Abstract
Automated image acquisition, a custom analysis algorithm, and a distributed computing resource were used to add time as a third dimension to a quantitative trait locus (QTL) map for plant root gravitropism, a model growth response to an environmental cue. Digital images of Arabidopsis thaliana seedling roots from two independently reared sets of 162 recombinant inbred lines (RILs) and one set of 92 near isogenic lines (NILs) derived from a Cape Verde Islands (Cvi) × Landsberg erecta (Ler) cross were collected automatically every 2 min for 8 hr following induction of gravitropism by 90° reorientation of the sample. High-throughput computing (HTC) was used to measure root tip angle in each of the 1.1 million images acquired and perform statistical regression of tip angle against the genotype at each of the 234 RIL or 102 NIL DNA markers independently at each time point using a standard stepwise procedure. Time-dependent QTL were detected on chromosomes 1, 3, and 4 by this mapping method and by an approach developed to treat the phenotype time course as a function-valued trait. The QTL on chromosome 4 was earliest, appearing at 0.5 hr and remaining significant for 5 hr, while the QTL on chromosome 1 appeared at 3 hr and thereafter remained significant. The Cvi allele generally had a negative effect of 2.6–4.0%. Heritability due to the QTL approached 25%. This study shows how computer vision and statistical genetic analysis by HTC can characterize the developmental timing of genetic architectures.
Keywords: root gravitropism, QTL, natural variation, Arabidopsis
METHODOLOGIES for characterizing genomes and genotypes are more advanced in terms of their degree of automation and throughput than their phenotype counterparts. This imbalance hinders progress in mapping genotype to phenotype in model systems such as the Arabidopsis thaliana plant, the subject of the present study. Computer vision methodologies can help address this imbalance by enabling objective, accurate, and potentially automated measurements of size and shape captured in digital images (Spalding 2009). The Arabidopsis seedling root is well suited for this approach because it grows and responds well to stimuli in experimental scenarios that permit acquisition of high-quality images from which shape and size descriptors such as midlines, local curvature, and angles can be computationally extracted (Miller et al. 2007; Chavarria-Krauser et al. 2008; French et al. 2009). For example, Durham Brooks et al. (2010) used automated analysis of high-resolution images to measure the influence of seed size, seedling age, and media composition on the time course of root gravitropism, a dynamic growth response to a change in orientation with respect to the gravity vector (Blancaflor and Masson 2003; Morita 2010). Miller et al. (2010) used the same approach plus extended computational analysis to quantify a transient gravitropism phenotype and its precise time of onset in young roots of glr3.3 glutamate receptor mutants. Other researchers have taken advantage of automated image acquisition and analysis to measure shoot growth features over time (Zhang et al. 2012; Tisné et al. 2013) or to acquire rotational series of branching root systems growing in a transparent gel to capture a third spatial dimension instead of time (Iyer-Pascuzzi et al. 2010; Clark et al. 2011).
The degree of automation made possible by computational image analysis increases the feasibility of systematically acquiring high-resolution phenotype data from genetically structured populations for the purpose of large-scale statistical genetic studies. The benefits of accuracy and objectivity attending this approach increase the quality of the resulting phenotype data. Accordingly, automated or semiautomated image analysis is beginning to impact the mapping of quantitative trait loci in plants (Herridge et al. 2011; Moore et al. 2013; Tisné et al. 2013; Topp et al. 2013). The project presented here combines an ability to capture the time course of gravitropism in high resolution with the throughput needed to make mapping populations feasible subjects. Gravitropism was selected for study because it is a model that embodies perception and transduction of an external cue, hormonal control of growth, and adaptation to a new environmental state (Boonsirichai et al. 2002). These are fundamental physiological and developmental processes, now made accessible to statistical genetic analysis by novel application of automated computer vision and computation methodologies.
Materials and Methods
Germplasm
The Arabidopsis seed stocks used here were described in a recent machine-vision study of seed size and shape QTL (Moore et al. 2013). As in the previous report, recombinant inbred line 1 (RIL1) refers to a set of 162 F10 RILs derived from a cross between the Landsberg erecta (Ler) and Cape Verde Islands (Cvi) accessions of A. thaliana (Alonso-Blanco et al. 1998) donated to us by Patrick Masson, University of Wisconsin, and to the phenotype dataset obtained with those seeds. RIL2 refers to progeny of RIL1 produced in a random-designed and carefully controlled seed-bulking exercise to duplicate the population in a different maternal environment as described in Moore et al. (2013). NIL refers to 92 near isogenic lines created by introgression of various short regions of Cvi DNA into the Ler background (Keurentjes et al. 2007) and raised in the same manner as the RIL2 seeds, again as described in Moore et al. (2013). The plant populations or the phenotype datasets derived from them are referred to as RIL1, RIL2, or near isogenic line (NIL).
Plant culture
In a row on a Petri plate containing 1 mM KCl, 1 mM CaCl2, 5 mM 4-morpholineethanesulfonic acid, pH 5.7 with BIS-TRIS propane, and gelled with 1% agar, three seeds were sown ∼0.5 cm apart. Plates with seeds were placed at 4° for 2–4 days of stratification before being cultured vertically in a 22°-growth chamber under constant white light for 3 days, at which point the primary root was 2–8 mm in length.
Root imaging
An imaging system, consisting of 11 charge-coupled device cameras (Marlin F146B; Allied Vision Technologies) each equipped with a macro zoom lens (model NT59-157; Edmund Optics), was used to capture digital images of roots on as many Petri plates, each backlit with 880 nm radiation in a room maintained at ∼22°, essentially as previously described (Durham Brooks et al. 2010; Miller et al. 2010). Instructions on how to create a similar apparatus are posted at http://phytomorph.wisc.edu/hardware/fixed-cameras.php. To initiate the experiment, each Petri dish was held in a custom plate holder in front of a camera and rotated by 90°. Framing, focusing, and starting image acquisition, in that order, were performed within 20 sec of sample rotation. Images were acquired automatically every 2 min for 8 hr at a resolution of 100 pixels/mm, resulting in a 241-frame movie of the root gravitropic response. The resulting images showed the dark roots of the three seedlings on a plate, against a light background. A video of an example experiment is provided as Supporting Information, File S1. Because each response recording lasted 8 hr, two successive trials per camera were acquired within a given day to increase the throughput of phenotype acquisition. To ensure that the roots being compared were at similar growth stages, only those with initial lengths between 3 and 8 mm were used for QTL analysis. RIL1, RIL2, and NIL were phenotyped consecutively. Within each population, genotypes were selected for measurement according to a randomized order.
