Abstract
Many insects feed on only one or a few types of host. These host specialists often evolve a preference for chemical cues emanating from their host and develop mechanisms for circumventing their host’s defenses. Adaptations like these are central to evolutionary biology, yet our understanding of their genetics remains incomplete. Drosophila sechellia, an emerging model for the genetics of host specialization, is an island endemic that has adapted to chemical toxins present in the fruit of its host plant, Morinda citrifolia. Its sibling species, D. simulans, and many other Drosophila species do not tolerate these toxins and avoid the fruit. Earlier work found a region with a strong effect on tolerance to the major toxin, octanoic acid, on chromosome arm 3R. Using a novel assay, we narrowed this region to a small span near the centromere containing 18 genes, including three odorant binding proteins. It has been hypothesized that the evolution of host specialization is facilitated by genetic linkage between alleles contributing to host preference and alleles contributing to host usage, such as tolerance to secondary compounds. We tested this hypothesis by measuring the effect of this tolerance locus on host preference behavior. Our data were inconsistent with the linkage hypothesis, as flies bearing this tolerance region showed no increase in preference for media containing M. citrifolia toxins, which D. sechellia prefers. Thus, in contrast to some models for host preference, preference and tolerance are not tightly linked at this locus nor is increased tolerance per se sufficient to change preference. Our data are consistent with the previously proposed model that the evolution of D. sechellia as a M. citrifolia specialist occurred through a stepwise loss of aversion and gain of tolerance to M. citrifolia’s toxins.
Keywords: Drosophila, octanoic acid, adaptation, speciation, host specialization
HALF of all insects interact with plants (Grimaldi and Engel 2005). Most phytophageous insects, however, use only a few plant genera for food, mating, and oviposition (Bernays and Chapman 1994). Changes in host use can result in both new species and new adaptations (Ehrlich and Raven 1964; Janz 2011). For example, the evolution of a new host specialization may have contributed to the formation of new species in pea aphids (Acyrthosiphon) among others (Via 2001; Matsubayashi et al. 2010). Adapting to a new host can drive genetic and phenotypic change that is critical for isolating nascent species. In some cases, specialization has a price: Increased performance on the new host correlates with reduced performance on other hosts (Futuyma and Moreno 1988; Jaenike 1990; Fry et al. 1996; Scheirs et al. 2005; Via and Hawthorne 2005). This scenario poses a new challenge for the nascent specialist, as it must keep together alleles for finding and selecting the appropriate host (“preference”) along with those for utilizing that host (“performance,” e.g., physiologically adapting to that host’s secondary compounds or nutritional content; Jaenike 1990; Janz 2011). Theory suggests that a genetic correlation between the preference and performance alleles, such as caused by pleiotropy or genetic linkage, can overcome this problem and facilitate the switch to a new host (Lande 1979; Jaenike 1990; Fry et al. 1996; Janz 2011).
Until recently, evidence for this “genetic linkage” hypothesis in phytophagous insects has been mixed. Early genetic data in Drosophila by Jaenike (1986, 1987, 1989) suggested that oviposition preference and “settling” behavior are unlinked in Drosophila tripunctata, while Taylor and Condra (1983) found linkage between preference and performance in D. pseudoobscura. No linkage was found in other herbivorous species, such as Callosobruchus maculatus (southern cowpea weevil; Wasserman and Futuyma 1981), Colias philodice (butterfly; Tabashnik 1986), Papilionidae (swallowtail butterflies; Thompson 1988; Thompson et al. 1990), Chrysomelidae (leaf-feeding beetles; Keese 1996), Nilaparvata lugens (brown planthopper; Sezer and Butlin 1998a,b), and Oreina elongata (leaf beetle; Ballabeni and Rahier 2000). However, more recent QTL mapping data for aphids (Hawthorne and Via 2001; Caillaud and Via 2012; Sauge et al. 2012), and other genetic association studies in Euphydryas editha (Edith’s checkerspot butterfly; Ng 1988; Singer et al. 1988), Liriomyza sativae (leafminer fly; Via 1986), Phyllotreta nemorum (flea beetle; Nielsen 1996), and Papilio glaucus (eastern tiger swallowtail butterfly; Bossart 2003), suggest that some preference and performance alleles can be genetically linked.
A major concern with many of these genetic studies is their low resolution. QTL and marker-association studies produce candidate regions with large confidence intervals, increasing the chance that preference and performance alleles will overlap. Other studies simply infer genetic linkage due to the apparent heritability of host preference to well-performing offspring (e.g., Singer et al. 1988). Most finer resolution genetic studies of adaptive host specialization have focused on host preference or avoidance. Few studies have focused on the genetics of tolerance of a specific compound in the host plant because few species with obvious host adaptations have the requisite genetic tool kit needed to study these traits (except studies involving domesticated plants and agricultural pests, in which selection pressures are often different than in natural populations).
In contrast, D. sechellia’s specialization on Morinda citrifolia has both an obvious adaptation and a genetic tool kit. D. sechellia is endemic to Seychelles (Tsacas and Bachli 1981), specializes on the fruit produced by M. citrifolia (Louis and David 1986), and is closely related to D. simulans, a well-studied human commensal and habitat generalist. M. citrifolia contains octanoic acid (OA) (Legal et al. 1994), which is a fatty acid that is toxic to D. simulans and other insects but tolerated by D. sechellia (Rkha et al. 1991). OA typically comprises 58% of the volatile chemicals in a ripe M. citrifolia (Farine et al. 1996; Pino et al. 2010), which makes it the main toxic component of the fruit. D. sechellia also prefers M. citrifolia over other fruit for consumption and oviposition (Rkha et al. 1991; Legal et al. 1992; Matsuo et al. 2007). As a result of this adaptation, D. sechellia has limited competition for access to M. citrifolia and may be protected from predation (Jones 2005).
Prior work coarsely mapped several tolerance factors and identified some loci underlying the preference behavior (Jones 1998, 2001, 2004; Colson 2004; Matsuo et al. 2007; Earley and Jones 2011). As with other studies of host preference, the earlier work either did not assay tolerance or lacked the resolution needed to confidently test the genetic linkage hypothesis (e.g., Matsuo et al. 2007; Earley and Jones 2011). Moreover, because a specific gene involved in OA tolerance was not identified, the specific mechanism of OA tolerance in D. sechellia remains unknown.
In this study, we ultrafine map OA tolerance by genotyping independently derived recombinants using visible markers and a panel of molecular markers, along with a new phenotypic assay that provides reliable doses of OA vapor to flies without allowing them to directly contact the toxic chemical. We then measured the preference behavior of these recombinants in a test of the genetic linkage hypothesis. As preference and tolerance are not tightly linked, we reject the linkage hypothesis for this region. We hypothesize that the evolution of D. sechellia into an M. citrifolia specialist occurred through a stepwise, gradual gain of tolerance and loss of behavioral aversion to M. citrifolia’s toxins.
Materials and Methods
D. simulans/D. sechellia introgression lines
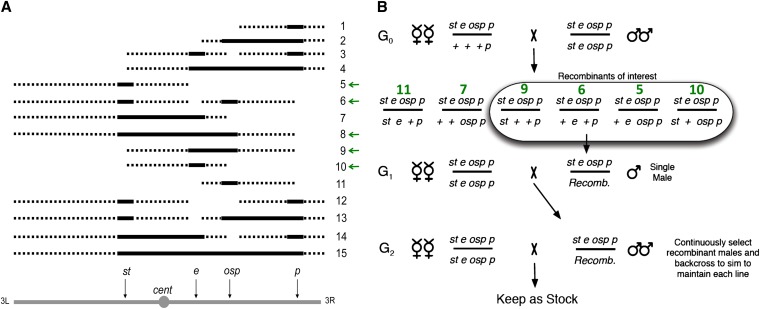
Jones (1998) identified a region harboring resistance alleles on chromosome arm 3R between two visible markers. As this interval had the greatest effect on resistance, we dissected it further by generating a set of D. sechellia/D. simulans introgression lines. We used these 15 original introgression lines (OILs) (Figure 1) to recombine elements of D. sechellia into the D. simulans background, using the Dsim\jv st e osp p mutant line (14021-0251.173, Drosophila Species Stock Center, University of California, San Diego) and D. sechellia S9 (M. Ashburner stock collection, Cambridge, UK). The presence or absence of a D. sechellia introgression was monitored with these recessive visible markers. Introgressed regions were present if the dominant wild-type phenotype was seen (from D. sechellia) rather than the recessive visible mutation (from D. simulans).
