | ||
| Pikine | ≥3ya | 2004 | [45] | ||
| Dakar | All | 2004 | [46] | ||
| Thies | All | 2007 | [47] | ||
| Dakar | All | 2009–10 | [48] | ||
| Central Senegal (3 districts : Mbour, Fatick, Bambey) and Southern Senegal(3 districts : Tambacounda, Velingara, Saraya) | <10 ya | 2009–11 | [49] | ||
| Dakar | all | 2010–2011 | Pradines, unpublished data | ||
| Mali | Kolle | <5 ya | 2002–03 | [50] | |
| Bougoula-Hameau | >6 mths | 2002–04 | [51] | ||
| Bancouma, Monteourou, Bandiagara, Faladie, Koulikoro Ba, Sirakoro-meg, Niena, Kolebougou, Markakoungo, Dimbal, Kafana, Siekorole, Toguel, M'pessoba Banamba, N'debougou | all | 2002–04 | [52] | ||
| Kangaba et kela | >6 mths | 2001–03 | [53] | ||
| Bandiagara, Faladje Kolle Pongenon | all | 2010 | Djimdé, unpublished data | ||
| Cote d’Ivoire | Anonkoua-koute (Abidjan), Ayamé, Dabakala | all | 2003–08 | Ako, unpublished data | |
| Bonoua and Samo | <5 ya | 2005 | [54] | ||
| Abidjan (2 districts: Yopougon and Adjamé) | Children | 2006 | [55] | ||
| Adzope | Children | 2007 | [56] | ||
| Cameroon | Maroua, Ndop, Bafoussam, Hévécam | <5 ya | 2000–01 | [57] | |
| Yaoundé | >12 ya | 2000–01 | [58] | ||
| Garoua, Yaounde, Mutengene | <5 ya | 2004–06 | [59] | ||
| Yaoundé, Mfou (suburb of Yaoundé) | All ages | 2005–08 | [60] | ||
| Pfdhfr 108 | Senegal | Dielmo, Sine Saloum | All | 1996–99 | [61] |
| Pikine, Tambacounda, Thies | ≥5 ya | 2000–03 | [62] | ||
| Dakar | 3–65 ya | 2002 | [44] | ||
| Dakar | <5 ya | 2006–08 | [63] | ||
| Dakar | all | 2009–10 | [47] | ||
| Mali | Tieneguebougou | 2–12 ya | 1996 | [64] | |
| Kidal | All | 1999 | [65] | ||
| Bandiagara | 5–15 ya | 2000 | [66] | ||
| Kolle | <5 ya | 2002–03 | [50] | ||
| Bongoula-Hameau | >6 mths | 2002–04 | [50] | ||
| Kolokani | <5 ya | 2006–07 | [67] | ||
| Cote d’Ivoire | Yopougon | <5 ya | 2000–01 | [68] | |
| Anonkoua-koute (Abidjan), Ayamé, Dabakala | 2003–08 | Ako, unpublished data | |||
| Bonoua and Samo | <5 y | 2005 | [54] | ||
| Abidjan (2 districts: Yopougon and Adjamé) | Children | 2006 | [55] | ||
| Adzope | Children | 2007 | [56] | ||
| Cameroon | Bertoua, Douala, Eseka, Yaounde | <5 ya | 1999 | [69] | |
| Bafoussam, Bertoua, Djoum, Garoua, Hevecam, Manjo, Maroua, Mengang, Ndop, Ngaoundere, Sangmelima, Yaounde | <10 ya | 1999–03 | [70] | ||
| Dschang, Fontem, Limbe, Nkambe | <10 ya | 2002–03 | [71] | ||
| Garoua, Mutengene, Yaounde | <5 ya | 2004–06 | [59] | ||
| Yaounde | ≥12 ya | 2001–05 | [72] | ||
| In vitro susceptibility test | |||||
| chloroquine | Senegal | Mlomp | 1996–98 | [73] | |
| Pikine, Dielmo, NDiop | All | 1996 | [74] | ||
| Dielmo, NDiop | 1997 | [75], [76] | |||
| Pikine | ≥5 ya | 2000 | [41] | ||
| Pikine | ≥18 ya | 2001 | [42] | ||
| Dakar | 2002 | [44] | |||
| Dakar | 2009–2010 | [77] | |||
Laboratory Analysis of Parasites
Molecular analysis
Two molecular markers, pfcrt 76 for CQ resistance and pfdhfr 108 for pyrimethamine resistance, were used in this study to compare the trends between travellers and field data. Although the presence of these two markers does not perfectly correlate with treatment failure, each is a good proxy of the intrinsic resistance of the parasite [12], [13]. They are used here as a proof of concept since they have been widely and consistently collected in both field studies and travellers surveillance over the period of interest. Due to the availability of travellers’ data, the time period of 2000–2011 was studied for pfcrt 76T, while the time period of 1996–2011 was used for pfdhfr 108N.
For molecular analyses of parasites from travellers, DNA was extracted from blood samples of P. falciparum, using the QIAamp DNA Mini Kit, Qiagen® before 2008 and the MagNA Pure LC DNA Isolation Kit I, Roche after 2008. PCR and subsequent allele-specific restriction analyses were performed to identify polymorphic codons of interest at the pfcrt 76 locus (Lys to Thr) and the pfdhfr 108 locus (Ser to Asn) [14].
For field studies, the genotyping methods differed slightly between studies. The detailed method for each study was described in the corresponding publication (see Table 2 for the references to the studies).
Only “pure” pfcrt 76T and pfdhfr 108N infections among the total number of samples tested were included to improve comparability of the allele prevalence calculated between studies. Indeed, genotyping methods for detecting mixed infections vary in sensitivity across studies. There is not clear evidence that patients living in endemic countries are more likely to carry mixed alleles (mutant and sensitive) than travellers returning from endemic countries as the number of mosquitoes’ bites is not the only factor to consider and the presence of mixed infections is possible after only one bite [15].
In vitro assay
For the in vitro susceptibility tests, only data from Senegal were analysed in this study because there were sufficient available data for parasites from both the travellers and within-country isolates over the complete period of interest.
