Abstract
The cyclin-dependent kinase inhibitor p27Kip1 has been suggested as a prognostic marker in prostate cancer. The aim of this study was to determine the clinical and prognostic role of p27 expression in hormone-naive prostate cancers. A tissue microarray containing samples from 4,699 prostate cancers with attached pathological, clinical follow-up and molecular data was analyzed for nuclear p27 expression by immunohistochemistry. p27 staining was negative in 18.6%, weak in 33.5%, moderate in 28.4% and strong in 19.5% of 3,701 interpretable cancer spots. Loss of p27 immunostaining was linked to tumors of low Gleason grade (P<0.0001) and ERG fusion-negative cancers (P<0.0001). p27 levels were not associated with other parameters, including tumor stage, nodal stage, preoperative prostate-specific antigen (PSA) levels, surgical margin status and cell proliferation (as measured by the Ki67 labeling index). p27 expression was also unrelated to clinical outcome in all cancers, as well as in the subsets of ERG fusion-positive and -negative cancers. Overall, the present data demonstrated that elevated p27 expression was often unrelated to prostate cancer phenotype. Furthermore, the lack of an effect of the p27 protein levels on PSA recurrence following radical prostatectomy indicated that factors other than p27 expression are likely to be the major determinants of prostate cancer recurrence. However, a subset of ERG-negative, low-grade tumors was frequently characterized by loss of p27, suggesting a role of this alteration for the development of these tumors.
Keywords: p27, ERG fusion, prostate cancer
Introduction
Prostate cancer is a significant health problem and a leading cause of cancer-associated mortality in males. Approximately 50% of males develop prostate cancer during their lifetime, and about half of these cancers become potential life-threatening disease requiring therapeutic intervention (1). Thus, molecular markers discriminating between indolent and aggressive forms of the disease are highly desired but currently lacking (2,3).
The expression levels of critical cell cycle regulators, such as cyclin dependent kinases (CDK) and their inhibitors, are frequently altered in cancers. The CDK inhibitor p27Kip1 has often been proposed as a prognostic marker in human cancers (4). p27 inhibits CDK2 and CDK4 and induces growth arrest. The observed deregulation of p27 expression in cancers is not primarily due to mutations, but is instead caused by ubiquitin-mediated proteasomal degradation (4). Reduced levels of p27 occur in several cancer types and are generally associated with poor prognoses. For example, loss of p27 has been revealed to be an independent prognostic factor in breast, colon and gastric carcinomas (4,5). Studies investigating the prognostic significance of p27 in prostate cancer have yielded conflicting results [reviewed in (6)]. A number of studies have proposed that tumors with lost or diminished p27 expression are more aggressive (7–16), while other studies could not confirm these data (17–22). As all previous studies on the prognostic relevance of p27 expression in prostate cancer have analyzed comparatively small tumor sets with a maximum of 130 patients, it is possible that the analysis of large cohorts may provide clearer results.
In the present study, a large tissue microarray (TMA) containing samples from 4,699 hormone naive prostate cancers, obtained from patients who had undergone radical prostatectomy, was used. The present data showed that the loss of p27 expression was correlated with ERG fusion-negative tumors, but did not identify a significant effect of p27 expression on prostate cancer phenotype or patient prognosis.
Materials and methods
Patients
A set of prostate cancer prognosis TMAs containing one 0.6-mm tissue core each from 4,699 consecutive radical prostatectomy specimens, obtained from patients undergoing surgery between 1992 and 2008 at the Department of Urology or the Martini Clinic at the University Medical Center Hamburg-Eppendorf (Hamburg, Germany), was used in the present study. The pathological and clinical data of the arrayed prostate cancers are shown in Table I. The composition of this TMA is described in detail in Table II. Clinical follow-up data were available for 4,203 patients and the median follow-up was 46.7 months, ranging between one and 219 months. None of the patients received neoadjuvant or adjuvant therapy prior to prostate-specific antigen (PSA) recurrence, which was the clinical endpoint of this study. The first PSA value ≥0.2 ng/ml following surgery was used to define the time of recurrence. Fluorescence in situ hybridization (FISH) data for ERG and immunohistochemical ERG (23) and Ki67 (24) results were available from previous studies. This study was approved by the ethics committee of the University Medical Center Hamburg-Eppendorf (Hamburg, Germany). Written informed consent was obtained from the patients.
Table I.