Figure 1.
Description of OIL genotypes and the cross to construct additional recombinants. (A) Diagram of original introgression lines (OILS). Solid black lines represent D. sechellia chromosome in D. simulans background. Dashed lines indicate the possible extent of the D. sechellia introgressions between visible markers. Four visible markers on chromosome 3 were used to categorize the OILs (scarlet, st; ebony, e; outspread, osp; pink, p). Introgression lines used for OA tolerance testing are denoted with green arrows (i.e., have a breakpoint between either st/e, e/osp, or both). (B) Diagram of fly cross to generate new introgression lines for OA tolerance assay. G0 females are from OIL 8 and males are pure D. simulans. The recombinants of interest (G1 males) were backcrossed to D. simulans females. The six recombinant genotypes (second line of cross) correspond to OILs numbered in green. Visible mutant phenotype was continually selected in males to preserve the haplotype without recombination. D. simulans background indicated by recessive mutations (st, e, osp, and p). Presence of D. sechellia introgression denoted by symbol for wild type (+). Recomb, the recombinant chromosome inherited from the G0 female.
The OILs were made by crossing D. sechellia females with the males of the D. simulans marker line. F1 hybrid females were then backcrossed to D. simulans males from the same line. The recessive mutations in the D. simulans background were then visible in some of the F2 recombinants. Individual females with the desired visible marker combinations were backcrossed to mutant D. simulans males for >20 generations to reduce the size of the D. sechellia introgression on chromosome 3 and eliminate D. sechellia contamination from the rest of the genome. The 15 OILs represent every combination of the four visible markers, with the presence of the D. sechellia introgression in each OIL indicated by a black line in Figure 1A. Each OIL was derived from a pool of many F2 females sharing the same marker phenotype, so each OIL “line” is actually a population. While male F1 D. simulans/D. sechellia hybrids are sterile, 20th generation introgression males are fertile. To stably maintain the D. sechellia introgressions of each OIL, we backcrossed 20th generation OIL males to D. simulans females. The D. sechellia introgressions were maintained as heterozygotes.
Generation of two “gold standard” lines and large panel of recombinant lines
The OILs were tested for OA tolerance (data not shown) and it was determined that OIL 10 flies could be used to generate high and low gold standard lines. These lines, called “high 10” and “low 10,” contain a D. sechellia introgression spanning the e locus, but exhibited different responses to OA exposure. We assayed many OIL 10 males and used three high and three low tolerance flies, respectively, to create high 10 and low 10. They were used to calibrate further tolerance assays.
Preliminary data indicated the OA tolerance locus was near e. We used OIL 8 to generate a huge population of individual recombinants in this region with unknown tolerance (Figure 1B). OIL 8 contains a large introgression spanning st, e, and osp loci. Three highly tolerant OIL 8 males were backcrossed to D. simulans females to create the line “high 8.” To generate individual recombinant flies (Figure 2), we crossed high 8 females to D. simulans males and collected male offspring with introgressions at st and osp but not e (OIL 6), only at e (OIL 10), at e and osp (OIL 9), and only at st (OIL 5). These recombinants had a breakpoint between e and a neighboring marker (either st or osp). We gave each line an arbitrary number followed by a hyphen and the number of the OIL to which it was phenotypically identical (e.g., 197-6 is an OIL 6 line). Initially, we generated 36 new recombinant lines to validate the OA tolerance assay (called the 36 “unknown” lines; see Supporting Information, File S1). Once it was clear that the assay was viable, we created another 700 recombinant lines and genotyped/phenotyped them as described below to finely map the OA tolerance locus.
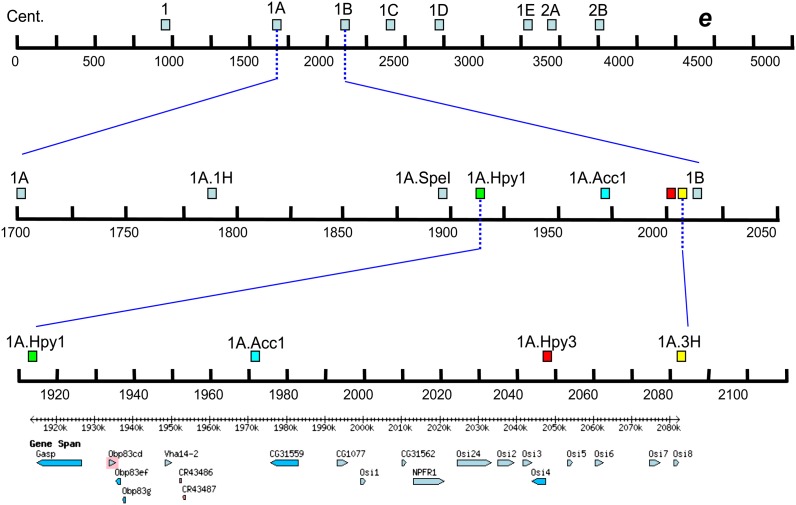
Figure 2.
Map of CAPS markers used to narrow OA tolerance region on chromosome arm 3R. The dashed and solid blue lines indicate zooming in (note the new scale on each level). The light blue boxes on the top level are markers using EcoRI spanning the region on 3R between the centromere (Cent.) and ebony (e). Between the first and second levels, the focus becomes the region between markers 1A and 1B on the second level. The marker names ending in H are for HindIII; SpeI for SpeI; Hyp for HypCH4V; and Acc1 for Acc65I. The markers shaded with light blue are outside of the region of interest. The third level represents the ∼170-kb region between markers 1A.Hpy1 and 1A.3H. These markers are color coded for easy identification between second and third levels. The bottom panel shows the names and positions of the 18 candidate genes (FlyBase). Scale for chromosome position: ×1000.
Genotyping using CAPS
We genotyped recombinant lines using cleaved amplified polymorphic sequence (Konieczny and Ausubel 1993). Primers were designed to amplify both D. simulans and D. sechellia sequences that contained polymorphic restriction cut sites. PCR amplicons from D. sechellia sequence had an intact restriction site, whereas D. simulans amplicons did not. Recombinant flies with an introgression between flanking CAPS sites were propagated, and recombinant offspring underwent further CAPS genotyping with increasingly finemapped CAPS marker sites (Figure 2). Overall, 700 unique recombinant males were genotyped, using Acc65I, EcoRI, HindIII, HpyCH4IV, and SpeI.
Genomic DNA was extracted from males at each generation using a single fly purification method. Briefly, a single male fly was frozen at −80° and then homogenized with a pipette tip in a “squishing buffer” (10 mM Tris-HCL pH 8.2, 1 mM EDTA, 25 mM NaCl). To this mixture, 1 μl of 0.2 mg/ml of Proteinase K was added and incubated at 37° for 20 min and then inactivated at 95° for 2 min. The resulting DNA was PCR ready.
Genomic positions listed herein are the D. melanogaster positions from D. melanogaster Gene Models/Evidence Release 5 (FlyBase 1999) identified using a syntenic alignment with D. simulans and D. sechellia.
Octanoic acid tolerance assay
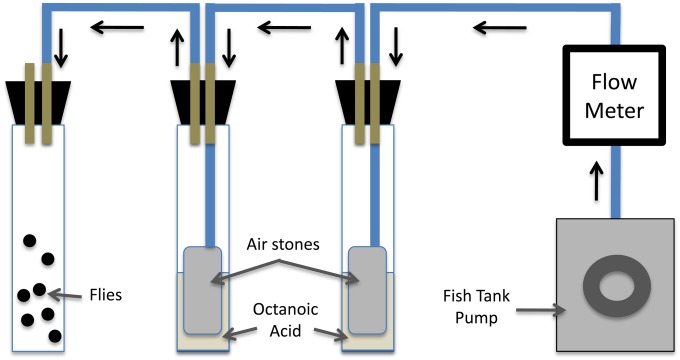
Tolerance to OA in D. sechellia, D. simulans, and hybrid recombinants was assayed using a vapor delivery system (Figure 3). A fish tank pump, regulated by a flow meter, pushed air through plastic tubing submerged in a tube of liquid OA at ∼2.2 liter/min, followed by a second tube of OA, and finally into a third tube with flies (additional details are provided in File S1). To ensure full OA saturation, air was pumped for at least 1 hr before fly testing. The entire apparatus was in a fume hood with full light and ambient temperature (20–25°).