For susceptibility tests of parasites from travellers, the following methods were used. The batches of plates were validated on the CQ-susceptible 3D7 reference strain and the CQ-resistant W2 reference strain using the standard 42-hour 3H-hypoxanthine uptake inhibition method in controlled atmospheric conditions in the incubator (5% CO2, 10% O2 and 85% N2) [16], [17]. The isotopic microtests were performed, aliquoting 200 µl/well of the suspension of fresh parasitized erythrocytes into 96-well plates pre-dosed with CQ. Radioactivity incorporated by the parasites was measured using a scintillation counter. The CQ susceptibility was calculated as the 50% inhibitory concentration (IC50) of CQ of the isolates tested [18], [19]. The drug concentration that inhibited 50% of parasite growth (IC50) was estimated by using nonlinear regression to fit an inhibitory sigmoid Emax model [20]. The In Vitro Analysis and Reporting Tool (IVART) enabled the transformation, standardization and analysis of the data [21].
For within-country surveys, the in vitro methods differed between studies and were described in the publications, which are referenced in Table 2. However, the measurement of the drug susceptibility of fresh P. falciparum parasites was mainly performed by isotopic assays using the 3H-hypoxanthine uptake inhibition method. The in vitro CQ susceptibility was determined by a P. falciparum Lactate DeHydrogenase (pLDH) ELISA assay in four studies.
Statistical Analysis
Sample size calculation
In order to select eligible countries with enough data per year for significant molecular analysis, a sample size calculation was first performed. The basic comparison of the trends used a simple logistic regression model. The prevalence of isolates from traveller samples that carried a mutant allele (Pt) or from studies on field samples (Pf) was the metric used. In the models: S.
where X is the time covariate and a the intercept, the null hypothesis of equal temporal slopes was tested [22]. That is,
A two sided t-test with a test significance level of α = 0.05, a power of 1−β = 0.80 and an effect size of δ = 0.15 was used. The total sample size required for showing a significant difference between the slopes St and Sf was n = 642 isolates for each data type (field and travellers data) per country.
Logistic regression
For the molecular analysis, a logistic regression model with time as a linear covariate was fitted to the prevalence of the mutant isolates (separately, for the pfcrt 76 and pfdhfr 108 data) for the travellers and field studies, for each country. Given the probability of the mutant isolates, the observed number of mutant isolates in each year was assumed to be binomially distributed. The estimated slope of the fitted logistic regression curve for the travellers and field data, the 95% confidence intervals for the slopes and whether the slopes differ significantly from each other are presented (see figures in the results section).
For the in vitro susceptibility analysis, a Generalized Linear Model (GLM) with a log-link function was fitted to the travellers and field data for the period 2000–2011. The slopes of the changes in CQ susceptibility for the two datasets were assessed to determine whether they differed significantly from null (0) and whether they differed significantly from each other.
Software
All statistical analyses were performed using Stata version 11 for Windows (Stata Corp, College Station, TX, USA) and R version 2.10 (R – project).
Results
Four African countries had sufficient numbers of field and traveller derived isolates to allow meaningful comparisons between the two populations: Senegal, Mali, Cote d’Ivoire and Cameroon. The characteristics for the field studies are summarised in Table 2 for each publication. A total of 23 studies were included for pfcrt 76 analysis over the 2000–2011 period, mainly from Senegal; 21 studies for pfdhfr 108 analysis for 1996–2010 and 8 studies from Senegal for the in vitro analysis over the same period. The characteristics of the patients differed between studies regarding the population age and the study settings (urban or rural area) but this heterogeneity was observed for the four countries.
The median patient age for travellers experiencing malaria after their return to France was 31 years, with 79% older than 15 and 61% of the travellers had visited friends and relatives in endemic countries for more than one month. Only 38% reported prophylaxis intake during their travel and most patients presented with uncomplicated malaria (95%) (Table 3). No differences among these characteristics were observed among the four countries except for gender; a majority of travellers to Senegal and Mali were male.
Table 3. Characteristics of travellers with malaria returning from Senegal, Mali, Cote d’Ivoire and Cameroon and reported in France during the period from 2000 to 2011.
| Travellers | Senegal (n = 1,993)* | Mali (n = 2,372)* | Cote d’Ivoire (n = 4,778 )* | Cameroon (n = 3,272)* |
| Median age (year) [Min-Max] | 30 [0–94] | 31 [0–76] | 30 [0–83] | 33 [0–87] |
| Gender ratio (Male/Female) | 2.47 | 2.20 | 1.40 | 1.15 |
| Chemoprophylaxis | ||||
| Yes n (%) | 746 (38) | 959 (41) | 1,955 (41) | 1,048 (32) |
| Duration of stay | ||||
| ≤2 weeks n (%) | 218 (13) | 152 (8) | 457 (12) | 439 (16) |
| 2–4 weeks n (%) | 356 (21) | 361 (18) | 1,150 (30) | 868 (33) |
| 1–3 months n (%) | 699 (41) | 928 (48) | 1,221 (32) | 679 (25) |
| >3 months n (%) | 428 (25) | 498 (26) | 1,021 (26) | 688 (26) |
| Purpose of travel | ||||
| Tourism n (%) | 322 (18) | 251 (12) | 514 (12) | 397 (13) |
| Visit friends and relatives n (%) | 1,108 (61) | 1,520 (71) | 2,482 (58) | 1,738 (59) |
| Severe malaria** | ||||
| Yes n (%) | 136 (7) | 123 (5) | 225 (5) | 177 (5) |
Numbers may not add to totals because of missing information.
Severe malaria are cases of imported malaria that fulfilled at least one criteria of the WHO clinical and laboratory classification of severity [78].
Between 2000–2011, 2,874 P. falciparum positive isolates were collected from travellers for analysis of the pfcrt 76 allele prevalence and 3,351 isolates for analysis of the pfdhfr 108 allele prevalence between 1996–2011. Between 1996 and 2011, 305 fresh blood samples were collected from travellers, and tested in Paris or Marseille to measure susceptibility to CQ in vitro.