Pathological and clinical data of the arrayed prostate cancers.
| No. of patients | ||
|---|---|---|
|
|
||
| Data | Study cohort on TMA (n=4699) | Biochemical relapse among categories (n=904) |
| Follow-up (months) | ||
| Mean | 56.6 | - |
| Median | 46.7 | - |
| Age (years) | ||
| <50 | 126 | 22 |
| 50–60 | 1399 | 227 |
| 61–70 | 2596 | 520 |
| >70 | 337 | 84 |
| Pretreatment PSA (ng/ml) | ||
| <4 | 666 | 77 |
| 4–10 | 2559 | 376 |
| 10–20 | 894 | 269 |
| >20 | 289 | 159 |
| pT category (AJCC 2002) | ||
| pT2 | 3010 | 272 |
| pT3a | 926 | 286 |
| pT3b | 489 | 307 |
| pT4 | 42 | 39 |
| Gleason grade | ||
| ≤3+3 | 1761 | 125 |
| 3+4 | 2055 | 450 |
| 4+3 | 512 | 261 |
| ≥4+4 | 135 | 68 |
| pN category | ||
| pN0 | 2317 | 580 |
| pN+ | 151 | 111 |
| Surgical margin | ||
| Negative | 3634 | 573 |
| Positive | 810 | 324 |
Numbers do not always add up to 4,699 in the various categories due to cases with missing data. TMA, tissue microarray; PSA, prostate-specific antigen; pT, primary tumor; AJCC, American Joint Committee on Cancer; pN, regional lymph node.
Table II.
Clinicopathological features of study cohort, associations with p27 IHC and subsets of ERG-negative and -positive cancers.
| p27 IHC result (all cancers/ERG-negative/ERG-positive) | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Parameter | n all | n evaluable | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | P-value |
| All cancers | 4699 | 3701 (1876/1709) | 18.6 (27.8/7.5) | 33.5 (36.9/30.3) | 28.4 (22.9/34.7) | 19.5 (12.4/27.5) | |
| Tumor stage | |||||||
| pT2 | 3019 | 2338 (1260/994) | 20.1 (29.5/7.0) | 32.9 (36.0/29.5) | 27.9 (22.6/34.7) | 19.2 (11.9/28.8) | 0.0185 (0.0758/0.2009) |
| pT3a | 946 | 778 (342/420) | 15.8 (23.1/8.1) | 33.3 (39.8/28.8) | 29.1 (22.0/35.2) | 21.9 (15.2/27.9) | |
| ≥pT3b | 527 | 461 (216/231) | 15.0 (22.2/8.7) | 37.5 (38.0/37.2) | 29.5 (26.4/32.0) | 18.0 (13.4/22.1) | |
| Gleason grade | |||||||
| ≤3+3 | 1765 | 1279 (675/552) | 25.5 (37.9/9.2) | 32.1 (35.1/29.7) | 25.9 (20.3/32.6) | 16.5 (6.7/28.4) | <0.0001 (<0.0001/0.5422) |
| 3+4 | 2074 | 1715 (823/894) | 15.0 (23.0/6.6) | 34.5 (38.9/30.5) | 29.3 (23.9/34.9) | 21.2 (14.2/28.0) | |
| 4+3 | 528 | 435 (229/193) | 14.9 (19.7/7.8) | 34.0 (37.6/30.1) | 30.1 (23.1/38.3) | 20.9 (19.7/23.8) | |
| ≥4+4 | 166 | 145 (88/51) | 8.3 (10.2/3.9) | 33.1 (30.7/37.3) | 34.5 (33.0/35.3) | 24.1 (26.1/23.5) | |
| Lymph node metastasis | |||||||
| N0 | 2359 | 1837 (916/877) | 16.2 (23.9/7.4) | 34.0 (37.7/30.7) | 29.5 (24.5/34.9) | 20.3 (14.0/27.0) | 0.9015 (0.1647/0.6316) |
| N+ | 174 | 150 (73/73) | 14.0 (17.8/8.2) | 34.0 (32.9/35.6) | 30.7 (26.0/35.6) | 21.3 (23.3/20.6) | |
| Preoperative PSA level (ng/ml) | |||||||
| <4 | 672 | 493 (217/247) | 17.0 (26.7/7.7) | 30.0 (33.2/27.1) | 31.9 (26.3/36.4) | 21.1 (13.8/28.8) | 0.002 (0.1787/0.0023) |
| 4–10 | 2585 | 2060 (1039/964) | 17.9 (27.6/6.2) | 33.7 (38.0/30.0) | 27.3 (20.1/34.1) | 21.0 (13.8/29.8) | |
| 10–20 | 909 | 734 (413/303) | 21.4 (30.0/9.2) | 33.8 (35.6/31.0) | 30.0 (24.7/38.3) | 14.9 (9.7/21.5) | |
| >20 | 311 | 249 (134/107) | 18.1 (20.9/13.1) | 39.8 (39.6/41.1) | 24.1 (26.1/21.5) | 18.1 (13.4/24.3) | |
| Surgical margin | |||||||
| Negative | 3665 | 2866 (1473/1299) | 18.5 (27.5/7.3) | 33.7 (37.1/30.3) | 27.9 (22.9/34.0) | 19.8 (12.6/28.5) | 0.6104 (0.9984/0.3255) |
| Positive | 844 | 687 (328/340) | 18.5 (27.5/8.5) | 32.9 (36.9/30.6) | 30.3 (22.8/37.1) | 18.3 (12.8/23.9) | |
IHC, immunohistochemistry; PSA, prostate-specific antigen.