Tip angle measurement
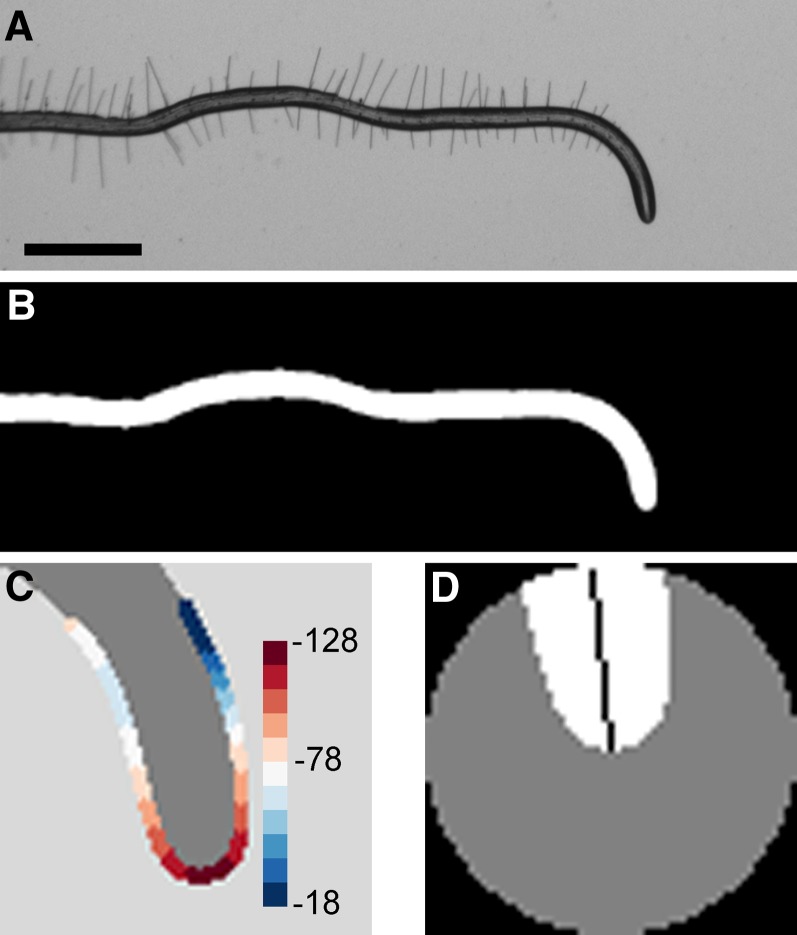
The camera firmware that controlled acquisition also automatically saved the images to a local disk array. A file synchronization protocol nightly copied the images to disks housed in the Department of Computer Sciences from where they were automatically read and processed for tip angle measurements on the University of Wisconsin Center for High-Throughput Computing (CHTC) grid, which is managed and scheduled by the HTCondor software tool (Thain et al. 2005). The custom algorithm that measured tip angle at each time point in a time series of frames was implemented in Matlab. The algorithm first binarized the grayscale images to isolate the roots from the background (Figure 1, A and B). Erosion and pruning produced a continuous, branchless skeleton of the root. The set of pixels forming the boundary of the root apex was located within an 81 × 81 pixel patch of the binarized root image centered at the terminus of the skeleton. The local curvature at each position of the boundary was determined by continuous wavelet transformation of a parameterized trace of the boundary pixels. The point of maximum curvature was labeled the root tip (Figure 1C). The largest circle fit within a 60 × 60 pixel patch centered at the root tip was used to mask (set to zero) a portion of the root apex large enough that the tip angle could be equated with the first principal component determined by principal components analysis (Figure 1D). The angle of each root tip in each frame was thus determined, stored in comma-separated value files, and automatically copied to the local server one day after the experiment was performed. The large majority of trials were successfully processed automatically, resulting in 8–20 independent time course measurements for each genotype. The most common problems encountered were blurry images caused by excessive condensation forming on the lid of the Petri plate and two roots colliding during the experiment. In all, 2123, 2325, and 2536 roots were successfully imaged and analyzed in the RIL1, RIL2, and NIL populations, respectively.
Figure 1.
Root tip angle determined by image analysis. (A) Grayscale image of a representative root undergoing a growth response to a change in the gravity vector, or gravitropism. Bar, 1 mm. (B) Binarized image of the responding root. (C) Curvature values are calculated at each boundary point along the root apex. The point of highest curvature is taken to be the root tip. Color legend indicates curvature values in mm−1. (D) A patch centered at the root tip is subjected to principal components analysis. The first eigenvector (black line) determines the tip angle relative to the horizon.
Stepwise QTL analysis using HTCondor
To add a high-resolution time axis to a QTL map, stepwise QTL analyses (Manichaikul et al. 2009) were performed at each of the 241 time points using tip angle as the phenotype. The approach uses a penalized LOD score, with the LOD score for a multiple-QTL model (the log10 likelihood ratio comparing the given model to the null model with no QTL) penalized by the complexity of the model with separate penalties for main effects and pairwise interactions derived by a permutation test with a nominal significance level of 5% in the context of a two-dimensional, two-QTL genome scan. The approach uses a penalty on the main effects, Tm, and two different penalties on interactions, TiH (heavy) and TiL (light), and considers the structure of a QTL model to assign these penalties; see Manichaikul et al. (2009).
Performing 241 separate multiple-QTL analyses presented computational challenges especially because, to establish the penalties for each time point, 25,000 permutations of the phenotype against the genotype were performed in the context of a two-dimensional, two-QTL genome scan, using Haley–Knott regression (Haley and Knott 1992). The calculations were performed with R/qtl (Broman et al. 2003), an add-on package for the R statistical software (R Development Core Team 2013). A directed acyclic graph (DAG) created in DAGman (Couvares et al. 2007) structured the permutation tests into five per job and the HTCondor software tool was used to execute the 1.2 million separate compute jobs (5000 jobs per time point × 241 time points) in parallel on the CHTC or the Open Science Grid (OSG) distributed computing resources. After the results of each permutation test were automatically collated, the DAG triggered automatic model selection and QTL processing, again performed using R/qtl (Broman et al. 2003). Model fitting was performed in the manner described in Moore et al. (2013).