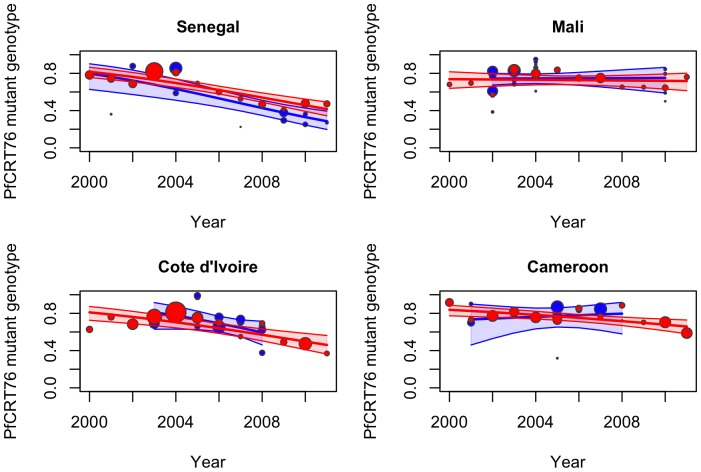
Figure 2 and Table 4 summarize the temporal trends in the prevalence of the pfcrt 76T mutant isolates (associated with CQ resistance) in each of the targeted countries. The prevalence of the pfcrt 76T mutant genotype significantly decreased for travellers between 2000 and 2011 in Senegal (St = −0.17, p<10−3), Cote d’Ivoire (St = −0.15, p<10−3) and less dramatically in Cameroon (St = −0.09, p<10−3). However, over that same period, no overall decrease was observed in isolates from Mali (St = −0.01, p = 0.72).
Figure 2. Observed data, fitted model (by logistic regression) and 95% confidence interval (shaded area) for the prevalence of the pfcrt 76 mutant isolates from 2000 to 2011 for travellers (red) and field studies (blue) for A-Senegal, B-Mali, C-Cote d’Ivoire and D-Cameroon.
Each data point represents the prevalence of resistant isolates per year for travellers’ data and per study for field studies, where the size of the circle is proportional to the number of isolates in the sample.
Table 4. Comparison between travellers and field data for the pfcrt 76 and pfdhfr 108 molecular markers and for the CQ in vitro susceptibility in Senegal.
| Country | Travellers Slope [95% CI*] | Field Study Slope [95% CI] | p-value** | |
| Pfcrt 76 | Senegal | −0.167 [−0.219; −0.115] | −0.208 [−0.312; −0.105] | 0.575 |
| Mali | −0.009 [−0.082; 0.063] | 0.005 [−0.106; 0.116] | 0.885 | |
| Cote d’Ivoire | −0.146 [−0.215; −0.078] | −0.215 [−0.463; 0.032] | 0.578 | |
| Cameroon | −0.090 [−0.146; −0.033] | 0.050 [−0.220; 0.321] | 0.264 | |
| Pfdhfr 108 | Senegal | 0.117 [0.088; 0.147] | 0.148 [0.088; 0.209] | 0.386 |
| Mali | 0.182 [0.124; 0.240] | 0.119 [0.086; 0.152] | 0.116 | |
| Cote d’Ivoire | 0.083 [0.052; 0.113] | 0.132 [−0.025; 0.289] | 0.484 | |
| Cameroon | 0.213 [0.115; 0.311] | 0.130 [−0.155; 0.415] | 0.753 | |
| CQ in vitro analysis | Senegal | −0.050 [−0.085; −0.015] | −0.028 [−0.059; 0.002] | 0.264 |
CI = confidence interval,
The p-value indicates whether the fitted slopes for travellers data and field studies were significantly different from each other.
After comparing the slopes of the trends for the isolates from travellers (St) and from locally studied parasites (Sf), no significant differences were observed between 2000 and 2011 in Senegal (St = −0.17 versus Sf = −0.21, p = 0.58), Mali (St = −0.01 versus Sf = 0.01, p = 0.89), Cote d’Ivoire (St = −0.15 versus Sf = −0.22, p = 0.58) and Cameroon (St = −0.09 versus Sf = 0.05, p = 0.26) (Table 4, Figure 2). After performing a power calculation on the four previous tests, the probabilities of rejecting the null hypothesis, St = Sf, when it is false, were between 92 and 96%. These data derived from studies of parasites from returning travellers reflected accurately the trends of the prevalence of molecular markers of CQ resistance that were occurring in the countries in which the travellers acquired their malaria.
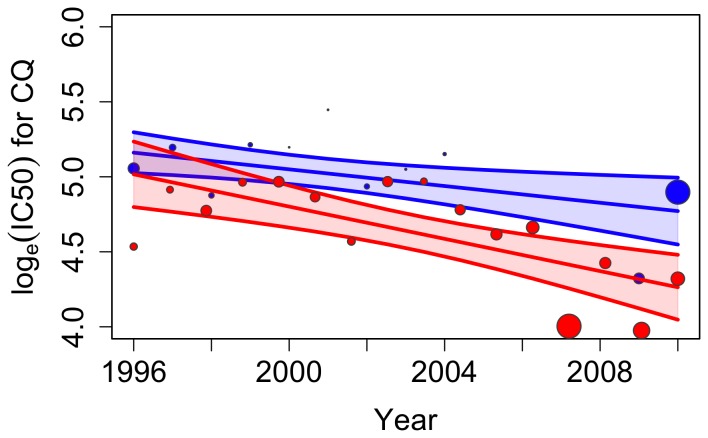
Changes in CQ susceptibility were also assessed using the in vitro response of isolates. From 1996 to 2011 the geometric mean of the IC50 for CQ of the isolates tested in vitro decreased in isolates from travellers and those studied in Senegal (Table 4, Figure 3). The geometric means of the IC50 values measured for the isolates from travellers were lower than those measured in Senegal. However, the slopes showing the trends did not differ significantly (St = −0.05 versus Sf = −0.03, p = 0.26) with a power of 94%. In this case, as well, the data gathered from travellers was an accurate reflection of the trend among parasite populations in the country of origin.
Figure 3. Observed data, fitted model (by Generalized Linear Model) and 95% confidence interval (shaded area) for the in vitro CQ response (IC50) isolates from 1996 to 2011 for travellers (red) and field studies (blue) from Senegal.
Each data point represents the ln (mean IC50) per year for travellers’ data and per study for field studies, where the size of the circle is proportional to the number of isolates in the sample.
The increase of the molecular marker pfdhfr 108N has been commonly associated with an increase of pyrimethamine resistance for more than fifteen years [23]. When this parameter was compared between travellers and field-derived isolates, a significant increase in the pfdhfr 108N genotype was observed in all 4 countries over the period from 1996–2011: Senegal (St = 0.12, p<10−3), Mali (St = 0.18, p<10−3), Cote d’Ivoire (St = 0.08, p<10−3) and Cameroon (St = 0.21, p<10−3) (Table 4, Figure 4). For this comparison as well, no significant difference was observed in the trends of the molecular marker pfdhfr 108N when data from travellers (St) and field-derived (Sf) isolates were compared for samples taken between 1996 and 2011: Senegal (St = 0.12 versus Sf = 0.15, p = 0.39), Mali (St = 0.18 versus Sf = 0.12, p = 0.12), Cote d’Ivoire (St = 0.08 versus Sf = 0.13, p = 0.48) and Cameroon (St = 0.21 versus Sf = 0.13, p = 0.75) (Table 4, Figure 3). The powers of the four comparative analyses ranged between 91 and 97%.