Immunohistochemistry (IHC)
Freshly cut TMA sections were used for immunostaining. Slides were deparaffinized and exposed to heat-induced antigen retrieval for 5 min in an autoclave at 121°C and pH 9.0. p27 IHC was performed using a monoclonal antibody (1:50; DCS72; Calbiochem, Darmstadt, Germany). An EnVision™ system (Dako, Glostrup, Denmark) was used to visualize the immunostaining. Nuclear p27 staining was evaluated according to the following scoring system. The staining intensity (0, 1+, 2+ and 3+) and the fraction of positive tumor cells were recorded for each tissue spot. A final score was built from these two parameters as follows: Negative, staining intensity of 0; weak, staining intensity of 1+ in ≤70% of tumor cells or staining intensity of 2+ in ≤30% of tumor cells; moderate, staining intensity of 1+ in >70% of tumor cells, staining intensity of 2+ in >30% but ≤70% of tumor cells or staining intensity of 3+ in ≤30% of tumor cells; strong, staining intensity of 2+ in >70% of tumor cells or staining intensity of 3+ in >30% of tumor cells.
Statistical analysis
For the statistical analysis, JMP 8.0 software (SAS Institute, Inc., Cary, NC, USA) was used. Contingency tables were calculated to study associations between the p27 score and clinico-pathological variables. The χ2 (likelihood ratio) test was used to identify significant associations. Kaplan-Meier curves were generated for PSA recurrence-free survival. The log-rank test was applied to test the significance of differences between stratified survival functions. Cox proportional hazards regression analysis was performed to test the statistical independence and significance between pathological, molecular and clinical variables.
Results
Technical issues
A total of 4,699 hormone-naive cancers were analyzed for p27 expression. The analysis was successful in 3,701 tumors and failed in 998 cases (21.2%) due to lack of tissue spots or absence of unequivocal cancer cells in the p27-stained TMA section. Data were available for genomic ERG rearrangement (by FISH; n=2,336), ERG expression (by IHC; n=4,266) and Ki67 labeling index (by IHC; n=3,711) from previous studies (22,23).
p27 expression in prostate cancer
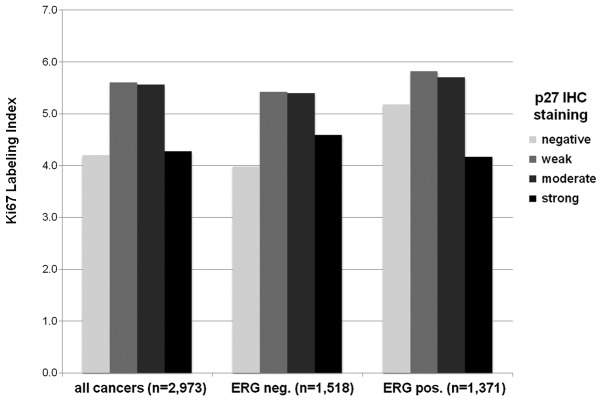
Examples of p27 immunostaining are shown in Fig. 1. Strong p27 expression was observed in tissue spots containing normal prostate tissue and was predominantly identified in the nucleus, with weak diffuse cytoplasmic staining, and was almost undetectable in stromal cells. By contrast, negative or weak nuclear p27 expression was observed in 52.1% of the 3,701 interpretable tumor spots. Expression was categorized as negative in 18.6% (n=689), weak in 33.5% (n=1,239), moderate in 28.4% (n=1,052) and strong in 19.5% (n=721) of the tumor spots analyzed. Associations between the p27 expression score and tumor phenotype are shown in Table II. Loss of p27 was correlated with low Gleason grade; there was a decrease in the fraction of p27-negative tumors from Gleason ≤3+3 (25.5%) to Gleason ≥4+4 (8.3%) (P<0.0001). This was paralleled by an increase in the fraction of tumors with strong p27 staining from 16.5% in Gleason ≤3+3 to 24.1% in Gleason ≥4+4. All data are summarized in Table II. The levels of p27 staining were unrelated to other parameters, including nodal stage, surgical margin status and cell proliferation index as measured by the Ki67 labeling index (Fig. 2). A significant P-value was observed for the association between the p27 score and tumor stage (P=0.0185). However, we did not consider this association to be true, as the fraction of tumors with negative, weak, moderate or strong p27 scores was almost identical in the different tumor stages.