Figure 3.
Diagram of OA tolerance test setup. OA vapor flows in the direction of the black arrows through plastic tubing (thick blue lines) and is bubbled through air stones twice to ensure saturation before reaching the flies. The OA/air stones and flies are housed in glass vials sealed with black stoppers. The stoppers have holes for glass tubing to pass through (brown lines), which are attached to the plastic tubing. The chamber containing flies is vented.
Flies were collected 4–7 days posteclosion with light CO2 anesthesia no fewer than 4 days pretest. Between 10 and 60 flies were dumped in the test chamber and every 2 min the number of “knocked-down” (KD) flies were counted, up to a total of 30 min. Typically, OA exposure induces neurotoxin-like symptoms in flies: frantic whole-body movement, leg and wing twitching, and finally KD, where flies either invert their body or collapse while upright. When needed, we tapped the test chamber to distinguish tolerant flies at rest vs. KD flies.
Data analysis
Cumulative KD counts within a line were calculated as a proportion (no. flies KD/no. flies total) then logit transformed, and a linear model compared these values against log-transformed time of OA exposure (0–30 min at 2-min intervals). “Knock down 50%” (KD50) was calculated as the time at which 50% of the flies in a given assay were knocked down (R library MASS; R Development Core Team 2012). To determine the influence of genotype, mutant phenotype, fly test chamber density, air flow rate, and sex on KD50 values, we constructed a linear model and performed an ANOVA. Significant differences in KD50 between lines and sexes were calculated using Welch’s t-test. All data have been deposited in Dryad (http://doi.org/10.5061/dryad.rp6gt).
Morinda tolerance assay
Five recombinant lines were tested for tolerance to ripe Morinda (high 8, high 10, low 10, 335-6, and 197-6) and pure D. sechellia and D. simulans. Naturally fallen fruit from M. citrifolia trees in a climate-controlled greenhouse were stored in plastic bags and used within 3 days. A pea-sized fruit pulp (no seeds) was spread across the top of a 60-mm Pyrex Petri dish. Individual flies were aspirated into this Petri dish and then placed in a growth chamber (25°, 60% humidity). After 30 min, flies were observed for KD every 10 min up to a maximum of 60 min (most flies survived to 30 min). “Survivorship” was measured as the first block of time during which knockdown was noticed. If no KD was observed at 60 min, that fly was scored as 70 and the test ended. Wilcoxon rank sum test compared differences between D. simulans and recombinant lines (Holm corrected P-value).
Behavior experiments
F1 backcrossed flies aged 2–10 days posteclosion were subjected to a behavioral assay as in Dworkin and Jones (2009) and Earley and Jones (2011). Briefly, flies of mixed sex were introduced without anesthesia into the assay chamber (2L glass beaker, Fisher). Within the chamber were two glass milk bottles, open on the top, containing either control or test food (22 ml diH20, 4.1 g instant fly media 4-24; Carolina Biological Supply), where test food was identical to control food save for acid inoculate (0.2% OA and 0.06% hexanoic acid, HA, v/v). The two food types were otherwise similar in color, texture, and water content. Cheese cloth was rubber banded over the entire chamber to prevent escape, and the chamber was placed in a growth chamber (25°, 60% humidity) overnight. The next day, flies located within each milk bottle were counted and sexed. A response index (RI) was calculated: RI = (no. flies in test bottle − no. in control bottle)/(no. in test bottle + no. in control bottle). This index was logit transformed to perform parametric tests. Data were analyzed using all-by-all t-tests with a false discovery rate (FDR) of 0.05 (R Development Core Team 2012), and a general linear model (GLM) (normal, model: RI = line + sex + line × sex + err; SAS Institute, Cary, NC). This assay captures fly settling behavior, which likely includes both positional preference and ovipositional preference—prior work suggests that they give qualitatively similar results (Earley and Jones 2011). Flies that chose a medium generally do not switch to the alternate medium (data not shown).
The recessive effect of introgressed loci was tested by selfing F1 flies to create F2 that segregated introgressed regions in both heterozygous (D. simulans/D. sechellia) and homozygous (D. sechellia/D. sechellia) states. Any F2 flies expressing the recessive D. simulans markers (no introgression) were removed. F2 flies were then pooled and tested in the same way as F1.
Results
New high-throughput assay for volatile fatty acid tolerance
We developed and validated a new apparatus for measuring adult tolerance to OA and other volatile fatty acids. Adult foraging is important in host preference for some phytophagous insect species, such as the grass miner Chromatomyia nigra (Scheirs et al. 2005), the chrysomelid Altica carduorum (Scheirs et al. 2005), and L. trifolii (Scheirs et al. 2005). These species prefer to oviposit and feed on host plants best suited for adult performance. As Scheirs et al. (2005) point out, several studies that only consider larval performance also suggest that adult performance may have been affected by host quality (e.g., Karowe 1990; Herr and Johnson 1992; Lu and Logan 1994). Because adult D. sechellia tend to feed on fresh M. citrifolia (Tsacas and Bachli 1981), which can be toxic even to D. sechellia larvae (Rkha et al. 1991), we believe that adult performance, rather than larval, is a key component of this adaptation.
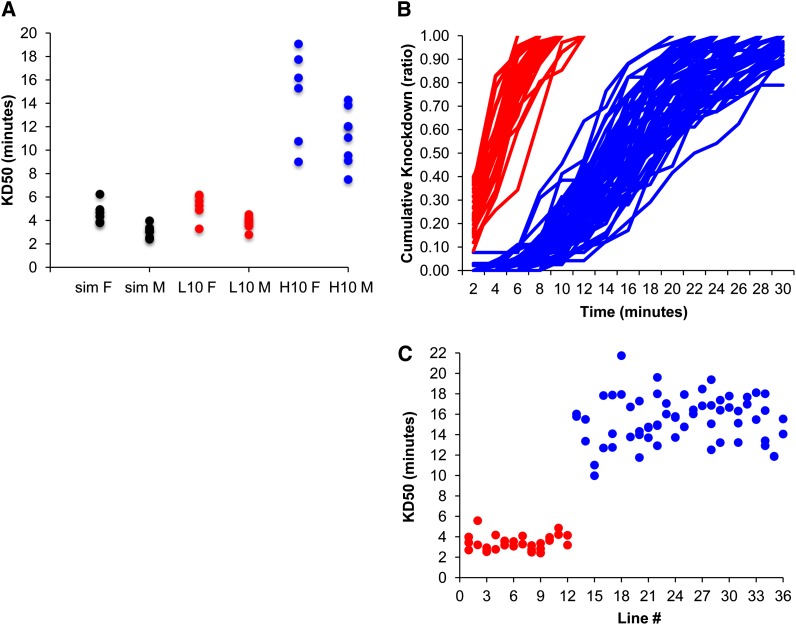
The high 10 and low 10 gold standard lines were used to initially validate our new “vapor” assay. We found a significant, and repeatable, difference in tolerance when high 10 males and females were compared to their respective sexes for low 10 and D. simulans, but not between the latter two lines for each sex (P-values in Table 1; KD50s plotted in Figure 4A; Figure S1). D. simulans was used as the low tolerance control, while D. sechellia was not knocked down after 3 consecutive hours of exposure. We used Welch’s t-test because the variances for the high and low 10 male and the high and low 10 female comparisons were not equal (Bartlett test P-values = 0.0079 and 0.0034, respectively).
Table 1. Welch’s t-test P-values for comparisons of KD50 between lines and sexes.
| Female |
Male |
|||||
|---|---|---|---|---|---|---|
| Line | D. sim | High 10 | Low 10 | D. sim | High 10 | Low 10 |
| Female | ||||||
| D. sim | — | <0.0001 | 0.3324 | 0.0068 | — | — |
| High 10 | — | — | <0.0001 | — | 0.0072 | — |
| Low 10 | — | — | — | — | — | 0.0229 |
| Male | ||||||
| D. sim | — | — | — | — | <0.0001 | 0.0635 |
| High 10 | — | — | — | — | — | <0.0001 |
| Low 10 | — | — | — | — | — | — |
Significant P-value <0.05 are shown in boldface type; D. sim, D. simulans; KD50, time that 50% of flies are knocked down.
Figure 4.