Figure 4. Observed data, fitted model (by logistic regression) and 95% confidence interval (shaded area) for the prevalence of the pfdhfr 108 mutant isolates from 1996 to 2011 for travellers (red) and field studies (blue) for A-Senegal, B-Mali, C-Cote d’Ivoire and D-Cameroon.
Each data point represents the prevalence of resistant isolates per year for travellers’ data and per study for field studies, where the size of the circle is proportional to the number of isolates in the sample.
Thus, all three measures of changes in prevalence of molecular markers and in vitro parasites resistance to CQ and pyrimethamine demonstrate that information from parasites imported by travellers was an accurate measure of the changes in parasites within the 4 countries studied.
Discussion
This study suggests that the surveillance of travellers may be used for monitoring antimalarial drug resistance in endemic countries. The proof of concept was demonstrated using the prevalence of two molecular markers, pfcrt 76 and pfdhfr 108, and in vitro susceptibility for CQ. In this study, no significant difference between the trends of antimalarial drug resistance for travellers’ and field data were observed over more than 10 years. A decrease of the prevalence of the pfcrt 76T mutant genotype was observed over a period of 10 years in travellers returning from Senegal, Cote d’Ivoire and Cameroon, whilst this prevalence remains stable in Mali. An increase of mutant genotype isolates for pfdhfr 108 was observed in the four countries of West and Central Africa. The in vitro CQ susceptibility results supported the molecular results for Senegal. The trend in pfcrt 76 is downward while the trend in pfdhfr 108 is upward. The fact that screening travellers was able to detect temporal trends in opposite directions strengthens the proof of concept significantly.
Sustainable, reliable and systematic monitoring of drug efficacy is needed for tracking resistance [24]. Monitoring antimalarial drug resistance is based on clinical assessment and biological assays as part of a clinical trial [25]. Since the emergence of resistance to CQ, and then later to SP, capacities to conduct such monitoring in endemic countries have substantially improved, but remain very heterogeneous. In particular regions, human and/or technical resources are limited and, as such, conducting a clinical trial for the purpose of surveillance has competed with other high priorities that Ministries of Health must contend with and has not been systematically conducted.
Previous studies highlight the usefulness of travellers’ surveillance as an early warning detection system for emergence or re-emergence of communicable diseases [26], [27], [28]. Travellers’ surveillance has proven in the past to be an effective early alert system for detecting the emergence of CQ resistance (Table 1).
One strength of using travellers as a sentinel system of resistance is that detection of clinical therapeutic failure due to resistance is facilitated in this non immune population with a low risk of re-infection. Moreover, the French Malaria Reference Centre use standardized methods for prospectively collecting reliable information.
This study does have several limitations. First, due to the complexity of collecting laboratory data systematically, consistently and over a long period of time in both populations, travellers and field studies, only four countries and two molecular markers have been used in this proof of concept. Second, precise information regarding the location of infection within each country could not be collected for P. falciparum infected travellers returning from endemic countries. However, travellers did not visit all parts of a country and they were more likely to frequent particular places such as touristic and/or, or business-oriented locations. The reported information was highly dependent on factors such as the areas that were visited, the period of travel, migration history and the political context in endemic areas. Of course, these factors can also impact information on exact locations where patients acquire their infections within the country, as well. Perhaps more importantly, the travellers in this work were not representative of the native population in that their baseline characteristics differ, including age, immune status and parasitemia before treatment. Finally, especially for the field studies, different approaches were used for determining the molecular markers of resistance and the in vitro susceptibility for CQ. The heterogeneity between methods is encouraging WWARN to standardise approaches and to develop common tools like IVART [21].
However, these limitations do not diminish the clarity of the outcome presented here. Surveillance of parasites from travellers provided an accurate picture of events occurring in the field. This does not suggest that this approach should replace studies conducted in endemic countries. Rather, information from travellers can be used as an additional surveillance system.
Given the utility, surveillance of travellers can be useful in tracking resistance to ACTs, as well. Currently, only the response to the long-acting partner drugs, can be assessed, but if putative molecular markers are defined, tracking of resistance to the artemisinin component can also be added. The collaboration between Ministries of Health in endemic countries and the malaria reference centres in non-endemic countries for sharing and validating collected information should be reinforced and facilitated.
Due to the length of time between a field study and the publication of results, data collected from imported cases may be available in a more timely manner and, as such, could be used for early alert of emerging resistance. The complexity of the available tools for assessing drug efficacy and monitoring resistance highlights the importance of a standardized and coordinated approach. The follow-up of imported cases in several non-endemic countries should also enable the collaborators to track the evolution of resistance to antimalarial drugs at an international scale and thus provide novel information of value to policy makers.
The goal of this work was to validate the use of international traveller surveillance systems, for detecting the emergence of antimalarial drug resistance and for following resistance trends where local information is not otherwise available and/or sufficient. Easy access to reproducible and standardized data should be implemented. The existing health international, European or American institutions (WHO, European Centre for Disease Prevention and Control (ECDC), US Centres for Disease Control and Prevention (CDC Atlanta) and the different networks for infectious diseases surveillance in travellers (TropNet Europe, EuroTravNet, GeoSentinel) should be used for facilitating the coordination and data sharing between national surveillance systems [26], [27], [28].
Conclusions
This study has not shown different trends in antimalarial drug resistance between travellers and field studies. An international travellers’ database can be used as an additional surveillance system to assess and monitor the emergence of drug resistance in endemic areas where information is limited.
Acknowledgments
We thank Prof Carol Sibley for critical reading of the manuscript. The authors would like to thank Sandie Menard and Antoine Berry for generously providing additional information and data from their previous publication. The authors would like to thank Fabrice Legros (deceased) for his contribution to the study.