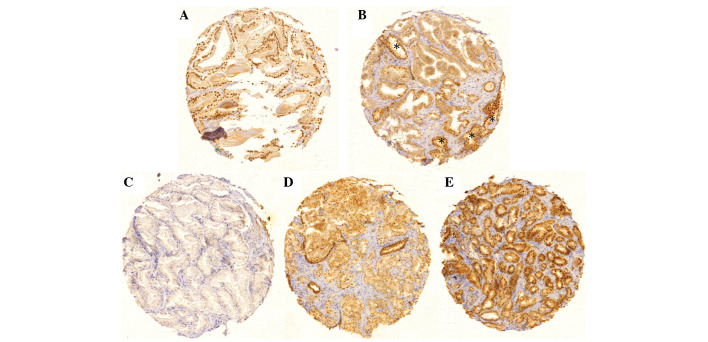
Figure 1.
Representative images of p27 immunostaining. (A) Positive nuclear staining in a spot harboring non-neoplastic prostate epithelium; (B) loss of p27 staining in neoplastic epithelium, but not in normal epithelium (*); (C) negative staining in prostate cancer; (D) moderate positive staining in prostate cancer; (E) strong positive staining in prostate cancer.
Figure 2.
Association of p27 staining score with Ki67 labeling index (Ki67Li) in all prostate cancers, and in ERG fusion-negative (ERG neg.) and -positive (ERG pos.) prostate cancers.
Association of p27 with ERG gene breaks and ERG protein expression, and the clinical significance of this
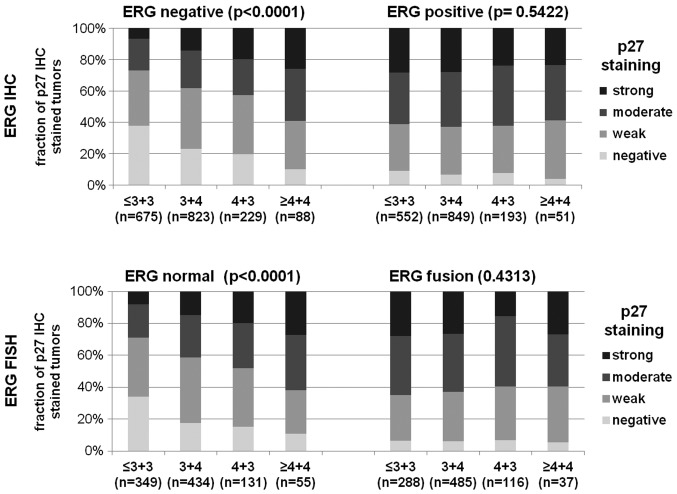
The availability of ERG data from a previous study (23) made it possible to search for associations between ERG and p27, and compare p27 scores and prostate cancer phenotypes, in separate subsets of ERG fusion-positive and -negative cancers. Loss of p27 staining was associated with ERG fusion-negative cancers. This was true for immunohistochemical detection of ERG protein expression and FISH detection of ERG breaks (P<0.0001 for both; Fig. 3). Loss of p27 expression was observed in 7.6% of all ERG fusion-positive tumors compared with 27.8% in ERG fusion-negative tumors, as revealed by ERG IHC (Fig. 3A). Consistent with this result, p27 expression was considered to be negative in only 6.5% of ERG fusion-positive cancers compared with 22.8% in ERG fusion-negative cancers, as revealed by ERG FISH (Fig. 3B). Notably, subset analysis showed that the loss of p27 was correlated with low Gleason grades only in ERG fusion-negative cancers (P<0.0001) and not in ERG-positive tumors (P=0.5422, Fig. 4) by IHC. This significant association in ERG fusion-negative tumors was demonstrated by FISH analysis of ERG rearrangement (P<0.0001; Fig. 4). Other parameters did not show significant associations in ERG-negative or -positive cancers, including tumor stage, nodal stage and surgical margin status (data not shown).
Figure 3.
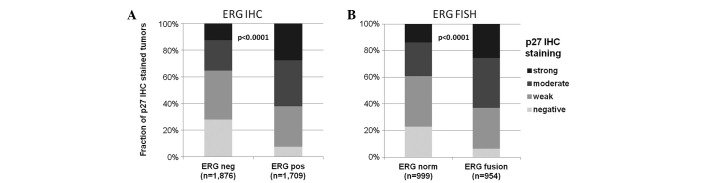
Association of p27 immunohistochemical staining and ERG fusion status. Comparison of p27 staining scores in ERG-positive and -negative prostate cancers. ERG fusion status was determined by either (A) immunohistochemistry (IHC) or (B) fluorescence in situ hybridization (FISH).
Figure 4.
Association of p27 immunohistochemical staining and Gleason grade in ERG fusion-negative and -positive prostate cancers by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) analysis.
No associations were observed between the various p27 staining scores and patient prognosis in all cancers, nor in the subsets of ERG fusion-positive or -negative cancers (Fig. 5).
Figure 5.