OA tolerance for gold standard and 36 unknown lines. (A) KD50 of all replicates for females (labeled “F”) and males (“M”), separately, of D. simulans (sim, black), low 10 (L10, red), and high 10 (H10, blue). Plot of the (B) cumulative distribution of knockdown and (C) KD50 for all replicates of the 36 lines of unknown tolerance. The lines/dots clustered into two distinct groups. The blue lines/dots have been classified as high tolerance and the red as low tolerance. None of the lines/dots classified as high tolerance had replicates that clustered with the low tolerance lines and vice versa. Line numbers (1–36) in C correspond to the “Line #”. column in Table S2 (mean KD50s).
Our data show that knockdown is highly reproducible at a given flow rate, with higher flow rates resulting in more rapid knockdown (Figure S2). Fly density had no effect (see File S1). The assay could reliably and repeatedly classify 36 “unknown” lines for resistance (Figure 4, B and C; File S1). These lines were also used to test whether the visible genetic markers affected tolerance. While the visible marker ebony affected tolerance, this effect was generally weak and background specific (e.g., the same marker did not always have the same effect across lines; File S1) and did not correlate with the number of markers in the genetic background.
High 10/low 10 gold standard lines show differential tolerance by sex
In all lines tested for male/female differences (high 10, low 10, and D. simulans; Table 1), the females were more tolerant to OA exposure than males. The percentage of cumulative knockdown over time of all individual replicates for high 10, low 10, and D. simulans are plotted by sex in Figure S3 (mean KD50s in Table S1).
The difference in tolerance by sex could be due to a different mechanism for tolerance in females than in males. Jones (1998) found the effects to be of different magnitudes between the sexes for every region he studied, along with an epistatic interaction between all three major chromosomes and one between markers y and f on the X chromosome in females, but not males. However, he did find effects for females in every region in which he found effects for males and explained the epistasis involving the X chromosome as possibly due to the X being hemizygous. Our results showed a significantly higher tolerance in females than males, but both sexes in highly tolerant lines exhibited significantly higher tolerances than their respective low tolerance counterparts. As the difference between the low and high tolerance lines was much larger than that between the sexes, it seems for this particular locus that the mechanism involved in OA tolerance is the same for both sexes. Instead it is likely that females were more tolerant due to their larger size, although we cannot specifically rule out that females have a differences in their tolerance mechanism compared to males.
Fine mapping of tolerance using recombinant line screening with CAPS markers
After validating the tolerance assay with the high/low 10 and 36 unknown lines, we focused on mapping tolerance factors in the region of interest. Using a marker panel consisting of 47 CAPS markers, we screened 700 independent D. simulans/D. sechellia recombinants. Twenty-four of these new recombinants helped reduce the target region by several hundred kilobases. Seven recombinants had a breakpoint that helped define the final 18-gene region (i.e., had a boundary marker adjacent to it). We preserved the haplotypes of these seven “boundary lines” for further testing. The boundary lines clustered into clear low and high tolerance groups, similar to the 36 unknown lines, with a significant difference between the KD50s of the lines from each group (Welch’s P-value = 4.385e-04; Figure S4). The final boundary markers of the region containing the tolerance locus are on 3R at positions 1,913,252, defined by 335-6 (low tolerance), 697-6 (low), 505-10 (high), 525-10 (high), and 553-10 (high), and 2,082,441, defined by 197-6 (high) and 725-6 (high).
Formally, these effects could be a byproduct of the particular lines used in this introgression, the species chosen, or the hybrid background produced by this introgression. However, earlier work used a variety of different D. simulans and D. sechellia backgrounds and all were qualitatively similar (Amlou et al. 1998b; Jones 1998, 2001). Alternatively, D. simulans could be the outlier instead of D. sechellia. Analysis of D. mauritiana, a susceptible sister species of D. sechellia and D. simulans, shows that in this region D. mauritiana alleles tend to be more D. simulans-like than D. sechellia-like or none of the three species appears to be a strong outlier (Table S6 and Figure S5), suggesting that D. simulans alleles in this region were likely not influencing tolerance any more than D. mauritiana alleles would have been.
Tolerance locus confirmed using fresh M. citrifolia fruit
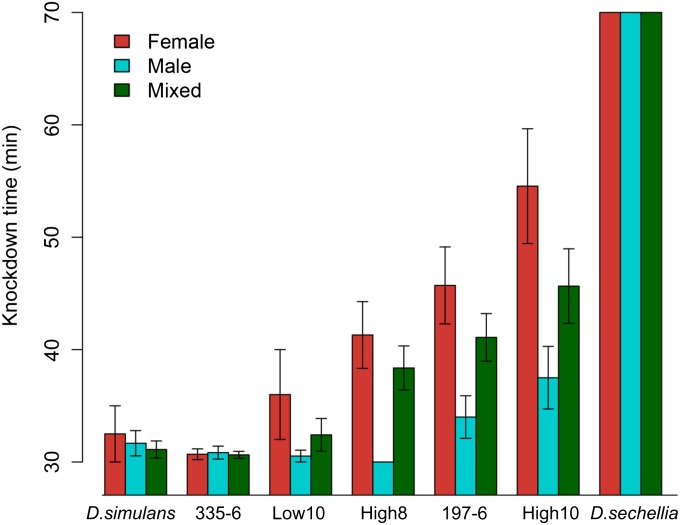
Consistent with our vapor-based tolerance assay, the M. citrifolia tolerance assay showed clear differences between the D. simulans background line (Dsim\jv st e osp p) and several of the recombinant lines. D. simulans had a mean (±SE) KD of 31.67 ± 3.89 min in males and 32.50 ± 5 min in females, which is close to the minimum KD time allowed (i.e., 30 min) in the experiment. A D. sechellia line (synA) remained upright for the entire 70 ± 0 min (maximum time allowed) in both males and females.
Lines that were tolerant to pure OA were generally also tolerant to Morinda fruit. In a mixed sex analysis comparing individual lines to their D. simulans background (Wilcoxon rank sum test), lines high 8, high 10, and 197-6 were all significantly more tolerant than D. simulans, whereas low 10 and 335-6 were not (Table 2). However, this difference appears to be driven by higher tolerance in females compared to males across all lines (P < 0.001, Wilcoxon; Figure 5, Table 2). This was particularly pronounced in the high tolerance lines. High 8 males were knocked down at 30 ± 0 min, while the females had a mean of 50 ± 4.71 min. The 197-6 males were down by 34 ± 1.89 min, while the females went down at 45.71 ± 3.42 min. High 10 males were down by 37.5 ± 2.7 min and females by 54.5 ± 5.1. This pattern was also seen in one of the low tolerance lines, low 10, where males averaged 30.53 ± 0.53 min and females averaged 36 ± 4.00 min. The 335-6 group was the exception, with the males going down at 30.83 ± 0.58 min and the females similarly at 30.69 ± 0.48 min. Of the highly tolerant recombinant lines, only 197-6 and high 10 had males stay upright beyond the initial 30-min check. Thus, the mixed sex results were mostly driven by the female tolerance.
Table 2. Wilcox rank-sum test (unpaired) P-values* comparing differences in Morinda tolerance between lines and D. simulans background.
| Line | Females | Males | Mixed |
|---|---|---|---|
| High10 | 0.035 | 0.076 | <<0.001 |
| High8 | 0.25 | 0.063 | 0.03 |
| 197-6 | 0.10 | 0.76 | 0.0019 |
| Low10 | 0.86 | 0.32 | 0.96 |
| 335-6 | 0.27 | 0.48 | 0.16 |
P-values are estimates because of rank ties between lines and D. simulans background. Bold type indicates significance at P < 0.05.
Figure 5.
M. citrifolia tolerance assay. Introgression flies from high and low tolerant lines were assayed for tolerance of ripe M. citrifolia fruit. Individual flies were exposed, and knockdown was observed between 30 and 60 min every 10 min. Flies used: D. simulans (susceptible background of introgression lines), D. sechellia (SynA tolerant line), high 8, low 10, 335-6, 197-6, and high 10 (recombinant lines and high and low tolerant lines).
Final tolerance region includes 18 genes, including Obp and Osiris families
A list of the 18 genes found in the ∼170 kb tolerance region on 3R is in Table S3. Two families of genes are represented: a cluster of three odorant-binding proteins (Obp) and nine Osiris genes. The three other named genes are Gasp, Vha14-2, and NPFR1. Of the 18 genes, only three remain unnamed.