Heat maps of profile logarithm of odds (LOD) scores were used to visualize the evidence for QTL in the context of a multiple-QTL model. This is analogous to a display technique used in Zeng et al. (2000): Each QTL was considered separately, and its position was allowed to vary, keeping all other QTL fixed at their estimated positions. For each possible position of the QTL under test, the LOD score comparing that multiple-QTL model to the model with the given QTL omitted was calculated.
Source code, raw and processed data
All of the raw images, custom image analysis computer code, quantified phenotype data, R/qtl input files, and reduced QTL results are available for download at http://phytomorph.wisc.edu/download/. These resources enable repetition of the analyses presented here but, probably more importantly, they enable computer vision scientists to extract additional traits from the image sets and statisticians to develop new methods for mapping phenotypes quantified with high time resolution.
Results
The gravitropism dataset
The biological material studied here was a set of 162 recombinant inbred lines of A. thaliana derived from a cross between the Cvi and Ler ecotypes (Alonso-Blanco et al. 1998) and a set of 92 near isogenic lines containing introgressions of Cvi DNA in the Ler background. The seed stocks used were the same previously studied by image analysis to determine the size distributions and genetic architectures of length, width, and area traits (Moore et al. 2013). The phenotype data are time series of root tip angles extracted from sequences of grayscale digital images automatically acquired at 2-min intervals during gravitropism and batch processed on a HTCondor-managed high-throughput computing grid (Thain et al. 2005). Figure 2A shows five images of a representative root undergoing gravitropism for 8 hr. Each of the 6984 separate trials in this study consists of 241 frames that form a time series. The average tip angle responses of the two parental ecotypes and three example members of the mapping population are shown in Figure 2B. Regardless of genotype, all roots reoriented by approximately 90° within 8 hr, although the time courses differed significantly. The set of phenotype data obtained for this set of recombinant inbred lines is referred to as RIL1. A separate rearing and harvest of the recombinant inbred lines including the parental accessions and a repeat of the phenotype measurements from the next generation of seedlings produced the RIL2 dataset. In general, RIL2 responses were slower than RIL1 responses despite the genetic identity of the plants (Figure 2C). The same measurements were performed on the near isogenic lines, the seeds of which were generated in the manner of the second RIL generation, to produce the NIL dataset (Figure 2D).
Figure 2.
Quantifying root tip angle during gravitropism generates a time-course phenotype. (A) Representative series of images of a root responding to gravity at 2-hr intervals. At the beginning of the experiment, seedlings were rotated 90° such that the root tip was approximately horizontal to the gravity vector. By 8 hr after rotation, the root had grown to reorient its tip parallel to the direction of gravity. Bar, 0.5 mm. (B–D) Tip angle development during gravitropism in the RIL1, RIL2, and NIL datasets. The response of the Cvi parental line is shown by a black line, Ler by an orange line, and the other lines indicate the responses of representatives of the population of inbred lines (labeled A, B, and C). Vertical bars indicate the standard error of the mean. For RIL1, n = 27, 28, 10, 10, and 18 for Cvi, Ler, RIL1-A, RIL1-B, and RIL1-C. For RIL2, n = 14, 18, 18, 11, and 12 for Cvi, Ler, RIL2-A, RIL2-B, and RIL2-C. For NIL, n = 32, 33, and 20 for NIL-A, NIL-B, and NIL-C.
Analysis of variance
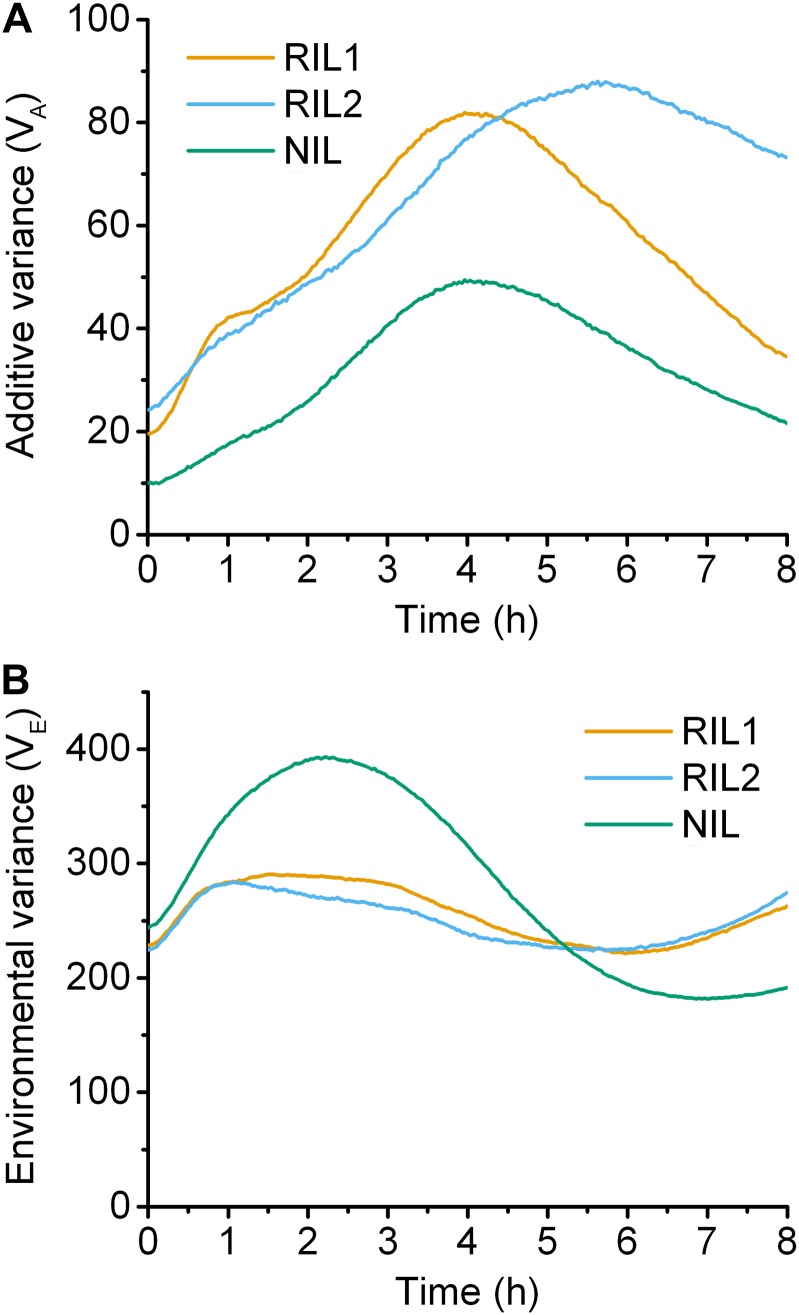
The variance attributed to additive genetic variation and environmental variance in RIL1, RIL2, and NIL are plotted in Figure 3. Additive genetic variance was initially low in all populations but increased as the gravitropic response progressed, peaking 4 to 6 hr after gravistimulation (Figure 3A). Environmental contribution to total phenotypic variance exceeded the variance attributed to additive genetic variation, developing mostly during the first few hours of the response, especially in NIL (Figure 3B). These analyses of variance are based on the differences between the average responses of genetically distinct lines.