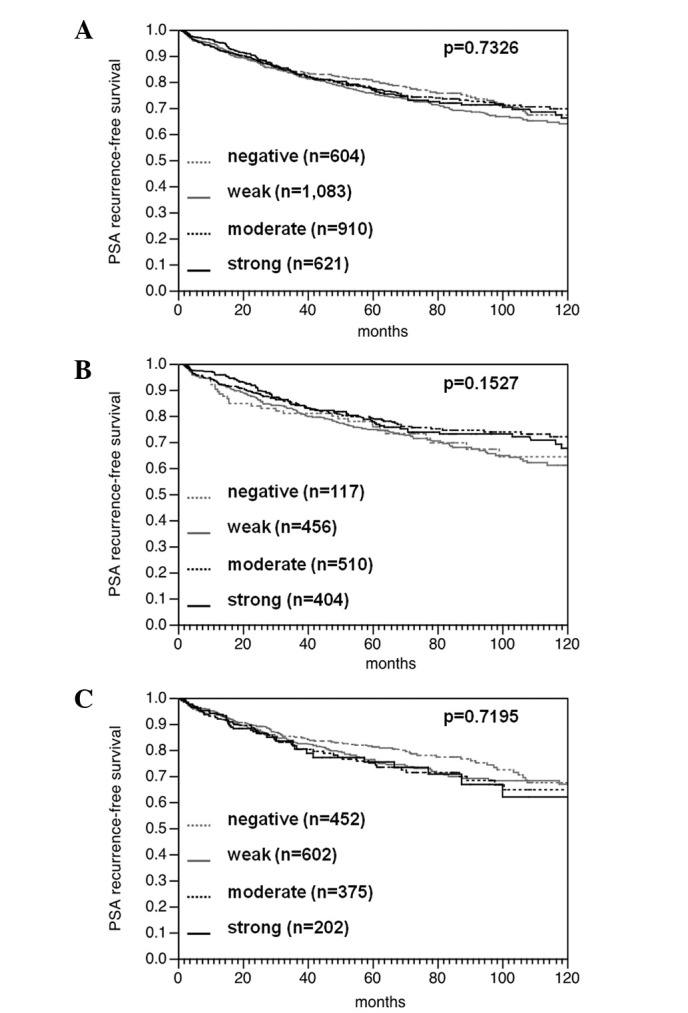
Prostate-specific antigen (PSA) recurrence-free survival stratified for p27 staining score in (A) all prostate cancers, as compared with subsets of (B) ERG fusion-negative and (C) ERG fusion-positive prostate cancers.
Discussion
The prognostic value of p27 expression in prostate cancer has been discussed with contrary conclusions in a number of previous studies [reviewed in (6)]. In the present study, >4,000 prostate cancers with long-term clinical data were analyzed in a TMA format. This database enabled assessment of the effect of p27 separately and in the two major molecular subgroups of prostate cancer, as defined by the ERG fusion status. The data revealed negative and weak p27 staining scores in 18.6 and 33.5% of samples, respectively, and showed that the loss of p27 expression was associated with ERG-negative cancers. The frequency of tumors with reduced p27 expression was within the range of previous studies, which reported absent or reduced p27 expression in 16–68% of cancers (6,12,13). The present results clearly demonstrated that the loss of p27 expression had no prognostic significance in radically operated prostate cancers. In particular, there was no clear difference with respect to PSA recurrence between strongly p27-positive and -negative cancers, neither when tumors were jointly analyzed, nor in subsets of ERG fusion-negative and -positive cancers. This lack of prognostic relevance is concordant with data from a number of other studies that were also unable to identify a prognostic effect of reduced p27 expression (17–22). Other studies have demonstrated that prostate cancers with a loss of or diminished p27 expression have particularly poor prognoses (7–11,13–15,17,25). It is possible that these controversial data are partly attributable to sampling issues, as these studies have all analyzed relatively small patient sets comprising between 86 and up to 130 prostate cancers. The present data, together with published findings, clearly argue against a clinically relevant impact of p27 expression on prostate cancer development and progression. This hypothesis is also consistent with the lack of association between p27 expression and tumor cell proliferation, as well as the observation that p27 knockout results in only a moderate increase in prostate epithelial proliferation and altered differentiation (15,26).
In the present study, the loss of p27 expression was significantly associated with ERG fusion-negative prostate cancers. The association with ERG was demonstrated by two independent approaches, namely IHC and FISH. This finding largely excluded the possibility of an artificial association caused by false negative IHC for both parameters in a subset of tissues that may have tissue damage compromising immunoreactivity. Previous studies have already demonstrated that there are considerable molecular differences between ERG fusion-positive and -negative prostate cancers (27–29). A number of studies have demonstrated gene expression profiles that were markedly distinctive between ERG fusion-positive and -negative cancers (30–32). At the level of specific genes and pathways, it is now well accepted that ERG interacts and modulates several tumor-relevant pathways involving AR, C-MYC, NKX3.1 and PTEN, all of which are more frequently identified to be altered in ERG fusion-positive prostate cancer, compared with ERG fusion-negative prostate cancer (27,28,33). The present study revealed that the loss of p27 protein expression is another feature associated with ERG-negative tumors.