This article has been submitted on behalf of the National Malaria Reference Centre Study Group
Ahmed Aboubacar (Institut de parasitologie et de pathologie tropicale, Strasbourg, France), Patrice Agnamey (Service de Parasitologie, CHU Amiens, France), Adela Angoulvant (Service Parasitologie, APHP Bicêtre, Paris, France), Patricia Barbut (Laboratoire de Biologie Médicale, CH Longjumeau, France), Didier Basset (Service Parasitologie, CHU Montpellier, France), Ghania Belkadi (Service Parasitologie, APHP Saint Antoine, Paris, France), Anne-Pauline Bellanger (Service Parasitologie, CHRU Jean Minjoz, Besançon, France), Dieudonné Bemba (Service Microbiologie, APHP Bondy-Jean Verdier, Paris, France), Françoise Benoit-Vical (Service Parasitologie, CHU Rangueil, Toulouse, France), Antoine Berry (Service de Parasitologie, CHU Rangueil, Toulouse, France), Marie-Laure Bigel (Laboratoire Biologie Médicale, CH François Quesnay, Mantes-La-Jolie, France), Julie Bonhomme (Service Microbiologie, CHU Caen, France), Françoise Botterel (Service de Parasitologie, APHP Henri Mondor, Créteil, France), Olivier Bouchaud (Service de Maladies Infectieuses et Tropicales, CHU Avicenne-Bobigny, Paris, France), Marie-Elisabeth Bougnoux (Service Microbiologie, APHP Necker, Paris, France), Patrice Bourée (Service Parasitologie, APHP Bicêtre, Paris, France), Laurent Bret (Service Microbiologie, CHR Orleans, France), Pierre Buffet (Service Parasitologie, APHP Pitié-Salpétrière, Paris, France), Bernadette Buret (Laboratoire Biologie Médicale, CH Niort, France), Sylviane Chevrier (Service Parasitologie, CHU Rennes, France), Bernadette Cuisenier (Service Parasitologie, CHU Dijon, France), Martin Danis (Service de Parasitologie, APHP Pitié-Salpétrière, Paris, France), Marie-Laure Dardé (Service Parasitologie, CHU Limoges, France), Ludovic De Gentile (Service de médecine tropicales, CHU Angers, France), Jean-Marie Delarbre (Service Microbiologie, CH Mulhouse, France), Pascal Delaunay (Service Parasitologie, CHU Nice, France), Anne Delaval (Service Microbiologie, CHG Aulney-sous-Bois, France), Jean Delmont (Service Médecine Tropicale et Infectieuse, Hôpital Nord, Marseille, France), Guillaume Desoubeaux (Service Parasitologie, CHRU de Tours, France), Michel Develoux (Service Paraistologie, APHP Saint Antoine, Paris, France), Jean Dunand (Service Microbiologie et Hygiene, APHP Ambroise Paré, Paris, France), Rémy Durand (Service Parasitologie, APHP Avicenne-Bobigny, Paris, France), Odile Eloy (Service Parasitologie, CH Versailles, France), Nathalie Fauchet (Service Microbiologie, CHI Créteil, France), Bernard Faugere (Service Parasitologie, APHM Timone, Marseille, France), Albert Faye (Service Pédiatrie, APHP Robert Debré, Paris, France), Pierre Flori (Service Parasitologie, CHU Saint-Etienne, France), Chantal Garabedian (Laboratoire de Maladies Infectieuses et Hygiène, CH Pays d’Aix, Aix-en-Provence, France), Françoise Gay-Andrieu (Service Parasitologie, CHU Nantes, France), Nadine Godineau (Service Microbiologie, CHG Delafontaine, St Denis, France), Pascal Houzé (Laboratoire Biochimie, APHP Saint Louis, Paris, France), Sandrine Houzé (Service Parasitologie, APHP Bichat-Claude Bernard, Paris, France), Houria Ichou (Service Microbiologie et Hygiène, APHP Louis Mourier, Paris, France), Laurence Lachaud (Service Microbiologie, CHU Nîmes, France), Magalie Lefevre (Laboratoire de Biologie Médicale, CH Creil, France), Gwenaël Le Moal (Service Maladies Infectieuses, CHU Poitiers, France), Marie Machouart (Service Parasitologie, CHU Nancy, France), Denis Malvy (Service Parasitologie, CHU Bordeaux, France), Sophie Matheron (Service Maladies Infectieuses et Tropicales, APHP Bichat-Claude Bernard, Paris, France), Danièle Maubon (Service Parasitologie, CHU Grenoble, France), Bruno Megarbane (Service de Réanimation Médicale et Toxicologique, APHP Lariboisière, Paris, France), Sandie Menard (Service de Parasitologie, CHU Rangueil, Toulouse, France), Laurence Million (Service Parasitologie, CHRU Jean Minjoz, Besançon, France), Muriel Mimoun-Aiach (Service Microbiologie, APHP Trousseau, Paris, France), Philippe Minodier (Service Pédiatrie, APHM Hôpital Nord, Marseille, France), Gilles Nevez (Service Parasitologie, CHU Brest, France), Philippe Parola (Service Maladies Infectieuses et Tropicales APHM Hôpital Nord, Marseille, France), Daniel Parzy (Service Parasitologie, IMTSSA, Marseille, France), Olivier Patey (Service Maladies Infectieuses et Tropicales, CHI Villeneuve Saint Georges, France), Pierre Patoz (Laboratoire Biologie Médicale, CH Tourcoing, France), Pascale Penn (Service Microbiologie, CH Le Mans, France), Alice Perignon (Service de maladies infectieuses et tropicales, APHP Pitié-Salpetrière, Paris, France), Stéphane Picot (Service Parasitologie, CHU Lyon, France), Jean-Etienne Pilo (Service de Santé et des Armées Maladie Infectieuses et Tropicales, HIA Begin, Saint Mandé, France), Isabelle Poilane (Service Microbiologie, APHP Bondy Jean Verdier, Paris, France), Denis Pons (Service Microbiologie, CHU Clermont Ferrand, France), Christophe Rapp (Service de Santé et des Armées Maladie Infectieuses et Tropicales, HIA Begin, Saint Mandé, France), Marie-Catherine Receveur (Service Médecine, CHU Bordeaux, France), Claudine Sarfati (Service Parasitologie, APHP Saint-Louis Lariboisière, Paris, France), Yaye Senghor (Service Parasitologie, APHP Bobigny Avicenne, Paris, France), Jean-Yves Siriez (Service Pédiatrie, APHP Robert Debré, Paris, France), Nicolas Taudon (Service Parasitologie, IMTSSA, Marseille, France), Marc Thellier (Service Parasitologie, APHP Pitié-Salpétrière, Paris, France), Maxime Thouvenin (Service Microbiologie, CH Troye, France), Dominique Toubas (Service Parasitologie, CHU Reims, France)
Funding Statement