The Ka/Ks ratios (Li 1993) from the D. simulans and D. sechellia lineages, as well as nonsynonymous sites and other sequence information, for the 18 genes are summarized in Table S4 and Table S5 (also see File S1). There is not a strong signature of positive selection at any of these loci. Only 7 of the 17 genes in D. simulans had nonsynonymous changes (41%), while 13 had such changes in D. sechellia (76%).
Earlier work contrasted levels of transcription of these genes across species, tissues, and treatments (Kopp et al. 2008; Dworkin and Jones 2009). None of the Osiris family genes showed differential expression. Across animal bodies, only CG31562 (a CHK kinase-like protein of unknown function) was significantly different between D. simulans and D. sechellia (D. sechellia expresses approximately one-third as much as D. simulans). In heads, Obp83cd was also significantly down in D. sechellia compared to D. simulans (approximately one-half), but not differentially expressed in antennae. Obp83ef was differentially expressed in antennae, but the difference between D. simulans and D. sechellia was substantially less than the difference between D. simulans and D. melanogaster, suggesting that expression of this gene is evolutionarily labile and the change is not associated with the shift to M. citrifolia. In an experiment looking at differential changes in gene expression associated with exposure to OA and HA, the gene CG1077 was weakly induced (∼1.5 times).
Tolerance alleles do not affect host preference behavior
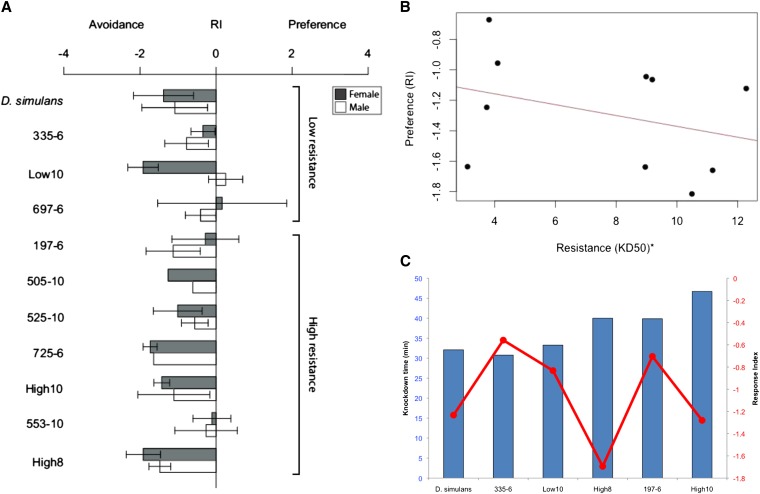
D. sechellia exhibits strong preference for M. citrifolia’s fruit and its constituent fatty acids, OA and HA. These compounds, in contrast, are highly aversive to D. simulans. To test whether the tolerance conferred by the 18-gene region or the genes within this region affected behavior, we measured the preference of seven recombinant lines using our established preference assay (Earley and Jones 2011). Most lines did not differ from the D. simulans background control (Figure 6A; pairwise t-test and GLM, P > 0.05). The exceptions, high 8 (both males and females, P = 0.017) and low 10 (males only, P = 0.011), behaved in the opposite of expectation—lower tolerance resulted in less aversion. Similarly, the trend was for flies with higher tolerance to avoid the OA medium (Figure 6B), although this trend was not significant (P = 0.3506). To improve power, we pooled high and low tolerance lines and compared preference behavior between the two groups (replicates: 55 high, 26 low; 4993 flies). There was no effect for either sex and the trend was in the opposite of expectation (mean RIhigh = −1.49; mean RIlow = −1.21; main effect P = 0.148, sex P = 0.157, interaction P = 0.207). Similarly, there was no difference between the pooled high or low lines and the D. simulans control (high, P = 0.408; low, P = 0.121). We confirmed for a subset of lines that this pattern was consistent for tolerance to M. citrifolia fruit (Figure 6C). These data suggest that the increased tolerance conferred by the introgressed D. sechellia region is not sufficient to change behavior and excludes the possibility of an additive or dominant acting preference locus in this interval.
Figure 6.
Tolerance alleles in the 18-gene region do not affect behavior in heterozygotes. (A) We measured the settling behavior of several introgression lines, as well as high/low 10 and D. simulans (also called “sim-marked”). The response index (RI) describes the degree of aversion (negative values) or preference (positive values). Most lines were not significantly different from controls, except for high 8 males and females, and low 10 males. Neither (B) OA tolerance nor (C) M. citrifolia fruit tolerance were positively correlated with OA preference.
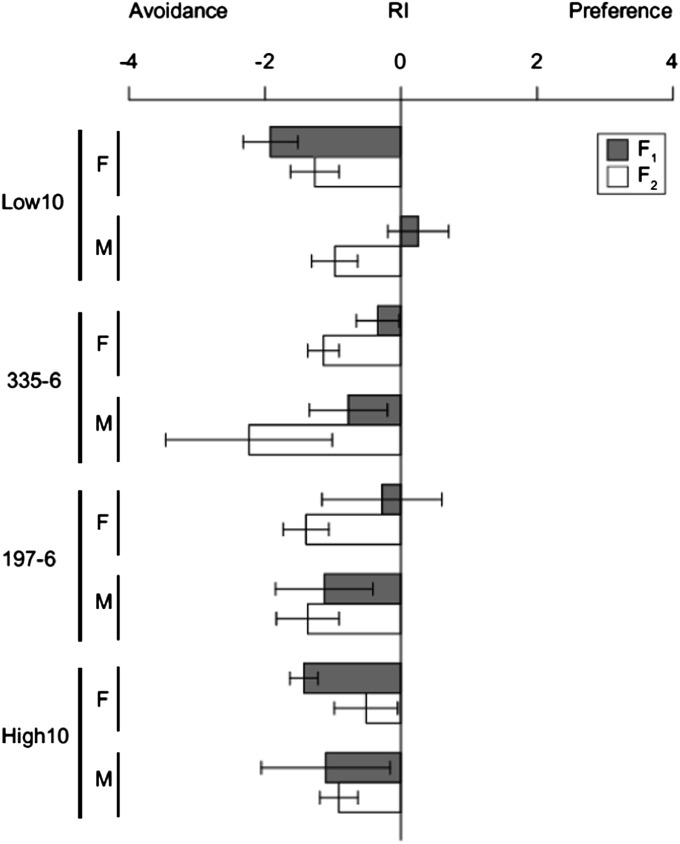
We assayed a subset of lines for a recessive preference factor linked to the tolerance factors. From the lines showing highest (high 10, 197-6) and lowest OA tolerance (low 10, 335-6), we sibmated F1 flies to create F2 progeny, segregating introgressions in homozygous (D. sechellia/D. sechellia) and heterozygous (D. simulans/D. sechellia) states, removing F2’s that had no introgression (D. simulans/D. simulans). If a recessive D. sechellia preference factor existed within an introgression line, then homozygous flies for that factor should have manifested higher OA preference and shifted F2 behavior relative to a population of fully heterozygous introgressions. We pooled homozygote and heterozygote F2’s and found no significant difference in their behavior compared to fully heterozygous F1 flies (Figure 7).
Figure 7.
D. sechellia recessive alleles have no effect on behavior, as shown by the plot of the response index (RI). To test the effect of recessive alleles from D. sechellia on preference behavior, we selfed F1 low (low 10 and 335-6) and high (high 10 and 197-6) tolerant introgression lines to make F2 progeny. Flies possessing both homozygous and heterozygous D. sechellia introgressions were pooled. Their behavior was not different from purely heterozygous F1. Females (“F”) and males (“M”) were assayed for all four lines.
Discussion
Many insects feed on only one or a few types of host. Genetic linkage between alleles contributing to host preference and alleles contributing to host usage, such as tolerance of secondary compounds, has been suggested to facilitate the evolution of new host specializations. We used a forward genetic approach and a novel assay to see whether this type of genetic correlation contributed to the evolution of the host specialization in D. sechellia. We isolated a ∼170-kb region on 3R, harboring 18 genes that contains at least one locus affecting OA tolerance, a critical element of D. sechellia’s adaptation to the toxic fruit of its host plant, M. citrifolia. While the D. sechellia introgression conferred OA tolerance in a D. simulans background, it had little to no effect on host-seeking behavior.