In the present study, loss of p27 was associated with tumors of low Gleason grade. This is in contrast to numerous earlier studies that either did not identify a significant correlation (10,11,14,18,21,22) or even reported inverse associations (7,8,12,16,19,20,25). Of note, the association with low-grade cancers was limited to the subset of ERG fusion-negative tumors. This finding suggests a distinct role for p27 loss in ERG fusion-positive and -negative cancers. In ERG fusion-positive cancers, the fraction of tumors with p27 loss was low and unrelated to the Gleason grade, suggesting that neither tumor differentiation nor progression are dependent on p27 levels. By contrast, in ERG fusion-negative cancers, there was a noticeably large fraction (40%) of low-grade cancers harboring p27 loss. This fraction significantly decreased to ~10% in high-grade (Gleason ≥4+4) cancers. It may be hypothesized that the loss of p27 favors the development of cancers that are characterized by low malignant potential.
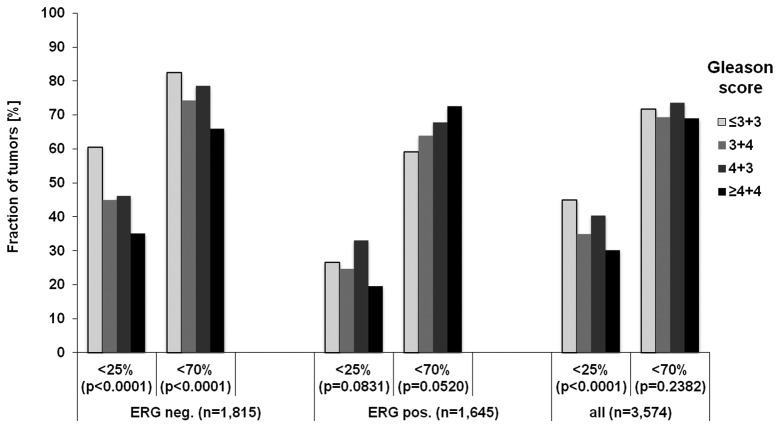
The majority of earlier studies analyzing p27 in prostate cancer were performed prior to the discovery of ERG fusions in 2005 (33), and later studies did not consider the ERG status (20–22). It is possible that variable fractions of ERG fusion-negative or -positive cancers may obscure this association, particularly if only small sample sizes are analyzed. In addition, it cannot be excluded that the various criteria applied to define loss or low p27 expression may have a significant effect on the outcome in different studies. For example, Vlachostergios et al observed reduced p27 expression in 86% of cancers defined as <70% of tumor cells staining positive for p27 (21). By contrast, Tsihlias et al defined low expression as <25% of tumor cells staining positive for p27 (7). To estimate the effect of different scoring systems on the associations tested, these scores were rebuilt in the present dataset according to the criteria used by Vlachostergios et al(21) and Tsihlias et al(7). Both positive and negative associations between the loss of p27 and Gleason grades based on the ERG status, as well as the scoring system used, were observed (Fig. 6). In the present study, an established scoring system based on the staining intensity and the fraction of tumor cells, which has been successfully used in numerous previous studies, was used to detect associations between molecular markers and tumor phenotype or patient prognosis (34,35). Such a predefined scoring system has the advantage that it includes the important information of staining intensity and enables an unbiased, standardized analysis. However it may not be optimally suited for establishing potential diagnostic thresholds.
Figure 6.
Association of p27 loss and Gleason grade in ERG fusion-negative (ERG neg.) and -positive (ERG pos.) prostate cancers, as well as in all prostate cancers, according to the criteria used by Tsihlias et al (<25%) and Vlachostergios et al (<70%) (6,20).
The present data also demonstrated the power of large-scale TMAs for the evaluation of biomarkers. Although the use of just one 0.6-mm TMA spot per donor tissue typically results in a loss of 20–30% of data points due to lack of cancer in individual TMA spots, the number of interpretable tissues remains sufficient for high-power statistical analysis in large TMAs. The ‘one core per donor tissue’ approach has the important advantage that the same amount of tissue (one spot) is analyzed per patient. TMAs containing two or more samples per tumor suffer from the statistical problem whereby individual samples are not always interpretable, resulting in patient subgroups with one, two, three and perhaps more interpretable samples, which in turn leads to higher positivity rates in patients with more interpretable tissue samples (36). In various earlier studies, large-scale prostate cancer TMAs that included >3,000 patients were used to demonstrate the prognostic role of p53, HER2, EGFR, Ki67LI, 8p deletion and PSMA (23,24,35,37–39). The large number of cases included in prostate cancer TMAs increasingly enables the selective analysis of relevant, molecularly defined subsets, such as ERG-positive prostate cancers.