This study was supported in part by a grant for doctoral studies to MG from the Doctoral Network of the École des Hautes Études en Santé Publique, Rennes, France and a grant for CNRpaludisme from Institut National de Veille Sanitaire, St Maurice, France. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, et al. (2012) Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379: 1960–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, et al. (2009) Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, et al. (2008) Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359: 2619–2620. [DOI] [PubMed] [Google Scholar]
- 4. Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, et al. (2002) Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418: 320–323. [DOI] [PubMed] [Google Scholar]
- 5. Roper C, Pearce R, Nair S, Sharp B, Nosten F, et al. (2004) Intercontinental spread of pyrimethamine-resistant malaria. Science 305: 1124. [DOI] [PubMed] [Google Scholar]
- 6. Morens DM, Folkers GK, Fauci AS (2004) The challenge of emerging and re-emerging infectious diseases. Nature 430: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO (2011) Global Plan for Artemisinin Resistance Containment (GPARC). Geneva: World Health Organization. 88 p. [Google Scholar]
- 8.WHO (2010) Global report on antimalarial drug efficacy and drug resistance: 2000–2010. Geneva: World Health Organization. 116 p. [Google Scholar]
- 9. MacPherson DW, Gushulak BD, Baine WB, Bala S, Gubbins PO, et al. (2009) Population mobility, globalization, and antimicrobial drug resistance. Emerg Infect Dis 15: 1727–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO (2010) International Travel and Health. Geneva: World Health Organization. 264 p. [Google Scholar]
- 11. Sabatinelli G, Ejov M, Joergensen P (2001) Malaria in the WHO European Region (1971–1999). Euro Surveill 6: 61–65. [PubMed] [Google Scholar]
- 12. Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, et al. (2001) A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344: 257–263. [DOI] [PubMed] [Google Scholar]
- 13. Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, et al. (2002) Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis 185: 380–388. [DOI] [PubMed] [Google Scholar]
- 14. Durand R, Jafari S, Vauzelle J, Delabre JF, Jesic Z, et al. (2001) Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol Biochem Parasitol 114: 95–102. [DOI] [PubMed] [Google Scholar]
- 15. Jafari S, Le Bras J, Bouchaud O, Durand R (2004) Plasmodium falciparum clonal population dynamics during malaria treatment. J Infect Dis 189: 195–203. [DOI] [PubMed] [Google Scholar]
- 16. Desjardins RE, Canfield CJ, Haynes JD, Chulay JD (1979) Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother 16: 710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaddouri H, Nakache S, Houze S, Mentre F, Le Bras J (2006) Assessment of the drug susceptibility of Plasmodium falciparum clinical isolates from africa by using a Plasmodium lactate dehydrogenase immunodetection assay and an inhibitory maximum effect model for precise measurement of the 50-percent inhibitory concentration. Antimicrob Agents Chemother 50: 3343–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basco L (2007) Field Application of In Vitro Assays for the Sensitivity of Human Malaria Parasites to Antimalarial Drugs. Geneva: World Health Organization. 202 p. [Google Scholar]
- 19. Pradines B, Hovette P, Fusai T, Atanda HL, Baret E, et al. (2006) Prevalence of in vitro resistance to eleven standard or new antimalarial drugs among Plasmodium falciparum isolates from Pointe-Noire, Republic of the Congo. J Clin Microbiol 44: 2404–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Nagard H, Vincent C, Mentre F, Le Bras J (2011) Online analysis of in vitro resistance to antimalarial drugs through nonlinear regression. Comput Methods Programs Biomed 104: 10–18. [DOI] [PubMed] [Google Scholar]
- 21. Woodrow CJ, Dahlstrom S, Cooksey R, Flegg JA, Le Nagard H, et al. (2013) High-throughput analysis of antimalarial susceptibility data by the WorldWide Antimalarial Resistance Network (WWARN) in vitro analysis and reporting tool. Antimicrob Agents Chemother 57: 3121–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armitage P (1980) Statistical Methods in Medical Research. Oxford: Blackwell Scientific Publications.
- 23. Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, et al. (1997) Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis 176: 1590–1596. [DOI] [PubMed] [Google Scholar]
- 24. Vestergaard LS, Ringwald P (2007) Responding to the challenge of antimalarial drug resistance by routine monitoring to update national malaria treatment policies. Am J Trop Med Hyg 77: 153–159. [PubMed] [Google Scholar]
- 25. Sibley CH, Barnes KI, Watkins WM, Plowe CV (2008) A network to monitor antimalarial drug resistance: a plan for moving forward. Trends Parasitol 24: 43–48. [DOI] [PubMed] [Google Scholar]
- 26. Guerin PJ, Grais RF, Rottingen JA, Valleron AJ (2007) Using European travellers as an early alert to detect emerging pathogens in countries with limited laboratory resources. BMC Public Health 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilson ME (2003) The traveller and emerging infections: sentinel, courier, transmitter. J Appl Microbiol 94 Suppl: 1S–11S [DOI] [PubMed] [Google Scholar]
- 28.Gautret P, Cramer JP, Field V, Caumes E, Jensenius M, et al.. (2012) Infectious diseases among travellers and migrants in Europe, EuroTravNet 2010. Euro Surveill 17. [PubMed]
- 29. Fogh S, Jepsen S, Effersoe P (1979) Chloroquine-resistant Plasmodium falciparum malaria in Kenya. Trans R Soc Trop Med Hyg 73: 228–229. [DOI] [PubMed] [Google Scholar]
- 30. Kean BH (1979) Chloroquine-resistant falciparum malaria from Africa. JAMA 241: 395. [PubMed] [Google Scholar]