Figure 3.
Genetic and environmental contributions to variance. (A) The proportion of the phenotypic variance attributed to the additive genetic variation. (B) The environmental contribution to the phenotypic variance. Orange, blue, and green lines indicate the RIL1, RIL2, and NIL populations, respectively.
Time-resolved QTL map
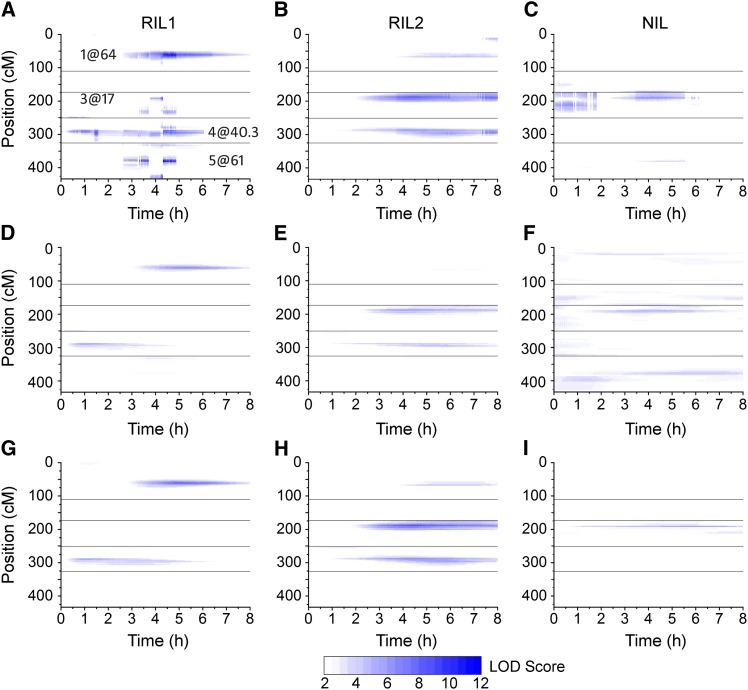
The mean tip angle at each of the 241 time points for each genotype was used as the phenotype in 241 separate and independent multiple-QTL modeling analyses. The result was a highly time-resolved QTL map for each of the datasets (Figure 4, A–C). LOD score is shown on a color-intensity scale as a function of time (abscissa) and genome position (ordinate). In RIL1 (Figure 4A), a total of eight loci, with a maximum of six loci at any given time, were detected during the 8-hr response. Two loci in particular were present during a large portion of the response. The locus on chromosome 4 at 40.3 cM affected variation in the response from ∼0.5 to 6 hr past gravistimulation, and the other on chromosome 1 at 64 cM contributed from ∼3 to 8 hr. Some QTL were more limited in duration. For example, shortly after 3 hr, a QTL appeared on chromosome 3 at 44 cM and lost influence at 5 hr. This particular QTL illustrates a problematic feature of treating the phenotype at one time point independent of the next, namely that a QTL can abruptly disappear or shift position due to data at adjacent time points being best fit by different QTL models.
Figure 4.
Time course of QTL development. Magnitude of LOD score is displayed as color intensity as a function of time. (A–C) Profile LOD scores for models selected by stepwise QTL analyses, considering each time point individually. For RIL1, (Tm, TiH, TiL) = (2.58, 3.41, 1.53); for RIL2, (Tm, TiH, TiL) = (2.56, 3.44, 1.63); and for NIL, (Tm, TiH, TiL) = (2.75, 2.40, 0.67). (D–F) Single-QTL analysis results from RIL1, RIL2, and NIL populations. Threshold for significance of single QTL is 1.91 for RIL1, 1.99 for RIL2, and 1.91 for NIL. (G–I) Profile LOD scores for the model selected by stepwise QTL analyses using the SLOD criterion. For RIL1, (Tm, TiH, TiL) = (1.91, 2.41, 1.10); for RIL2, (Tm, TiH, TiL) = (1.99, 2.62, 1.51); and for NIL, (Tm, TiH, TiL) = (1.91, 2.40, 1.66). The position of the loci on the y-axis is shown as the cumulative position of the five Arabidopsis chromosomes. Horizontal lines indicate chromosome breaks. The x-axis shows time in hours since onset of gravistimulation. All coordinates whose LOD scores were not significant are shown in white. Darker blue colors indicate higher support for the loci.
RIL2 supported the selection of three loci as genetic contributors to variation in the gravitropism response (Figure 4B). Two loci contributed from 2 to 8 hr after gravistimulation, with the third locus on chromosome 3 at 17 cM arising later to influence the response between 4 and 8 hr after the start of the experiment. A locus on chromosome 4 at 40 cM was identified and displayed a similar time course in RIL1 and RIL2 (Figure 4B).
Figure 4C shows that three or possibly four loci were statistically significant in NIL during the early and middle parts of the response. The locus on chromosome 3 at 17 cM detected throughout the first 6 hr of the response was also featured prominently in RIL2, and in the “alternate” model centered around 4 hr in RIL1. A second locus on chromosome 5 at 55 cM, influential from 3.5 to 5.5 hr after gravistimulation, was also apparent over the same time period in RIL1. NIL was the only dataset to support a QTL on chromosome 2, located at 40 cM, which contributed to the variation in the response during the first hour.