M. citrifolia assay validates OA apparatus and methodology
To screen the thousands of flies needed for introgression mapping of tolerance loci, we developed a new assay for volatile fatty acid resistance that mimics the toxic effects of the fruit. Our analysis revealed that exposure to M. citrifolia fruit is quantitatively similar to the OA assay results, in terms of consistency in the lines that exhibited high and low tolerance behavior. However, the mixed sex results for lines exhibiting high tolerance to M. citrifolia were driven by tolerance of the females (i.e., the males showed lower tolerance levels). Unfortunately, we do not know the concentration of OA in either the actual vapor of the OA assay or in the fruit itself (there is considerable variation among fruits and across ripening stages; Legal et al. 1992; Pino et al. 2010). Also, it is likely the fruit has a higher concentration of OA than the maximum our pump can produce. The consistently high tolerance of the females in both experimental setups suggests that the same tolerance mechanism is being assayed in both. Some of the variability among the sexes may reflect the larger size of the females. Experimentally, the high variability among lines in the M. citrifolia assay suggests that using the OA vapor methodology may be sensitive enough to detect moderate-to-weak effect loci undetectable using fruit.
Tolerance region harbors several candidate loci
Of the 18 genes in this region, two gene families represented two-thirds of the total and only three remain unnamed. A cluster of nine Osiris genes was present, along with three Obps. None of these 18 genes, however, showed a strong signature of positive selection that may be expected for a gene contributing to D. sechellia adaptation to its host. Likewise, gene expression data did not strongly implicate any one locus (but see Obp83cd below).
Osiris genes:
According to Dorer et al. (2003), the Osiris gene family is clustered at the Triplo-lethal locus in D. melanogaster. All have endoplasmic reticulum signal peptides, may be integral to the plasma membrane, may have important housekeeping functions, and are highly dosage sensitive. In addition, their linkage and sequences are unusually highly conserved, as seen in Anopheles gambiae (Dorer et al. 2003). While none of these genes can be ruled out, it seems unlikely that their functionality can be appreciably altered without dire consequences to the individual.
Odorant-binding proteins:
Obp83cd, Obp83ef, and Obp83g are members of a family of odorant binding proteins (OBPs) involved in olfactory perception, although their function is not fully understood (Vieira et al. 2007). OBPs are water soluble and exist in the aqueous lymph surrounding odorant receptors in the chemosensory sensilla of insects. They enhance the solubility of hydrophobic odorants by binding to them and transporting them through the extracellular lymph to the dendritic membrane of neurons (Whiteman and Pierce 2008). Obps are typically found in olfactory tissues, but expression analysis shows that they are not limited to them (Pelosi et al. 2006). Besides transporting odorants, it has been suggested that Obps may also act as scavengers, removing toxic odorant molecules to prevent damage to cells (Steinbrecht 1998; Blomquist and Vogt 2003). The possible involvement of Obps in OA tolerance is consistent with the hypothesized link between Obp chemical detection and detoxification.
Obp83cd (Galindo and Smith 2001) and Obp83ef (Galindo and Smith 2001; Kopp et al. 2008) are significantly down-regulated in the head and up-regulated in the antennae, respectively. Obp83cd is expressed in the labellum (Galindo and Smith 2001), while Obp83ef is expressed in the antennae and other nonspecific tissues (Galindo and Smith 2001; Kopp et al. 2008). Intriguingly, Obp83cd shows a species-specific pattern of expression: It has reduced expression in D. sechellia relative to D. simulans (Dworkin and Jones 2009).
As with most genes, the regulatory regions of these Obps are not well characterized. The Regulatory Element Database for Drosophila v3.0 (Gallo et al. 2011) indicated that the regulatory regions for both Obp83cd and Obp83ef are 3 kb upstream of the start codons. We aligned these upstream regions, using D. melanogaster as the outgroup. For Obp83cd, there were 34 changes in D. sechellia, along with a 43-bp deletion. Obp83ef had 28 changes in D. sechellia. REDfly did not specify transcription factor binding sites, but any one of these upstream changes could alter the regulation of these genes in D. sechellia. No regulatory information was available for Obp83g.
Obp83cd and Obp83g have a D. sechellia Ka/Ks higher than the mean for the region, while Obp83ef has a Ka/Ks that is much lower than the mean. D. simulans alleles for Obp83cd and Obp83g have no nonsynonymous changes, while D. sechellia alleles have five and two, respectively. Of the genes that do not end prematurely or contain frameshifts, Obp83cd has the largest Ka (0.0076) of the D. sechellia alleles in this region. Obp83ef has one nonsynonymous change in both species, but a much higher Ka/Ks in D. simulans (0.6249 vs. 0.1073). Coding changes may not be involved in OA tolerance, but if they are, these genes are candidates with at least one amino acid change each.
Obps have previously been associated with OA avoidance behavior (Dworkin and Jones 2009), M. citrifolia preference (Matsuo et al. 2007), and host plant preference in Drosophila (Kopp et al. 2008), but not to tolerance of the normally toxic effects of OA experienced by most Drosophila species. While we are unable to eliminate the other 15 genes, the three Obps are our strongest candidates for future analysis.
No evidence for genetic linkage between preference and tolerance loci in this region
A positive genetic correlation between the preference and performance alleles due to pleiotropy or genetic linkage can facilitate the evolution of a new host specialization. This 18-gene region spans only ∼170,000 bp and has an estimated recombination rate of 0.28 cM. These genes are therefore tightly linked. However, we find no evidence that harboring the D. sechellia tolerance alleles in this region causes the flies to behave in a more D. sechellia-like manner. Indeed the high 8 line, which spans a much larger interval (up to 23.3 cM on chromosome 3, although the actual boundaries have not been mapped), does not appear to be significantly different from the controls and is marginally more aversive than some low tolerance lines. Previous work noted that D. sechellia preference for OA was recessive to D. simulans aversion (Higa and Fuyama 1993; Amlou et al. 1998a). We tested for recessive effect loci on high and low tolerance lines by selfing F1 hybrids to segregate F2 progeny with pooled homozygous and heterozygous D. sechellia introgressions (removing any F2 with recessive homozygous D. simulans markers—hence, no introgression). If recessive OA preference loci exist within these introgressions, then F2 flies should exhibit increased preference relative to F1 flies. We did not see this pattern (Figure 7).
The limited genetic resolution of the earlier studies that suggested linkage between preference and tolerance potentially biased these studies toward overlap between QTL for preference and performance. Our data suggest that if these studies had higher resolution—and thus smaller confidence intervals—the apparent linkage between preference and performance may be reduced.
In the current study we have a different concern: We are only looking at a single locus and the critical early association between behavior and tolerance could have evolved elsewhere in the genome. Recent work, however, has identified strong preference factors on chromosome 2, which has the weakest effect on tolerance (Jones 1998; Matsuo et al. 2007; Dworkin and Jones 2009). Similarly, the X chromosome, which has at least two tolerance factors, has no effect on preference behavior (Jones 1998, 2001, 2004; Earley and Jones 2011). Furthermore, the tolerance region on 3R was previously shown to be one of the two largest contributors to resistance (the other locus is flanking; Jones 1998). Preferring the toxic host without tolerance alleles in this region of 3R would be deleterious. Together these data suggest that linkage between preference and tolerance factors did not play a major role in the evolution of D. sechellia’s specialization.
There are several examples of genetically unlinked development of host preference and performance. Earlier work by Jaenike (1989) ruled out tight linkage between preference and performance in D. tripunctata. The same was found in other herbivorous species (see Introduction). Theoretically, the quickest way to achieve speciation through host specialization is if host preference and performance each have a simple genetic architecture and are tightly linked to each other (Fry 2003). Jaenike (1987) posited that it is unlikely for linkage disequilibrium to establish a genetic correlation between preference and performance in a system with more than a few interacting alleles. More likely, pleiotropy would explain such a linkage. (In the case of D. sechellia, we also eliminate this possibility.) Assortative mating on the new host plant may allow unlinked evolution of preference and performance during sympatric divergence (Diehl and Bush 1989). This scenario would likely require a few new alleles of large effect, which are seen in D. sechellia’s adaptation to Morinda fruit, but also may have resulted in a fitness gap between the old and new hosts. In this case, the deleterious effects of the new host on the performance of the unadapted fly may have been overcome by reduced competition on the new host (Wallace 1968; Berlocher and Feder 2002), since OA is toxic to D. sechellia’s sibling species. Alternatively, since five or more loci may be involved in OA tolerance (i.e., a polygenic architecture), it is possible that part or most of the speciation process occurred gradually in allopatry (Templeton 1981).