- 31. Moran JS (1983) Failure of chloroquine prophylaxis in Plasmodium falciparum in Zaire. Lancet 2: 171–172. [DOI] [PubMed] [Google Scholar]
- 32. Le Bras J, Decazes JM, Deloron P, Verdier F, Modai J (1984) R-II chloroquine-resistant falciparum malaria from Burundi. Trans R Soc Trop Med Hyg 78: 410–411. [DOI] [PubMed] [Google Scholar]
- 33. Le Bras J, Coulaud JP, Bricaire F, Le Bras M, Roue R, et al. (1985) Chloroquine-resistant falciparum malaria in the Congo. Lancet 2: 1071. [DOI] [PubMed] [Google Scholar]
- 34. Sansonetti PJ, Lebras C, Verdier F, Charmot G, Dupont B, et al. (1985) Chloroquine-resistant Plasmodium falciparum in Cameroon. Lancet 1: 1154–1155. [DOI] [PubMed] [Google Scholar]
- 35. Olsen VV, Jensen T, Jorgensen M (1984) Chloroquine-resistant Plasmodium falciparum malaria from Angola. Lancet 1: 1462–1463. [DOI] [PubMed] [Google Scholar]
- 36. Neequaye J, Coene J, Taelman H, Wéry M, Greenberg AE, et al. (1986) In vivo chloroquine-resistant falciparum malaria in western Africa. Lancet 1() 2. [PubMed] [Google Scholar]
- 37. Le Bras J, Hatin I, Bouree P, Coco-Cianci O, Garin JP, et al. (1986) Chloroquine-resistant falciparum malaria in Benin. Lancet 2: 1043–1044. [DOI] [PubMed] [Google Scholar]
- 38. Hellgren U, Ardal OK, Lebbad M, Rombo L (1987) Is chloroquine-resistant Plasmodium falciparum malaria emerging in Senegal or The Gambia? Trans R Soc Trop Med Hyg 81: 728. [DOI] [PubMed] [Google Scholar]
- 39. Charmot G, Le Bras J, Doury JC, Baudon D, Roue R, et al. (1988) Chloroquine-resistant Plasmodium falciparum malaria from Ivory Coast. Trans R Soc Trop Med Hyg 82: 392–393. [DOI] [PubMed] [Google Scholar]
- 40. Chabasse D, De Gentile L, Ligny C, Le Bras J, Rialland X, et al. (1988) Chloroquine-resistant Plasmodium falciparum in Mali revealed by congenital malaria. Trans R Soc Trop Med Hyg 82: 547. [DOI] [PubMed] [Google Scholar]
- 41. Thomas SM, Ndir O, Dieng T, Mboup S, Wypij D, et al. (2002) In vitro chloroquine susceptibility and PCR analysis of pfcrt and pfmdr1 polymorphisms in Plasmodium falciparum isolates from Senegal. Am J Trop Med Hyg 66: 474–480. [DOI] [PubMed] [Google Scholar]
- 42. Sarr O, Myrick A, Daily J, Diop BM, Dieng T, et al. (2005) In vivo and in vitro analysis of chloroquine resistance in Plasmodium falciparum isolates from Senegal. Parasitol Res 97: 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bertin G, Ndam NT, Jafari-Guemouri S, Fievet N, Renart E, et al. (2005) High prevalence of Plasmodium falciparum pfcrt K76T mutation in pregnant women taking chloroquine prophylaxis in Senegal. J Antimicrob Chemother 55: 788–791. [DOI] [PubMed] [Google Scholar]
- 44. Henry M, Diallo I, Bordes J, Ka S, Pradines B, et al. (2006) Urban malaria in Dakar, Senegal: chemosusceptibility and genetic diversity of Plasmodium falciparum isolates. Am J Trop Med Hyg 75: 146–151. [PubMed] [Google Scholar]
- 45. Sarr O, Ahouidi AD, Ly O, Daily JP, Ndiaye D, et al. (2008) Mutations in PFCRT K76T do not correlate with sulfadoxine-pyrimethamine-amodiaquine failure in Pikine, Senegal. Parasitol Res 103: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bob NS, Diop BM, Renaud F, Marrama L, Durand P, et al. (2010) Parasite polymorphism and severe malaria in Dakar (Senegal): a West African urban area. PLoS One 5: e9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ndiaye D, Patel V, Demas A, LeRoux M, Ndir O, et al. (2010) A non-radioactive DAPI-based high-throughput in vitro assay to assess Plasmodium falciparum responsiveness to antimalarials–increased sensitivity of P. falciparum to chloroquine in Senegal. Am J Trop Med Hyg 82: 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wurtz N, Fall B, Pascual A, Diawara S, Sow K, et al. (2012) Prevalence of molecular markers of Plasmodium falciparum drug resistance in Dakar, Senegal. Malar J 11: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ndiaye M, Faye B, Tine R, Ndiaye JL, Lo A, et al. (2012) Assessment of the Molecular Marker of Plasmodium falciparum Chloroquine Resistance (Pfcrt) in Senegal after Several Years of Chloroquine Withdrawal. Am J Trop Med Hyg 87: 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tekete M, Djimde AA, Beavogui AH, Maiga H, Sagara I, et al. (2009) Efficacy of chloroquine, amodiaquine and sulphadoxine-pyrimethamine for the treatment of uncomplicated falciparum malaria: revisiting molecular markers in an area of emerging AQ and SP resistance in Mali. Malar J 8: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Djimde AA, Fofana B, Sagara I, Sidibe B, Toure S, et al. (2008) Efficacy, safety, and selection of molecular markers of drug resistance by two ACTs in Mali. Am J Trop Med Hyg 78: 455–461. [PubMed] [Google Scholar]
- 52. Djimde AA, Barger B, Kone A, Beavogui AH, Tekete M, et al. (2010) A molecular map of chloroquine resistance in Mali. FEMS Immunol Med Microbiol 58: 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wele M, Djimde AA, Guindo A, Beavogui AH, Traore IZ, et al. (2011) High frequency of PfCRT 76T in two Malian villages and its prevalence in severe relative to non-severe malaria. Acta Trop 119: 11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ako BA, Offianan AT, Johansson M, Penali LK, Nguetta SP, et al. (2012) Molecular analysis of markers associated with chloroquine and sulfadoxine/pyrimethamine resistance in Plasmodium falciparum malaria parasites from southeastern Cote-d'Ivoire by the time of Artemisinin-based Combination Therapy adoption in 2005. Infect Drug Resist 5: 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Djaman JA, Ahibo H, Yapi FH, Bla BK, Ouattara L, et al. (2010) Molecular Monitoring of Falciparaum Malaria Isolates in Côte d’Ivoire: Genetic Markers (dhfr-ts, dhps, pfcrt, pfmdr-1) for Antimalarial-Drugs Resistance. Eur J Sci Research 40: 10. [Google Scholar]