A function-valued approach
The analysis described above adjusted for the search across QTL models but not for the search across time points. Another shortcoming is that the selected model occasionally abruptly changed between adjacent time points despite the phenotype following a smooth curve across time. A simple but effective approach to integrating information across time points was devised to accommodate the high time resolution in the phenotype data generated by the automated image analysis. The method used a model comparison criterion analogous to that of Manichaikul et al. (2009). It was initiated by performing a genome scan, or single-QTL analysis, at each time point to generate a 2D matrix of LOD scores (genomic positions × time points). Figure 4, D–F shows the results of these single-QTL analyses in the three populations. For RIL1, strong support was obtained for loci on chromosomes 1 and 4 acting at different times (Figure 4D), while for RIL2, loci affecting this response were predicted on chromosomes 3 and 4 (Figure 4E). The locus on chromosome 4 was predicted to be at ∼40.3 cM in both RIL1 and RIL2. The NIL population showed some support for a locus on the proximal end of chromosome 4 and some support for loci on chromosomes 3 and 5 (Figure 4F). The next step in the method was to calculate the maximum LOD (MLOD) and the average LOD (SLOD) at each genomic position across time. Significance thresholds were obtained by a permutation test (Churchill and Doerge 1994), using the corresponding SLOD or MLOD statistic. SLOD exhibits higher power to detect QTL with effects across a large time interval, while MLOD exhibits higher power to detect QTL with large effects over a narrow interval. The MLOD or SLOD statistics were used to derive a penalized LOD criterion, which multiple-QTL analyses sought to maximize in a stepwise search, starting with the first QTL at the position with the highest MLOD or SLOD score. Additional QTL were added to the model if the significance threshold was met. After selecting the QTL model with the highest penalized LOD score, profile LOD scores were derived to evaluate the evidence and localization of each QTL, evaluating each time point in the 8-hr response, individually: The position of each QTL was varied, one at a time, and at each location for a given QTL, we derived a LOD score comparing the multiple-QTL model to the model with the given QTL omitted. Figure 4, G–I presents these profile LOD scores, based on the model identified with the stepwise QTL analysis using the SLOD statistic. In RIL1, a two-QTL model was identified using the SLOD statistic, with loci on chromosomes 1 and 4 (Figure 4G), and a two-QTL model with evidence of epistasis was predicted using the MLOD statistic, with the same QTL on chromosome 4 and an additional one on the distal end of chromosome 3 (data not shown but presented for download). For RIL2, the SLOD statistic predicted a three-QTL model, with loci on chromosomes 1, 3, and 4 (Figure 4H), while the MLOD statistic gave a four-QTL model, with two loci on chromosome 1 and one locus on chromosomes 3 and 4 (data not shown but presented for download). In the NIL dataset, the SLOD statistic resulted in a two-QTL model with both loci on chromosome 3, at 16.4 and 39.7 cM (Figure 4I). Analysis of this population using the MLOD statistic found both the two QTL on chromosome 3, as well as an additional locus on chromosome 2 at 81.3 cM (data not shown but presented for download). None of the loci identified by the above methods were found in a QTL study of Arabidopsis root skewing (the degree to which a seedling root axis deviates from the gravity vector when grown on a vertically maintained agar surface) that utilized the same germplasm (Vaughn and Masson 2011).
Heritability
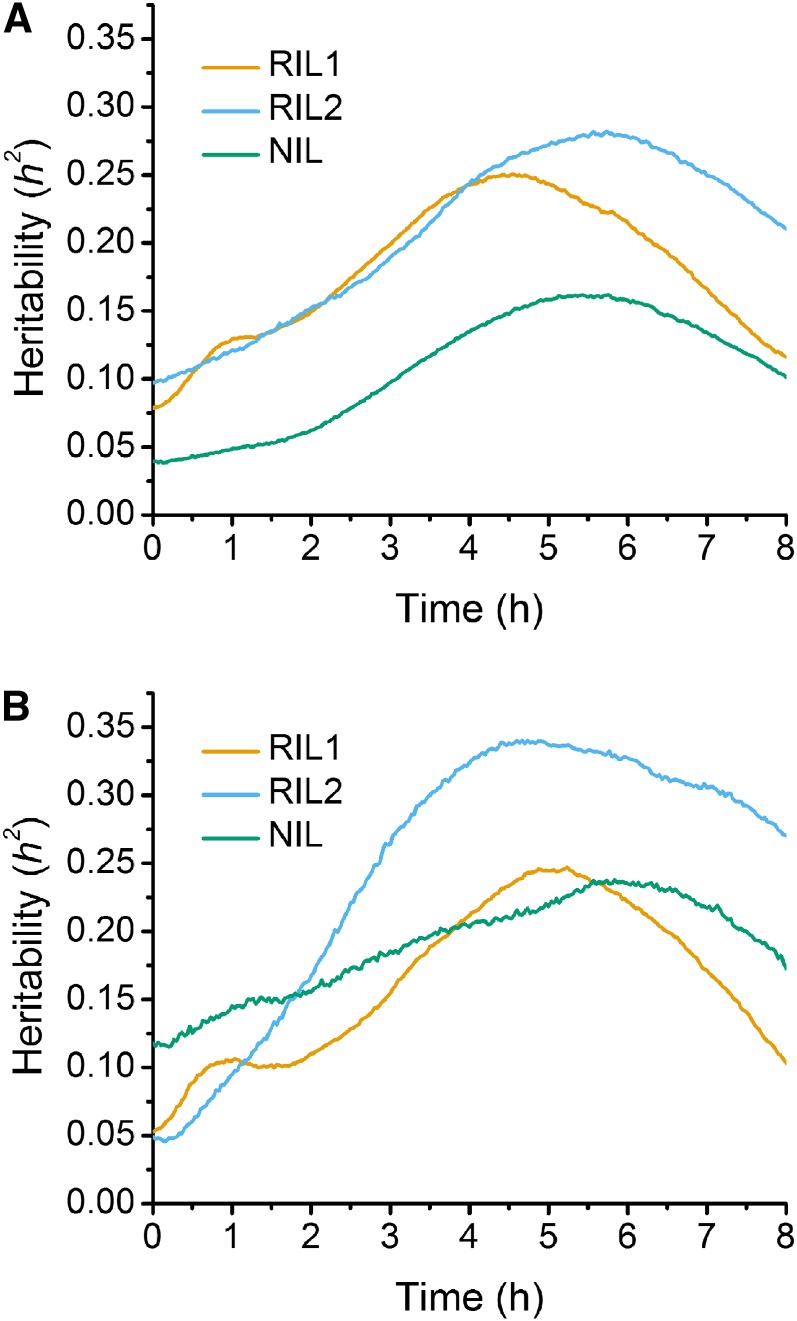
Analysis of variance was performed to calculate the heritability (h2) due to the identified QTL across time in each population, using both the stepwise QTL analysis results at individual time points as well as the results achieved via stepwise QTL analysis using the SLOD statistic. For RIL1, h2 ranged from a low of 10%, occurring at the initiation of the gravitropic stimulus, to a high of 27% around 6 hr postgravistimulation using trait data from each time point (Figure 5A). Analysis of RIL1 with the SLOD statistic showed a similar range in h2, from 5% to a high of 33% (Figure 5B). The heritability calculated from the RIL2 population was somewhat lower than RIL1, ranging from 8–24% and 5–23% for analysis at each time point and via the SLOD statistic, respectively (Figure 5, A and B). For the NIL population, the measured heritability was less than that of the RIL1 and RIL2 populations, with estimates of 3–15% from individual time point data (Figure 6A) and 12–22% from the SLOD statistic analysis (Figure 5B). The lower heritability of NIL can probably be attributed to the lesser amount of genetic variation present in these lines.