Evolution of D. sechellia’s host specialization likely occurred in a stepwise manner
D. sechellia’s loss of OA avoidance and its development of preference for M. citrifolia seemed to derive in part from the elimination of related Obps. Dworkin and Jones (2009) found that Obp56e had a premature stop codon in D. sechellia and that D. melanogaster showed reduced avoidance of M. citrifolia when Obp56e was knocked down. Matsuo et al. (2007) found a 4-bp insertion upstream of Obp57e in the D. sechellia allele affecting preference, which prevented expression when heterozygous within a D. melanogaster Obp57e deficiency line. If loss of functional Obps occurred first in the evolution of D. sechellia’s specialization on M. citrifolia, it would certainly have produced large selection pressure toward the development of tolerance on the flies that were coming into contact with the fruit.
Loss-of-function mutations are usually recessive, as they often result in the elimination or reduction of protein expression or of nonfunctional protein structures. However, Jones (1998) concluded that at least five loci involved in D. sechellia tolerance were dominant. Similarly, pesticide resistance factors are usually dominant or codominant (Ottea and Plapp 1984; Roush and McKenzie 1987; Houpt et al. 1988; ffrench-constant et al. 1993; Roush 1993). This asymmetry in dominance may reflect the different genetic mechanisms through which these two traits evolved. Loss of avoidance can be achieved by the loss or reduction of a sensory response, such as through pseudogenization of Obps. In fact, McBride (2007) discovered that D. sechellia harbors an unusually large number of pseudogenized chemosensory genes relative to D. simulans or D. melanogaster, even when accounting for D. sechellia’s small population size. Toxin resistance, in contrast, often requires increased expression of existing detoxification genes or gain of a new physiological mechanism. Both cases may result in additive-to-completely dominant phenotypes. This pattern may imply that during the early genetic steps of the evolution of a new specialization—or any adaptation—that “loss” of an ancestral trait may readily evolve from common recessive nulls segregating in the standing genetic variation. In contrast, dominant phenotypes associated with new traits and gain-of-function alleles may involve more new mutations and other relatively rare alleles, as these dominant alleles are expected to be at a lower frequency in the ancestral population (Orr and Betancourt 2001).
Supplementary Material
Acknowledgments
We thank the anonymous reviewers for their comments. We also thank Karna Mital for his contributions and Daniel Matute for his helpful advice. This work was supported by the National Science Foundation (NSF) DEB-0212686 (C.D.J.), NSF-Graduate Research Fellowship Program (E.J.E.), the National Institute of Diabetes and Digestive and Kidney Diseases grant P30DK056350 to the University of North Carolina Nutrition Obesity Research Center, and National Institutes of Health (NIH) R01 GM058686 and NIH ARRA R01 supplement GM058686-08S1.
Footnotes
Communicating editor: K. Nichols
Literature Cited
- Amlou M., Moreteau B., David J. R., 1998a Genetic analysis of Drosophila sechellia specialization: oviposition behavior toward the major aliphatic acids of its host plant. Behav. Genet. 28: 455–464 [DOI] [PubMed] [Google Scholar]
- Amlou M., Moreteau B., David J. R., 1998b Larval tolerance in the Drosophila melanogaster species complex toward the two toxic acids of the D. sechellia host plant. Hereditas 129: 7–14 [DOI] [PubMed] [Google Scholar]
- Ballabeni P., Rahier M., 2000. Performance of leaf beetle larvae on sympatric host and non-host plants. Entomol. Exp. Appl. 97: 175–181 [Google Scholar]
- Berlocher S. H., Feder J. L., 2002. Sympatric speciation in phytophagous insects: Moving beyond controversy? Annu. Rev. Entomol. 47: 773–815 [DOI] [PubMed] [Google Scholar]
- Bernays E. A., Chapman R. F., 1994. Host-Plant Selection by Phytophagous Insects. Chapman and Hall, New York [Google Scholar]
- Blomquist G. J., Vogt R. G., 2003. Insect Pheromone Biochemistry and Molecular Biology: The Biosynthesis and Detection of Pheromones and Plant Volatiles. Elsevier Academic Press, London [Google Scholar]
- Bossart J., 2003. Covariance of preference and performance on normal and novel hosts in a locally monophagous and locally polyphagous butterfly population. Oecologia 135: 477–486 [DOI] [PubMed] [Google Scholar]
- Caillaud M. C., Via S., 2012. Quantitative genetics of feeding behavior in two ecological races of the pea aphid, Acyrthosiphon pisum. Heredity 108: 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson I., 2004. Drosophila simulans’ response to laboratory selection for tolerance to a toxic food source used by its sister species D-sechellia. Evol. Ecol. 18: 15–28 [Google Scholar]
- Diehl S. R., Bush G. L., 1989. The Role of Habitat Preference in Adaptation and Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Dorer D. R., Rudnick J. A., Moriyama E. N., Christensen A. C., 2003. A family of genes clustered at the Triplo-lethal locus of Drosophila melanogaster has an unusual evolutionary history and significant synteny with Anopheles gambiae. Genetics 165: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I., Jones C. D., 2009. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia. Genetics 181: 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley E. J., Jones C. D., 2011. Next-Generation Mapping of Complex Traits with Phenotype-Based Selection and Introgression. Genetics 189: 1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich P. R., Raven P. H., 1964. Butterflies and plants: a study in coevolution. Evolution 18: 586–608 [Google Scholar]
- Farine J. P., Legal L., Moreteau B., LeQuere J. L., 1996. Volatile components of ripe fruits of Morinda citrifolia and their effects on Drosophila. Phytochemistry 41: 433–438 [Google Scholar]
- ffrench-Constant R. H., Steichen J. C., Rocheleau T. A., Aronstein K., Roush R. T., 1993. A single-amino acid substitution in a gamma-aminobutyric-acid subtype-a receptor locus is associated with cyclodiene insecticide resistance in Drosophila populations. Proc. Natl. Acad. Sci. USA 90: 1957–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase , 1999. The FlyBase database of the Drosophila Genome Projects and community literature. The FlyBase Consortium. Nucleic Acids Res. 27: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry J. D., 2003. Multilocus models of sympatric speciation: Bush vs. Rice vs. Felsenstein. Evolution 57: 1735–1746 [DOI] [PubMed] [Google Scholar]
- Fry J. D., Einsohn S. L. H., Ackay T. F. C. M., 1996. The contribution of new mutations to genotype-environment interaction for fitness in Drosophila melanogaster. Evolution 50: 2316–2327 [DOI] [PubMed] [Google Scholar]
- Futuyma D. J., Moreno G., 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19: 207–233 [Google Scholar]
- Galindo K., Smith D. P., 2001. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 159: 1059–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo S. M., Gerrard D. T., Miner D., Simich M., Des Soye B., et al. , 2011. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 39: D118–D123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi D., Engel M. S., 2005. Evolution of Insects. Cambridge University Press, Cambridge, UK [Google Scholar]
- Hawthorne D. J., Via S., 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412: 904–907 [DOI] [PubMed] [Google Scholar]
- Herr J. C., Johnson M. W., 1992. Host plant preference of Liriomyza sativae (Diptera: Agromyzidae) populations infesting green onion in Hawaii. Environ. Entomol. 21: 1097–1102 [Google Scholar]
- Higa I., Fuyama Y., 1993. Genetics of food preference in Drosophila sechellia. I. Responses to food attractants. Genetica 88: 129–136 [DOI] [PubMed] [Google Scholar]
- Houpt D. R., Pursey J. C., Morton R. A., 1988. Genes-controlling malathion resistance in a laboratory-selected population of Drosophila melanogaster. Genome 30: 844–853 [DOI] [PubMed] [Google Scholar]
- Jaenike J., 1986. Genetic complexity of host-selection behavior in Drosophila. Proc. Natl. Acad. Sci. USA 83: 2148–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J., 1987. Genetics of oviposition-site preference in Drosophila-Tripunctata. Heredity 59: 363–369 [DOI] [PubMed] [Google Scholar]