In summary, the results of the present study have demonstrated that p27 expression was lost in a considerable fraction (~20%) of prostate cancers, but had no overall effect on patient prognosis and cancer progression. Differences may exist, however, in the biology of p27 in ERG fusion-negative and -positive cancers.
Acknowledgements
The authors would like to thank Christina Koop, Julia Schumann, Sünje Seekamp and Inge Brandt for their technical assistance.
References
- 1.Cooperberg MR, Cowan J, Broering JM, Carroll PR. High-risk prostate cancer in the United States, 1990–2007. World J Urol. 2008;26:211–218. doi: 10.1007/s00345-008-0250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samaratunga H, Epstein JI. What is the molecular pathology of low-risk prostate cancer? World J Urol. 2008;26:431–436. doi: 10.1007/s00345-008-0260-5. [DOI] [PubMed] [Google Scholar]
- 3.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsihlias J, Kapusta L, Slingerland J. The prognostic significance of altered cyclin-dependent kinase inhibitors in human cancer. Annu Rev Med. 1999;50:401–423. doi: 10.1146/annurev.med.50.1.401. [DOI] [PubMed] [Google Scholar]
- 5.Wander SA, Zhao D, Slingerland JM. p27: a barometer of signaling deregulation and potential predictor of response to targeted therapies. Clin Cancer Res. 2011;17:12–18. doi: 10.1158/1078-0432.CCR-10-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanson GP, Quinn D. Using molecular markers to help predict who will fail after radical prostatectomy. Prostate Cancer. 2011;2011:290160. doi: 10.1155/2011/290160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsihlias J, Kapusta LR, DeBoer G, et al. Loss of cyclin-dependent kinase inhibitor p27Kip1 is a novel prognostic factor in localized human prostate adenocarcinoma. Cancer Res. 1998;58:542–548. [PubMed] [Google Scholar]
- 8.Vis AN, Noordzij MA, Fitoz K, Wildhagen MF, Schröder FH, van der Kwast TH. Prognostic value of cell cycle proteins p27(kip1) and MIB-1, and the cell adhesion protein CD44s in surgically treated patients with prostate cancer. J Urol. 2000;164:2156–2161. [PubMed] [Google Scholar]
- 9.Vis AN, van Rhijn BW, Noordzij MA, Schröder FH, van der Kwast TH. Value of tissue markers p27(kip1), MIB-1, and CD44s for the pre-operative prediction of tumour features in screen-detected prostate cancer. J Pathol. 2002;197:148–154. doi: 10.1002/path.1084. [DOI] [PubMed] [Google Scholar]
- 10.Yang RM, Naitoh J, Murphy M, et al. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol. 1998;159:941–945. [PubMed] [Google Scholar]
- 11.Revelos K, Petraki C, Gregorakis A, et al. p27(kip1) and Ki-67 (MIB1) immunohistochemical expression in radical prostatectomy specimens of patients with clinically localized prostate cancer. In Vivo. 2005;19:911–920. [PubMed] [Google Scholar]
- 12.Guo Y, Sklar GN, Borkowski A, Kyprianou N. Loss of the cyclin-dependent kinase inhibitor p27(Kip1) protein in human prostate cancer correlates with tumor grade. Clin Cancer Res. 1997;3:2269–2274. [PubMed] [Google Scholar]
- 13.Kuczyk M, Machtens S, Hradil K, et al. Predictive value of decreased p27Kip1 protein expression for the recurrence-free and long-term survival of prostate cancer patients. Br J Cancer. 1999;81:1052–1058. doi: 10.1038/sj.bjc.6690806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuczyk MA, Bokemeyer C, Hartmann J, et al. Predictive value of altered p27Kip1 and p21WAF/Cip1 protein expression for the clinical prognosis of patients with localized prostate cancer. Oncol Rep. 2001;8:1401–1407. doi: 10.3892/or.8.6.1401. [DOI] [PubMed] [Google Scholar]
- 15.Cordon-Cardo C, Koff A, Drobnjak M, et al. Distinct altered patterns of p27KIP1 gene expression in benign prostatic hyperplasia and prostatic carcinoma. J Natl Cancer Inst. 1998;90:1284–1291. doi: 10.1093/jnci/90.17.1284. [DOI] [PubMed] [Google Scholar]
- 16.Cote RJ, Shi Y, Groshen S, et al. Association of p27Kip1 levels with recurrence and survival in patients with stage C prostate carcinoma. J Natl Cancer Inst. 1998;90:916–920. doi: 10.1093/jnci/90.12.916. [DOI] [PubMed] [Google Scholar]
- 17.Cheville JC, Lloyd RV, Sebo TJ, et al. Expression of p27kip1 in prostatic adenocarcinoma. Mod Pathol. 1998;11:324–328. [PubMed] [Google Scholar]