Figure 5.
(A) Heritability calculated via results from stepwise QTL analysis at individual time points. (B) Heritability based on the chosen SLOD model from each population. Orange, blue, and green lines indicate the RIL1, RIL2, and NIL populations, respectively.
Figure 6.
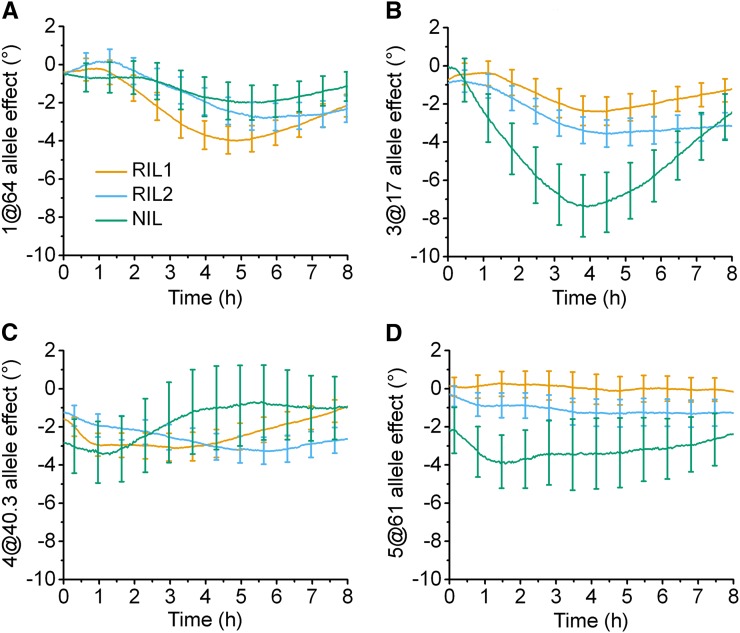
Time-dependent allele effects on root tip angle during gravitropism. The four QTL found by stepwise QTL analyses at each time point that were common to at least two populations were added to a model and the contribution of each locus was estimated. Positive values indicate that substitution of a Cvi allele at the indicated locus increases the tip angle trait, while a negative value corresponds to the Ler allele increasing the trait value. The 95% confidence interval for the degree of the effect is displayed as error bars. (A) Chromosome 1 at 64 cM. (B) Chromosome 3 at 17 cM. (C) Chromosome 4 at 40.3 cM. (D) Chromosome 5 at 61 cM.
A four-QTL model
Independent analysis of the three datasets identified some shared and some unique QTL. In an effort to consolidate the results in a single analysis, loci at chromosome 1 at 64 cM (1@64 cM), 3@17 cM, 4@40.3 cM, and 5@61 cM, identified as contributors by stepwise QTL analysis at each time point (Figure 4, A–C), were used to create a four-QTL model that was evaluated in each of the three datasets. Figure 6 shows plots of the effect on the tip angle resulting from replacement of the Ler allele with the Cvi allele at the indicated positions, i.e., the Cvi allele effect, as inferred from the four-QTL model fit separately to each of the datasets. Figure 6A shows that the Cvi allele at the 1@64 cM locus was estimated to have a negative time-dependent effect on the root tip angle in all three populations. The LOD scores for this position (Figure 4, A–C) indicate strong support for the negative effect of this allele in RIL1 and RIL2, peaking around 5 hr. Locus 3@17 cM was also estimated to have a negative effect on root tip angle, this time with NIL providing strong support, peaking about 4 hr into the response (Figure 6B). The other two loci, 4@40.3 cM and 5@61 cM, also showed a negative Cvi allele effect but with inconsistent (Figure 6C) or little (Figure 6D) time dependence.
Discussion
Gravitropic signaling initiates in specific cells of the root cap (Hashiguchi et al. 2013), resulting in a redistribution of indole-3-acetic acid (IAA) flowing back shootward such that a growth-inhibiting level of this auxin accumulates on the lower side of the root (Spalding 2013). Slowing of cell expansion on the lower side drives downward bending, which is first detectable within ∼15 min (Lewis et al. 2007; Miller et al. 2007). The response time course in Arabidopsis seedling roots depends on factors such as seedling age, the size of the seed from which the seedling emerged, nutrient conditions (Durham Brooks et al. 2010), and even on the previous generation’s culture environment (Elwell et al. 2011) but, generally speaking, the bending speed increases during the first 2 hr, peaks, then gradually slows as the tip angle approaches the new vertical. The present study captured the time course of this process across genetically structured populations of Arabidopsis seedlings so that the dynamics of its genetic architecture could be determined.
High-throughput computing advantages matched technical needs of temporal QTL study
Automated image acquisition and feature extraction were important contributors to the feasibility of this study but were not sufficient for success. Also necessary was the establishment of an automated high-throughput computing (HTC) workflow. HTCondor software enabled batch processing of raw images and statistical genetic modeling of the quantified feature by matching jobs and available resources in a distributed computing environment. Obtaining the results presented in Figure 4, A–C alone entailed launching 21,700 jobs that consumed ∼5000 central processor unit hours. Such work would probably not have been feasible in the absence of such a computing infrastructure. Data quality was also increased by adopting HTC because the degree of throughput and automation increased the feasibility of larger sample sizes.
Comparison of the two methods of QTL analysis
The first of the two methods of searching for significant QTL used in this study relied on automated stepwise QTL selection of a model based on tip angle data at a single time point, while the second method combined information from all time points to choose a multiple-QTL model. The first method is conceptually simpler, treating each time point independently and separately as a new trait to map. However, especially in the case of RIL1, the independent treatment of time points produced occasional discontinuities along the time axis as the selected QTL model jumped between alternatives. The alternate QTL positions, for example ∼4 hr on chromosomes 3 and 5 (Figure 4A), could reflect genes that contribute to the response but without sufficient strength to force their selection in each independent modeling exercise. In the second method, the function-valued approach, one model is chosen to represent the entire response, leading to improved power to detect QTL and better separation of linked QTL. The second approach is also more conservative as it seeks to control for the search across time points as well as the search across QTL models. When applied to the two RIL datasets, both methods detected the QTL on chromosomes 1, 3, and 4 (Figure 4). Replacement of Ler with Cvi alleles at these loci had a negative effect on the root tip angle (Figure 6) as did the Cvi allele at the weakly supported QTL on chromosome 5, which was only detected with the first method and only in RIL1 (Figure 4A).