- 56. Ouattara L, Bla KB, Assi SB, Yavo W, Djaman JA (2010) PFCRT and DHFR-TS Sequences for Monitoring Drug Resistance in Adzopé Area of Côte d’Ivoire After the Withdrawal of Chloroquine and Pyrimethamine. Tropical Journal of Pharmaceutical Research 9: 8. [Google Scholar]
- 57. Basco LK, Ndounga M, Ngane VF, Soula G (2002) Molecular epidemiology of malaria in Cameroon. XIV. Plasmodium falciparum chloroquine resistance transporter (PFCRT) gene sequences of isolates before and after chloroquine treatment. Am J Trop Med Hyg 67: 392–395. [DOI] [PubMed] [Google Scholar]
- 58. Basco LK (2002) Molecular epidemiology of malaria in Cameroon. XIII. Analysis of pfcrt mutations and in vitro chloroquine resistance. Am J Trop Med Hyg 67: 388–391. [DOI] [PubMed] [Google Scholar]
- 59. Mbacham WF, Evehe MS, Netongo PM, Ateh IA, Mimche PN, et al. (2010) Efficacy of amodiaquine, sulphadoxine-pyrimethamine and their combination for the treatment of uncomplicated Plasmodium falciparum malaria in children in Cameroon at the time of policy change to artemisinin-based combination therapy. Malar J 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Menard S, Morlais I, Tahar R, Sayang C, Mayengue PI, et al. (2012) Molecular monitoring of Plasmodium falciparum drug susceptibility at the time of the introduction of artemisinin-based combination therapy in Yaounde, Cameroon: implications for the future. Malar J 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Noranate N, Durand R, Tall A, Marrama L, Spiegel A, et al. (2007) Rapid dissemination of Plasmodium falciparum drug resistance despite strictly controlled antimalarial use. PLoS One 2: e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ndiaye D, Daily JP, Sarr O, Ndir O, Gaye O, et al. (2005) Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in Senegal. Trop Med Int Health 10: 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Faye B, Ndiaye M, Ndiaye JL, Annie A, Tine RC, et al. (2011) Prevalence of molecular markers of Plasmodium falciparum resistance to sulfadoxine-pyrimethamine during the intermittent preventive treatment in infants coupled with the expanded program immunization in Senegal. Parasitol Res 109: 133–138. [DOI] [PubMed] [Google Scholar]
- 64. Doumbo OK, Kayentao K, Djimde A, Cortese JF, Diourte Y, et al. (2000) Rapid selection of Plasmodium falciparum dihydrofolate reductase mutants by pyrimethamine prophylaxis. J Infect Dis 182: 993–996. [DOI] [PubMed] [Google Scholar]
- 65. Djimde AA, Dolo A, Ouattara A, Diakite S, Plowe CV, et al. (2004) Molecular diagnosis of resistance to antimalarial drugs during epidemics and in war zones. J Infect Dis 190: 853–855. [DOI] [PubMed] [Google Scholar]
- 66. Thera MA, Sehdev PS, Coulibaly D, Traore K, Garba MN, et al. (2005) Impact of trimethoprim-sulfamethoxazole prophylaxis on falciparum malaria infection and disease. J Infect Dis 192: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dicko A, Sagara I, Djimde AA, Toure SO, Traore M, et al. (2010) Molecular markers of resistance to sulphadoxine-pyrimethamine one year after implementation of intermittent preventive treatment of malaria in infants in Mali. Malar J 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Djaman JA, Mazabraud A, Basco L (2007) Sulfadoxine-pyrimethamine susceptibilities and analysis of the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum isolates from Cote d'Ivoire. Ann Trop Med Parasitol 101: 103–112. [DOI] [PubMed] [Google Scholar]
- 69. Basco LK, Ndounga M, Tejiokem M, Ngane VF, Youmba JC, et al. (2002) Molecular epidemiology of malaria in Cameroon. XI. Geographic distribution of Plasmodium falciparum isolates with dihydrofolate reductase gene mutations in southern and central Cameroon. Am J Trop Med Hyg 67: 378–382. [DOI] [PubMed] [Google Scholar]
- 70. Tahar R, Basco LK (2006) Molecular epidemiology of malaria in Cameroon. XXII. Geographic mapping and distribution of Plasmodium falciparum dihydrofolate reductase (dhfr) mutant alleles. Am J Trop Med Hyg 75: 396–401. [PubMed] [Google Scholar]
- 71. Mbacham WF, Evehe MSB, Netongo PM, Ali IM, Nfor NE, et al. (2009) Mutations within folate metabolising genes of Plasmodium falciparum in Cameroon. African Journal of Biotechnology 8: 6. [Google Scholar]
- 72. McCollum AM, Basco LK, Tahar R, Udhayakumar V, Escalante AA (2008) Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob Agents Chemother 52: 4089–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Brasseur P, Guiguemde R, Diallo S, Guiyedi V, Kombila M, et al. (1999) Amodiaquine remains effective for treating uncomplicated malaria in west and central Africa. Trans R Soc Trop Med Hyg 93: 645–650. [DOI] [PubMed] [Google Scholar]
- 74. Pradines B, Tall A, Parzy D, Spiegel A, Fusai T, et al. (1998) In-vitro activity of pyronaridine and amodiaquine against African isolates (Senegal) of Plasmodium falciparum in comparison with standard antimalarial agents. J Antimicrob Chemother 42: 333–339. [DOI] [PubMed] [Google Scholar]
- 75. Pradines B, Tall A, Rogier C, Spiegel A, Mosnier J, et al. (2002) In vitro activities of ferrochloroquine against 55 Senegalese isolates of Plasmodium falciparum in comparison with those of standard antimalarial drugs. Trop Med Int Health 7: 265–270. [DOI] [PubMed] [Google Scholar]
- 76. Pradines B, Tall A, Ramiandrasoa F, Spiegel A, Sokhna C, et al. (2006) In vitro activity of iron-binding compounds against Senegalese isolates of Plasmodium falciparum. J Antimicrob Chemother 57: 1093–1099. [DOI] [PubMed] [Google Scholar]
- 77. Fall B, Diawara S, Sow K, Baret E, Diatta B, et al. (2011) Ex vivo susceptibility of Plasmodium falciparum isolates from Dakar, Senegal, to seven standard anti-malarial drugs. Malar J 10: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.WHO (2013) Management of severe malaria: A practical handbook. Geneva: World Health Organization. 90 p. [Google Scholar]