- 18.Erdamar S, Yang G, Harper JW, et al. Levels of expression of p27KIP1 protein in human prostate and prostate cancer: an immunohistochemical analysis. Mod Pathol. 1999;12:751–755. [PubMed] [Google Scholar]
- 19.Halvorsen OJ, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res. 2003;9:1474–1479. [PubMed] [Google Scholar]
- 20.Nassif AE, Tambara Filho R. Immunohistochemistry expression of tumor markers CD34 and P27 as a prognostic factor of clinically localized prostate adenocarcinoma after radical prostatectomy. Rev Col Bras Cir. 2010;37:338–344. doi: 10.1590/s0100-69912010000500006. [DOI] [PubMed] [Google Scholar]
- 21.Vlachostergios PJ, Karasavvidou F, Kakkas G, et al. Lack of prognostic significance of p16 and p27 after radical prostatectomy in hormone-naïve prostate cancer. J Negat Results Biomed. 2012;11:2. doi: 10.1186/1477-5751-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu TT, Wang JS, Jiaan BP, et al. Role of p21(WAF1) and p27(KIP1) in predicting biochemical recurrence for organ-confined prostate adenocarcinoma. J Chin Med Assoc. 2007;70:11–15. doi: 10.1016/S1726-4901(09)70294-1. [DOI] [PubMed] [Google Scholar]
- 23.Minner S, Enodien M, Sirma H, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–5888. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 24.Minner S, Jessen B, Stiedenroth L, et al. Low level HER2 overexpression is associated with rapid tumor cell proliferation and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:1553–1560. doi: 10.1158/1078-0432.CCR-09-2546. [DOI] [PubMed] [Google Scholar]
- 25.Thomas GV, Schrage MI, Rosenfelt L, et al. Preoperative prostate needle biopsy p27 correlates with subsequent radical prostatectomy p27, Gleason grade and pathological stage. J Urol. 2000;164:1987–1991. [PubMed] [Google Scholar]
- 26.Mukai M, Dong Q, Hardy MP, Kiyokawa H, Peterson RE, Cooke PS. Altered prostatic epithelial proliferation and apoptosis, prostatic development, and serum testosterone in mice lacking cyclin-dependent kinase inhibitors. Biol Reprod. 2005;73:951–958. doi: 10.1095/biolreprod.105.040980. [DOI] [PubMed] [Google Scholar]
- 27.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sreenath TL, Dobi A, Petrovics G, Srivastava S. Oncogenic activation of ERG: A predominant mechanism in prostate cancer. J Carcinog. 2011;10:37. doi: 10.4103/1477-3163.91122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–439. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 30.Setlur SR, Mertz KD, Hoshida Y, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jhavar S, Brewer D, Edwards S, et al. Integration of ERG gene mapping and gene-expression profiling identifies distinct categories of human prostate cancer. BJU Int. 2009;103:1256–1269. doi: 10.1111/j.1464-410X.2008.08200.x. [DOI] [PubMed] [Google Scholar]
- 32.Brase JC, Johannes M, Mannsperger H, et al. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-β signaling. BMC Cancer. 2011;11:507. doi: 10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller J, Ehlers A, Burkhardt L, et al. Loss of p(Ser2448)-mTOR expression is linked to adverse prognosis and tumor progression in ERG-fusion-positive cancers. Int J Cancer. 2012;132:1333–1340. doi: 10.1002/ijc.27768. [DOI] [PubMed] [Google Scholar]
- 35.Minner S, Wittmer C, Graefen M, et al. High level PSMA expression is associated with early PSA recurrence in surgically treated prostate cancer. Prostate. 2011;71:281–288. doi: 10.1002/pros.21241. [DOI] [PubMed] [Google Scholar]
- 36.Tennstedt P, Köster P, Bruchmann A, et al. The impact of the number of cores on tissue microarray studies investigating prostate cancer biomarkers. Int J Oncol. 2012;40:261–268. doi: 10.3892/ijo.2011.1216. [DOI] [PubMed] [Google Scholar]
- 37.El Gammal AT, Bruchmann M, Zustin J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- 38.Schlomm T, Iwers L, Kirstein P, et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21:1371–1378. doi: 10.1038/modpathol.2008.104. [DOI] [PubMed] [Google Scholar]
- 39.Schlomm T, Kirstein P, Iwers L, et al. Clinical significance of epidermal growth factor receptor protein overexpression and gene copy number gains in prostate cancer. Clin Cancer Res. 2007;13:6579–6584. doi: 10.1158/1078-0432.CCR-07-1257. [DOI] [PubMed] [Google Scholar]