Similarities and differences between RIL1 and RIL2
Independent rearings of the same set of recombinant inbred lines created the seed stocks used to produce the RIL1 and RIL2 datasets. Therefore, differences between the two datasets cannot be due to genetic differences. Instead, environmental differences during the production of the seed lots are the most probable causes of the differences between RIL1 and RIL2 root responses. Variation in seed size and shape within the two stocks were previously measured by image analysis and subjected to statistical genetic analysis. The seeds used to generate RIL2 were found to be significantly larger than those that generated RIL1. Such maternal environment effects on Arabidopsis seed size are known to affect next-generation growth of seedlings including root gravitropism (Durham Brooks et al. 2010; Elwell et al. 2011). Therefore, the QTL on chromosome 3 that is apparent in RIL2 (Figure 4H) but only weakly and briefly or not at all in RIL1 (Figure 4, A and G) may be a locus that affects gravitropism only in certain seed-size contexts. However, none of the seed size or shape QTL previously identified (Moore et al. 2013) using the same seed stocks overlapped with the root tip angle QTL mapped here.
Candidate genes
The 1.5-LOD support intervals calculated for each QTL permitted consideration of whether or not any corresponded to genes known to encode components of the gravitropism mechanism, such as PIN2 auxin transporters (Chen et al. 1998; Müller et al. 1998), determinants of starch content in the sedimenting statoliths (Caspar and Pickard 1989; Kiss et al. 1989), and plastid outer envelope proteins (Stanga et al. 2009), among others (Morita 2010; Baldwin et al. 2013).
The confidence interval of QTL 1@61 spans a region approximately from gene AT1G21945 to gene AT1G22420. One gene within this region is AUXIN UP-REGULATED F-BOX PROTEIN2 (AUF2), which functions with the related AUF1 protein to regulate root growth responses to the hormones cytokinin and auxin (Zheng et al. 2011). Mutating AUF1 and AUF2 impairs both of the major auxin transport streams in the root and makes root growth more sensitive to auxin transport inhibitors (Zheng et al. 2011). Natural variation at this genetic locus could potentially affect the gravitropism time course.
The confidence interval for QTL 3@15 spans from AT3G23980 to AT3G24400. No gene within this region is known to function in gravitropism, hormonal control of root growth, or other processes that could support a hypothesis about how natural variation within it could affect the phenotype measured here.
The confidence interval for QTL 4@40.3 extends from approximately AT4G15080 to AT4G15396 and includes ABCG30, a gene predicted to encode a membrane protein belonging to the ATP-binding cassette (ABC) superfamily of transporters (Verrier et al. 2008). Members of the B subfamily of ABC transporters have been linked to IAA transport (Noh et al. 2001) and subsequently shown to affect gravitropism (Noh et al. 2003; Lewis et al. 2007), but only relatively recently have members of the G subfamily been connected to auxin transport. Specifically, ABCG36 and ABCG37 have a subcellular localization and an apparent transport activity that leads to efflux from roots of indole-3-butyric acid (Strader and Bartel 2009; Růžička et al. 2010), a compound that has intrinsic auxinic activity and is believed to be a precursor of IAA (Strader and Bartel 2011). ABCG30, formerly known as PLEIOTROPIC DRUG RESISTANCE 2 (PDR2), resides near ABCG36 and ABCG37 in the ABCG phylogenetic tree, and it is expressed specifically in roots (van den Brule and Smart 2002). Expression in roots and a close sequence relationship with membrane proteins now believed to function in auxin homeostasis makes ABCG30 an interesting candidate for the cause of phenotypic variation associated with QTL 4@40.3.
The genes within the confidence interval surrounding QTL 5@61, covering loci AT5G16770 to AT5G17330, include MERISTEM-DEFECTIVE (MDF), which encodes a protein in the SART-1 family. MDF is expressed throughout the plant and mediates root, shoot, and flower development (Casson et al. 2009). Messenger RNA levels of PIN2 and PIN4 are lower in mdf mutants, affecting the ability of the root to achieve a normal auxin maximum in the meristem, which may be how MDF functions to maintain meristematic activity (Casson et al. 2009). The influence of MDF on meristem activity does not make it a strong candidate for the causative gene at this QTL because cell division is not expected to play an important part during an 8-hr gravitropic response. However, the effect of MDF on PIN2 expression makes it a very interesting candidate of causation. PIN2 mediates the shootward flow of auxin, which is an important component of the mechanism that creates differential growth between the upper and lower sides of the root during gravitropism (Spalding 2013). Natural variation in MDF may cause natural variation in PIN2 expression, and thereby affect the strength of the shootward auxin transport stream and the tropic responses that depend on it.
Adding a highly resolved time dimension to an analysis of quantitative trait loci posed some significant technical challenges met here by an emphasis on automated data acquisition and HTC for data analysis and modeling, but it also generates significant new insights and perspectives on the genetic architecture of the process that gives rise to a trait, rather than a trait itself. With image analysis becoming more common in studies of plant biology, in scenarios ranging from confocal microscopes to whole plants (Spalding and Miller 2013), time may be an axis on many more QTL profiles in the future.
Supplementary Material
Acknowledgments
This work was supported by grant IOS-1031416 from the National Science Foundation Plant Genome Research Program to E.P.S. and by National Institutes of Health grant GM074244 to K.W.B.