- Jaenike J., 1989. Genetic population structure of Drosophila tripunctata: patterns of variation and covariation of traits affecting resource use. Evolution 43: 1467–1482 [DOI] [PubMed] [Google Scholar]
- Jaenike J., 1990. Host specialization in phytophagous insects. Annu. Rev. Ecol. Syst. 21: 243–273 [Google Scholar]
- Janz N., 2011. Ehrlich and Raven revisited: mechanisms underlying codiversification of plants and enemies. Annu. Rev. Ecol. Evol. Syst. 42(42): 71–89 [Google Scholar]
- Jones C. D., 1998. The genetic basis of Drosophila sechellia’s resistance to a host plant toxin. Genetics 149: 1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. D., 2001. The genetic basis of larval resistance to a host plant toxin in Drosophila sechellia. Genet. Res. 78: 225–233 [DOI] [PubMed] [Google Scholar]
- Jones C. D., 2004. Genetics of egg production in Drosophila sechellia. Heredity 92: 235–241 [DOI] [PubMed] [Google Scholar]
- Jones C. D., 2005. The genetics of adaptation in Drosophila sechellia. Genetica 123: 137–145 [DOI] [PubMed] [Google Scholar]
- Karowe D. N., 1990. Predicting host range evolution: colonization of Coronilla varia by Colias philodice (Lepidoptera: Pieridae). Evolution 44: 1637–1647 [DOI] [PubMed] [Google Scholar]
- Keese M. C., 1996. Feeding responses of hybrids and the inheritance of host-use traits in leaf feeding beetles (Coleoptera: Chrysomelidae). Heredity 76: 36–42 [Google Scholar]
- Konieczny A., Ausubel F. M., 1993. A procedure for mapping Arabidopsis mutations using codominant ecotype-specific pcr-based markers. Plant J. 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kopp A., Barmina O., Hamilton A. M., Higgins L., McIntyre L. M., et al. , 2008. Evolution of gene expression in the Drosophila olfactory system. Mol. Biol. Evol. 25: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1979. Quantitative genetic-analysis of multivariate evolution, applied to brain–body size allometry. Evolution 33: 402–416 [DOI] [PubMed] [Google Scholar]
- Legal L., David J. R., Jallon J. M., 1992. Toxicity and attraction effects produced by Morinda citrifolia fruits on the Drosophila melanogaster complex of species. Chemoecology 3: 125–129 [Google Scholar]
- Legal L., Chappe B., Jallon J. M., 1994. Molecular-basis of Morinda-Citrifolia (L): toxicity on Drosophila. J. Chem. Ecol. 20: 1931–1943 [DOI] [PubMed] [Google Scholar]
- Li W. H., 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36: 96–99 [DOI] [PubMed] [Google Scholar]
- Louis J., David J. R., 1986. Ecological specialization in the Drosophila-Melanogaster species subgroup: a case-study of Drosophila-Sechellia. Acta Oecologica-Oecologia Generalis 7: 215–229 [Google Scholar]
- Lu W. H., Logan P., 1994. Genetic-variation in oviposition between and within populations of Leptinotarsa decemlineata (Coleoptera, Chrysomelidae). Ann. Entomol. Soc. Am. 87: 634–640 [Google Scholar]
- Matsubayashi K. W., Ohshima I., Nosil P., 2010. Ecological speciation in phytophagous insects. Entomol. Exp. Appl. 134: 1–27 [Google Scholar]
- Matsuo T., Sugaya S., Yasukawa J., Aigaki T., Fuyama Y., 2007. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 5: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C. S., 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc. Natl. Acad. Sci. USA 104: 4996–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D., 1988. A novel level of interactions in plant–insect systems. Nature 334: 611–612 [Google Scholar]
- Nielsen, J. K., 1996 Intraspecific variability in adult flea beetle behaviour and larval performance on an atypical host plant, pp. 160–162 in Proceedings of the 9th International Symposium on Insect-Plant Relationships Springer-Verlag, New York. [Google Scholar]
- Orr H. A., Betancourt A. J., 2001. Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottea J. A., Plapp F. W., 1984. Glutathione S-transferase in the housefly: biochemical and genetic changes associated with induction and insecticide resistance. Pestic. Biochem. Physiol. 22: 203–208 [Google Scholar]
- Pelosi P., Zhou J. J., Ban L. P., Calvello M., 2006. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63: 1658–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino, J. A., E. Marquez, C. E. Quijano, and D. Castro, 2010 Volatile compounds in noni (Morinda citrifolia L.) at two ripening stages. Ciencia e Tecnologia de Alimentos 30: 183–187.
- R Development Core Team , 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rkha S., Capy P., David J. R., 1991. Host plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetic analysis. Proc. Natl. Acad. Sci. USA 88: 1835–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush R. T., 1993. Occurrence, genetics and management of insecticide resistance. Parasitol. Today 9: 174–179 [DOI] [PubMed] [Google Scholar]
- Roush R. T., McKenzie J. A., 1987. Ecological genetics of insecticide and acaricide resistance. Annu. Rev. Entomol. 32: 361–380 [DOI] [PubMed] [Google Scholar]
- Sauge M. H., Lambert P., Pascal T., 2012. Co-localisation of host plant resistance QTLs affecting the performance and feeding behaviour of the aphid Myzus persicae in the peach tree. Heredity 108: 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheirs J., Jordaens K., De Bruyn L., 2005. Have genetic trade-offs in host use been overlooked in arthropods? Evol. Ecol. 19: 551–561 [Google Scholar]
- Sezer M., Butlin R., 1998a The genetic basis of oviposition preference differences between sympatric host races of the brown planthopper (Nilaparvata lugens). Proc. R. Soc. Lond. B Biol. Sci. 265: 2399–2405 [Google Scholar]
- Sezer M., Butlin R. K., 1998b The genetic basis of host plant adaptation in the brown planthopper (Nilaparvata lugens). Heredity 80: 499–508 [Google Scholar]
- Singer M. C., Ng D., Thomas C. D., 1988. Heritability of oviposition preference and its relationship to offspring performance within a single insect population. Evolution 42: 977–985 [DOI] [PubMed] [Google Scholar]
- Steinbrecht R. A., 1998. Odorant-binding proteins: expression and function. Olfaction and Taste Xii 855: 323–332 [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E., 1986. Evolution of Host Plant Utilization in Colias Butterflies. Evolutionary Genetics of Invertebrate Behavior, pp. 173–184 Plenum, New York [Google Scholar]
- Taylor C. E., Condra C., 1983. Resource partitioning among genotypes of Drosophila pseudoobscura. Evolution 37: 135–149 [DOI] [PubMed] [Google Scholar]
- Templeton A. R., 1981. Mechanisms of speciation: a population genetic approach. Annu. Rev. Ecol. Syst. 12: 23–48 [Google Scholar]
- Thompson J. N., 1988. Evolutionary ecology of the relationship between oviposition preference and performance of offspring in phytophagous insects. Entomol. Exp. Appl. 47: 3–14 [Google Scholar]
- Thompson J. N., Wehling W., Podolsky R., 1990. Evolutionary genetics of host use in swallowtail butterflies. Nature 334: 148–150 [Google Scholar]
- Tsacas L., Bachli G., 1981. Drosophila sechellia. n. sp., huitieme espece du sous-groupe melanogaster des iles Seychelles (Diptera, Drosophilidae). Rev. Fr. Entomol. 3: 146–150 [Google Scholar]
- Via S., 1986. Genetic covariance between oviposition preference and larval performance in an insect herbivore. Evolution 40: 778–785 [DOI] [PubMed] [Google Scholar]
- Via S., 2001. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 16: 381–390 [DOI] [PubMed] [Google Scholar]
- Via S., Hawthorne D. J., 2005. Back to the future: genetic correlations, adaptation and speciation. Genetica 123: 147–156 [DOI] [PubMed] [Google Scholar]
- Vieira F. G., Sanchez-Gracia A., Rozas J., 2007. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 8: R141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B., 1968. Polymorphism, population size, and genetic load, pp. 87–108 in Population Biology and Evolution, edited by Lewontin R. C. Syracuse University Press, Syracuse, NY [Google Scholar]
- Wasserman S. S., Futuyma D. J., 1981. Evolution of host plant utilization in laboratory populations of the southern cowpea weevil, Callosobruchus maculatus Fabricius (Coleoptera: Bruchidae). Evolution 35: 605–617 [DOI] [PubMed] [Google Scholar]
- Whiteman N. K., Pierce N. E., 2008. Delicious poison: genetics of Drosophila host plant preference. Trends Ecol. Evol. 23: 473–478 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.