Footnotes
Communicating editor: K. Nichols
Literature Cited
- Alonso-Blanco C., Peeters A. J. M., Koornneef M., Lister C., Dean C., et al. , 1998. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Baldwin K. L., Strohm A. K., Masson P. H., 2013. Gravity sensing and signal transduction in vascular plant primary roots. Am. J. Bot. 100: 126–142 [DOI] [PubMed] [Google Scholar]
- Blancaflor E. B., Masson P. H., 2003. Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol. 133: 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsirichai K., Guan C., Chen R., Masson P. H., 2002. Root gravitropism: an experimental tool to investigate basic cellular and molecular processes underlying mechanosensing and signal transmission in plants. Annu. Rev. Plant Biol. 53: 421–447 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Caspar T., Pickard B. G., 1989. Gravitropism in a starchless mutant of Arabidopsis. Implications for the starch-statolith theory of gravity sensing. Planta 177: 185–197 [PubMed] [Google Scholar]
- Casson S. A., Topping J. F., Lindsey K., 2009. MERISTEM-DEFECTIVE, an RS domain protein, is required for the correct meristem patterning and function in Arabidopsis. Plant J. 57: 857–869 [DOI] [PubMed] [Google Scholar]
- Chavarria-Krauser A., Nagel K. A., Palme K., Schurr U., Walter A., et al. , 2008. Spatio-temporal quantification of differential growth processes in root growth zones based on a novel combination of image sequence processing and refined concepts describing curvature production. New Phytol. 177: 811–821 [DOI] [PubMed] [Google Scholar]
- Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., et al. , 1998. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. T., MacCurdy R. B., Jung J. K., Shaff J. E., McCouch S. R., et al. , 2011. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 156: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvares P., Kosar T., Roy A., Weber J., Wenger K., 2007. Workflow in Condor, Springer-Verlag, New York [Google Scholar]
- Durham Brooks T. L., Miller N. D., Spalding E. P., 2010. Plasticity of Arabidopsis root gravitropism throughout a multidimensional condition space quantified by automated image analysis. Plant Physiol. 152: 206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell A. L., Gronwall D. S., Miller N. D., Spalding E. P., Brooks T. L., 2011. Separating parental environment from seed size effects on next generation growth and development in Arabidopsis. Plant Cell Environ. 34: 291–301 [DOI] [PubMed] [Google Scholar]
- French A., Ubeda-Tomas S., Holman T. J., Bennett M. J., Pridmore T., 2009. High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol. 150: 1784–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y., Tasaka M., Morita M. T., 2013. Mechanism of higher plant gravity sensing. Am. J. Bot. 100: 91–100 [DOI] [PubMed] [Google Scholar]
- Herridge R. P., Day R. C., Baldwin S., Macknight R. C., 2011. Rapid analysis of seed size in Arabidopsis for mutant and QTL discovery. Plant Methods 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A. S., Symonova O., Mileyko Y., Hao Y., Belcher H., et al. , 2010. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 152: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J. J., Bentsink L., Alonso-Blanco C., Hanhart C. J., Blankestijn-De Vries H., et al. , 2007. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J. Z., Hertel R., Sack F. D., 1989. Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177: 198–206 [PubMed] [Google Scholar]
- Lewis D. R., Miller N. D., Splitt B. L., Wu G., Spalding E. P., 2007. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis Multidrug Resistance-Like ABC transporter genes. Plant Cell 19: 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Moon J. Y., Sen S., Yandell B. S., Broman K. W., 2009. A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics 181: 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N. D., Parks B. M., Spalding E. P., 2007. Computer-vision analysis of seedling responses to light and gravity. Plant J. 52: 374–381 [DOI] [PubMed] [Google Scholar]
- Miller N. D., Durham Brooks T. L., Assadi A. H., Spalding E. P., 2010. Detection of a gravitropism phenotype in glutamate receptor-like 3.3 mutants of Arabidopsis thaliana using machine vision and computation. Genetics 186: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. R., Gronwall D. S., Miller N. D., Spalding E. P., 2013. Mapping quantitative trait loci affecting Arabidopsis thaliana seed morphology features extracted computationally from images. G3 (Bethesda) 3: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M. T., 2010. Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 61: 705–720 [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Galweiler L., Tanzler P., Huijser P., et al. , 1998. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B., Murphy A. S., Spalding E. P., 2001. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B., Bandyopadhyay A., Peer W. A., Spalding E. P., Murphy A. S., 2003. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423: 999–1002 [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2013 R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Růžička, K., L. C. Strader, A. Bailly, H. Yang, J. Blakeslee et al., 2010 Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc. Natl. Acad. Sci. USA 107: 10749–10753. [DOI] [PMC free article] [PubMed]
- Spalding E. P., 2009. Computer vision as a tool to study plant development, pp. 317–326 in Plant Systems Biology, edited by Belostotsky D.A. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Spalding E. P., 2013. Diverting the downhill flow of auxin to steer growth during tropisms. Am. J. Bot. 100: 203–214 [DOI] [PubMed] [Google Scholar]
- Spalding E. P., Miller N. D., 2013. Image analysis is driving a renaissance in growth measurement. Curr. Opin. Plant Biol. 16: 100–104 [DOI] [PubMed] [Google Scholar]
- Stanga J. P., Boonsirichai K., Sedbrook J. C., Otegui M. S., Masson P. H., 2009. A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol. 149: 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Bartel B., 2009. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Bartel B., 2011. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain D., Tannenbaum T., Livny M., 2005. Distributed computing in practice: the Condor experience. Concurr. Comput. 17: 323–356 [Google Scholar]
- Tisné S., Serrand Y., Bach L., Gilbault E., Ben Ameur R., et al. , 2013. Phenoscope: an automated large-scale phenotyping platform offering high spatial homogeneity. Plant J. 74: 534–544 [DOI] [PubMed] [Google Scholar]
- Topp C. N., Iyer-Pascuzzi A. S., Anderson J. T., Lee C. R., Zurek P. R., et al. , 2013. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc. Natl. Acad. Sci. USA 110: E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brule S., Smart C. C., 2002. The plant PDR family of ABC transporters. Planta 216: 95–106 [DOI] [PubMed] [Google Scholar]
- Vaughn L. M., Masson P. H., 2011. A QTL study for regions contributing to Arabidopsis thaliana root skewing on tilted surfaces. G3 (Bethesda) 1: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier P. J., Bird D., Burla B., Dassa E., Forestier C., et al. , 2008. Plant ABC proteins: a unified nomenclature and updated inventory. Trends Plant Sci. 13: 151–159 [DOI] [PubMed] [Google Scholar]
- Zeng Z. B., Liu J. J., Stam L. F., Kao C. H., Mercer J. M., et al. , 2000. Genetic architecture of a morphological shape difference between two Drosophila species. Genetics 154: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hause R. J., Borevitz J. O., 2012. Natural genetic variation for growth and development revealed by high-throughput phenotyping in Arabidopsis thaliana. G3 (Bethesda) 2: 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. H., Miller N. D., Lewis D. R., Christians M. J., Lee K. H., et al. , 2011. AUXIN UP-REGULATED F-BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiol. 156: 1878–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.